Abstract
Topoisomerases are essential enzymes that solve topological problems arising from the double-helical structure of DNA. As a consequence, one should have naively expected to find homologous topoisomerases in all cellular organisms, dating back to their last common ancestor. However, as observed for other enzymes working with DNA, this is not the case. Phylogenomics analyses indicate that different sets of topoisomerases were present in the most recent common ancestors of each of the three cellular domains of life (some of them being common to two or three domains), whereas other topoisomerases families or subfamilies were acquired in a particular domain, or even a particular lineage, by horizontal gene transfers. Interestingly, two groups of viruses encode topoisomerases that are only distantly related to their cellular counterparts. To explain these observations, we suggest that topoisomerases originated in an ancestral virosphere, and that various subfamilies were later on transferred independently to different ancient cellular lineages. We also proposed that topoisomerases have played a critical role in the origin of modern genomes and in the emergence of the three cellular domains.
INTRODUCTION
DNA topoisomerases (hereafter referred to as topoisomerases) are molecular magicians that are absolutely essential to solve topological problems arising from the double-helical structure of DNA (1,2). Type I topoisomerases (Topo I) introduce transient DNA single-stranded breaks in order to change the topological linking number of a double-stranded DNA molecule by steps of one, whereas type II topoisomerases (Topo II) introduce transient double-stranded breaks and change the linking number by steps of two. Although topological problems are obvious with circular DNA genomes, they also occur with linear DNA, as indicated by the requirement of topoisomerases in eukaryotes, and in viruses with long linear genomes (from around 100 kb to 1 Mb). From such consideration, one should have naively expected to find in all cells two homologous topoisomerases descendant from two ancestral topoisomerases present in the Last Universal Common Ancestor (LUCA), which was usually assumed to contain a DNA genome (3). An intelligent designer would have probably invented only one ubiquitous Topo I and one ubiquitous Topo II to facilitate the task of future biochemists. The reality turned out to be quite different, and more interesting. As in the case of other enzymes working with DNA, such as DNA polymerases, the distribution of topoisomerases families and sub-families among modern organisms is not congruent with the universal tree of life based on 16S rRNA sequence comparison (with the trinity Archaea, Bacteria and Eukarya). This is a challenging observation, since the phylogenies of many other important cellular proteins (universal ribosomal proteins, large RNA polymerase subunits, components of the protein-secretion system, ATP syntheses), as well as whole genome phylogenies (based on various methods) follow the tripartite rule.
The phylogenomics of various topoisomerase families and sub-families have been recently reviewed in details (4,5). Here, we will summarize this topic, while adding some important updates, and focus on the evolutionary puzzle of topoisomerase origin [for details on the phylogenetic analyses discussed here, see (5)]. Considering the importance of topoisomerases in cellular life, an important assumption will be that their differential distribution may have had a critical role in shaping the variety of modern genomes and even possibly in shaping modern cellular organisms. Another important aspect is that viruses, here defined as capsid-encoding organisms (6) should be considered on an equal footing with cells (ribosome-encoding organisms) when discussing the origin and early evolution of DNA genomes (hence of DNA topoisomerases). With these considerations in mind, we will first discuss the taxonomic distribution of topoisomerases in the cellular world, within the different domains, trying to clearly distinguish topoisomerases that were already present at the emergence of each domain from those that were introduced later on in a particular domain by lateral gene transfer. We will consider that a topoisomerase was present in the last common ancestor of a particular domain when it is present today in most members of this domain (covering its phylogenetic diversity), whereas we will conclude that there was lateral gene transfer when the enzyme is only present in some members of the domain and branches within another domain in phylogenetic trees. We will then briefly discuss viral topoisomerases and their possible relationships with their cellular counterparts. Finally, we will briefly discuss the evolutionary relationships between topoisomerases and other proteins involved in DNA metabolism in order to get possible clues about their origin. To set the stage for these discussions, we will first review the nomenclature and diversity of the different topoisomerase families [for excellent reviews on the biological roles and mechanism of topoisomerases, see (7,8)].
THE WORLD OF TOPOISOMERASES
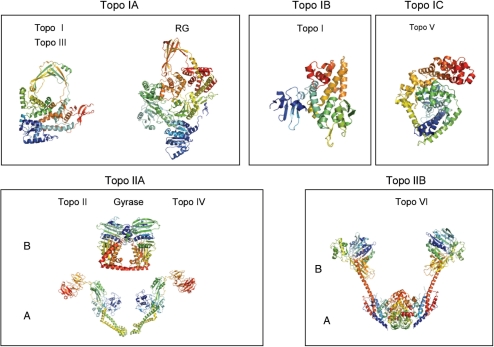
Topoisomerases were first classified according to their mechanistic features (cleavage of either one or two DNA strands for Topo I and Topo II, linking in 5′ or 3′ of the DNA break for Topo IA and IB, respectively). Now that sequences and structural data are available for all known classes of topoisomerases, it is wise to adopt a ‘natural classification’ based on their evolutionary relationships. Such natural classification recognizes five distinct families of topoisomerases, each one corresponding to homologous proteins that originated from five distinct ancestral enzymes. Type I topoisomerases are divided into three families, IA, IB and IC, whereas Type II topoisomerases are divided into two families IIA and IIB (Figure 1). The Topo I families are unrelated to each others and to Topo II, except for the presence of common protein domains that they share with other enzyme families. Hence, Topo IA, Topo IIA and Topo IIB share the so-called Toprim domain (for topoisomerase-primase) which is also present in bacterial primases and several nuclease families (9). The B subunits of Topo IIA and IIB are homologous and contains an ATP-binding domain, the Bergerat fold, also present in the chaperone Hsp90, the proteins of the MutL family and histidine-kinases (10,11). However, their A subunits are evolutionary unrelated. All Type I topoisomerases are monomeric, whereas all Type IIB topoisomerases exhibit a 2-fold symmetry, being either homodimers (some Topo IIAs), heterodimers (most Topo IIAs and all IIBs) or heterotrimers (T4 Topo IIA). The subunits of Topo IIA and IIB have been named A and B, and some DNA topoisomerases are also named according to the timing of their discovery (Topo I, II, III, IV, V, VI) which may be quite confusing for newcomers in the field. Fortunately, all Topo Is have been labelled by chance with odd numbers (I, III, V), whereas all Topo IIs have been labelled with even numbers (II, IV, VI) (Table 1). Finally, a few topoisomerases have been given specific names, such as ω protein for bacterial Topo IA (the first topoisomerase to be discovered), gyrase (to emphasize the negative supercoiling activity of this Topo IIA) and reverse gyrase (to emphasize the positive supercoiling activity of this atypical Topo IA). The distribution of the various families and subfamilies of topoisomerases in the universal tree of life does not follow a general rule (Table 1, Figure 2), some families being widely distributed among the three domains of life but others being restricted to one domain or, in an extreme case, to a single species (Topo IC). The general conclusion is that the topoisomerase I and II activities originated several times independently in different protein families, and that their distribution in modern organisms does not follow mechanistic rules, but is a product of the particular history of these organisms.
Figure 1.
The topoisomerase families represented by one or two crystal structures of each families: Topoisomerase IA. (a) Structure of full-length topoisomerase I from T. Maritima in monoclinic crystal form (PDB entry 2GAJ) (93), and (b) reverse gyrase from A. Fulgidus (PDB entry 1GKU) (94); topoisomerase IB: crystal structure of D. Radiodurans topoisomerase IB (PDB entry 2F4Q) (95); topoisomerase IC: crystal structure of topoisomerase V (61 Kda Fragment) (PDB entry 2CSD) (52); topoisomerase IIA. (a) Crystal structure of E. Coli topoisomerase IV ParE 43kda subunit complexed with Adpnp (PDB entry 1S16) (96) and (b) structure of the full-length E. Coli ParC subunit (PDB entry 1ZVU) (97); topoisomerase IIB: crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms (PDB entry 2ZBK) (98). Each structures were download from the Protein Data Bank: http://www.rcsb.org/pdb (99), and the figures were generated in PyMOL available at http://www.pymol.org/, with each protein chains coloured differently as rainbow.
Table 1.
Distribution of DNA topoisomerases
| Bacteria | Archaea | Eukarya | Virus | ||
|---|---|---|---|---|---|
| Topo IA | Topo I (ω–like protein) | X | (X)T | (X)T (Mimivirus) | |
| Topo III | X | X | X | ||
| reverse gyrase | (X)T | (X)? | |||
| Topo IB | Topo IB | X | X | X | X (Poxviruses) (X)T (Mimivirus) |
| Topo IC | Topo V | X (Methanopyrus) | |||
| Topo IIA | gyrase | X | (X)T | (X)T | |
| Topo IV | (X)? | ||||
| Topo II | X | X (T4 and NCLDV) | |||
| Topo IIB | Topo VI | (X)T | X | (x)? |
X, present; X, already present in the last common ancestor of the domain; (X)?, different hypotheses (present in the last common ancestor or ancient gene transfer); (X)T, clear case of lateral gene transfer.
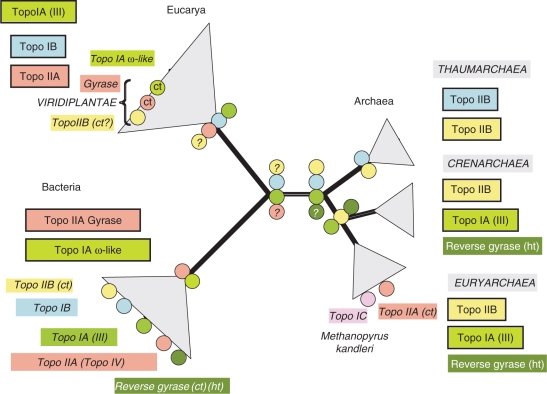
Figure 2.
Phylogenomic distribution of cellular topoisomerases. The universal tree of cellular life is unrooted and the Archaea divided into three phyla according to (39). The name of the various families and subfamilies of topoisomerases are within framed coloured boxes when the enzyme was most likely already present in the last common ancestor of this domain. In that case, they are symbolized by coloured circles at the nodes corresponding to the domain ancestors. A question mark indicates an uncertainty. The name of the various families and subfamilies of topoisomerases that were probably transferred from another cellular domain or from viral families are in italic and within unframed coloured boxes. Question marks indicate that it's unclear if the enzyme was present at the indicated node. Ct, means that theses enzymes were clearly transferred from another cellular domain, ht, thermophiles or hyperthermophiles.
BACTERIAL TOPOISOMERASES
Bacteria are one of the three cellular domains and should not be confused with prokaryotes (a very misleading term) or with primitive cells (12). The domain Bacteria is probably the most successful of the three cellular domains. Bacteria have adapted successfully to all possible biotopes (except the most hyperthermophilic ones, i.e. above 95°C) by inventing a huge variety of metabolisms and ways of life. They have been at the origin of atmospheric oxygen, of mitochondria, and of chloroplasts. The majority of bacteria have a relatively simple structure (they lack a nucleus and cellular organelles). However, some bacteria, such as Planctomycetales, have intracytoplasmic membranes. In the case of the species Gemmata obscuriglobus, this membrane entirely surrounds the bacterial chromosome, producing a nuclear-like body reminiscent of the eukaryotioc nucleus (13). Bacteria are presently divided into a large number of phyla (from 20 to 30 depending on the authors) based on 16S rRNA phylogeny, and more than 500 complete bacterial genomes sequences are now available in public databases. Unfortunately, the relationships between the various bacterial phyla have not yet been sorted out, despite some interesting data obtained from whole-genome tree (super-tree) analyses (14) or the identification of the bacterial superphylum PVC including Planctomycetales, Verrucomicrobiales and Chlamydiae (15). Early claims based on 16S rRNA analyses that hyperthermophilic bacteria correspond to ancient lineages have been challenged, because the high GC content of rRNA from hyperthermophiles might have artificially group these organisms at the base of the bacterial tree (16,17).
All bacterial genomes contain at least two topoisomerase coding genes, a Topo IA related to the Escherichia coli ω protein and a gyrase (Topo IIA). These two proteins were therefore likely already present in the last common bacterial ancestor (Figure 2). Bacterial Topo I (ω-like) and gyrase have been transferred from Cyanobacteria to plants Viridiplantae, via the chloroplast. Later on, the plant gyrase, still active in the chloroplast, was also targeted to plant mitochondria (18). Bacterial gyrases have been also transferred to several Euryarchaea (one of the three main archaeal phyla), as indicated by the branching of archaeal gyrases within the gyrase bacterial tree (5).
Gyrases are very special Topo IIAs that introduce negative supercoils into DNA (19). As a consequence, the genome of organisms containing gyrase is negatively supercoiled (19–22). Gyrase is essential for bacterial life and is the target of several key antibacterial drugs (quinolones and coumarins). In Bacteria, the intracellular level of DNA supercoiling is regulated by the opposite activities of gyrase and of the ω protein (the latter only relaxes negative supercoils) (23). Importantly, the ATP-dependent activity of gyrase allows coupling of the level of intracellular supercoiling to the intracellular ATP/ADP ratio and thus to the physiological state of the cell (24). This directly links the overall metabolic state of the cell to the gene expression network, because the activity of various promoters is differentially affected by supercoiling (some being activated by negative supercoiling, others repressed, still others untouched) (25). Considering the importance of gyrase in cell physiology and the advantage of supercoiling in gene regulation, it is tempting to suggest that the acquisition of a gyrase (and of the associated Topo IA ω protein) has been one of the crucial event in the emergence of the domain Bacteria, providing to these organisms a critical advantage over competitors in natural selection.
In addition to gyrase, many bacteria contain another Topo IIA named Topo IV. Like DNA gyrase, Topo IV is a heterotetramer composed of two different subunits, ParE (homologous to GyrB) and ParC (homologous to GyrA). Although Topo IV closely resembles gyrase in structure and sequence similarity, it lacks the gyrase activity. This correlates with differences in a small region (the GyrA box) of the C-terminal domain (CTD) of the two enzymes. The CTD of gyrase is indeed responsible for the positive wrapping of the DNA around the enzyme: a crucial step in the supercoiling reaction, leading to sign-inversion of the DNA bound to the enzyme following strand passage [(26) and references therein]. In E. coli, Topo IV is a more powerful decatenase than gyrase and is more efficient in relaxing positive superturns (27). The redundance of the bacterial Topo IIA gene thus corresponds to a division of labour: gyrase is being involved in the control of DNA supercoiling and Topo IV in the relaxation of positive supercoils during the elongation step of transcription and the decatenation of the two daughter chromosomes at the end of DNA replication. Many bacteria contain only one Topo IIA which must combine the activities performed by gyrase and Topo IV in E. coli. These lonely Topo IIAs all probably have gyrase activity since they group with bona fide gyrases in phylogenetic trees and contain the GyrA box; however, one of them, the unique Topo IIA of the hyperthermophile Aquifex aeolicus, is probably an exception, since its does not harbour a canonical GyrA box (28).
Gyrases can be easily transformed into Topo IV by mutations in the GyrA box (26). It is therefore likely that Topo IV evolved from a bona fide gyrase, following either duplication of a gyrase coding gene, or introduction of a second gyrase gene in a bacterial genome (or in the genome of a bacterial ancestor) by lateral gene transfer. Since gyrase sequences branch within Topo IV sequences in phylogenetic trees of the Topo IIA family (4,5), it is possible that the transformation of a gyrase into a Topo IV occurred before the divergence of modern bacterial phyla, and that Topo IV was later on lost many times independently in several phyla. Alternatively, if Topo IVs evolved more rapidly (with less constraints) than gyrases, it might have only originated after the divergence of bacterial phyla, and its basal position in the bacterial part of the Topo IIA phylogenetic tree might result from an artefact of phylogenetic reconstruction (long branch attraction by the outgroup sequences). Phylogenetic analyses presently do not allow choosing between these two hypotheses, probably because of a lack of phylogenetic signal (5). It is also still not clear if all Topo IVs are monophyletic in relation to gyrase or if Topo IV-like proteins (negatively defined by the lack of gyrase activity) originated several times independently in the bacterial domain.
Hyperthermophilic bacteria and some moderately thermophilic ones contain a reverse gyrase (Topo IA) in addition to a gyrase (29,30). This enzyme, which produces positive supercoiling, is probably of archaeal origin. It has been shown that a plasmid present in a strain of the hyperthermophilic bacterium Thermotoga maritima, which contains both gyrase and reverse gyrase, is negatively supercoiled (21). This indicates that gyrase activity predominates over the reverse gyrase one in cells containing the two enzymes (see also an archaeal example below).
A few bacterial phyla (Proteobacteria, Firmicutes, Bacteroides) contain a second Topo IA, in addition to the ω-like Topo IA protein. This protein has been called Topo III because it was the third topoisomerase discovered in E. coli. The bacterial Topo III does not seem to play a role in the control of DNA topology, but cooperates in E. coli with the RecQ helicase to resolve Holliday junctions and converging replication forks, suppressing cross-over formation and stabilizing stalled replication forks (31). The scattered distribution of Topo III among bacterial phyla suggests that this enzyme was not present in the last common ancestor of all Bacteria (Figure 2). Bacterial-like Topo III are also encoded by several plasmids, suggesting that the bacterial enzymes may be ultimately of plasmid (viral) origin (5).
Finally, several bacteria from diverse phyla (mainly Proteobacteria and Actinobacteria, but also Thermus/Deinococcus) encode a Type IB topoisomerase (5,32). These bacterial enzymes were recently discovered thanks to the large-scale sequencing of bacterial genomes. They are much shorter than their eukaryotic or archaeal counterparts (see below) and resemble more to Topo IB encoded by poxviruses (32). The bacterial and viral enzymes form two clearly distinct monophyletic groups in a phylogenetic tree of the Topo IB family (5). Considering that Topo IBs are missing in many major bacterial phyla, such as Firmicutes, Planctomycetes and Cyanobacteria, and exhibit a scattered distribution in others, they were probably not present in the last common bacterial ancestor (Figure 2). The dramatic differences between these enzymes and their archaeal or eukaryal counterparts also argue against the idea that bacterial Topo IBs were acquired from an archaeon or a eukaryote. Finally, the existence of a group of viral enzymes with unique features suggests that bacterial Topo IB originated from another group of viral Topo IB for which we have presently no known member (they might be all extinct or not yet discovered).
ARCHAEAL TOPOISOMERASES
Archaea superficially resemble Bacteria (they have no nucleus or intracellular organelles, a compact genome, and coupled transcription and translation). However, they are very different in terms of 16S RNA and most aspects of their molecular biology [for recent reviews on Archaea, see (33,34)]. Archaea were once considered to be mainly composed of methanogens (strict anaerobes producing methane) and a disparate collection of extremophilic organisms (halophiles, thermoacidophiles and hyperthermophiles). However, this view has changed with the recent discovery that soils and aquatic environments are populated by ubiquitous groups of Archaea that seems to play a major role in the nitrogen cycle (35). As Bacteria, Archaea therefore exhibit a great variety of lifestyles and metabolisms. Although Archaea coexist with Bacteria in many biotopes (such as our intestine) they seem to be less successful than Bacteria, except in harsh environments. Hence, whereas Bacteria predominate in the upper layer of the ocean, Archaea become more abundant below 100 m in the ocean column or below 1 m in marine sea floor when resources become scarce (36). Similarly, whereas Bacteria dominate in moderately hot springs, Archaea become dominant at higher temperatures and they are the only organisms that have succeeded to colonize biotopes with temperatures above 95°C (up to 113°C). It has been recently proposed that Archaea are better adapted than Bacteria to extreme low energy conditions because of the lower permeability of their membranes, whereas Bacteria are more efficient in dynamic and rich environment (37).
Archaea can be divided into three major phyla based on ribosomal protein phylogeny and comparative genomics, the Euryarchaea, which exhibit a broad variety of phenotypes (halophiles, methanogens, moderate thermoacidophiles and hyperthermophiles), the Crenarchaea, which only include hyperthermophiles and thermoacidophiles, and the Thaumarchaea, which group psychrophilic, mesophilic and possibly thermophilic ammonium oxidizers (Figure 2). The division between Euryarchaea and Crenarchaea was recognized early on based on 16S rRNA sequence analyses (38), whereas the recognition of Thaumarchaea (formerly grouped with Crenarchaeota) was only recently recognized based on phylogenetic analysis of ribosomal proteins and comparative genomic analyses (39). Interestingly, Archaea resemble Eukarya much more than Bacteria at the molecular level. This is especially striking in the case of DNA replication, since archaeal proteins involved in this process only have eukaryotic homologues. Interestingly, there are both differences and similarities between the archaeal and the eukaryotic topoisomerase sets.
Nearly all Archaea contain a Topo II of the B family, known as Topo VI (10), except thermoacidophilic Euryarchaeota of the order Thermoplasmatales, which have a bacterial gyrase instead. The most parsimonious explanation of these observations is that Topo VI was already present in the most recent archaeal ancestor (Figure 2) and was replaced in Thermoplasmatales by DNA gyrase. Topo VI is the only known topoisomerase that can relax positive superturns in Euryarchaeota and Crenarchaeota (this task can also be done by a Topo IB in Thaumarchaea, see below). Topo VI is therefore likely essential in these Archaea both to remove positive superturns induced in transcription and replication and for chromosome segregation (decatenation). Topo VI has no gyrase activity and therefore can only control DNA topology by its relaxation activity. One can wonder if the lack of gyrase activity in most Archaea could partly explain why they seem to be less successful than Bacteria in ‘normal’ biotopes. In that context, it should be very interesting to compare Archaea containing a gyrase to those lacking this enzyme. As previously mentioned, several Archaea, all members of the phylum Euryarchaea, have indeed acquired a bacterial gyrase. These include all genera of halophiles (Haloarchaea), all mesophilic methanogens of the order Methanosarcina, Thermoplasmatales and hyperthermophiles of the genus Archaeoglobus. In Haloarchaea and Methanosarcina, the genes encoding the two gyrase genes (GyrA and GyrB) are adjacent to those encoding the two Topo VI subunits (Topo VIA and Topo VIB) indicating that both Topo II are probably co-regulated (40). Plasmids are negatively supercoiled in gyrase-containing Archaea (20,21) including Archaeoglobus, which contains both gyrase and reverse gyrase (22). In contrast, archaea lacking gyrase have relaxed intracellular plasmids (41). This indicates that, when present, gyrase determines the intracellular DNA topological state in archaea, as in bacteria. Accordingly, the lateral gene transfer and fixation of a gyrase gene in an organism previously lacking this activity should have produced a dramatic change in its intracellular topology. This means that the selective advantage obtained by acquiring a gyrase gene should have been expressed immediately, despite the burden created by the necessity for the cell to adapt itself to a drastic modification of the topological state of the DNA.
Surprisingly, although archaeal Topo VI (Topo IIB) and bacterial Topo IV (Topo IIA) have similar activities in vitro (relaxation of both positive and negative superturns), the bacterial Topo IV has not yet been detected in Archaea (50 genome sequenced) and the archaeal Topo VI has only been detected in the genomes of three bacteria (although more than 500 bacterial genomes have now been sequenced). This suggests that these enzymes interact with different specific partners in each domain and cannot easily complement each other and/or transfers are rarely fixed in the absence of a strong positive selection pressure.
Beside Topo VI, the most widespread topoisomerase in Archaea is a Topo IA, which is present in one or two copies in all Euryarchaea and Crenarchaea, but absent in the two Thaumarchaea whose sequences are available (39). The most parsimonious explanation of this observation is that Topo IA was present in the last common ancestor of all Archaea and was lost in Thaumarchaea (Figure 2). The archaeal Topo IA is more similar in term of sequence to bacterial and eukaryotic Topo III than to bacterial Topo IA of the ω-type, and we will therefore call it Topo III thereafter (5). Phylogenetic analysis has revealed the existence of several subgroups of archaeal Topo III whose positions in a phylogenetic tree of the family does not match the traditional archaeal phylogeny based on ribosomal proteins (5). Several Euryarchaea even contain Topo III of different subgroups (5). It is possible that several subfamilies of Topo III were already present in the last common archaeal ancestor, and that some of them were later on lost in various archaeal groups. Alternatively, several subgroups of Topo III may have originated by gene duplication in the archaeal domain and, with some of them exchanged later on within the Archaea via lateral gene transfer. The resolution of Topo III phylogeny is presently not sufficient to discriminate between these two possibilities (5). Although the role of these archaeal Topo III is unknown, it is reasonable to think that they are involved in DNA recombination, as it has been shown for their bacterial and eukaryal counterparts.
In additon to Topo III, all hyperthermophilic archaea contain another Topo IA, reverse gyrase [for recent reviews see (42,43)]. Reverse gyrase is a very unusual DNA topoisomerase, formed by the fusion of a helicase-like domain of the SFII superfamily N-terminal to a bona fide Topo IA g (44). Reverse gyrase can introduce positive supercoiling in the presence of ATP and is present not only in all hyperthermophiles, either archaea or bacteria, but also in some moderately thermophilic bacteria (45). In contrast, reverse gyrase has never been found in mesophiles, suggesting a strong link between this enzyme and life at high temperature (46). A role of reverse gyrase in adaptation to high temperature was later confirmed by the thermosensitive phenotype of a reverse gyrase knock-out mutant (47).
Unrooted phylogenies based on the complete reverse gyrase gene (with much more positions available) indicate that archaeal and bacterial reverse gyrases are very similar, suggesting that these enzymes did not diverged from an ancestral reverse gyrase present in the last common ancestor of Archaea and Bacteria, but that it appeared in one of the two domains and was later on transferred into the other (48). Strikingly, the archaeal reverse gyrase tree can be nicely superimposed to the tree of hyperthermophilic archaea based on ribosomal proteins, suggesting that reverse gyrase was present in the last common ancestor of Euryarchaeota and Crenarchaeota. In contrast, the bacterial reverse gyrase tree is not congruent with classical bacterial trees: bacterial reverse gyrases are divided in two subgroups and bacteria from the same phylum can have reverse gyrases of different subgroups. These data suggest that reverse gyrase originated in the archaeal domain and was later on transferred twice independently to different bacteria. Reverse gyrase might have been already present in the last common archaeal ancestor, and later on lost in Thaumarchaea, or it might have appeared after the divergence of Thaumarchaea, but before the split between Euryarchaeota and Crenarchaeota.
For a long time, there was no known archaeal Topo IB. However, the two recently sequenced genomes of the Thaumarchaea Cenarchaeum symbiosum and Nitrosopumilus maritimus contain a Topo IB gene (and surprisingly no Topo IA) (48). This Topo IB strikingly resembles by its size and sequence signatures the eukaryotic Topo IB, and phylogenetic analysis shown that these archaeal Topo IB branch as sister group of eukaryotic Topo IB in a Topo IB tree, away from Poxviruses and bacterial Topo IB (48). This strongly suggests that Topo IB was present in the last common ancestor of Archaea and Eukarya and, consequently, in the last common ancestor of Archaea. The finding of a Topo IB gene in Thaumarchaea also supports the rooting of the archaeal tree between Thaumarchaea and other Archaea, inferred from ribosomal protein phylogeny (39). The most parsimonious explanation for the phylogenomic distribution of Topo IB is now that Topo IB has been lost once, in a lineage common to Crenarchaea and Euryarchaea.
Finally, a very atypical topoisomerase, Topo V, was discovered 15 years ago in one particular species, Methanopyrus kandleri, a hyperthermophilic methanogen that can grow up to 110°C (49,50). Topo V is formed by the fusion of a N-terminal topoisomerase domain with a C-terminal DNA repair domain with apurinic/apyrimidic lyase activity. This domain contains 24 helix–hairpin–helix (HhH) DNA-binding motifs arranged in 12 tandem (HhH)2 domains. The M. kandleri Topo V was previously classified as a Topo IB, because it forms a transient covalent linkage in 3′-end of the DNA break and it relaxes both negative and positive superturns in the absence of magnesium. Furthermore, both Topo IB and Topo V relaxe DNA via events that release multiple DNA turns, employing a constrained swiveling mechanism (51). However, the resolution of the topoisomerase domain of Topo V has shown that this protein exhibits a new fold and, consequently, is not homologous to Topo IB (52). As a consequence, it has been suggested that Topo V should be considered the prototype for a new DNA topoisomerase family, Topo IC (5). A recent BLAST search in the nr database, as well as in the environmental database, indicates that the M. kandleri Topo V is still an orphan protein, despite the increasing number of sequenced genomes and environmental sequences available.
From this survey of the distribution and phylogeny of topoisomerases in the archaeal domain, one can conclude that the last common ancestor of all Archaea harboured a Topo IIB (Topo VI), at least one Topo IA and one Topo IB (Figure 2). However, it lacked the gyrase activity and its associated ω-like relaxing enzyme, indicating that, in contrast to its bacterial counterpart, it was not able to use DNA supercoiling to regulate its intracellular DNA topology in response to environmental changes. This could explain why Bacteria have been more successful in exploiting rich and versatile biotopes. Nevertheless, in contrast to Bacteria, some ancient Archaea acquired a reverse gyrase that allowed its descendants to exploit high temperature biotopes that were out of reach for Bacteria. Later on, some Archaea acquired a bacterial gyrase that probably allow them to compete more efficiently with bacteria and possibly to acquire a ‘bacterial lifestyle’. Indeed, most gyrase-containing Archaea are mesophiles with large genomes that exhibit a great metabolic versatility and adaptability to environmental changes.
EUKARYAL TOPOISOMERASES
The domain Eukarya includes ribosome-encoding organisms that originated from a common ancestor already containing an elaborate nucleus, multiple RNA polymerases and complex molecular machinery such as the spliceosome. It is also widely assumed that the ancestor of modern eukaryotes already harboured a mitochondrion that originated from an alpha-proteobacterium and that the few modern amitochondriate eukaryotes have lost mitochondria. Eukarya are presently divided into six major divisions that diverged early on (53). With few exceptions, most eukaryotes that have been investigated for their molecular biology (and in particular for their topoisomerases) belong to only two of these six divisions, the Opisthokonta, that includes animals and fungi, and the Viridiplantae, that include land plants, as well as green and red algaea. In addition to mitochondria, all members of the division Viridiplantae and several members of other divisions contain chloroplasts that originated from cyanobacteria by primary endosymbiosis (Viridiplantae) or by secondary or tertiary endosymbiosis.
Most eukaryotes contain at least three topoisomerases, one Topo IA, one Topo IB and one Topo IIA (Figure 2). The eukaryotic Topo IA, which is called Topo III, is indeed more similar in sequence to archaeal and bacterial Topo III than to bacterial ω-like Topo IA. As in Bacteria, eukaryotic Topo III are clearly involved in the resolution of Holliday junction in order to prevent excessive recombination. For this task, Topo III cooperate with RecQ-like helicases, such as sgs or Bloom's syndrome helicases, and other protein factors, such as Rmi1 [see (54) and references therein]. In mammals, the Topo III gene has been duplicated, leading to the existence of two isoforms of the protein, Topo IIIα and Topo IIIβ. The Topo IIIβ seems to be mainly involved in the resolution of Holliday junctions that occurs during meiotic recombination (55). The ubiquity of Topo III in the eukaryotic domain strongly suggests that the last common ancestor of all eukaryotes already contained a Topo IA of this subfamily (Figure 2).
The eukaryotic Topo IB (often simply called eukaryotic Topo I) was first described as a swivelase or an untwisting enzyme, since it can relax positive superturns. It seems to be the main enzyme involved in the relaxation of positive superturns that accumulate in front of the replication forks and transcription bubbles (although these tasks could be a priori performed by the eukaryotic Topo IIA as well). In mammals, the gene encoding the Topo IB has been duplicated, and one of the two copies has been targeted to mitochondria (56). Interestingly, the mitochondrial version has been streamlined, losing most of the long N-terminal extension characteristic of the nuclear eukaryotic Topo IB. As previously mentioned, eukaryotic Topo IBs are very similar to archaeal Topo IBs, indicating that a Topo I was most likely present in the last common ancestor of Archaea and Eukarya (Figure 2).
The eukaryotic Topo IIA is a homodimer, each monomer corresponding to the fusion of an homologue of GyrB(ParE) protein with an homologue of the bacterial GyrA(ParC) protein. Also, although eukaryotic Topo IIAs resemble bacterial Topo IVs in terms of activity, they are much more divergent from Topo IVs than Topo IVs are from DNA gyrases. In particular, the CTD of eukaryotic Topo IIA is not homologous to the CTD of bacterial Topo IIA.
Eukarya of the division Viridiplantae contain, in addition to the classical eukaryotic Topo IIA, a Topo IIB closely related to archaeal Topo VI (57) (Figure 2). In plants, this Topo IIB is essential for endoreduplication, a polyploidization process that is responsible for the enlargement of plant cells. This enlargement is essential for plants to reach their normal sizes (Topo IIB mutants are dwarfs) (58,59). In addition to the homologues of the archaeal Topo VI A and B subunits, genetic analyses have shown that plant Topo VIs probably also contain two small subunits, BIN4 and RHL1 (60,61). Another recently described putative plant Topo VI subunit, MIDGET (62), appears to be identical to BIN4 except for the first 31 N-terminal amino acids (unpublished observation). Interestingly, BIN4 and RHL1 exhibit some sequence similarities with the CTD of eukaryotic Topo IIA, suggesting that Topo IIA and B in plants may interact with similar partners.
All Eukarya contain a protein, called Spo11, which is a homologue of the archaeal Topo VI A subunit. This protein, which performs the cleavage-religation reaction required for strand passage in topoisomerases, promotes the formation of chromosome double-stranded breaks that trigger meiotic recombination (10,63). This surprising observation draws an unexpected link between Archaea and the origin of sex in Eukarya. Plants contain two Spo11 paralogues involved in meiosis in addition to the paralogue (called Spo11-3) involved in the formation of the plant Topo IIB. The A subunits of eukaryotic Topo IIB and all Spo11 homologues form a monophyletic clade, clearly separated from Archaeal Topo VI in phylogenetic trees of the Topo IIB A subunits (5). It is therefore unclear if a Topo IIB was present in the last common ancestor of Archaea and plants and later on lost in many eukaryotic divisions, or if an archaeal Topo VI was transferred to eukaryotes at the origin of the Viridiplantae division (64,65). Beside a Topo IIB of possible archaeal origin, Viridiplantae also contain a Topo IA and a DNA gyrase of bacterial origin. Viridiplantae are therefore the organisms with the best equipment in topoisomerases, since they harbour topoisomerases from all known families, except Topo IC. Some protists also contain a DNA gyrase of bacterial origin. The presence of a gyrase in Plasmodium and other pathogenic protists (whose ancestors once harboured a chloroplast) is especially interesting, considering the possibility of targeting this enzyme for therapeutic purposes.
VIRAL DNA TOPOISOMERASES
Viruses have been considered for a long time (and are still considered by many biologists) as by products of biological evolution, i.e. fragments of cellular genomes (plasmids or transposons) that have escaped from the control of ancestral cells (either prokaryote or eukaryotes), acquired a capsid, and became autonomous infectious parasites. They were often not even considered as biological entities. This view has radically changed in the last years, following dramatic discoveries, such as novel viruses with unique morphotypes in the archaeal domain (66) or the giant mimivirus with a genome three times as big as the genome of a mycoplasma (67). New hypotheses have been proposed about the nature of viruses, their origin and their roles in biological evolution (68–70). Viruses are now considered by several authors as bona fide living organisms (6) and it has been proposed to focus on the intracellular stage of the viral infection (the viral factory) instead of the virion, to get a better understanding of their nature (69). Viruses are presently the most abundant living entities (71) and are also very ancient (72,73). The discovery of homologous DNA replication and capsid proteins in viruses infecting cells from different domains suggests indeed that the formation of major viral lineages predated the divergence of Archaea, Bacteria and Eukarya (68,72,73). It is likely that viruses have played a major role in shaping the history of modern cells, since they have probably always outnumbered their cellular hosts and since transfers of genes from viruses to cells have been probably always a major source of cellular proteins (73). To put viral nomenclature in line with modern evolutionary theories, it has been suggested to replace the ancient dichotomic nomenclature, viruses versus bacteriophages (that mirrored the eukaryote/prokaryote dichotomy) by a new one following the nomenclature proposed by Woese and colleagues for the three cellular domains, bacterioviruses, for virus infecting Bacteria, archaeoviruses, for virus infecting Archaea and eukaryoviruses, for virus infecting Eukarya (66). It has also been proposed to elevate the taxonomic status of viruses to the same level as cells by dividing the living world in two classes of organisms, those encoding capsids, and those encoding ribosomes (6).
Many DNA viruses encode their own DNA polymerases, as well as a variety of other enzymes involved in DNA replication, repair or recombination. In particular, two completely unrelated families of DNA viruses with large DNA genomes (>100 kb and up to 1.2 Mb) encode their own topoisomerases. Most bacterioviruses of the T4 superfamily and many eukaryotic viruses of the NCLDV (Nucleo-Cytoplasmic Large DNA Viruses) superfamily encode a Topo IIA, whereas Poxviruses (a subfamily of NCLDV) additionally encode a Topo IB. The Topo IIA encoded by T4-like bacterioviruses form a distinct monophyletic group in the Topo IIA phylogenetic tree, very well separated from their bacterial and eukaryotic homologues (5) (Table 1). Strikingly, the bacteriovirus Topo IIAs are no more related to bacterial than to eukaryotic Topo IIAs. Furthermore, an insertion common to the bacteriovirus and eukaryotic Topo IIA clearly indicates that the T4 enzyme cannot be derived from the bacterial enzymes (73). Topo IIAs from NCLDV are closely related to eukaryotic Topo IIA, both in term of structure (they are homodimers) and sequence (they form a monophyletic group with eukaryotic Topo IIA in the Topo IIA tree). However, they do not group specifically with their respective hosts (for instance Poxviruses with animals), but branch instead at the base of the eukaryotic Topo IIA tree, either as outgroup of eukaryotic sequences or intermixed with Topo II from various protists (5). These data suggest ancient transfers of Topo IIA genes between NCLDV and eukaryotes. Although the direction of these transfers cannot be rigorously determined, we believe that transfers from viruses to cells are more likely.
The Poxviruses Topo IB harbour a specific short N-terminal domain, instead of the longer N-terminal domain found in their eukaryotic and thaumarchaeal homologues (32) suggesting that this viral enzyme was not acquired from a eukaryotic host. In fact, Poxviruses Topo IB group together with bacterial Topo IB in a Topo IB tree, suggesting that the bacterial Topo IB could be of viral origin (5).
Curiously, the recently described giant mimivirus, a relative of Poxviruses in the NCLDV family, encodes a Topo IB and a Topo IA (5,74) (Table 1), which both group with their bacterial counterparts, in the respective phylogenetic trees (5). These enzymes were thus probably acquired by lateral gene transfer, from a bacterium that was ingested by a protist infected by mimivirus (5,74).
ORIGIN OF TOPOISOMERASES
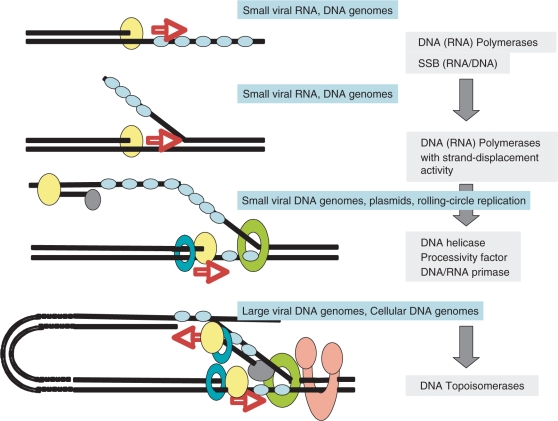
Topoisomerases could have originated by combining protein modules previously involved in RNA metabolism, such as RNA-binding proteins, RNA endonucleases or RNA ligases. Alternatively, they could have evolved from protein modules that were already working with DNA, if the first steps in the evolution of DNA genomes occurred in the absence of any topoisomerase activity, i.e. before the emergence of long double-stranded DNA genomes. Two arguments favour the latter hypothesis: first, whereas RNA polymerases and RNA-binding proteins are obvious candidates to be direct ancestors of DNA polymerases and single-stranded DNA-binding proteins, ‘RNA topoisomerases’ that could be direct ancestor of DNA topoisomerases are unknown. Secondly, it is likely that double-stranded DNA genomes with complex DNA-replication mechanisms (i.e. concurrent symmetric DNA replication) were preceded by single-stranded or even short double-stranded DNA genomes replicated by simpler mechanisms, such as asymmetric DNA replication, and/or rolling circle (RC) replication (75) (Figure 3). These simple systems probably did not require topoisomerases, as it is still the case for their modern counterparts (the RC replication of some replicons require supercoiled DNA, hence gyrase activity, but only for the recognition step of the initiator protein). If this scenario is correct, topoisomerases probably originated when more complex DNA genomes (long linear or circular DNA molecules) were selected in the course of evolution, together with more elaborate replication machineries. DNA polymerases should have originated first (from RNA polymerases) followed by single-stranded DNA-binding proteins (from RNA-binding proteins) to increase the processivity of the DNA-replication process. Various DNA specific nucleases and possibly DNA ligases should have originated shortly thereafter, to eliminate template strands, maturate DNA genomes and/or destroy foreign DNA in the on-going competition between viruses and cells. Later on, the replication of double-stranded DNA would have required first the invention of DNA helicases, and finally topoisomerases, once the size of DNA genomes became such that topological problems had to be solved (Figure 3). This scenario is supported by the fact that all known viruses encoding topoisomerases have large DNA genomes and elaborated multi-enzymatic systems for DNA replication.
Figure 3.
Hypothetical and schematic scenario for the evolution of the elongation step of RNA and DNA replication, from simple to complex, with the progressive ‘invention’ of enzymes specifically involved in genome replication. Steps 1 and 2, asymetric replication (one strand at a time), observed in organisms with single or double-stranded RNA or DNA genomes. A complementary strand is first synthesized and will serve of template for the synthesis of the new strand (step 2). These steps require more and more processive polymerases and single-stranded DNA or RNA-binding proteins. Step 3, partially symmetric replication (one strand starts to be replicated before the first one has been fully replicated). This introduces the notion of leading and lagging strands and requires the recruitment of a primase activity possibly previously only used in the initiation step (together with other mechanisms such as tRNA priming and protein priming). Steps 2 and 3 can be progressively improved by the introduction of helicases and processivity factors (clamp-like) to help the polymerase. Step 4, symetric replication with two polymerases and the primase activity linked to the helicase activity. The formation of short fragments on the lagging strand require the intervention of other proteins (nuclease/ligases) which have been omitted for clarity. This step can be improved by coupling the two polymerases in a physical complex and by rotating the lagging strand by 180° to allow concurrent replication of the two strands. At this stage, topoisomerases are required to replicate long linear genomes or circular genomes. All steps in that scenario are observed today in the viral world. Steps 1 and 2 in both RNA and DNA viruses, step 3 only in DNA viruses and step 4 in both DNA viruses with large genomes (including concurrent replication) and in cellular organisms.
The evolution of DNA-replication machineries from simple to complex might have occurred in primitive DNA cells, following the transition from RNA to DNA genomes. However, an alternative possibility is that this evolution occurred in ancestral DNA viruses that still infected RNA cells. This would explain why there is no congruence between topoisomerase and cellular phylogenies, as well as the existence of specific viral families of topoisomerases. As previously mentioned, topoisomerases are not the only DNA enzymes whose phylogenomic distribution do not fit with the topology of the universal tree of life [see (76) for the case of DNA polymerases]. In general, DNA-replication proteins encoded by DNA viruses are not specifically related to functional analogues of their cellular hosts. In most cases, they form specific viral subfamilies only distantly related to their cellular counterparts, or they are strictly virus specific (70,73 and references therein). To explain these observations, one of us (PF) as well as Koonin and coworkers, have suggested that most viral DNA-replication proteins originated in an ancestral virosphere of DNA viruses that predated modern cells with DNA genomes (68,70,76). This led to the idea that DNA itself could have appeared first in a primitive virosphere of RNA viruses infecting RNA cells (77). In that model, the first individual with a DNA genome (a virus) was selected because chemical modifications of RNA into DNA produced a direct selective advantage for this virus in bypassing cellular defence mechanisms directed against RNA. If this hypothesis is correct, the first viruses with DNA genomes might have contain simple genomes formed by single-stranded or short linear double-stranded DNA molecules. Enzymes involved in DNA replication, DNA recombination and repair, might have then originated step-by-step in the ancestral virosphere, in coordination with the evolution of more and more complex viral DNA-replication mechanisms (75) (Figure 3).
Two observations indeed suggest an evolutionary connection between topoisomerases and the viral world. The most compelling is the evolutionary relationships between Topo IB and virally encoded tyrosine recombinases (78). Topo IBs belong to a huge superfamily of tyrosine recombinases, suggesting that they evolved from viral encoded integrases. Another is that viral Rep proteins that initiate rolling-circle replication exhibit type IA-like DNA topoisomerase activities (79,80). The relaxases encoded by conjugative plasmids and some restriction endonuclease or DNA ligases (many of them also encoded by viruses) are also mechanistically related to Topo I (81,82). The emergence of a topoisomerase from such enzymes was probably easy, since it has been shown that the restriction endonuclease NaeI can be transformed from an endonuclease to a topoisomerase by a single amino-acid substitution (82).
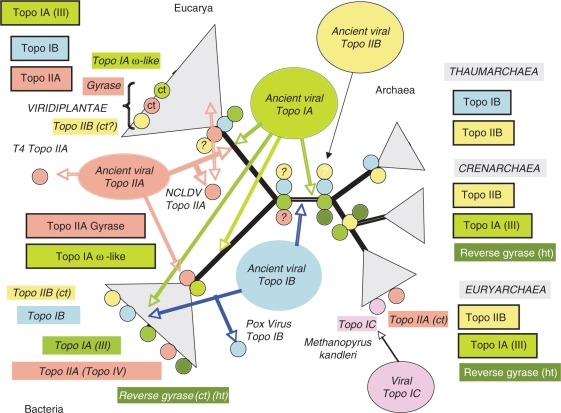
In the viral scenario for the origin of topoisomerases, one can imagine that the different families of Topo I (Topo IA, IB, IC and possibly others) have thus originated from different integrases, endonucleases, ligases, or endonuclease-ligases involved in the replication of various viruses and/or plasmids. To explain the present distribution of topoisomerase families and subfamilies, one should posit that a specific subfamily of Topo IB was transferred to the common archaeal/eukaryal lineage, before the divergence of these two domains, whereas another subfamily was transferred later on to Proteobacteria (and even later to other bacterial phyla by lateral gene transfers between bacteria). Eukaryotic and archaeal Topo III, as well as bacterial Topo IA (ω-like) were probably transferred to Eukarya, Archaea and Bacteria before the emergence of the last common ancestor of each of these domains (i.e. in the branch leading from LUCA to these domains) (Figure 4). Topo IA of the Topo III subfamily were transferred later on in Bacteria (i.e. after diversification of the bacterial phyla). Although Topo IAs are nearly universal (only missing in Thaumarchaea), it is unclear if a Topo IA was already present in LUCA, since the phylogenetic tree of the Topo IA family is not congruent with the universal tree of life. Indeed, the bacterial (ω-like) Topo IA is sister group of archaeal and bacterial reverse gyrases, whereas the bacterial Topo III is sister group of archaeal and eukaryal Topo III (5). This odd phylogeny could be better explained by independent viral origins of different Topo IA subfamilies in different domains than by classical ‘cellular-only’ evolutionary scenario.
Figure 4.
Hypothetical scenario in which cellular topoisomerases originated from an ancestral pool of viral topoisomerases already diversified into five families and various subfamilies. The arrows indicate the direction of transfers from viruses to cellular lineages. Drawing of the universal tree and colour symbols are as in Figure 2.
If various Topo I families (and subfamilies) originated independently in several ancestral lineages of DNA viruses, one can suspect that some of them were never transferred to cells and remain to be discovered. Topo V of M. kandleri might be just such a viral-specific enzyme (50), recently introduced in the Methanopyrus lineage, and discovered by chance, because a curious biochemist decided to investigate all topoisomerases present in a newly discovered archaeon available in his laboratory (49).
Viral type II topoisomerases should have originated after viral Topo I, i.e. when the evolution of long linear or circular DNA genomes started to raise problems for the segregation of viral genomes. Considering their structure, Topo II should have emerged from the association of proteins that were previously working independently (4). Topo IIA and IIB thus should have evolved from three proteins: the ancestors of the A subunits of Topo IIA and IIB, and those of the B subunits common to Topo IIA and IIB (4,8). Before being topoisomerase subunits, these proteins should have been selected to play another role in ancient cells or viruses. It would be interesting to check the possibility that individual Topo II subunits still play additional roles in modern cells, besides their role as Topo II components. In fact, we already know that an eukaryotic homologue of the A subunit of Topo IIB (Spo11) is a double-stranded endonuclease with a role in genetic recombination (10,63). It is usually assumed that Spo11 was recruited from an archaeal Topo IIB to work in meiosis. However, it is possible that the endonuclease activity of Spo11 originated first, and was later recruited to become a Topo II subunit. The ancestral Topo II A subunits (involved in cleavage and religation) might have been previously a Topo I, a DNA ligase and/or an endonuclease, whereas the ancestral Topo II B subunit appears to be an ATP-driven molecular machine recruited several time independently to drive large scale conformational changes in various molecular motors (83).
We presently know four Topo IIA subfamilies, one in Bacteria, one in T4-like bacterioviruses, one in Poxviruses and one in Eukarya. We suspected that all of them are also of viral origin and were transferred in cellular organisms when the segregation of cellular chromosome became a critical issue (Figure 4). The unusual Topo IIA of Mycobacterium smegmatis (84), which branches in between Bacteria and T4 bacterioviruses in the Topo IIA phylogenetic tree (5), could be the stand-alone member of a fifth subfamily of viral origin recently introduced in this mycobacterium.
The presence of two distinct versions of homologous Topo IIA in Bacteria and Eukarya could suggests that this enzyme was already present in LUCA and later on lost in Archaea. However, in the viral scenario for the origin of topoisomerases, Topo IIA might have been introduced independently in Bacteria and Eukarya by two different viral lineages (Figure 4). In that case, the virus that introduced Topo IIA in Eukarya was probably related to NCLDV. As we have seen previously, NCLDV indeed encode Topo IIA specifically related to the eukaryotic enzymes. Later on, some eukaryotic Topo IIA might have been displaced by Topo II from other NCLDV, explaining the complex phylogenetic pattern of viral and cellular lineages at the base of the eukaryotic Topo IIA tree. The viral origin of Topo IIB, and its transfer in the branch leading to Archaea (or to the common ancestor of Archaea and Bacteria) could also explain the odd distribution of this topoisomerase (Figure 4).
Several authors have now considered the possibility that LUCA was still a member of the RNA genomes world, or at least that the modern DNA-replication machinery present in modern cells has not yet been invented at the time of LUCA (85–88). This would explain the existence of two non-homologous sets of the three major DNA-replication proteins (DNA polymerase, primase, helicase): one specific to Bacteria, the other common to Archaea and Eukarya. The hypothesis of an independent transfer of Topo IIA and IIB in different cellular domains (implying the absence of Topo II activity in LUCA) is in agreement with this idea (4). It is tempting to suggest that the elaborate mechanism of DNA replication present in modern cells was finalized first in the ancestral virosphere. This mechanism is characterized by the simultaneous and rapid replication of the two DNA strands by two DNA polymerases that are physically associated (with the lagging strand wrapping by 180°C along the DNA-replication axis) in order to move in the same direction. This sophisticated way to efficiently replicate double-stranded DNA molecules does not only occur in cells but probably also in all DNA viruses with large genomes such as the T4 and T7 bacterioviruses, Herpesviridae or NCLDV. An appealing hypothesis is that the transfers of such elaborate DNA-replication mechanisms from viruses to cells triggered the transition from RNA to DNA cells, providing immediately a tremendous selective advantage to DNA cells over RNA cells. If such transfer occurred two or three times independently, the first DNA cells may have been the direct ancestors of modern cellular domains (87). Alternatively, it has been suggested that viruses predated cellular organisms and that DNA enzymes originated in an acellular RNA/protein world before being transferred into the first cellular organisms [(70), see also (89) for a recent review on the first steps of cellular evolution].
In the viral scenario for the origin of topoisomerases, gyrase and reverse gyrase could have also originated first in the viral world, or alternatively later on from cellular topoisomerases (Figure 4). In any case, the appearance of these new activities should have had a profound influence in shaping the bacterial and the archaeal domains. As previously proposed, gyrase may have give a decisive advantage to bacteria to outcompete other ancestral cell lineages, explaining the incredible success of this domain. One could even suggest that the appearance of gyrase selected the individual at the origin of the bacterial domain (the last common bacterial ancestor). On the other hand, the appearance of reverse gyrase allowed archaea to become the first organisms to thrive at temperatures above 80°C and to occupy new biotopes that were previously out of reach for bacteria. Our love and long time interest for topoisomerases possibly leads us to speculate too much on the decisive roles that these enzymes might have played in shaping the modern biosphere. However, part of these speculations seems to be inevitable consequences of the critical roles that these proteins still play today and of their specific and unusual distributions patterns in the various ancestors of the three domains.
CONCLUSION
Our proposal that most topoisomerase families and subfamilies originated in the viral world predicts that Topo IC is only the tip of the iceberg, i.e. that many viruses and plasmids encode new families and subfamilies of topoisomerases that are still unknown. All viral and plasmid genomes sequences indeed contain a huge proportion of ORFans genes encoding proteins of unknown function (90). We predict that some of these ORFans encode new topoisomerases, as it was found recently that some encode new families of DNA polymerases (91). One should therefore promote in the future the systematic analysis of proteins encoded by ORFans present in viruses, plasmids or cellular genomes (the latter being possibly in many cases relics of integrated viruses (92). Another interesting line of research would be to start manipulating the topoisomerase content of cells to gain insights into their roles (past and present) in shaping genome and cells. It should be interesting for instance to introduce a thermophilic gyrase in a thermophile containing only reverse gyrase, or into a mesophilic archaea with none of these activities, to see how a cell copes with the arrival of an activity that dramatically alters the topology of its genome. It should be also interesting to transfer a Topo IV into an archaeon or an archaeal Topo III into a bacterium to understand why such transfer has never been successful in nature. The study of topoisomerases and DNA topology should greatly benefit from such approaches based on the new vision of the biosphere emerging from the work of modern evolutionists.
FUNDING
The work on DNA topoisomerases in our laboratory was funded by Association de la Recherche contre le Cancer (ARC). Funding for open access charge: ARC n° 4869.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Simonetta Gribaldo for critical reading of the manuscript.
REFERENCES
- 1.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Lazcano A, Guerrero R, Margulis L, Oró J. The evolutionary transition from RNA to DNA in early cells. J. Mol. Evol. 1988;27:283–290. doi: 10.1007/BF02101189. [DOI] [PubMed] [Google Scholar]
- 4.Gadelle D, Filée J, Buhler C, Forterre P. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003;3:232–242. doi: 10.1002/bies.10245. [DOI] [PubMed] [Google Scholar]
- 5.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;4:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Raoult D, Forterre P. Redefining viruses: lessons from Mimivirus. Nat. Rev. Microbiol. 2008;4:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 7.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;6:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 8.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L, Leipe DD, Koonin EV. Toprim a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 11.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 12.Pace NR. Time for a change. Nature. 2006;441:289. doi: 10.1038/441289a. [DOI] [PubMed] [Google Scholar]
- 13.Fuerst JA. Intracellular compartmentation in planctomycetes. Annu. Rev. Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- 14.Daubin V, Gouy M, Perrière G. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 2002;12:1080–1090. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin. Biotechnol. 2006;17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Forterre P. A hot topic: the origin of hyperthermophiles. Cell. 1996;85:789–792. doi: 10.1016/s0092-8674(00)81262-3. [DOI] [PubMed] [Google Scholar]
- 17.Brochier C, Philippe H. Phylogeny: a non-hyperthermophilic ancestor for bacteria. Nature. 2002;417:244. doi: 10.1038/417244a. [DOI] [PubMed] [Google Scholar]
- 18.Wall MK, Mitchenall LA, Maxwell A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl Acad. Sci. USA. 2004;101:7821–7826. doi: 10.1073/pnas.0400836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gellert M, O'Dea MH, Itoh T, Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl Acad. Sci. USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sioud M, Possot O, Elie C, Sibold L, Forterre P. Coumarin and quinolone action in archaebacteria: evidence for the presence of a DNA gyrase-like enzym. J. Bacteriol. 1988;170:946–953. doi: 10.1128/jb.170.2.946-953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guipaud O, Marguet E, Noll KM, de la Tour CB, Forterre P. Both DNA gyrase and reverse gyrase are present in the hyperthermophilic bacterium Thermotoga maritima. Proc. Natl Acad. Sci. USA. 1997;94:10606–10611. doi: 10.1073/pnas.94.20.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Garcia P, Forterre P, Van der Oost J, Erauso G. Plasmid pGS5 from the hyperthermophilic archaeon Archaeoglobus profundus is negatively supercoiled. J. Bacteriol. 2000;182:4998–5000. doi: 10.1128/jb.182.17.4998-5000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruss GJ, Manes SH, Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982;31:35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 24.Drlica K. Control of bacterial DNA supercoiling. Mol. Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 25.Dorman CJ. DNA supercoiling and bacterial gene expression. Sci. Prog. 2006;89(Pt 3–4):151–166. doi: 10.3184/003685006783238317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramlinger VM, Hiasa H. The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J. Biol. Chem. 2006;281:3738–3742. doi: 10.1074/jbc.M511160200. [DOI] [PubMed] [Google Scholar]
- 27.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guipaud O, Forterre P. DNA gyrase from Thermotoga maritima. Methods Enzymol. 2001;334:162–171. doi: 10.1016/s0076-6879(01)34465-8. [DOI] [PubMed] [Google Scholar]
- 29.Bouthier de la Tour C, Portemer C, Huber R, Forterre P, Duguet M. Reverse gyrase in thermophilic eubacteria. J. Bacteriol. 1991;173:3921–3923. doi: 10.1128/jb.173.12.3921-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brochier-Armanet C, Forterre P. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea. 2006;2:83–93. doi: 10.1155/2006/582916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol. Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogh BO, Shuman S. A poxvirus-like type IB topoisomerase family in bacteria. Proc. Natl Acad. Sci USA. 2002;99:1853–1858. doi: 10.1073/pnas.032613199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrett R, Klenk HP. Oxford: Blackwell Pub; 2007. Archaea: Evolution, Physiology, and Molecular Biology. [Google Scholar]
- 34.Cavicchioli R. Archaea: Molecular and Cellular Biology. Washington, DC: ASM Press; 2007. [Google Scholar]
- 35.Nicol GW, Schleper C. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol. 2006;14:207–212. doi: 10.1016/j.tim.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Lipp JS, Morono Y, Inagaki F, Hinrichs KU. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature. 2008;454:991–994. doi: 10.1038/nature07174. [DOI] [PubMed] [Google Scholar]
- 37.Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- 38.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 40.Berthon J, Cortez D, Forterre P. Genomic context analysis in Archaea suggests previously unrecognized links between DNA replication and translation. Genome Biol. 2008;9:71. doi: 10.1186/gb-2008-9-4-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charbonnier F, Forterre P. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J. Bacteriol. 1994;176:1251–1259. doi: 10.1128/jb.176.5.1251-1259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nadal M. Reverse gyrase: an insight into the role of DNA-topoisomerases. Biochimie. 2007;89:447–455. doi: 10.1016/j.biochi.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 43.D'Amaro A, Rossi M, Ciaramella M. Reverse gyrase: an unusual DNA manipulator of hyperthermophilic organisms. Ital. J. Biochem. 2007;56:103–109. [PubMed] [Google Scholar]
- 44.Confalonieri F, Elie C, Nadal M, de La Tour C, Forterre P, Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc. Natl Acad. Sci. USA. 1993;90:4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- 46.Brochier-Armanet C, Forterre P. Widespread distribution of archaeal reverse gyrase in thermophilic bacteria suggests a complex history of vertical inheritance and lateral gene transfers. Archaea. 2007;2:83–93. doi: 10.1155/2006/582916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atomi H, Matsumi R, Imanaka T. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 2004;186:4829–4833. doi: 10.1128/JB.186.14.4829-4833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brochier-Armanet C, Gribaldo S, Forterre P. 2008. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biology Direct, 2008 December 23. doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slesarev AI, Stetter KO, Lake JA, Gellert M, Krah R, Kozyavkin SA. DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature. 1993;364:735–737. doi: 10.1038/364735a0. [DOI] [PubMed] [Google Scholar]
- 50.Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragón A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl Acad. Sci. USA. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brinkmann H, Philippe H. The diversity of eukaryotes and the root of the eukaryotic tree. Adv. Exp. Med. Biol. 2007;607:20–37. doi: 10.1007/978-0-387-74021-8_2. [DOI] [PubMed] [Google Scholar]
- 54.Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific ‘dissolvasome’? Trends Biochem. Sci. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 55.Kwan KY, Moens PB, Wang JC. Infertility and aneuploidy in mice lacking a type IA DNA topoisomerase III beta. Proc. Natl Acad. Sci. USA. 2003;100:2526–2531. doi: 10.1073/pnas.0437998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Pommier Y. Mitochondrial Topoisomerase I Sites in the Regulatory D-Loop Region of Mitochondrial DNA. Biochemistry. 2008;47:11196–203. doi: 10.1021/bi800774b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartung F, Puchta H. Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene. 2001;271:81–86. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- 58.Hartung F, Angelis KJ, Meister A, Schubert I, Melzer M, Puchta H. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 2002;12:1787–1791. doi: 10.1016/s0960-9822(02)01218-6. [DOI] [PubMed] [Google Scholar]
- 59.Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 2002;12:1782–1786. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- 60.Sugimoto-Shirasu K, Roberts GR, Stacey NJ, McCann MC, Maxwell A, Roberts K. RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc. Natl Acad. Sci. USA. 2005;102:18736–18741. doi: 10.1073/pnas.0505883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breuer C, Stacey NJ, West CE, Zhao Y, Chory J, Tsukaya H, Azumi Y, Maxwell A, Roberts K, Sugimoto-Shirasu K. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell. 2007;19:3655–3656. doi: 10.1105/tpc.107.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirik V, Schrader A, Uhrig JF, Hulskamp M. MIDGET unravels functions of the Arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell. 2007;19:3100–3110. doi: 10.1105/tpc.107.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 64.Huang J, Gogarten JP. Ancient horizontal gene transfer can benefit phylogenetic reconstruction. Trends Genet. 2006;22:361–366. doi: 10.1016/j.tig.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 65.Malik SB, Ramesh MA, Hulstrand AM, Logsdon JM. Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol. Biol. Evol. 2007;24:2827–2841. doi: 10.1093/molbev/msm217. [DOI] [PubMed] [Google Scholar]
- 66.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view, Nat. Rev. Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 67.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 68.Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 69.Claverie JM. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koonin EV, Senkevitch TG, Dolja VV. The ancient Virus World and evolution of cells. Biol. Direct. 2006;1:19. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suttle CA. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 72.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 73.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 74.Benarroch D, Claverie JM, Raoult D, Shuman S. Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria. J. Virol. 2006;80:314–321. doi: 10.1128/JVI.80.1.314-321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forterre P, Filée J, Myllykallio H. Origin and evolution of DNA and DNA replication machineries. In: Ribas L, editor. The Genetic Code and the Origin of Life. Landes Bioscience / Eurekah.com, Georgetown, Tex.; 2004. pp. 145–168. [Google Scholar]
- 76.Filée J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 77.Forterre P. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 2002;5:525. doi: 10.1016/s1369-5274(02)00360-0. [DOI] [PubMed] [Google Scholar]
- 78.Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 79.Koepsel RR, Murray RW, Rosenblum WD, Khan SA. The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc. Natl Acad. Sci. USA. 1985;82:6845–6849. doi: 10.1073/pnas.82.20.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marsin S, Marguet E, Forterre P. Topoisomerase activity of the hyperthermophilic replication initiator protein Rep75. Nucleic Acids Res. 2000;28:2251–2255. doi: 10.1093/nar/28.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pansegrau W, Schröder W, Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 82.Jo K, Topal MD. DNA topoisomerase and recombinase activities in Nae I restriction endonuclease. Science. 1995;267:1817–1820. doi: 10.1126/science.7892605. [DOI] [PubMed] [Google Scholar]
- 83.Thomsen ND, Berger JM. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol. Microbiol. 2008;69:1071–1090. doi: 10.1111/j.1365-2958.2008.06364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jain P, Nagaraja V. An atypical type II topoisomerase from Mycobacterium smegmatis with positive supercoiling activity. Mol. Microbiol. 2005;58:1392–1405. doi: 10.1111/j.1365-2958.2005.04908.x. [DOI] [PubMed] [Google Scholar]
- 85.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain. Proc. Natl Acad. Sci. USA. 2006;103:3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poole AM, Logan DT. Modern mRNA proofreading and repair: clues that the last universal common ancestor possessed an RNA genome? Mol. Biol. Evol. 2005;22:1444–1455. doi: 10.1093/molbev/msi132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Forterre P, Gribaldo S. The origin of modern terrestrial life. HFSP J. 2007;1:156–168. doi: 10.2976/1.2759103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yin Y, Fischer D. Identification and investigation of ORFans in the viral world. BMC Genomics. 2008;9:24. doi: 10.1186/1471-2164-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 92.Daubin V, Lerat E, Perrière G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hansen G, Harrenga A, Wieland B, Schomburg D, Reinemer P. Crystal structure of full length topoisomerase I from Thermotoga maritima. J. Mol. Biol. 2006;358:1328–1340. doi: 10.1016/j.jmb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 94.Rodríguez AC, Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Patel A, Shuman S, Mondragón A. Crystal structure of a bacterial type IB DNA topoisomerase reveals a preassembled active site in the absence of DNA. J. Biol. Chem. 2006;281:6030–6037. doi: 10.1074/jbc.M512332200. [DOI] [PubMed] [Google Scholar]
- 96.Bellon S, Parsons JD, Wei Y, Hayakawa K, Swenson LL, Charifson PS, Lippke JA, Aldape R, Gross CH. Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase. Antimicrob. Agents Chemother. 2004;48:1856–1864. doi: 10.1128/AAC.48.5.1856-1864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 98.Graille M, Cladière L, Durand D, Lecointe F, Gadelle D, Quevillon-Cheruel S, Vachette P, Forterre P, van Tilbeurgh H. Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms. Structure. 2008;16:360–370. doi: 10.1016/j.str.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 99.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]