Abstract
The genetic basis for the Ara-C resistance of CCRF-CEM Ara-C/8C leukemia cells was investigated. DNA sequencing revealed that these cells expressed an equilibrative nucleoside transporter 1 (ENT1) with a single missense mutation resulting in glycine to arginine replacement (G24R). To test the importance of this residue, additional G24 mutants were created and examined for [3H]-uridine and [3H]-Ara-C uptake. Both a G24E and G24A mutant showed reduced ENT1-dependent activity. An EGFP-tagged G24R ENT1 displayed plasma membrane localization even though it was unable to bind [3H]-NBMPR, an ENT1-specific inhibitor. These results define G24 as critical amino acid for ENT1 nucleoside uptake and suggest that mutations in TM1 may provide a mechanism for Ara-C resistance in CCRF-CEM Ara-C/8C cells.
Keywords: equilibrative nucleoside transporter, AraC, drug resistance, missense mutation, leukemia cells
Introduction
Cytarabine (Ara-C), a pyrimidine analog, is a conventional anti-cancer chemotherapeutic commonly used for the treatment of acute myeloid leukemia. Ara-C is a hydrophilic molecule that achieves intracellular penetration via nucleoside transporter proteins. Once inside the cell, this agent exerts cytotoxic effects on proliferating cancer cells by the direct inhibition of DNA polymerases and the arrest of DNA synthesis. Although Ara-C treatment often induces partial or complete remission of cancerous tissue, many patients eventually develop resistance [1]. Thus, understanding the mechanisms of resistance is important for the development of effective anti-cancer therapies.
CCRF-CEM Ara-C/8C (Ara-C/8C) cells, a nucleoside transport-deficient T cell leukemia cell line, were isolated by single cell cloning and resistance to Ara-C; these cells demonstrate cross-resistance to gemcitabine and 2′, 3′-dideoxycytidine [2,3]. Despite these observations, the molecular defect responsible for the loss of nucleoside transport in these cells was not elucidated.
The equilibrative nucleoside transporters (ENTs) are a Na+-independent class of nucleoside transporters (SLC29) responsible for the uptake of a large number of nucleosides and nucleoside analogs [4]. This includes purine and pyrimidine nucleosides and analogs such as gemcitabine, Ara-C, and fludarabine [5–7]. Four distinct members of this class of proteins have been identified (ENT1-4) [4]. ENT1 and ENT2 are the best-characterized members of this family; ENT1 is selectively inhibited by nitrobenzylthioinosine (NBMPR), whereas both ENT1 and ENT2 are inhibited by dipyridimole and dilazep [8]. In comparison, ENT3 is distributed mainly within endomembranes [9] and studies suggest ENT4 may function as a monoamine/organic anion transporter [10,11].
ENTs share a common 11-transmembrane (TM) helix topology [12] and structure–function studies suggest that amino acid residues within TM 3-6 may be involved in nucleoside binding [13,14]. For example, site-directed mutagenesis of human ENT1 (hENT1) expressed in S. cerevisiae demonstrated that glycine 179 in TM5 was required for uridine transport and sensitivity to NBMPR whereas glycine 184 may partially determine targeting of the transporter to the plasma membrane [13]. Similarly, glycine 154 was reported to be important for nucleoside transport and sensitivity to the inhibitors NBMPR, dipyridamole and dilazep [14]. However, recent studies suggest that amino acids in transmembrane domains other than 3-6 may also be critical for the transport of nucleoside substrates and their analogs [15]. Specifically methionine 33 in TM1 and isoleucine 429 in the TM11 of hENT1, hENT2, and C. elegans ENT1 were reported to be required for nucleoside transport and may contribute to the binding of dipyridamole [15]. A report by Paproski et al. demonstrated that tryptophan 29 within TM1 was important for inhibitor binding and hENT1s containing mutations of W29 have altered nucleoside transport kinetics [16].
In our study we identified a single point mutation at glycine 24 in TM1 of hENT1 in Ara-C/8C cells. To investigate the importance of G24, we developed hENT1s with different point mutations of glycine 24 and expressed these mutants in PK15 cells, a nucleoside transport-deficient cell line. The mutant hENT1s were defective in transport activity as measured by [3H]-uridine uptake and inhibitor binding. Thus, mutation of glycine 24 in hENT1 may explain the loss of nucleoside transport and resistance to Ara-C and gemcitabine in Ara-C/8C cells.
Materials and Methods
Cell culture and reagents
The nucleoside transport-deficient swine epithelial cell line PK15-NTD (PK15) was generously provided by Dr. Chung-Ming Tse (Johns Hopkins University School of Medicine) and was maintained as described previously [17]. The CCRF-CEM cell line was obtained from the ATCC (Rockville, Md.) and the CCRF-CEM Ara-C/8C cell line, an Ara-C resistant cell line derived from the CCRF-CEM cell line, was kindly provided by Dr. Buddy Ullman (Oregon Health Sciences University). CCRF-CEM and Ara-C/8C cells were maintained as described previously [2]. NBMPR (6-[(4-nitrobenzyl) thio]-9-(β-D-ribofuranosyl) purine) was purchased from Sigma (St. Louis, MO). [3H]-NBMPR (specific activity: 22.0 Ci/mmol), [5, 6-3H]-Uridine (specific activity: 35–50 Ci/mmol), and [5-3H]-Ara-C (cytosine-β-D-arabinofuranoside, specific activity: 15–30 Ci/mmol) were obtained from Moravek Biochemicals (Brea, CA).
Total RNA extraction and cDNA synthesis
Total RNAs were extracted from CCRF-CEM and Ara-C/8C cells with Trizol reagent (Invitrogen; Carlsbad, CA) following the manufacturer’s instruction. Two micrograms of total RNA was used for first-strand cDNA synthesis using oligo (dT) as primers and SuperScript II RNase H− Reverse Transcriptase (SuperScript™ First-Strand Synthesis System for RT-PCR, Invitrogen).
PCR and DNA sequencing
One tenth of the resulting first-strand cDNA was then used for PCR amplification with High Fidelity pfu DNA polymerase (Invitrogen). The primers for amplifying the entire coding sequence of hENT1 were 5′-CCGCTCGAGATGACAACCAGTCACCTCAG-3′ (sense primer) and 5′-AGACTCGAGTCACACAATTGCCCGGAACAGG-3′ (antisense primer). PCR products were cloned into the TOPO blunt cloning vector (Invitrogen) and expressed in DH5α E. coli. (Invitrogen). Plasmids were submitted for automatic sequencing (UNC-CH Genome Analysis Facility, http://152.19.68.152/gafsite/Main.asp).
Construction of expression vectors and transfection
hENT1 cDNA was subcloned into the pcDNA 3.1 HisC expression vector (Invitrogen). Construction of the G24 hENT1 mutant expression vectors was performed using the Quickchange Site-directed Mutagenesis kit (Stratagene; Cedar Creek, TX) with primers specific to hENT1; G24R sense primer (5′-CTTCATGCTGGGTCTGAGAACGCTGCTCCCGTGG-3′) and antisense primer (5′ CCACGGGAGCAGCGTTCTCAGACCCAGCATGAAG 3′), G24A sense primer (CTTCATGCTGGGTCTGGCAACGCTGCTCCCGTGG) and antisense primer (CCACGGGAGCAGCGTTGCCAGACCCAGCATGAAG), and G24E sense primer (CTTCATGCTGGGTCTGGAAACGCTGCTCCCGTGG) and antisense primer (CCACGGGAGCAGCGTTTCCAGACCCAGCATGAAG) each confer a missense mutation at base pair 72. Wt and G24R hENT1s were subcloned into the EGFP-C3 expression vector (Clontech, Mountain View, CA) for confocal microscopy. Expression vectors were introduced into PK15 or HeLa cells by Lipofectamine transfection (Invitrogen) according to the manufacturer’s instruction.
[3H]-uridine and [3H]-Ara-C uptake assays
The uptake of [3H]-uridine was measured in CEM cell lines or transfected PK15 cells exactly as described previously [18]. The same method was used to measure [3H]-Ara-C uptake with the substitution of cold cytidine for uridine used to stop the reaction. The protein concentration of each sample was quantitated using Coomassie Protein Assay Reagent (Pierce; Rockford, IL).
Confocal Microscopy
pEGFP-C3 constructs were transfected into HeLa cells. This cell type was used to obtain high transfection efficiency and ease for microscopy. Twenty-four hours post-transfection images were captured using a 63X oil immersion objective on a Zeiss LSM 510 Meta confocal microscope (Thornwood, NY). Image capture was resolved using a 488 nm argon laser.
Isolation of membranes and [3H]-NBMPR binding assay
Crude cell membranes from the CEM cells were prepared as described previously with minor modification [8]. Briefly, approximately 5×107 cells were washed three times with room temperature phosphate-buffered saline (PBS), suspended in 500 μl of ice-cold 5 mM Na2HPO4 buffer (pH 8.0), and sonicated to completely lyse the cells. After a 30 min incubation on ice, the cell membranes were washed twice (40,000 × g centrifugation for 40 min) with ice-cold 5 mM Na2HPO4 buffer and suspended in 200 μl of the ice-cold 5 mM Na2HPO4 buffer. Binding assays were performed at room temperature in 10 mM Tris (pH 7.1) containing 0.01% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) (w/v) to prevent non-specific binding of the radioligand. Incubations were initiated by adding an aliquot of 500 μg cell membrane to room temperature 5 mM Na2HPO4 (final vol. 1 ml) containing 10 nM [3H]-NBMPR and were terminated after 45 min by dilution with 5 ml of ice-cold 10 mM Tris (pH 7.1) followed by rapid filtration through Whatman GF/B filters which were then washed once with 5 ml of ice-cold 10 mM Tris (pH 7.1). Radioactivity was measured by liquid scintillation using Scintisafe™ Econo-2 scintillation fluid (Fisher Scientific, Pittsburgh, PA) on a LS6500 Multi-purpose Scintillation Counter (Beckman Coulter, Fullerton, CA). Non-specific binding of [3H]-NBMPR was determined in the presence of 20 μM unlabelled NBMPR.
Results
Functional loss of hENT1 activity of Ara-C/8C cells is independent of mRNA expression
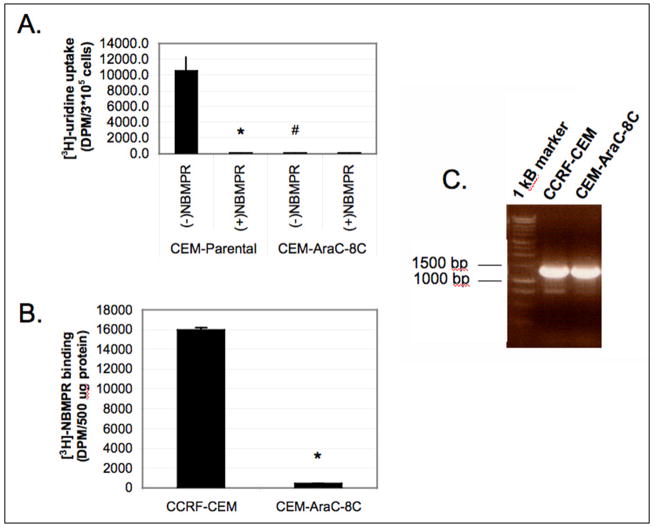
The nucleoside transport-deficient Ara-C/8C cells were isolated as a cell line highly resistant to Ara-C and gemcitabine [2,3]. Comparison of [3H]-uridine uptake in parental resistant Ara-C/8C cells showed that the Ara-C/8C were completely deficient in uridine uptake (Fig. 1A). [3H]-Uridine transport was abolished with the ENT1 specific inhibitor, NBMPR in the parental CCRF-CEM cells demonstrating that hENT1 activity accounts for the majority of uridine uptake in the CEM cell lines. In comparison, the binding of [3H]-NBMPR to crude membrane preparations of Ara-C/8C cells was almost completely abolished (Fig. 1B) suggesting that functional hENT1 protein was absent from Ara-C/8C cells.
Figure 1. Comparison of [3H]-uridine uptake, [3H]-NBMPR binding, and mRNA expression between CCRF-CEM cells and Ara-C/8C cells.
A) 3x105 CCRF-CEM or Ara-C/8C cells (CEM-AraC-8C) cells were incubated with [3H]-uridine in sodium-free transport buffer for 5 min in the presence or absence of 1.0 μM NBMPR, a selective inhibitor of hENT1, as described in Materials and Methods. Data points represent sample mean +/− SD in duplicate (N=4; *p<0.01 different from –NBMPR, #p<0.01 different from parental cell line). B) [3H]-NBMPR binding was performed as described in Materials and Methods. Data points represent mean +/− SD from samples run in triplicate (N=2; *p<0.01). C) RT-PCR analysis of hENT1 mRNA expression was determined as described in Materials and Methods. Representative gel comparing mRNA levels in the parental CCRF-CEM cell line and CCRF-CEM-AraC-8C cell line.
hENT1 mRNA expression was determined using RT-PCR. As shown in Fig. 1C, Ara-C/8C cells expressed similar levels of hENT1 mRNA as compared to the parental CCRF-CEM cells. This observation suggests that the defect in nucleoside uptake was independent of hENT1 expression.
Identification of a point mutation in hENT1 from Ara-C/8C cells that disrupts transport activity
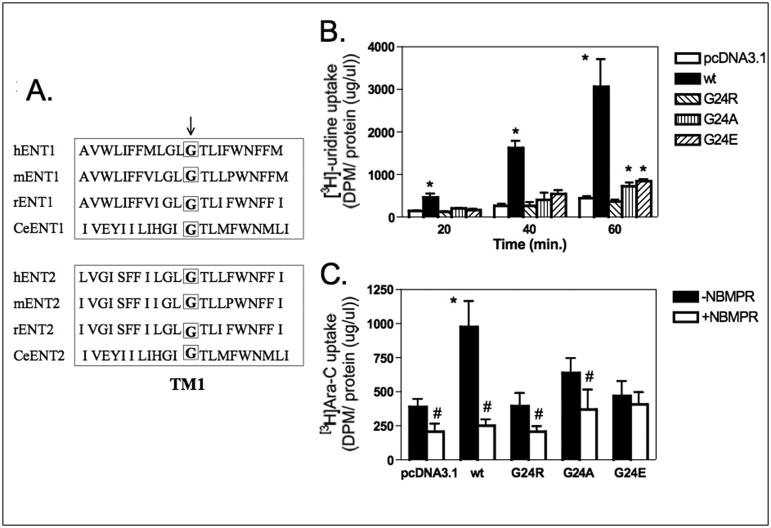
We next determined whether hENT1 from Ara-C/8C cell line contained mutations that would explain a loss of function. Full-length hENT1 from Ara-C/8C cells was amplified by PCR and sequenced. Analysis of the hENT1 sequence from these cells revealed a single nucleotide missense mutation resulting in the substitution of arginine for glycine at amino acid position 24 (GGA to AGA). Complete sequence analysis of hENT1 from Ara-C/8C cells did not reveal any additional mutations. G24 is located in the TM1 domain of hENT1 and is conserved amongst species and ENT2 isoforms (Fig. 2A).
Figure 2. Substitution of G24 in hENT1 disrupts nucleoside and Ara-C transport activity.
A) Cross-species comparison of amino acids 13-33 of ENT1 and ENT2. Arrow denotes G24. B) Wt hENT1 and G24A, G24E, and G24R hENT1 mutants were expressed separately in PK15 cells (6×104) and [3H]-uridine uptake was performed for 20, 40, and 60 minutes as described in Materials and Methods. Average transport by wt hENT1 was approx. 4.19 pmol/min/mg protein. Data points represent mean +/− SD of samples run in quadruplicate (N=1; *p<0.01 different from control). C) Wt hENT1 and G24A, G24E, and G24R hENT1 mutants were expressed separately in PK15 cells (6×104) and [3H]-Ara-C uptake was performed for 2 hours in the presence or absence of 1μM NBMPR as described in Materials and Methods. Average transport by wt hENT1 was approx. 2.08 pmol/min/mg protein. Data points represent mean +/− SD of samples run in quadruplicate (N=1; *p<0.01 different from control, #p<0.01 different from –NBMPR).
We created several point mutations of G24; substitution of alanine and glutamate were performed to determine the effect of amino acid size and charge on hENT1 function. Wild-type (wt) hENT1 and G24A, G24E, and G24R hENT1 mutants were transfected into nucleoside transport-deficient PK15 cells and [3H]-uridine uptake was measured as an index of ENT1 activity. As shown in Fig. 2B, G24R hENT1 was deficient in [3H]-uridine transport compared to wt hENT1. hENT1 proteins with a mutation of G24 to alanine or glutamate displayed only partial transport activity when compared to wt hENT1 (Fig. 2B). Similarly, the G24R hENT1 mutant displayed a deficiency in [3H]-Ara-C uptake; G24A hENT1 displayed partial uptake when compared to wt hENT1 whereas G24E hENT1 was uptake-deficient (Figure 2C). These data further indicated that glycine 24 is important for hENT1 activity and mutation of G24 severely reduced transport activity.
The G24R hENT1 mutant maintains plasma membrane localization but is deficient in NBMPR binding
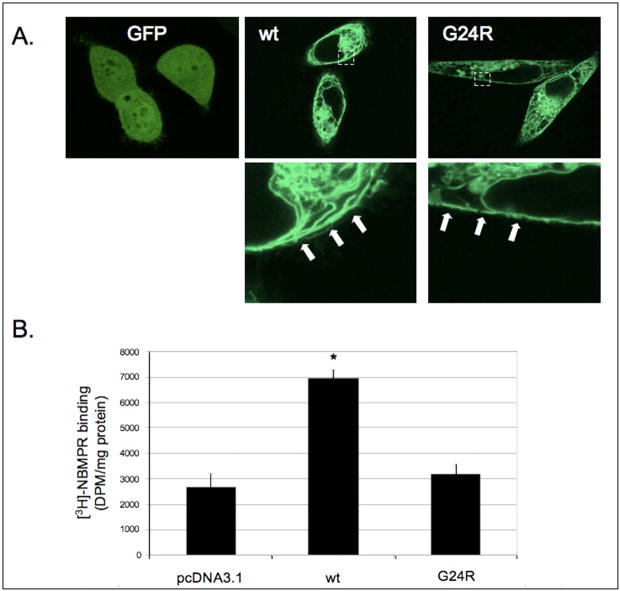
To determine if the G24R hENT1 mutant was localized to the plasma membrane as expected, wt and G24R hENT1s were subcloned into the pEGFP-C3 vector. Transfection of HeLa cells showed that the G24R hENT1 mutant exhibited predominantly membranous expression, similar to that observed with wt hENT1 (Fig. 3A). These data demonstrated that the G24 mutation did not alter protein maturation or plasma membrane localization.
Figure 3. G24R hENT1 mutant lacks [3H]-NBMPR binding even though plasma membrane expression is maintained.
A) Wt and G24R hENT1 expression was observed using confocal microscopy. Image fields are representative of expression. Arrows denote approximate plasma membrane location. B) Wt hENT1 and G24R hENT1 were transfected separately into PK15 cells. Forty-eight hours post-transfection cells were lysed, the membranes were isolated and [3H]-NBMPR binding to the isolated membrane was determined as described in Materials and Methods. Data points represent the mean +/− SD (N=3; *p<0.01 different from control).
To investigate whether the G24R hENT1 mutant was sensitive to inhibitor binding, [3H]-NBMPR binding assays were conducted with crude cell membrane preparations from PK15 cells expressing either wt or G24R hENT1. [3H]-NBMPR has selective affinity for ENT1 and inhibitor binding to cells expressing wt hENT1 was observed (Fig. 3B). In contrast, cells expressing the G24R hENT1 mutant did not display any detectable specific [3H]-NBMPR binding suggesting that the G24R mutation affected both substrate and inhibitor recognition.
Discussion
In this study we provide evidence for a novel missense mutation in hENT1 that conferred resistance to nucleoside uptake in an Ara-C resistant cell line of CCRF-CEM cells. Our results demonstrate that a single point mutation (G24R) in TM1 results in the generation of an inactive hENT1. G24 is an amino acid that is highly conserved between ENT isoforms and amongst species, suggestive of its importance to hENT1 function. Substitution of G24 with amino acids that vary in size and charge confirmed the importance of G24 in ENT1 recognition of substrates. Moreover, G24 may be an important contact for inhibitor recognition since G24R mutants of hENT1 did not bind [3H]-NBMPR. Partial [3H]-Ara-C transport was observed with the G24A mutation, which may be due to the similarity of the amino acids. Importantly, our data suggest that differences in hENT1 G24 mutant activity were not due to differential expression but due to a functional difference since the wt and G24R hENT1 proteins had dissimilar [3H]-uridine transport/[3H]-NBMPR binding ratios (2.54±0.10 vs. 0.67±.02). Thus these studies are the first to identify this conserved amino acid as an important determinant for nucleoside recognition and uptake.
The G24R point mutation in hENT1 may affect several processes related to the functional activity of hENT1 including substrate binding, protein folding or targeting of hENT1 to the plasma membrane. Topology predictions (SOSUI program; http://bp.nuap.nagoya-u.ac.jp/sosui/) suggest that G24R hENT1 is capable of targeting to plasma membrane similarly to the wt hENT1. This prediction is supported by our data showing similar membranous localization of EGFP-tagged wt and G24R hENT1. Our studies also show that G24R hENT1 is incapable of binding the inhibitor NBMPR. This may result from a disruption of the substrate-binding site since NBMPR, a purine analog, is expected to bind at or near the substrate-binding pocket [13,14]. Recent studies have shown that amino acids in close proximity (M33, W29) are important to hENT1 nucleoside transport and inhibitor binding [15,16]. Based on these observations, we speculate that the G24R substitution at this position similarly interferes with binding of nucleosides and nucleoside analogues to hENT1. Thus, TM1 may be a topologically-sensitive portion of hENT1 making the proper orientation of TM1 necessary for protein function.
ENT-mediated transport of nucleoside analogs is often the rate-limiting step for drug-induced cell cytotoxicity [6]. Thus, it is not surprising that studies have found a positive correlation between the expression of hENT1 and cancer cell sensitivity to the nucleoside analogs Ara-C [19] and gemcitabine [20]. The reduction in hENT1 expression is a common mechanism for resistance to antimetabolite treatment of cancer [7,21]. In addition, a recent study using an Ara-C-resistant CCRF-CEM cell line reported that genetic mutations of hENT1 that alter mRNA splicing and protein translation provide mechanisms for resistance to drug treatment [22].
In conclusion, these studies provide new information on the specific role of G24 in the TM1 domain of ENT1 in nucleoside recognition and uptake. Expression of a G24R hENT1 mutant provides the first molecular explanation for the loss of hENT1-dependent nucleoside uptake in the Ara-C/8C cells and indicates that mutation of this amino acid could result in resistance to Ara-C treatment.
Acknowledgments
This work was supported by NIH grant RO1-GM069976 to L.M.G.
Abbreviations
- ENT1
equilibrative nucleoside transporter 1
- AraC
1-β-D-arabinofuranosylcytosine
- NBMPR
nitrobenzylmercaptopurine ribonucleoside (6-[(4-nitrobenzyl) thiol]-9-β-D-ribofuranosyl purine)
- EGFP
enhanced green fluorescent protein
- ddC
2′, 3′-dideoxycytidine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cripe LD. Adult acute leukemia. Curr Probl Cancer. 1997;21(1):1–64. doi: 10.1016/s0147-0272(97)80006-2. [DOI] [PubMed] [Google Scholar]
- 2.Ullman B, Coons T, Rockwell S, McCartan K. Genetic analysis of 2′,3′-dideoxycytidine incorporation into cultured human T lymphoblasts. J Biol Chem. 1988;263:12391–96. [PubMed] [Google Scholar]
- 3.Ullman B. Dideoxycytidine metabolism in wild type and mutant CEM cells deficient in nucleoside transport or deoxycytidine kinase. Adv Exp Med Biol. 1989;253B:415–20. doi: 10.1007/978-1-4684-5676-9_61. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–43. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 5.Molina-Arcas M, Marce S, Villamor N, Huber-Ruano I, Casado FJ, Bellosillo B, Montserrat E, Gil J, Colomer D, Pastor-Anglada M. Equilibrative nucleoside transporter-2 (hENT2) protein expression correlates with ex vivo sensitivity to fludarabine in chronic lymphocytic leukemia (CLL) cells. Leukemia. 2005;19:64–8. doi: 10.1038/sj.leu.2403582. [DOI] [PubMed] [Google Scholar]
- 6.Pastor-Anglada M, Molina-Arcas M, Casado FJ, Bellosillo B, Colomer D, Gil J. Nucleoside transporters in chronic lymphocytic leukaemia. Leukemia. 2004;18:385–93. doi: 10.1038/sj.leu.2403271. [DOI] [PubMed] [Google Scholar]
- 7.Takagaki K, Katsuma S, Kaminishi Y, Horio T, Nakagawa S, Tanaka T, Ohgi T, Yano J. Gene-expression profiling reveals down-regulation of equilibrative nucleoside transporter 1 (ENT1) in Ara-C-resistant CCRF-CEM-derived cells. J Biochem. 2004;136:733–40. doi: 10.1093/jb/mvh180. [DOI] [PubMed] [Google Scholar]
- 8.Hammond JR. Interaction of a series of draflazine analogues with equilibrative nucleoside transporters: species differences and transporter subtype selectivity. Naunyn Schmiedebergs Arch Pharmacol. 2000;361:373–82. doi: 10.1007/s002100000214. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin SA, Yao SY, Hyde RJ, Ng AM, Foppolo S, Barnes K, Ritzel MW, Cass CE, Young JD. Functional characterization of novel human and mouse equilibrative nucleoside transporters (hENT3 and mENT3) located in intracellular membranes. J Biol Chem. 2005;280:15880–887. doi: 10.1074/jbc.M414337200. [DOI] [PubMed] [Google Scholar]
- 10.Engel K, Wang J. Interaction of organic cations with a newly identified plasma membrane monoamine transporter. Mol Pharmacol. 2005;68:1397–1407. doi: 10.1124/mol.105.016832. [DOI] [PubMed] [Google Scholar]
- 11.Zhou M, Xia L, Engel K, Wang J. Molecular determinants of substrate selectivity of a novel organic cation transporter (PMAT) in the SLC29 family. J Biol Chem. 2007;282:3188–95. doi: 10.1074/jbc.M609421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundaram M, Yao SY, Ingram JC, Berry ZA, Abidi F, Cass CE, Baldwin SA, Young JD. Topology of a human equilibrative, nitrobenzylthioinosine (NBMPR)-sensitive nucleoside transporter (hENT1) implicated in the cellular uptake of adenosine and anti-cancer drugs. J Biol Chem. 2001;276:45270–75. doi: 10.1074/jbc.M107169200. [DOI] [PubMed] [Google Scholar]
- 13.SenGupta DJ, Lum PY, Lai Y, Shubochkina E, Bakken AH, Schneider G, Unadkat JD. A single glycine mutation in the equilibrative nucleoside transporter gene, hENT1, alters nucleoside transport activity and sensitivity to nitrobenzylthioinosine. Biochemistry. 2002;41:1512–19. doi: 10.1021/bi015833w. [DOI] [PubMed] [Google Scholar]
- 14.SenGupta DJ, Unadkat JD. Glycine 154 of the equilibrative nucleoside transporter, hENT1, is important for nucleoside transport and for conferring sensitivity to the inhibitors nitrobenzylthioinosine, dipyridamole, and dilazep. Biochem Pharmacol. 2004;67:453–58. doi: 10.1016/j.bcp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Visser F, Baldwin SA, Isaac RE, Young JD, Cass CE. Identification and mutational analysis of amino acid residues involved in dipyridamole interactions with human and Caenorhabditis elegans equilibrative nucleoside transporters. J Biol Chem. 2005;280:11025–34. doi: 10.1074/jbc.M410348200. [DOI] [PubMed] [Google Scholar]
- 16.Paproski RJ, Visser F, Zhang J, Tackaberry T, Damaraju V, Baldwin SA, Young JD, Cass CE. Mutation of Trp 29 of human equilibrative nucleoside transporter 1 alters affinity for coronary vasodilator drugs and nucleoside selectivity. Biochem J. 2008 doi: 10.1042/BJ20080074. [DOI] [PubMed] [Google Scholar]
- 17.Ward JL, Sherali A, Mo ZP, Tse CM. Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. Ent2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J Biol Chem. 2000;275:8375–81. doi: 10.1074/jbc.275.12.8375. [DOI] [PubMed] [Google Scholar]
- 18.Huang M, Wang Y, Cogut SB, Mitchell BS, Graves LM. Inhibition of nucleoside transport by protein kinase inhibitors. J Pharmacol Exp Ther. 2003;304:753–60. doi: 10.1124/jpet.102.044214. [DOI] [PubMed] [Google Scholar]
- 19.Gati WP, Paterson AR, Larratt LM, Turner AR, Belch AR. Sensitivity of acute leukemia cells to cytarabine is a correlate of cellular es nucleoside transporter site content measured by flow cytometry with SAENTA-fluorescein. Blood. 1997;90:346–53. [PubMed] [Google Scholar]
- 20.Marce S, Molina-Arcas M, Villamor N, Casado FJ, Campo E, Pastor-Anglada M, Colomer D. Expression of human equilibrative nucleoside transporter 1 (hENT1) and its correlation with gemcitabine uptake and cytotoxicity in mantle cell lymphoma. Haematologica. 2006;91:895–902. [PubMed] [Google Scholar]
- 21.Lu X, Gong S, Monks A, Zaharevitz D, Moscow JA. Correlation of nucleoside and nucleobase transporter gene expression with antimetabolite drug cytotoxicity. J Exp Ther Oncol. 2002;2:200–12. doi: 10.1046/j.1359-4117.2002.01035.x. [DOI] [PubMed] [Google Scholar]
- 22.Cai J, Damaraju VL, Groulx N, Mowles D, Peng Y, Robins MJ, Cass CE, Gros P. Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer Res. 2008;68:2349–57. doi: 10.1158/0008-5472.CAN-07-5528. [DOI] [PubMed] [Google Scholar]