Abstract
How do proteins accomplish folding during early evolution? Theoretically the mechanism involves the selective stabilization of the native structure against all other competing compact conformations in a process that involves cumulative changes in the amino acid sequence along geological timescales. Thus, an evolved protein folds into a single structure at physiological temperature, but the conformational competition remains latent. For natural proteins such competition should emerge only near cryogenic temperatures, which places it beyond experimental testing. Here, we introduce a designed monomeric miniprotein (FSD-1ss) that within biological temperatures (330–280 K) switches between simple fast folding and highly complex conformational dynamics in a structurally degenerate compact ensemble. Our findings demonstrate the physical basis for protein folding evolution in a designed protein, which exhibits poorly evolved or primordial folding. Furthermore, these results open the door to the experimental exploration of primitive folding and the switching between alternative protein structures that takes place in evolutionary branching points and prion diseases, as well as the benchmarking of de novo design methods.
Keywords: energy landscape, glassy dynamics, molecular evolution, protein design, protein folding
Polypeptide chains with random sequences reflecting natural amino acid compositions are expected to form compact states in aqueous solution to minimize hydrophobic solvent exposure. Because it is impossible to simultaneously satisfy all of the potential interactions in the polypeptide chain, these compact states are structurally degenerate and, in analogy to random heteropolymers (1), should exhibit glassy conformational dynamics. The trademarks of such dynamics are highly stretched exponential decays and a superArrhenius temperature-dependent relaxation rate that experiences an abrupt slow down below a characteristic temperature (here termed Tg) (2). Naturally evolved protein domains, however, fold into unique 3D structures efficiently and with simple exponential kinetics (3). How natural selection drives the transformation from random polypeptides to well-evolved folding proteins is one of the major unresolved fundamental questions in modern biochemistry.
A widely held view has been to assume a thermodynamic criterion, namely that natural folding only requires the native structure to be the lowest energy state in the vast protein conformational space. The thermodynamic criterion has led to relative success in ab initio prediction of native structures (4), as well as in computer-guided de novo protein design (5). However, it has been recently highlighted that artificial proteins catalogued as successful designs may exhibit complex folding kinetics (6). In an alternative mechanism inspired by statistical mechanics the role of early evolution is to produce natively biased (7) or funneled (2) folding energy landscapes by selecting amino acid sequences that maximize the stability gap between the native and all other compact structures (8). Thus, an evolved natural protein domain folds with simple exponential kinetics to a unique structure at biological temperatures (i.e., slightly below its thermal denaturation midpoint, Tm), whereas the heteropolymer-like regime emerges only at very low temperature (Tg ≪ Tm) (2).
This physical mechanism for the evolution of protein folding has been thoroughly studied in computer simulations of simplified protein models (9). The big challenge is to test the mechanism experimentally. Such a test would entail resolving the switch from simple folding kinetics into a unique 3D structure to glassy dynamics in a structurally degenerate ensemble upon decreasing temperature. Theoretical estimates set Tm/Tg ratios of ≈1.6 (10) or higher (11) for natural proteins. Given typical protein thermal stabilities (12), these estimates place Tg below the temperatures at which protein–water–glycerol mixtures form glasses (13) and thus beyond experimental reach. Accordingly, efforts to detect glassy dynamics slightly below the water freezing point in the 2-state folding protein L have failed (14). On the other hand, there have been reports of stretched kinetic decays in laser temperature-jump (15) and single-molecule force-clamp unfolding (16) experiments of other proteins.
A de Novo Designed Protein as a Model of Primordial Folding.
An alternative to natural proteins is to investigate the folding behavior of poorly evolved proteins, which by definition should have much smaller Tm/Tg (17). As a model of poorly evolved folding, we use here a small de novo designed protein. Particularly, we focus on a simple α + β scaffold corresponding to the natural zinc-finger domain Zif268 (Fig. 1Bottom Left), which was then used by Dahiyat and Mayo (18) as a starting point for the first computer-generated de novo designed protein (FSD-1) (Fig. 1 Bottom Center). FSD-1 was designed with the energy of the target structure as the only fitness criterion (18); it only shares 6 residues with Zif268 and has an unusually large content in aromatic residues. These features hint at a Tm/Tg below that of natural proteins. As another advantage, from its small size (28 residues), FSD-1 is expected to exhibit simple ultrafast folding kinetics (19). The problem is that the accessible temperature range for FSD-1 is very narrow due to its marginal thermal stability (Tm ≈ 295 K) (18).
Fig. 1.
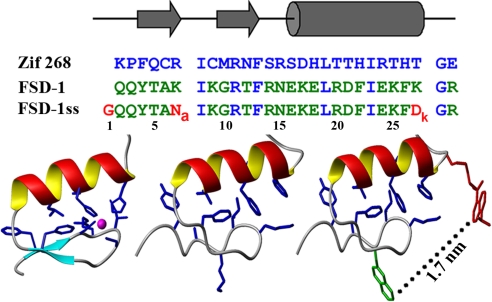
Amino acid sequences and structures of FSD-1ss and its predecessors. Sequences are lined up with the secondary structure assignment. The numbering scheme follows the FSD-1ss sequence, thus being shifted 1 position in the other 2 proteins. This numbering scheme is maintained throughout. The 3 structures are shown at the bottom (Zif268, FSD-1, and FSD-1ss from left to right) by using a ribbon backbone representation. Blue corresponds to critical core residues, green corresponds to Nal, and red corresponds to DnK in FSD-1ss. The magenta sphere shows the position of the zinc atom in Zif268.
For our experiments we introduced 2 nonnatural aromatic amino acids [naphthyl-alanine (Nal) and dansyl-lysine (DnK)] in the original sequence of FSD-1. The fluorescent aromatic amino acids were incorporated as a FRET pair to monitor protein compactness and were intrusively placed inside the protein structure (see Fig. 1 Bottom Right) rather than in floppy tails. The new version (FSD-1ss) is synthetic in 2 ways: Its amino acid sequence was designed de novo, and it includes nonnatural amino acids within its folded structure. The Nal–DnK pair placed in the FSD-1 structure should result in ≈90% FRET efficiency (i.e., with an Ro value of ≈2.5 nm, a k2 value of 2/3, and an ≈1.7-nm distance between probes) (see Fig. 1), providing a highly sensitive probe of protein conformational changes.
Natural-Like Folding Near the Denaturation Midpoint.
At low temperatures the FSD-1ss far-UV CD spectrum indicates a mix of α-helix, β-sheet, and coil (Fig. 2A), and FRET efficiency is ≈90% (Fig. 2C). The protein unfolds upon increasing temperature (Fig. 2A) and/or addition of urea (Fig. 2B), following typical sigmoidal curves with the broadness expected for natural proteins of similar size. The sigmoidicity is better appreciated in the curve derivative, which shows a clear peak at the midpoint (e.g., Fig. 2D). Both CD and FRET thermal denaturation experiments show a pronounced pretransition slope [Fig. 2C and supporting information (SI) Fig. S1]. FSD-1ss is very stable, requiring ≈6.5 M urea (Fig. 2B) or ≈355 K to be half-unfolded when monitored both by CD (Fig. S1) and FRET (Fig. 2D). Addition of urea decreases the Tm monotonically (Fig. 2D). Therefore, FSD-1ss exhibits equilibrium unfolding typical of small natural proteins and is much more stable than the parent FSD-1 (i.e., Tm ≈ 60 K higher).
Fig. 2.
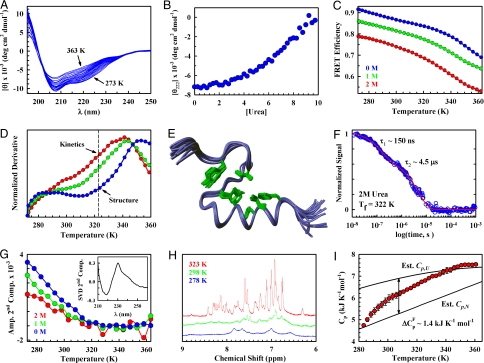
Folding and structural properties of FSD-1ss. (A) Equilibrium thermal unfolding by far-UV CD. (B) Equilibrium urea unfolding by CD at 222 nm. (C) Equilibrium thermal unfolding by FRET: 0 M (blue), 1 M (green), and 2 M (red) urea. (D) Derivative of the FRET equilibrium thermal unfolding curves shown in C. Color coding is as shown in C. (E) Superposition (backbone of residues 3–26) of the 20 lowest-energy NMR structures at 323 K. Core side chains are shown in green. (F) Relaxation decay after a ≈10 K ns T jump to 322 K final temperature in the presence of 2 M urea. The red curve is the best double exponential fit. (G) Singular value decomposition (SVD) of far-UV CD spectra as a function of temperature and urea. (Inset) The second component (Comp.). The main graph shows the second component's amplitude (Amp.) for 0, 1, and 2 M urea. Color coding is as shown in C. (H) One-dimensional NMR spectrum of the proton amide region: 323 K (red), 298 K (green), and 278 K (blue). (I) DSC thermogram in absolute heat capacity units showing the empirical estimates of the heat capacity baselines for the folded and unfolded states (SI Text). Error bars indicate the experimental errors of 4 runs.
We also determined the NMR structure of FSD-1ss at the highest temperature still below the unfolding transition (i.e., 323 K) (Fig. 2D). The NMR structure (PDB ID code 2K6R) shows the expected overall topology with an α-helix packed against 2 short strands connected by a loop (Fig. 2E). The structure is very well defined for a protein of such small size (Table S1). The superposition of the ensemble of 20 structures results in rmsd of 0.04 nm for the backbone heavy atoms of residues 3–26. Moreover, the side chains of the hydrophobic core residues overlay almost on top of each other (Fig. 2E). PROCHECK provides another indicator of the high structural quality with >90% of the residues in the most favorable regions of the Ramachandran plot and 0% in disallowed regions.
To determine the (un)folding kinetics of FSD-1ss near its Tm, we used a FRET nanosecond laser-induced temperature-jump instrument (20). Fig. 2F shows the folding relaxation of FSD-1ss after a T jump of ≈10 K to 323 K in the presence of 2 M urea. Under these conditions the protein is slightly below the Tm (see Fig. 2D), and thus the resulting relaxation is still dominated by the folding rate. The relaxation is very fast and can be well fit to a double exponential decay with relaxation times of 150 ns and 4.5 μs (Fig. 2F). Biphasic folding-relaxation decays with one phase in the hundreds of nanoseconds and another in the few microseconds are common in ultrafast folding proteins, such as the villin headpiece subdomain (21) or the albumin binding domain prb (22). In fact, 4.5 μs at 322 K place FSD-1ss at the top of the ultrafast folding chart (23). Thus, FSD-1ss exhibits natural-like simple ultrafast folding kinetics near its Tm.
Structural Rearrangements at Low Temperatures.
Below 320 K rather unusual behavior is observed. Far-UV CD indicates progressive increases in backbone secondary structure from 320 to 273 K (Fig. 2A). FRET efficiency also increases, suggesting more collapse (Fig. 2C). Moreover, the second component of CD singular value decomposition reveals changes in the tertiary environment of the Phe residues restricted to the 273- to 320-K range (Fig. 2G). The characteristic Phe negative band at ≈220- to 230-nm shifts to the blue as temperature decreases, indicating less solvent exposure. Such reorganization of the FSD-1ss protein core is reduced by addition of urea (Fig. 2G). Furthermore, the 1D NMR amide proton spectrum exhibits line broadening below 323 K. At 278 K the spectrum is so broadened that only a few signals can be detected (Fig. 2H). NMR line-broadening indicates structural reorganizations (i.e., large alterations in the electronic environment of amide protons) taking place in the 50- to 500-μs timescale. This behavior is entirely reversible and independent of protein concentration from 50 μM to 2 mM, indicating that FSD-1ss does not aggregate under our experimental conditions.
Differential scanning calorimetry (DSC) produces a broad thermogram without defined peaks (Fig. 2I). The thermogram shape is similar to that of apomyoglobin (24) but is also consistent with FSD-1ss's small size [little unfolding enthalpy (12)] and ultrafast folding [downhill regime (23)]. Furthermore, although broad, the FSD-1ss thermogram agrees very well with the empirical baselines for folded and unfolded states calculated from its sequence (Fig. 2I). This agreement allows the estimate of an average change in heat capacity on unfolding (ΔCp) of ≈1.4 kJ/mol. The thermogram also shows a heat-capacity shoulder in the same temperature interval as the changes in Phe tertiary environment (Fig. 2 I and G), consistent with the presence of conformational changes at low temperature.
The degree of NMR line-broadening shown in Fig. 2H is reminiscent of the onset of cold denaturation (25). In this case, however, cold denaturation is ruled out by other observations: (i) CD and FRET indicate that the protein becomes more structured and compact as temperature lowers; (ii) the low-temperature heat capacity is consistent with the empirical folded-state baseline (Fig. 2I) (SI Text); (iii) addition of urea eliminates the low-temperature changes rather than enhancing them (see Fig. 2G); and (iv) given ΔCp ≈ 1.4 kJ/mol and Tm ≈ 355 K, and assuming a standard enthalpy per residue of 2.7 kJ/mol at 333 K (12), cold denaturation should only occur below 220 K. Therefore, we can conclude that, below 320 K, FSD-1ss undergoes conformational transitions in a structurally degenerate compact ensemble. The structural changes involve both backbone and core rearrangements that produce large differences in amide proton chemical shifts, presumably because of strong ring-current effects from the many aromatic residues.
Glassy Conformational Dynamics.
We performed additional laser T jump experiments at lower temperatures. The first interesting observation is that the relaxation decay, after a T jump of ≈10 K to 310 K, is not single or double exponential but requires fitting to a stretched exponential function [i.e., exp((t/τ)β)] with τ ≈ 8 μs and β ≈ 0.55 (Fig. 3A). The degree of stretching in the relaxation increases as temperature decreases. This trend can be appreciated in Fig. 3A, which shows decays after ≈10-K jumps to final temperatures between 310 and 279 K together with best fits to single exponential decays for reference (thin black curves). Fits to a stretched exponential function are good at all temperatures (Fig. 3A, red curves) with a fitted β parameter that decreases to 0.33 at 279 K. At this low temperature the relaxation is highly stretched, extending for at least 5–6 time decades (Fig. 3A). A classical multiexponential approach requires >7 exponentials to produce fits of similar quality. This relaxation is in fact close to the superstretched structural relaxation of native myoglobin after CO photodissociation (26).
Fig. 3.
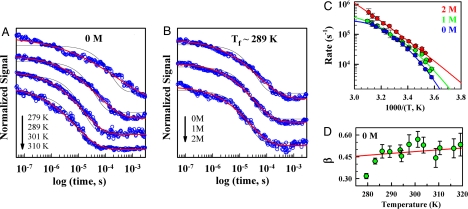
Glassy conformational dynamics in FSD-1ss. (A) Relaxation decays after ≈10-K jumps to final temperatures from 310 K (bottom trace) to 279 K (top trace) without urea. The red curves are best-stretched exponential fits and the thin black lines are best exponential fits for reference. (B) Decays as in A except ≈10-K jumps are to a final temperature of 289 K in 0, 1, and 2 M urea (from top to bottom). (C) Arrhenius plot of the inverse relaxation time obtained from stretched exponential fits: 0 M (blue), 1 M (green), and 2 M (red) urea. All data points correspond to folding conditions (<Tm). The curves are fits to a quadratic (superArrhenius) equation. Similar quality fits are obtained with a transition-state equation including an activation heat capacity term (SI Text). (D) Stretching parameter β as a function of temperature in the absence of urea. Error bars indicate fitting errors. The red curve is a fit to the Xie and Wolynes equation (30).
A strongly nonlinear temperature dependence is also apparent in the 4 decays shown in Fig. 3A with relaxation times of 8, 15, 50, and 330 μs for final temperatures from 310 K to 279 K in ≈10-K steps. The highly curved temperature dependence is evident in an Arrhenius plot (Fig. 3C, blue curve). Curved Arrhenius plots need to be interpreted with caution because the folding rate of 2-state folding proteins also shows curvature arising from the difference in heat capacity between native and folding transition states (27, 28). However, the curvature in FSD-1ss is much too pronounced to be accounted for by heat capacity changes. Fitting the 0 M data (Fig. 3C, blue curve) to a thermodynamic transition-state rate expression requires a folding activation heat capacity of 2.2 ± 0.22 kJ/(mol·K) (SI Text). This folding activation heat capacity is significantly larger than the total equilibrium change for FSD-1ss (Fig. 2I) and at least 3-fold larger than expected for 2-state folding [i.e., ≈40% of the equilibrium heat capacity change (28) or 0.56 kJ/(mol·K) for FSD-1ss]. The 2.2 kJ/(mol·K) value is indeed comparable with the folding activation heat capacity of the 3.7-times-larger protein FKBP-12 (29).
The addition of the chemical denaturant urea monotonically decreases the degree of relaxation stretching (see the decreasing deviations from single exponential fits in Fig. 3B), speeds up the relaxation rate in folding conditions (instead of the slowing down of 2-state folding), and drastically decreases the curvature of the Arrhenius plot (Fig. 3C, green and red curves). For example, the curvature at 2 M urea corresponds to only 0.4 ± 0.05 kJ/(mol·K) in folding activation heat capacity, more in line with the value expected for a 29-residue 2-state folder (28). The effects of urea in FSD-1ss are therefore incompatible with classical activated folding kinetics. In contrast, all of these observations are straightforwardly explained as a decrease in glassiness produced by the urea-induced smoothing of the rough FSD-1ss folding landscape.
Estimating the Roughness of the FSD-1ss Folding Landscape.
To estimate the roughness of the FSD-1ss folding landscape, we use the empirically determined stretching parameter β (Fig. 3D) and equation β = [1 + (δF/RT)2]−1/2 (30), where δF2 is the free-energy variance between adjacent conformations in the energy landscape. This calculation renders a roughness of (≈4.5 kJ/mol)2 for FSD-1ss in the absence of urea. Such an empirical estimate of landscape roughness provides a very useful benchmark for computer simulations, which could then be used to further investigate the relationship between glassy conformational dynamics and landscape roughness in atomistic simulations of FSD-1ss. It is also interesting to note that the experimental data follow well the theoretical relation at temperatures above 285 K but deviate quite apparently below (Fig. 3D). The deviation below 285 K can be interpreted as signaling the dynamic glass transition, resulting in an estimate of Tm/Tg of ≈1.3 for the synthetic superstable FSD-1ss miniprotein. A ratio of 1.3 is much lower than the estimates for natural proteins (11) and thus reminiscent of poorly evolved proteins.
Structural Basis for Degenerate Folding.
What are the structural determinants of FSD-1ss folding behavior at low temperature? Both FSD-1 (at 274 K) and FSD-1ss (at 323 K) have well-resolved NMR structures with little conformational heterogeneity in backbone and core side chains (e.g., Fig. 2E). Overall, the 2 structures are similar, but detailed inspection uncovers critical differences (Fig. 4A). The α-helix and several core side chains (F13, L19, and I23) (see Fig. 4A) superimpose on top of each other, whereas there are large changes in the N-terminal backbone region together with rotamer changes and spatial displacements of the other core side chains (Fig. 4A).
Fig. 4.
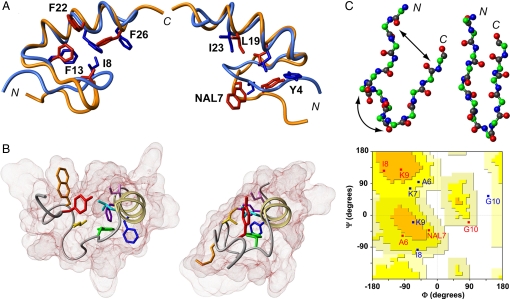
Structural basis for degenerate folding. (A) Superposition of FSD-1ss lowest-energy NMR structure at 323 K (red structures) and the averaged minimized FSD-1 structure at 275 K (blue structures) (18). The 2 views highlight different subsets of the main hydrophobic core residues. Numbering and sequences are as shown in Fig. 1B. (B) Comparison between FSD-1ss (Left) and FSD-1 (Right) structures. The backbone is shown as a ribbon, and the van der Waals surfaces are shown in shades of brown. The core side chains are colored according to the spectrum of light to indicate sequence order (from red to purple). (C) Comparison of backbone conformation in the N-terminal segment of FSD-1ss (Upper Left) and FSD-1 (Upper Right), by using a ball and sticks representation. The arrows highlight the structural differences. (Lower) Ramachandran plot showing the dihedral angle values of residues 6–10 in FSD-1ss (red) and FSD-1 (blue).
The most dramatic difference is an ≈180° flip of the side chain in position 7, Nal in FSD-1ss versus Lys in FSD-1 (see Fig. 1). In the FSD-1 structure, K7 is on the solvent-exposed face of the β-hairpin-like structure (Fig. 4B Right, orange residue). The highly hydrophobic character of Nal7, however, forces a backbone rearrangement to minimize its solvent exposure (Fig. 4B Left, orange residue), resulting in a nonoptimized structure. This finding is highlighted by comparison with the original FSD-1 structure. The protein core of FSD-1 is well-packed with all aromatics residues contained within the helix–hairpin interface, resulting in a prolate ellipsoidal structure (Fig. 4B Right). In contrast, the FSD-1ss structure is more open and spherical (Fig. 4B Left). The Nal7 flip opens the FSD-1ss core and pushes out the other aromatic residues (Fig. 4B; red, blue, and purple residues) to make room for the additional hydrophobe. Accessible surface-area calculations indicate that the real NMR structure of FSD-1ss buries more hydrophobic surface than its sequence modeled in the FSD-1 structure (Fig. 1 Right), explaining why it is indeed preferred. However, the overall gain comes at the expense of partially exposing F22 and F26 to the solvent. In other words, finding the best packing arrangement involves sacrificing the local environment of several hydrophobic residues.
The changes in core packing are accompanied by similarly drastic backbone rearrangements in the N-terminal region (Fig. 4C). Most notorious are the changes in the turn region connecting the 2 strands. For FSD-1, the first strand extends to K7, with the turn being placed at I8-K9. In FSD-1ss, the first strand is shorter and a turn-like conformation appears at A6-Nal7 to accommodate the Nal7 flip (see Fig. 4C Bottom). The shift in turn-register allows the formation of many hydrophobic contacts between Nal7 and I8 (observed as NOEs) but also displaces the 2 strands farther apart, impeding the formation of β-sheet-like hydrogen bonds (Fig. 4C Top Left). Therefore, there is backbone frustration in the N-terminal region of FSD-1ss, which cannot simultaneously satisfy the β-sheet hydrogen bonds and local hydrophobic contacts between Nal7 and I8.
Lessons for Protein Evolution and de Novo Design
Here we report the temperature-controlled switching of designed protein FSD-1ss from fast-folding into a single structure to glassy dynamics in a nonaggregating structurally degenerate compact ensemble. These results provide direct experimental demonstration of the inherent competition between the native and alternative structures during protein folding and evolution. Structural analysis indicates that the structural degeneration of FSD-1ss stems from a conflict between the backbone propensity to form hydrogen bonds and the efforts of different hydrophobic residues to maximize their insufficient packing. At low temperature these forces balance out, leading to a structurally degenerate ensemble, but as temperature rises the increasingly strong hydrophobic effect wins over selecting a single structure before unfolding takes place. Increasingly stretched exponential kinetics at low temperature have also been reported for short chains (8–12 residues) of phenylacetylene that form stacked helical structures in polar solvents (31), revealing striking similarities between the conformational behavior of poorly evolved proteins and much simpler synthetic homopolymers.
The folding properties of FSD-1ss have implications for protein design. De novo design strategies have frequently focused on matching amino acid sequences to a target backbone structure. It is interesting that substituting a single solvent-exposed Lys with Nal results in a more stable protein that, however, folds into a different structure. The result highlights the delicate interplay between stability and specificity when designing proteins and reinforces the importance of implementing negative selection during the design strategy. In this light, we could expect current successful de novo designs to exhibit more “evolved” folding than FSD-1ss (which contains intrusive nonnatural aromatic amino acids) but still less than natural proteins. This observation is consistent with the complex kinetics observed in α3d at low temperature (32) and in Top-7 (6), which are 2 proteins designed with entirely natural amino acid composition.
Finally, interpreting our results in evolutionary terms, we can conclude that FSD-1ss is analogous to a primordial or poorly evolved protein. Although FSD-1ss folds into a single structure and is very stable, the glassy regime is already present at physiological temperatures, making it inapt to perform classical biological functions. The narrow gap between folding and structural degeneration should also produce hypersensitivity to mutation (9). Turning FSD-1ss into a better-evolved or more “natural” protein involves decreasing the temperature at which the structurally degenerate glassy regime emerges (larger Tm/Tg). Our experiments suggest various strategies. For example, one could target the more stable structure (Fig. 4B Left) by reducing the bulkiness of some hydrophobic residues (e.g., F22 and F26) and retuning the sequence of residues 6–10 to decrease its local propensity to form the competing β-hairpin (Fig. 4C). Alternatively, one could target the original FSD-1 structure (Fig. 4B Right), removing Nal7 and further stabilizing the hairpin structure on its solvent-exposed face. The strong effect caused by Nal on FSD-1ss also offers some clues regarding the old question of what is the right hydrophobic balance to achieve protein folding. Nal is a 2-member-ring amino acid that is slightly bulkier and more hydrophobic than its aromatic natural counterparts (Phe, Tyr, and Trp), which could suggest that the 20-amino-acid natural alphabet has been carefully selected during early evolution to maintain the aromatic–hydrophobic protein content below a certain threshold, above which folding becomes a very hard design selection problem.
Therefore, FSD-1ss appears as an excellent model case to further explore the early stages of protein evolution via experiment. Moreover, FSD-1ss and FSD-1 fold into quite different structures (Fig. 4), with only 2 amino acid changes and, thus, offer a unique opportunity to investigate the physical transition from one native structure to another through sequence or environment manipulation. This investigation could be seen both as an experimental recreation of evolutionary branching points and as a simple model to mimic conformational switches such as those characteristic of prion diseases (33).
Experimental Procedures
Protein concentration was measured at 280 nm by using as a molar-extinction coefficient the sum of Nal (5,526 M−1·cm−1), DnK (1,571 M−1·cm−1), and tyrosine (1,280 M−1·cm−1). CD experiments were performed at a protein concentration of 50 μM in 20 mM citrate buffer, pH 5.0, at different urea concentrations. Equilibrium FRET experiments were performed at a protein concentration of 25 μM in 20 mM acetate buffer, pH 5.0. FRET efficiencies were determined from the donor fluorescence intensity by using a Nal sample as reference. DSC experiments were carried out in 20 mM acetate buffer, pH 5.0, as described previously (34). NMR samples were prepared in 90:10 (vol:vol) H2O/2H2O at pH 5.0. Nanosecond laser-induced temperature-jump experiments were performed as described in ref. 20. A more detailed methods description, the structure calculation procedures, and structure statistics are all included in SI Text, Figs. S1 and S2, and Table S1.
Supplementary Material
Acknowledgments.
This work has been supported by National Science Foundation Grant MCB-0317294 and Marie Curie Excellence Award MEXT-CT-2006-042334.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2K6R).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812108106/DCSupplemental.
References
- 1.Bryngelson JD, Wolynes PG. Intermediates and barrier crossing in a random energy-model (with applications to protein folding) J Phys Chem. 1989;93:6902–6915. [Google Scholar]
- 2.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding—A synthesis. Proteins Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SE. How do small single-domain proteins fold? Folding Des. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 4.Bonneau R, Baker D. Ab initio protein structure prediction: Progress and prospects. Annu Rev Biophys Biomol Struct. 2001;30:173–189. doi: 10.1146/annurev.biophys.30.1.173. [DOI] [PubMed] [Google Scholar]
- 5.Butterfoss GL, Kuhlman B. Computer-based design of novel protein structures. Annu Rev Biophys Biomol Struct. 2006;35:49–65. doi: 10.1146/annurev.biophys.35.040405.102046. [DOI] [PubMed] [Google Scholar]
- 6.Watters AL, et al. The highly cooperative folding of small naturally occurring proteins is likely the result of natural selection. Cell. 2007;128:613–624. doi: 10.1016/j.cell.2006.12.042. [DOI] [PubMed] [Google Scholar]
- 7.Go N. The consistency principle in protein structure and pathways of folding. Adv Biophys. 1984;18:149–164. doi: 10.1016/0065-227x(84)90010-8. [DOI] [PubMed] [Google Scholar]
- 8.Sali A, Shakhnovich EI, Karplus M. How does a protein fold? Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 9.Shakhnovich EI. Protein folding thermodynamics and dynamics: Where physics, chemistry, and biology meet. Chem Rev. 2006;106:1559–1588. doi: 10.1021/cr040425u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 11.Chan HS, Shimizu S, Kaya H. Cooperativity principles in protein folding. Methods Enzymol. 2004;380:350–379. doi: 10.1016/S0076-6879(04)80016-8. [DOI] [PubMed] [Google Scholar]
- 12.Robertson AD, Murphy KP. Protein structure and the energetics of protein stability. Chem Rev. 1997;97:1251–1267. doi: 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- 13.Angell CA. Formation of glasses from liquids and biopolymers. Science. 1995;267:1924–1935. doi: 10.1126/science.267.5206.1924. [DOI] [PubMed] [Google Scholar]
- 14.Gillespie B, Plaxco KW. Nonglassy kinetics in the folding of a simple single-domain protein. Proc Natl Acad Sci USA. 2003;97:12014–12019. doi: 10.1073/pnas.97.22.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabelko J, Ervin J, Gruebele M. Observation of strange kinetics in protein folding. Proc Natl Acad Sci USA. 1999;96:6031–6036. doi: 10.1073/pnas.96.11.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brujic J, Hermans RI, Walther KA, Fernandez JM. Single-molecule force spectroscopy reveals signatures of glassy dynamics in the energy landscape of ubiquitin. Nat Phys. 2006;2:282–286. [Google Scholar]
- 17.Wolynes PG. In: Protein Folding, Misfolding and Aggregation. Muñoz V, editor. Cambridge, UK: R Soc Chem; 2008. pp. 49–69. [Google Scholar]
- 18.Dahiyat BI, Mayo SL. De novo protein design: Fully automated sequence selection. Science. 1995;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 19.Naganathan AN, Muñoz V. Scaling of folding times with protein size. J Am Chem Soc. 2005;127:480–481. doi: 10.1021/ja044449u. [DOI] [PubMed] [Google Scholar]
- 20.Sadqi M, Lapidus LJ, Muñoz V. How fast is protein hydrophobic collapse? Proc Natl Acad Sci USA. 2003;100:12117–12122. doi: 10.1073/pnas.2033863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubelka J, Eaton WA, Hofrichter J. Experimental tests of villin subdomain folding simulations. J Mol Biol. 2003;329:625–630. doi: 10.1016/s0022-2836(03)00519-9. [DOI] [PubMed] [Google Scholar]
- 22.Vu DM, Myers JK, Oas TG, Dyer RB. Probing the folding and unfolding dynamics of secondary and tertiary structures in a three-helix bundle protein. Biochemistry. 2004;43:3582–3589. doi: 10.1021/bi036203s. [DOI] [PubMed] [Google Scholar]
- 23.Naganathan AN, Doshi U, Fung A, Sadqi M, Muñoz V. Dynamics, energetics, and structure in protein folding. Biochemistry. 2006;45:8466–8475. doi: 10.1021/bi060643c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griko YV, Privalov PL. Thermodynamic puzzle of apomyoglobin unfolding. J Mol Biol. 1994;235:1318–1325. doi: 10.1006/jmbi.1994.1085. [DOI] [PubMed] [Google Scholar]
- 25.Babu CR, van Hilser VJ, Wand AJ. Direct access to the cooperative substructure of proteins and the protein ensemble via cold denaturation. Nat Struct Biol. 2004;11:352–357. doi: 10.1038/nsmb739. [DOI] [PubMed] [Google Scholar]
- 26.Lim MH, Jackson TA, Anfinrud PA. Nonexponential protein relaxation—dynamics of conformational change in myoglobin. Proc Natl Acad Sci USA. 1993;90:5801–5804. doi: 10.1073/pnas.90.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveberg M, Tan YJ, Fersht AR. Negative activation enthalpies in the kinetics of protein-folding. Proc Natl Acad Sci USA. 1995;92:8926–8929. doi: 10.1073/pnas.92.19.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akmal A, Muñoz V. The nature of the free energy barriers to two-state folding. Proteins Struct Funct Bioinf. 2004;57:142–152. doi: 10.1002/prot.20172. [DOI] [PubMed] [Google Scholar]
- 29.Main ERG, Fulton KF, Jackson SE. Folding pathway of FKBP 12 and characterisation of the transition state. J Mol Biol. 1999;291:429–444. doi: 10.1006/jmbi.1999.2941. [DOI] [PubMed] [Google Scholar]
- 30.Xie X, Wolynes PG. Microscopic theory of heterogeneity and nonexponential relaxations in supercooled liquids. Phys Rev Let. 2001;86:5526–5529. doi: 10.1103/PhysRevLett.86.5526. [DOI] [PubMed] [Google Scholar]
- 31.Yang WY, Prince RB, Sabelko J, Moore JS, Gruebele M. Transition from exponential to nonexponential kinetics during formation of a nonbiological helix. J Am Chem Soc. 2000;122:3248–3249. [Google Scholar]
- 32.Zhu Y, et al. Ultrafast folding of alpha(3): A de novo designed three-helix bundle protein. Proc Natl Acad Sci USA. 2003;100:15486–15491. doi: 10.1073/pnas.2136623100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly JW. The environmental dependency of protein folding best explains prion and amyloid diseases. Proc Natl Acad Sci USA. 1998;95:930–932. doi: 10.1073/pnas.95.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzman-Casado M, Parody-Morreale A, Robic S, Marqusee S, Sanchez-Ruiz JM. Energetic evidence for formation of a pH-dependent hydrophobic cluster in the denatured state of Thermus thermophilus ribonuclease H. J Mol Biol. 2003;329:731–743. doi: 10.1016/s0022-2836(03)00513-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.