Summary
Progress in understanding the molecular pathogenesis of human myeloproliferative disorders (MPDs) has led to guidelines incorporating genetic assays with histopathology during diagnosis. Advances in flow cytometry have made it possible to simultaneously measure cell type and signaling abnormalities arising as a consequence of genetic pathologies. Using flow cytometry, we observed a specific evoked STAT5 signaling signature in a subset of samples from patients suspected of having juvenile myelomonocytic leukemia (JMML), an aggressive MPD with a challenging clinical presentation during active disease. This signature was a specific feature involving JAK-STAT signaling, suggesting a critical role of this pathway in the biological mechanism of this disorder and indicating potential targets for future therapies.
Significance
Recent advances have enabled simultaneous measurement of cell type and cell signals in primary populations using flow cytometry. This technique enables the question, "Can we track oncogenic cell populations from diagnosis through disease evolution via signaling?" Doing so in an era of using specific inhibitors against components of key signal transduction pathways will be necessary to assess treatment effects in human patients and adapt as cancer cells alter their signaling in response to these treatments. This work uses such an approach to follow patients over time and shows that disease status in juvenile myelomonocytic leukemia (JMML) -- at diagnosis, remission, relapse, and transformation -- is indicated by a subset of cells with an abnormal signaling profile.
Introduction
Myeloproliferative disorders (MPDs) are clonal malignancies characterized by overproduction of immature and mature myeloid cells showing organ infiltration. In particular, juvenile myelomonocytic leukemia (JMML) and chronic myelomonocytic leukemia (CMML) are characterized by malignant transformation in the stem cell compartment with clonal proliferation of progeny that variably retain the capacity to differentiate (Arico et al., 1997; Onida et al., 2002). Children suspected of having JMML often present with failure to thrive, fever, infection, splenomegaly, and a high white blood cell count with monocytosis. Current diagnostic criteria are imprecise and consist of major and minor requirements that are, in large part, based on excluding other conditions (Niemeyer et al., 1997). The major requirements include >1000 monocytes in peripheral blood (PB) (at a concentration of 1 × 109 cells/L), fewer than 20% bone marrow blasts, and absence of the t(9;22) or BCR/ABL fusion gene. Patients must also meet two of the minor criteria, including an elevated fetal hemoglobin for age, circulating myeloid precursors, a total white blood cell count of >10,000 (at a concentration of 10 × 109 cells/L), and in vitro hypersensitivity to granulocyte-macrophage colony stimulating factor (GM-CSF).
Extensive molecular data implicate genetic lesions that deregulate Ras signaling as key initiating events in JMML, with studies showing that 60% of patients harbor an oncogenic mutation in PTPN11, NRAS, or KRAS while another 15% have clinical Neurofibromatosis type 1 and/or demonstrate loss of the wildtype NF1 allele in their diseased bone marrow (Emanuel, 2004; Flotho et al., 2007). Patients with the myeloproliferative subtype of CMML exhibit NRAS, KRAS, and JAK2 mutations (Levine et al., 2005; Onida et al., 2002). A cellular characteristic of both JMML and CMML is the formation of abnormal numbers of granulocyte-macrophage colony-forming units (CFU-GM) in methylcellulose cultures containing sub-saturating concentrations of GM-CSF (Cambier et al., 1997; Emanuel et al., 1991), leading to prior suggestions that alterations downstream of the activated GM-CSF receptor collaborate to drive inappropriate cell growth and survival. One important primary signaling event post-binding of GM-CSF to the GM-CSF receptor is activation of the JAK/STAT pathway (Paukku and Silvennoinen, 2004). JAK2 trans-phosphorylates the β chain of the GM-CSF receptor, which creates docking sites for adapters and signal relay molecules resulting in activation of Ras and of downstream Ras effectors including ERK and S6 (Figure S1) (Irish et al., 2004; Kunz and Ibrahim, 2003; McCubrey et al., 2000; Rane and Reddy, 2002; Shuai and Liu, 2003).
CMML is an adult MPD that is clinically similar to JMML and shares certain genetic features such the frequent presence of RAS mutations (Onida et al., 2002). JAK2 mutations are rare in JMML and only slightly more common in CMML patients (Levine et al., 2005; Steensma et al., 2005; Zecca et al., 2007) whereas PTPN11 mutations are almost non-existent in CMML (Loh et al., 2005). Both JMML and CMML can progress to M4 or M5 acute myeloid leukemia (AML), which comprise the myelomonocytic (M4) and monocytic (M5) subtypes (Arico et al., 1997). Furthermore, somatic NRAS, KRAS and PTPN11 mutations occur frequently in the M4 and M5 subtypes of AML (Bacher et al., 2006; Loh et al., 2004).
Currently, it takes up to 3–4 weeks to confirm a suspected diagnosis of JMML with a CFU-GM assay. As early allogeneic hematopoietic stem cell transplant (HSCT) is the only potentially curative therapy for JMML (Locatelli et al., 2005), it is important to quickly and accurately diagnose these children in order to deliver appropriate therapy in a timely fashion. In addition, monitoring disease burden during treatment is challenging in patients with JMML due to imprecise clinical definitions of response. Current allele-specific PCR methodologies to detect minimal residual disease are only applicable to approximately 60% of patients (Archambeault et al., 2008). Importantly, because JMML and CMML exhibit considerable cellular heterogeneity, it has been difficult to elucidate the biologic features of cells that contribute to the cancer phenotype in vivo, and of precursor populations that might carry genetic lesions predisposing cells to an oncogenic fate.
Assays for identifying therapeutic agents and assessing efficacy in these patients based on the biochemical consequences of lesions in the GM-CSF and Ras signaling networks are few. Recent advances in flow cytometry however, have made it possible to simultaneously measure cell type and aberrant cell signals (Irish et al., 2006) arising as a consequence of these lesions. We used this approach to profile signaling at the single cell level (Irish et al., 2004; Van Meter et al., 2007), including molecules downstream of the GM-CSF receptor and molecules closely associated with Ras signaling, for the presence of primary JMML cells with altered signaling behavior that correlate with disease physiology. Our cohort of 52 samples included patients diagnosed with JMML, healthy individuals, infants with other MPDs, and children initially suspected of having JMML who were subsequently diagnosed with other disorders.
Results
A flow cytometry based signaling assay can be used to measure GM-CSF hypersensitivity
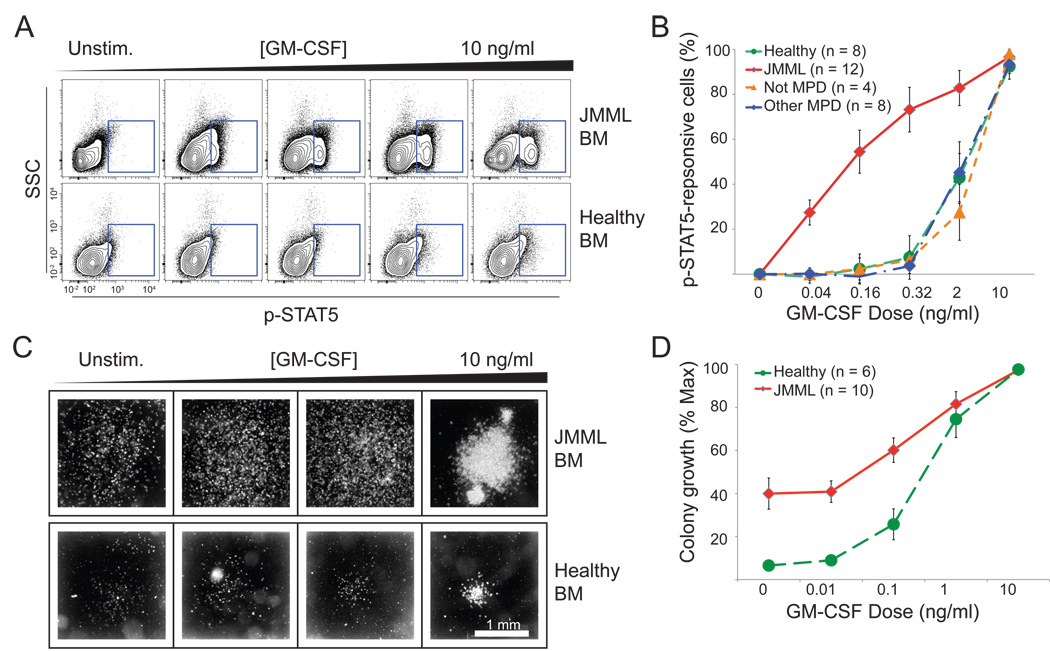
We used phospho-specific flow cytometry (Irish et al., 2004; Irish et al., 2006) after exposure to increasing concentrations of GM-CSF to interrogate evoked signaling responses in JMML cells. In a first test of GM-CSF induced phosphorylation of STAT5, we observed the dose-dependent appearance of a population of cells in a JMML bone marrow sample compared to normal healthy bone marrow (Figure 1A). This leukemia was also assessed via the traditional methylcellulose assay and exhibited hypersensitive colony formation (defined as clusters of >50 cells) at increasing concentrations of GM-CSF (Figure 1C and 1D), as previously described (Emanuel et al., 1991).
Figure 1. Hyperphosphorylation of STAT5 is seen in primary JMML cells at low concentrations of GM-CSF.
The diagnosis of JMML is confirmed through an in vitro CFU-GM assay showing colony formation at low concentrations of GM-CSF. Colonies form over 2 weeks at various doses of GM-CSF. (A) Phospho-specific flow cytometry based assay demonstrated that GM-CSF hypersensitivity can be measured via p-STAT5. Primary cells from JMML and normal bone marrow (BM) were stimulated at varying concentrations of GM-CSF. The JMML samples showed a subset of cells responding via p-STAT5 at low concentrations of GM-CSF. In (B), increase in p-STAT5 response to GM-CSF concentration was quantified as a function of maximal response. A higher percentage of p-STAT5+ cells were present at 0.16 and 0.32 ng/ml of GM-CSF concentrations in JMML (n = 12) samples than in healthy samples (n = 8), samples with NS or DS related TMD (n = 8) or non-MPD samples that were initially suspected to be JMML (n = 4). In (C) we show colonies formed in a JMML and a normal sample at increasing doses of GM-CSF. These are the same samples measured using phosphospecific flow cytometry in (A). (D) Colony formation from JMML patients (n=10) was quantified as a function of maximum colony growth, plotted over varying concentrations of GM-CSF and compared against growth from normal samples (n = 6). Error bars in (B) and (D) represent standard error of the mean (s.e.m).
We then investigated 11 additional JMML samples at diagnosis, and compared these leukemias to normal samples (n= 8), other childhood MPDs (these cases included 8 patients with Noonan syndrome/MPD (NS/MPD) or Down syndrome with transient myeloproliferative disorders (DS/TMD), and 4 children with an initial clinical suspicion of JMML (who were subsequently found to have another diagnosis). We observed an induced phosphorylated STAT5 (p-STAT5) population in the majority of JMML samples that were exposed to low levels of GM-CSF but not in the other samples interrogated. The data was quantified as a ratio of the percentage of positive cells compared to the unstimulated condition (Figure 1B).
p-STAT5 response to low doses of GM-CSF indicates JMML status
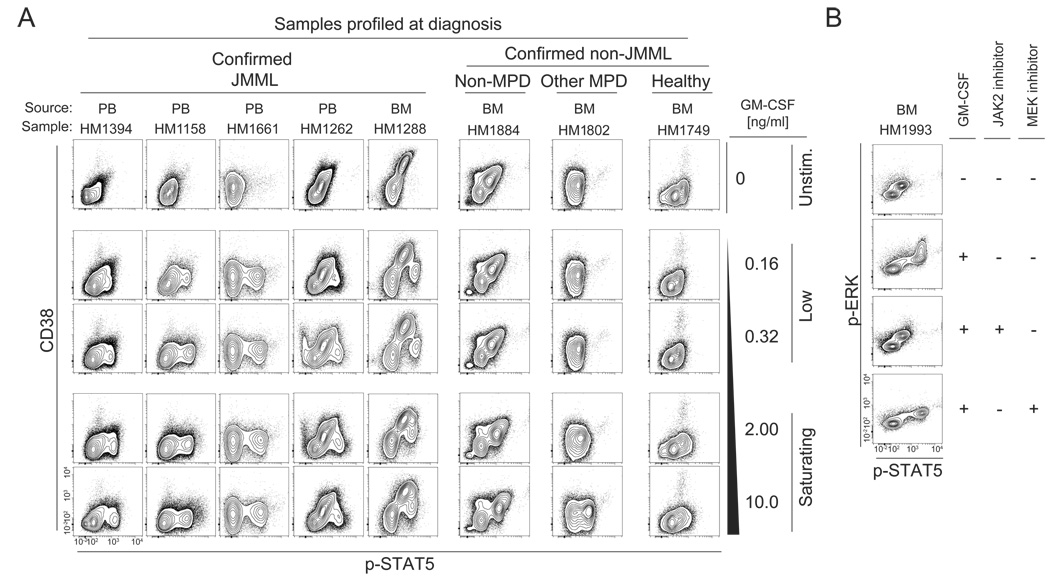
The combination of CD38 and p-STAT5 best stratified the GM-CSF hyper-responsive population, which was measurable in both peripheral blood and bone marrow samples. Representative samples from the patient cohort are shown in Figure 2A. The signature was present in both fresh and previously frozen primary samples (details about each sample are provided in Table S1).
Figure 2. The p-STAT5 responsive population can distinguish JMML from other myeloproliferative disorders of childhood at diagnosis.
(A) CD38 and p-STAT5 stratify the responsive population present in JMML peripheral blood and bone marrow samples at diagnosis. (B) Exposure to an oral JAK2 inhibitor, XL019, abrogated both the p-STAT5 and p-ERK response while exposure to a MEK inhibitor, CI-1040, only inhibited the p-ERK response.
p-STAT5 signaling cells in JMML samples are of myeloid origin and require JAK2 activity
Immunophenotyping revealed that the p-STAT5 responsive cells were of myeloid origin (CD33+, CD14+), CD34− and CD38low (Figure S2). The involvement of JAK-specific activation of the p-STAT5 response in these cells was first confirmed by exposing primary samples to a chemical JAK2 inhibitor for 30 minutes before GM-CSF stimulation at 10 ng/ml for 15 minutes (Figure S3). Similarly, exposing JMML cells to 5 µM of the oral JAK2 inhibitor XL019 (Exelixis) inhibited STAT5 and ERK phosphorylation in response to a saturating concentration of GM-CSF whereas the MEK inhibitor CI-1040 (Pfizer) failed to alter the p-STAT5 response despite inhibiting p-ERK (Figure 2B).
p-STAT5 response can be summarized using the 95th percentile
To visualize the dataset from all the patients studied, we calculated ratios of the 95th percentile of p-STAT5 activity to the unstimulated sample and displayed it in a heat map format (see Experimental procedures). Eleven of twelve JMML samples showed highly sensitive activation of p-STAT5 (at 0.32 ng/ml GM-CSF) as compared to lack of activation in the majority of the control specimens. Diagnostic samples from infants with other MPDs, including those with NS/MPD and DS/TMD did not demonstrate a hyper-responsive p-STAT5 population to GM-CSF (Figure 3A). In addition, samples obtained from children suspected of having JMML but subsequently found to have other disorders did not demonstrate the induced p-STAT5 population. The absence of the p-STAT5 signature in conditions that are phenotypically similar to JMML highlights the importance of the signaling disruptions that we observe specifically in JMML.
Figure 3. The p-STAT5 responsive population is indicative of active JMML.
(A) Clinically similar myeloproliferative disorders such as Noonan Syndrome/MPD and, Down Syndrome/TMD were distinguishable from JMML via phosphoflow cytometry. 11/12 JMML patient samples at diagnosis showed a GM-CSF hypersensitive population. 7/8 Normal BM samples did not have a GM-CSF hypersensitive population.
(B)Samples post diagnosis show a p-STAT5 response in patients who have relapsed or transformed into AML M4/M5. Samples in remission do not show the p-STAT5 response.
(C) The signature was also present in CMML (a similar disease to JMML that is diagnosed in adults) and AML M4/M5 (myeloid leukemia with myelomonocytic and monocytic differentiation).
(D) Samples sorted by the normalized p-STAT5 response at 0.32ng/ml. Samples >40 showed JMML activity (Signaling profiles 1 and 2). 8/12 JMML samples at diagnosis had a normalized p-STAT5 response >95 (Signaling profile 1).
Remiss. = Remission
PMAIO = PMA/Ionomyocin
Overall, 11/12 JMML diagnostic samples showed a GM-CSF hypersensitive population, 7/8 normal bone marrow samples did not show a GM-CSF hypersensitive population, 8/8 NS/MPD or DS-related TMD samples did not show the GM-CSF hypersensitive population, and 4 samples from patients that were suspected of having JMML but were later found to not have a MPD did not show the hypersensitive population (Figure 3A). Based on this analysis, the p-STAT5 diagnostic assay has a sensitivity of 91% and a specificity of 95%. An unsupervised grouping of these patients was calculated by normalizing the response of p-STAT5 (as measured by the 95th percentile) to GM-CSF concentrations within a patient (Figure 3D). Using this metric, samples with a response >40 showed a JMML-like evoked signaling pattern. Further information about these samples is available in Table S1. Elevated levels of p-ERK and basal activation of p-S6 were detected in some JMML samples, but these alterations were a less consistent feature of JMML than hypersensitive signaling at the level of p-STAT5 (Figures S4–S7).
p-STAT5 response to low doses of GM-CSF is detectable in CMML and AML M4/M5
We also analyzed evoked p-STAT5 responses in CMML and AML and found that all 5 samples from adults with CMML and 4 of 8 samples from patients with AML showed a hyper-responsive population (Figure 3C, S8 and S9). In AML, the presence of an aberrant p-STAT5 response to sub-therapeutic concentrations of GM-CSF correlated with the FAB M4/M5 morphologic subtype (n=4), and was not detected in non-M4/M5 AML samples (n=4). The eight AML samples were assessed for lesions in PTPN11, NRAS, KRAS, and FLT3. The genotype data was less predictive of the signaling phenotype than the cell type, as two patients with non-M4/M5 AML who were found to harbor a FLT3 ITD and one patient with an NRAS mutation did not exhibit the p-STAT5 signature. Further information is available in Tables S1 and S2.
Signaling response can indicate disease status
To determine if the signaling signature was maintained or altered over time, we followed two JMML patients through a series of samples obtained at various timepoints during therapy. In particular, patient HM1158 attained clinical remission, underwent matched-unrelated HSCT, and remains in remission. The hyper-responsive p-STAT5 population has been absent in his remission samples (Figure 4). In contrast, patient HM1394 was poorly responsive to chemotherapy, never achieved complete clinical remission, and relapsed shortly after undergoing a first matched-related HSCT. Following a second HSCT, the patient progressed to M4 AML. Over time, subsequent samples demonstrated an increasing p-STAT5 response at low concentrations of GM-CSF (Figure 4). To further demonstrate that the p-STAT5 population reflected clonal cells, genomic DNA was isolated from the responsive population after sorting on CD38 and p-STAT5 at 0.32 ng/ml of GM-CSF in a patient harboring a somatic NRASG12D mutation. These cells contained the NRAS mutation whereas DNA extracted from the patient’s T cells did not (data not shown).
Figure 4. The p-STAT5 responsive population disappears in remission and reappears during relapse.
Cells from JMML patients were profiled at diagnosis and later following a change in disease status. Both patients exhibited the characteristic JMML signature of hypersensitivity of STAT5 to 0.16 and 0.32 ng GM-CSF at diagnosis. Patient HM1394 responded poorly to upfront therapy, relapsed, and eventually transformed to AML M4. As the disease worsened, a greater percentage of cells were hyper-responsive. Likewise, patient HM1158 went into remission and no longer displayed the JMML signature.
Discussion
Based on the presence of Ras pathway mutations in JMML [reviewed in (Emanuel, 2004; Flotho et al., 2007)] the characteristic GM-CSF hypersensitivity of myeloid progenitors isolated from JMML patients in methylcellulose culture, and compelling data from Kras, Nf1, and Ptpn11 mutant mice linking genetic lesions found in JMML to MPD and GM-CSF hypersensitivity (Araki et al., 2004; Braun et al., 2004; Chan et al., 2004; Le et al., 2004; Mohi et al., 2005), we used phospho-specific flow cytometry to assay p-ERK and p-S6 levels in patient samples. The finding that a small proportion of CD33+, CD14+, CD38dim cells exhibited hyper-phosphorylation of p-STAT5 in response to subsaturating concentrations of GM-CSF was unexpected. Whereas studies of JMML patient samples and of mice lacking either Gmcsf or the βcommon chain of the murine GM-CSF receptor (Birnbaum et al., 2000; Kim et al., 2007) provide strong evidence that an aberrant response to GM-CSF is integral to pathogenesis of JMML, the role of STAT5 has not been explored.
Our data raise the intriguing possibility that Ras-GTP is upstream of JAK2-STAT5 activation in the aberrant response of JMML cells to GM-CSF. Ras pathway mutations might potentiate JAK/STAT signaling by stabilizing or directly activating the GM-CSF receptor or its associated signaling molecules. The SHP-2 phosphatase, which is deregulated by JMML-associated mutations, is recruited to phosphorylated tyrosine residues on the activated β subunit of the GM-CSF receptor, and is essential for efficient STAT5 activation in myeloid cells that are stimulated with interleukin 3(IL-3) (Yu et al., 2003). Ras localizes to activated receptor complexes, and elevated levels of Ras-GTP might, in turn, increase the degree and/or duration of JAK2 kinase activity.
We considered the possibility that hyperactivation of Ras/Raf/MEK/ERK signaling in JMML cells might overwhelm a negative regulatory molecule that normally suppresses GM-CSF induced JAK/STAT signaling. If so, inhibitors of this effector pathway would be expected to interfere with the hypersensitive response of STAT5 to GM-CSF. However, exposing primary JMML cells to CI-1040 failed to alter the p-STAT5 response despite inhibiting p-ERK. As a number of reports have shown that STAT5 is essential for establishing murine MPDs, including BCR/ABL positive chronic myelogenous leukemia (CML) and CMML (Baker et al., 2007; Cain et al., 2007; Ilaria and Van Etten, 1996; Paukku and Silvennoinen, 2004; Van Etten, 2004), understanding the biochemical mechanism underlying elevated p-STAT5 levels in myeloid malignancies with mutations in Ras signaling genes has therapeutic implications.
While highly concordant, we observed discrepancies between p-STAT5 activation in response to low concentrations of GM-CSF and a hypersensitive pattern of CFU-GM colony growth in three types of patients. This is perhaps not unexpected as the CFU-GM assay interrogates a population of cultured myeloid progenitors that form colonies after two weeks, whereas phospho-specific flow cytometry measures a specific biochemical response of a more mature monocytoid cell population to a burst of GM-CSF. It is also important to recall that a hypersensitive pattern of CFU-GM colony growth in methylcellulose is neither necessary nor sufficient to establish a diagnosis of JMML. With these caveats in mind, rare patients who showed differences between the two assays raise interesting questions that have implications for diagnosis and disease management.
Infants with NS/MPD are an interesting group. Whereas myeloid progenitors from these patients display hypersensitivity to GM-CSF in CFU-GM assays, we did not detect the p-STAT5 signature in our phospho-flow assay. Importantly, it is now recognized that, like the transient MPD seen in neonates with Down syndrome, the MPD that occurs in infants with NS usually resolves without treatment (Bader-Meunier et al., 1997; Kratz et al., 2005). Current management of these patients involves watchful waiting. Germline PTPN11 mutations encode weaker gain-of-function alleles than somatic leukemia-associated mutations by a number of criteria, including progenitor colony growth (Araki et al., 2004; Mohi et al., 2005; Schubbert et al., 2006). Based on these data, it is likely that phospho-flow cytometry provides a more specific readout of the rewired signaling networks found in JMML, an aggressive clonal myeloid malignancy, versus the transient MPD that occurs in patients with NS. An interesting question for future studies is if phospho-flow cytometry can prospectively identify those rare infants with NS/MPD who will manifest an aggressive clinical course.
We also encountered a child without NS (patient HM1753) who met diagnostic criteria for JMML early in life, but improved without treatment. When we evaluated him at 2 years of age, he demonstrated modest GM-CSF hypersensitivity but did not exhibit the phospho-flow signature. Molecular analysis revealed a NRASG13D mutation. Interestingly, other cases of JMML associated with somatic NRAS mutations have been reported to spontaneously clinically regress over time (Flotho et al., 2008; Matsuda et al., 2007); but have retained their RAS lesions. The molecular basis for this is unclear, but could involve the initiating NRAS mutation occurring in a cell lacking unlimited self-renewal potential (i.e. a non-stem cell). Alternatively, since Ras proteins with amino acid substitutions at codon 13 exhibit higher intrinsic GTPase activity than codon 12 mutant proteins (Ahmadian et al., 1999), it is possible that some mutant RAS alleles are insufficiently activated to cause aggressive JMML. This possibility is consistent with an infant with NS and a moderately activating germline KRAST58I mutation who showed spontaneous regression of a JMML-like MPD during the first year of life (Schubbert NG 2006), and with the NS/MPD paradigm discussed above. Importantly, our analysis of this patient with a NRASG13D mutation further suggest that a normal phospho-flow signature may identify children with NRAS and PTPN11 mutations who will have a benign clinical course and can be observed closely without aggressive treatment. Further studies of additional patients will be necessary to confirm this hypothesis.
The only case in our series with aggressive JMML who did not exhibit the characteristic GM-CSF/p-STAT5 phospho-flow signature harbored a KRASG12D mutation. In recent studies of additional cases, we have confirmed that the vast majority of children meeting the clinical criteria for JMML exhibit aberrant p-STAT5 activation in response to GM-CSF with the exception of a second patient with a KRASG12D mutation who did not show this signaling abnormality. Although these data need to be prospectively validated in a larger cohort study, they suggest that the KRASG12D, which is phenotypically aggressive in murine models (Braun et al., 2004; Chan et al., 2004), is less dependent upon JAK2/STAT5 signaling than other Ras pathway mutations. Interestingly, phospho-signaling analysis of c-kit+/lineagedim/− cells from KrasG12D mice with MPD did not reveal abnormal STAT5 activation by GM-CSF (Van Meter et al., 2007) (and data not shown). Our data therefore raise the intriguing possibility that myeloid malignancies with mutations in KRAS show differential activation of p-STAT5 (and perhaps other signaling molecules) than cells that express oncogenic NRAS or PTPN11.
A recent publication of WHO 2008 classification guidelines (Tefferi et al., 2007; Tefferi and Vardiman, 2008) suggests diagnostic approaches to combine molecular pathogenesis along with histology for both classic and atypical myeloproliferative disorders. Discoveries of somatic mutations in Ras signaling molecules have improved diagnostic capabilities for JMML but are still not universally applicable. In addition, following patients on therapy remains challenging in the absence of tractable markers. Our data imply that advances in proteomic and single-cell flow cytometry technologies will add to the genetic mapping of these disorders, allow tracking of rare cell populations that would be difficult to observe in bulk assay approaches (RNA expression or mass spectrometry) and allow us to measure specific activity at the protein level. We demonstrate the diagnostic value of such information in JMML and CMML and anticipate that further phospho-flow cytometry based assays will allow for direct measurements of the key signaling events required for disease maintenance. Finally, the results suggest that JMML, CMML, and M4/M5 AML are related entities in which hyperactive Ras and aberrant JAK2/STAT5 signaling are early or initiating events(Braun et al., 2004). As such, M4/M5 AML might be distinct from other subtypes of AML, in which aberrant transcription factor fusions such as PML-RARA and AML1-ETO likely represent primary leukemogenic events (Gilliland and Griffin, 2002). This has important therapeutic implications, as M4/M5 AML might be highly dependent on Ras and JAK2/STAT5 signaling, and therefore sensitive to inhibitors of these pathways. Revealing cell subpopulations associated with disease opens additional avenues for measuring minimal residual disease, assessing the biochemical effects of targeted therapies at the single cell level, and understanding drug action and mechanisms in diseases of heterogeneous origins and manifestations in diverse patient populations.
Experimental procedures
Sample Collection
Fresh bone marrow or peripheral blood samples were obtained from children suspected of having JMML or another myeloproliferative disorder related to a congenital syndrome, including NS or Down syndrome. In addition, the following hematopoietic tissues were analyzed in order to demonstrate the specificity of the JMML phosphoprotein signature: a) archived frozen bone marrow products from healthy sibling donors for hematopoietic stem cell transplants, b) fresh cord blood from term neonates, c) normal bone marrows taken from children suspected of having a metastatic solid tumor, d) diagnostic bone marrow or pheresis samples from children or adults with AML, e) archived bone marrow or peripheral blood samples from adult patients with chronic myelomonocytic leukemia (CMML). All samples were obtained with informed consent. This study has approval from the UCSF Committee on Human Research. Samples were collected in sodium heparin and mononuclear cells were isolated according to standard methods.
Frozen samples used in the study were cryopreserved in 90% FBS/10%DMSO. Supplementary Table 1 indicates fresh or frozen status of samples used for the phosphoflow analysis, however all colony assays were performed only on fresh material. Supplementary Table 2 lists cytogenetic information about the AML samples.
Genotyping
All patients referred for workup of a JMML diagnosis were genotyped for PTPN11 exon 3, 13, NRAS exon 1, 2, and KRAS exon 1, 2 according to previously published methodologies (Kalra et al., 1994; Loh et al., 2004; Meshinchi et al., 2003). Genomic DNA was prepared using PureGene reagents (Qiagen, Valencia, CA). Data was also collected for a family history of and World Health Organization criteria for NF1. The eight patients with AML were additionally genotyped for FLT3 ITD, FLTD835 according to previously published methodologies (Meshinchi et al., 2003; Zwaan et al., 2003).
CFU-GM assay
Mononuclear cells were isolated from fresh bone marrow samples and resuspended in Iscove’s Modified Dulbecco’s Medium (IMDM) + 2 % fetal bovine serum (FBS). Cells were suspended at a concentration of 200,000 cells per ml. 154 µL of the cell suspension was added to a tube with the following: 1.2 ml MethoCult H4230 methylcellulose (Cat. 04230, Stem Cell Technologies, Vancouver, British Columbia, 15 µL of 100 × Penicillin/Streptomycin, 30.8 µL of human GM-CSF titration (Cat. #300-03, PeproTech) diluted in water and IMDM to complete tube volume to 1.54 ml. The solution was vortexed for 15 seconds and rested for 15 minutes. 1.1 ml was plated into a 35×10 mm Petri dish (Cat. 351008, BD Falcon, Franklin Lakes, NJ) placed into a 150×15 mm Petri dish (Cat. 351058, BD Falcon) with another dish containing sterile water, and placed into an incubator at 37 degrees Celsius and 5% CO2.
After 14 days, plates were removed from the incubator and colonies (a cluster of 50 cells or more) were counted under a microscope at 40X magnification. Data is presented in supplementary table 1. Hypersensitive growth (+2) corresponds to ~50% of maximal growth whereas hypersensitive growth (+3) corresponds to maximal growth.
Cytokine stimulation and intracellular phospho-protein analysis using flow cytometry
Reagents used for flow cytometry included 16% paraformaldehyde (EM grade, Electron Microscopy Sciences Cat. #15710), methanol (Electron Microscopy Sciences Cat. #18510), StemSpan H3000 (Stem Cell Technologies #09800), and FACS Rehydration/Staining buffer (HBSS plus 4% FBS). Human GM-CSF (Peprotech #300-03), PMA (Sigma Technologies #P8139), and ionomycin (Sigma Technologies #I0634) were used to stimulate cells at concentrations indicated in the text. A chemical JAK2 inhibitor (Calbiochem, #420099, San Diego, CA), an oral JAK2 inhibitor XL019 (Exelixis, South San Francisco, CA) and an oral MEK inhibitor, CI-1040 (Pfizer Inc, NY, NY) were used as described. Antibodies used for phoshoprotein detection were p-STAT5 Alexa 647 (BD Biosciences #612599, 10 µL per sample), CD34-PerCPcy5.5 (BD Biosciences #347213, 7µL per sample), CD38-PECy7 (BD Biosciences #335790, 5 µL per sample), CD11b-Pacific Blue (BD Biosciences #558123, 8µL per sample), CD33 PE (BD Biosciences #347787, 5 µL per sample), CD3 Pacific Blue (BD Biosciences #558117) and CD14 APC Cy7 (BD Biosciences #557831, 5 µL per sample). The primary antibody against p-ERK (p-p44/42 MAPK) was obtained from Cell Signaling (#9101, used at 1:100). The FITC-labeled secondary antibody anti-rabbit IgG was from Jackson Immunoresearch (#711-096-152, used at 1:400).
The primary antibody against p-S6 was obtained from Cell Signaling (#4856, used at 1:50). The FITC-labeled secondary antibody anti-rabbit IgG was 711-096-152 from Jackson Immunoresearch (#711-096-152 used at 1:400).
Freshly isolated or defrosted mononuclear cells were suspended in pre-warmed StemSpan H3000 (StemCell Technologies Cat. #9800) at a concentration of 1–2 million cells/ml and rested at 37 degrees Celsius for 1 hour. The monocytic cell line, U937, which harbors a PTPN11 exon 3 mutation (178G>C, G60R) was used in each assay as a positive control.
Cells were transferred as 1 ml aliquots into flow cytometry test tubes (Falcon 2052; BD-Biosciences) and stimulated with various concentrations of human GM-CSF(Cat. #300-03, PeproTech) for 15 minutes. Cells were fixed by adding 100 µL of 16% paraformaldehyde (PFA; Electron Microscopy Sciences) at room temperature for 10 minutes. Cells were washed (centrifugation 1800 RPM for 5 minutes) twice with phosphate-buffered saline (PBS) and permeabilized by resuspension in 2 ml ice-cold 95% methanol for 10 minutes. Cells were stored in −20 degrees before staining for flow cytometry.
PFA-fixed, methanol-permeablilzed cells were rehydrated by adding 2 ml PBS and washed twice (centrifugation 2500 RPM for 5 minutes). Cells were resuspended in 500 µL FACS buffer (Hanks balanced salt solution containing 4% FBS; Hyclone) and incubated at 4 degrees Celsius for 2 hours.
Unconjugated primary antibodies were added at optimized concentrations (p-ERK 1:100 or p-S6 1:50) and incubated at room temperature for 45 mins. Samples were washed (centrifugation 2500 RPM for 5 min) once with FACS buffer. Secondary and directly conjugated antibodies were added at optimized concentrations and incubated in the dark at room temperature for 30 minutes. Samples were washed (centrifugation, 2500 RPM for 5 min) once with PBS and analyzed on a LSR II flow cytometer (BD Biosciences) equipped with 433 nm and 633 nm lasers.
Data collection and analysis
Data were collected using DIVA software (BD Biosciences) and analyzed using Cytobank, an open source flow cytometry storage and analysis application (Stanford University, Stanford CA). Samples that showed a p-ERK response to PMA were deemed viable and used for analysis.
For Figure 1B, the percent of p-STAT5 responsive cells was determined by drawing a gate using SSC and p-STAT5 to identifying the percent of p-STAT5+ cells and scaling the response such that the maximum percent of p-STAT5+ cells was equivalent to 100 and the percent of p-STAT5+ cells at unstimulated was equal to 0. Error bars represent standard error of the mean (s.e.m.)
For Figure 3, S6, and S7, the normalized p-STAT5 (or p-ERK or p-S6) response was calculated by transforming the raw data using the inverse hyperbolic sine, part of the biexponential class of functions used for digital flow cytometry data (Parks et al., 2006) and calculating a change in the 95th percentile of p-STAT5 response between a GM-CSF stimulated sample and its unstimulated/basal state. The algorithm is outlined as follows:
-
For each patient, stain and collect samples stimulated with GM-CSF at the following concentrations: Unstimulated, 0.04 ng/ml, 0.16ng/ml, 0.32 ng/ml, 2 ng/ml and 10 ng/ml
-
For each sample
Tranform the raw data: sinh−1(rawdata/150)
Identify live cells
Calculate the 95th percentile of p-STAT5 in the transformed space
Subtract this value from the 95th percentile of p-STAT5 of the unstimulated GM-CSF sample
-
For each patient
Calculate the range of p-STAT5 response to GM-CSF: max 95th percentile p-STAT5 difference – min 95th percentile p-STAT5 difference
Ignore patient if the range of response is <0.05
Normalize the change in p-STAT5 such that the unstimulated sample is equal to 0 and the max difference is equal to 100: difference in p-STAT5 / (range of response) *100
-
Supplementary Material
Acknowledgements
The authors wish to thank all of the patients, families, and referring physicians who contributed invaluable clinical information and samples to the investigators. We would also like to acknowledge investigators at MD Anderson Cancer Center who provided samples from CMML patients.
This work was supported by the NIH [NCI K22 CA113557 (M.L.L), NIH U54 CA119367 (G.P.N.), P30 CA82103 (M.L.L, N.F.), NIH N01 HV28183 (G.P.N, N.K.), NIH P01 CA34233 (G.P.N, N.K.) NIH PO1 CA108631 (M.L.L, K.M.S., D.S)], The Leukemia Lymphoma Society [LLS 7017-06 (G.P.N.), 2157-08 (M.L.L), 7019-04 (K.M.S.)], The V Foundation for Cancer Research, and the Frank A. Campini Foundation (M.L.L, K.M.S). M.L.L. is a Clinical Scholar of the Leukemia Lymphoma Society. J.M.I. is a Fellow of the Leukemia Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadian MR, Zor T, Vogt D, Kabsch W, Selinger Z, Wittinghofer A, Scheffzek K. Guanosine triphosphatase stimulation of oncogenic Ras mutants. Proc Natl Acad Sci U S A. 1999;96:7065–7070. doi: 10.1073/pnas.96.12.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, Neel BG. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- Archambeault S, Flores NJ, Yoshimi A, Kratz CP, Reising M, Fischer A, Noellke P, Locatelli F, Sedlacek P, Flotho C, et al. Development of an allele-specific minimal residual disease assay for patients with juvenile myelomonocytic leukemia. Blood. 2008;111:1124–1127. doi: 10.1182/blood-2007-06-093302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico M, Biondi A, Pui CH. Juvenile myelomonocytic leukemia. Blood. 1997;90:479–488. [PubMed] [Google Scholar]
- Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood. 2006;107:3847–3853. doi: 10.1182/blood-2005-08-3522. [DOI] [PubMed] [Google Scholar]
- Bader-Meunier B, Tchernia G, Miélot F, Fontaine JL, Thomas C, Lyonnet S, Lavergne JM, Dommergues JP. Occurrence of myeloproliferative disorder in patients with the Noonan syndrome. J Pediatr. 1997;130:885–889. doi: 10.1016/s0022-3476(97)70273-7. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene. 2007;26:6724–6737. doi: 10.1038/sj.onc.1210757. [DOI] [PubMed] [Google Scholar]
- Birnbaum RA, O'Marcaigh A, Wardak Z, Zhang YY, Dranoff G, Jacks T, Clapp DW, Shannon KM. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol Cell. 2000;5:189–195. doi: 10.1016/s1097-2765(00)80415-3. [DOI] [PubMed] [Google Scholar]
- Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, Rozmus J, Le Beau MM, Jacks TE, Shannon KM. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain JA, Xiang Z, O'Neal J, Kreisel F, Colson A, Luo H, Hennighausen L, Tomasson MH. Myeloproliferative disease induced by TEL-PDGFRB displays dynamic range sensitivity to Stat5 gene dosage. Blood. 2007;109:3906–3914. doi: 10.1182/blood-2006-07-036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier N, Baruchel A, Schlageter MH, Menot ML, Wattel E, Fenaux P, Chomienne C. Chronic myelomonocytic leukemia: from biology to therapy. Hematol Cell Ther. 1997;39:41–48. doi: 10.1007/s00282-997-0041-4. [DOI] [PubMed] [Google Scholar]
- Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, Shigematsu H, Johnson L, Akashi K, Tuveson DA, Jacks T, Gilliland DG. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel PD. Juvenile myelomonocytic leukemia. Curr Hematol Rep. 2004;3:203–209. [PubMed] [Google Scholar]
- Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77:925–929. [PubMed] [Google Scholar]
- Flotho C, Kratz C, Niemeyer CM. Targeting RAS signaling pathways in juvenile myelomonocytic leukemia. Curr Drug Targets. 2007;8:715–725. doi: 10.2174/138945007780830773. [DOI] [PubMed] [Google Scholar]
- Flotho C, Kratz CP, Bergstrasser E, Hasle H, Stary J, Trebo M, van den Heuvel-Eibrink MM, Wojcik D, Zecca M, Locatelli F, Niemeyer CM. Genotype-phenotype correlation in cases of juvenile myelomonocytic leukemia with clonal RAS mutations. Blood. 2008;111:966–967. doi: 10.1182/blood-2007-09-111831. author reply 967–968. [DOI] [PubMed] [Google Scholar]
- Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- Ilaria RL, Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6:146–155. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- Kalra R, Paderanga DC, Olson K, Shannon KM. Genetic analysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood. 1994;84:3435–3439. [PubMed] [Google Scholar]
- Kim A, Morgan K, Haszx DE, Wiesner SM, Lauchle JO, Geurts JL, Diers MD, Le DT, Kogan SC, Parada LF, et al. Beta common receptor inactivation attenuates myeloproliferative disease in Nf1 mutant mice. Blood. 2007;109:1687–1691. doi: 10.1182/blood-2006-05-025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz CP, Niemeyer CM, Castleberry RP, Cetin M, Bergstrasser E, Emanuel PD, Hasle H, Kardos G, Klein C, Kojima S, et al. The mutational spectrum of PTPN11 in juvenile myelomonocytic leukemia and Noonan syndrome/myeloproliferative disease. Blood. 2005;106:2183–2185. doi: 10.1182/blood-2005-02-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz M, Ibrahim SM. Molecular responses to hypoxia in tumor cells. Mol Cancer. 2003;2:23. doi: 10.1186/1476-4598-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Kong N, Zhu Y, Lauchle JO, Aiyigari A, Braun BS, Wang E, Kogan SC, Le Beau MM, Parada L, Shannon KM. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood. 2004;103:4243–4250. doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- Levine RL, Loriaux M, Huntly BJ, Loh ML, Beran M, Stoffregen E, Berger R, Clark JJ, Willis SG, Nguyen KT, et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood. 2005;106:3377–3379. doi: 10.1182/blood-2005-05-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli F, Nollke P, Zecca M, Korthof E, Lanino E, Peters C, Pession A, Kabisch H, Uderzo C, Bonfim CS, et al. Hematopoietic stem cell transplantation (HSCT) in children with juvenile myelomonocytic leukemia (JMML): results of the EWOG-MDS/EBMT trial. Blood. 2005;105:410–419. doi: 10.1182/blood-2004-05-1944. [DOI] [PubMed] [Google Scholar]
- Loh ML, Martinelli S, Cordeddu V, Reynolds MG, Vattikuti S, Lee CM, Wulfert M, Germing U, Haas P, Niemeyer C, et al. Acquired PTPN11 mutations occur rarely in adult patients with myelodysplastic syndromes and chronic myelomonocytic leukemia. Leuk Res. 2005;29:459–462. doi: 10.1016/j.leukres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Loh ML, Vattikuti S, Schubbert S, Reynolds MG, Carlson E, Lieuw KH, Cheng JW, Lee CM, Stokoe D, Bonifas JM, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004;103:2325–2331. doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Shimada A, Yoshida N, Ogawa A, Watanabe A, Yajima S, Iizuka S, Koike K, Yanai F, Kawasakix K, et al. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109:5477–5480. doi: 10.1182/blood-2006-09-046649. [DOI] [PubMed] [Google Scholar]
- McCubrey JA, May WS, Duronio V, Mufson A. Serine/threonine phosphorylation in cytokine signal transduction. Leukemia. 2000;14:9–21. doi: 10.1038/sj.leu.2401657. [DOI] [PubMed] [Google Scholar]
- Meshinchi S, Stirewalt DL, Alonzo TA, Zhang Q, Sweetser DA, Woods WG, Bernstein ID, Arceci RJ, Radich JP. Activating mutations of RTK/ras signal transduction pathway in pediatric acute myeloid leukemia. Blood. 2003;102:1474–1479. doi: 10.1182/blood-2003-01-0137. [DOI] [PubMed] [Google Scholar]
- Mohi MG, Williams IR, Dearolf CR, Chan G, Kutok JL, Cohen S, Morgan K, Boulton C, Shigematsu H, Keilhack H, et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 2005;7:179–191. doi: 10.1016/j.ccr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Niemeyer CM, Arico M, Basso G, Biondi A, Cantu Rajnoldi A, Creutzig U, Haas O, Harbott J, Hasle H, Kerndrup G, et al. Chronic myelomonocytic leukemia in childhood: a retrospective analysis of 110 cases. European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS) Blood. 1997;89:3534–3543. [PubMed] [Google Scholar]
- Onida F, Kantarjian HM, Smith TL, Ball G, Keating MJ, Estey EH, Glassman AB, Albitar M, Kwari MI, Beran M. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99:840–849. doi: 10.1182/blood.v99.3.840. [DOI] [PubMed] [Google Scholar]
- Parks DR, Roederer M, Moore WA. A new "Logicle" display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69:541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- Paukku K, Silvennoinen O. STATs as critical mediators of signal transduction and transcription: lessons learned from STAT5. Cytokine Growth Factor Rev. 2004;15:435–455. doi: 10.1016/j.cytogfr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Rane SG, Reddy EP. JAKs, STATs and Src kinases in hematopoiesis. Oncogene. 2002;21:3334–3358. doi: 10.1038/sj.onc.1205398. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- Steensma DP, Dewald GW, Lasho TL, Powell HL, McClure RF, Levine RL, Gilliland DG, Tefferi A. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both "atypical" myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005;106:1207–1209. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Thiele J, Orazi A, Kvasnicka HM, Barbui T, Hanson CA, Barosi G, Verstovsek S, Birgegard G, Mesa R, et al. Proposals and rationale for revision of the World Health Organization diagnostic criteria for polycythemia vera, essential thrombocythemia, and primary myelofibrosis: recommendations from an ad hoc international expert panel. Blood. 2007;110:1092–1097. doi: 10.1182/blood-2007-04-083501. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vardiman JW. Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2008;22:14–22. doi: 10.1038/sj.leu.2404955. [DOI] [PubMed] [Google Scholar]
- Van Etten RA. Mechanisms of transformation by the BCR-ABL oncogene: new perspectives in the post-imatinib era. Leuk Res. 2004;28 Suppl 1:S21–S28. doi: 10.1016/j.leukres.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, Nolan GP, Shannon K, Braun BS. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood. 2007;109:3945–3952. doi: 10.1182/blood-2006-09-047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WM, Hawley TS, Hawley RG, Qu CK. Catalytic-dependent and-independent roles of SHP-2 tyrosine phosphatase in interleukin-3 signaling. Oncogene. 2003;22:5995–6004. doi: 10.1038/sj.onc.1206846. [DOI] [PubMed] [Google Scholar]
- Zecca M, Bergamaschi G, Kratz C, Bergstrasser E, Danesino C, De Filippi P, Hasle H, Lisini D, Locatelli F, Pession A, et al. JAK2 V617F mutation is a rare event in juvenile myelomonocytic leukemia. Leukemia. 2007;21:367–369. doi: 10.1038/sj.leu.2404484. [DOI] [PubMed] [Google Scholar]
- Zwaan CM, Meshinchi S, Radich JP, Veerman AJ, Huismans DR, Munske L, Podleschny M, Hahlen K, Pieters R, Zimmermann M, et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: prognostic significance and relation to cellular drug resistance. Blood. 2003;102:2387–2394. doi: 10.1182/blood-2002-12-3627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.