Abstract
Purpose
Nance-Horan Syndrome (NHS) is an infrequent and often overlooked X-linked disorder characterized by dense congenital cataracts, microphthalmia, and dental abnormalities. The syndrome is caused by mutations in the NHS gene, whose function is not known. The purpose of this study was to identify the frequency and distribution of NHS gene mutations and compare genotype with Nance-Horan phenotype in five North American NHS families.
Methods
Genomic DNA was isolated from white blood cells from NHS patients and family members. The NHS gene coding region and its splice site donor and acceptor regions were amplified from genomic DNA by PCR, and the amplicons were sequenced directly.
Results
We identified three unique NHS coding region mutations in these NHS families.
Conclusions
This report extends the number of unique identified NHS mutations to 14.
Introduction
Nance-Horan Syndrome (NHS; OMIM 302350) is an infrequent and often overlooked X-linked disorder characterized by dense congenital cataracts, microphthalmia, and microcornea [1-3]. Affected individuals develop dental anomalies which include serrated incisal edges and barrel-shaped teeth, in which the incisal edge and the gingival margin are narrower than the cervix of the tooth. Distinctive facial features include anteverted and often simplex pinnae and a long face with a narrow mandible and a narrow nasal bridge. Some affected males also display developmental delay [4,5]. The phenotype of female carriers is less severe but includes corneal diameters ranged midway between normal and the affected males, and Y-sutural opacities which may occasionally affect vision even at younger ages [6]. The variability among carriers presumably results from differentially random X-inactivation.
The gene for NHS was mapped by several investigators to the Xp22.13 region [5,7-11]. The NHS gene was isolated and confirmed by independent laboratories [12,13]. Two additional studies [14,15] identified unique NHS gene mutations in four other independent NHS families. Previously, we identified five North American families with the unifying phenotypic characteristics of NHS [6,16]. Here we report the analyses of NHS gene mutation of these five families, bringing the total number of unique mutations to fourteen.
Methods
Clinical examination
Families exhibiting X-linked congenital cataract and microcornea were ascertained through the Medical Retina and Ocular Genetics Service of the Cullen Eye Institute at Baylor College of Medicine in Houston and by regional and national referral. A detailed family history and a pedigree were obtained through personal interviews and corroborated by medical and ophthalmological examination. Each subject or the responsible adult signed a Consent for Participation which was approved by the Institutional Review Board for Human Subject Research of the Baylor College of Medicine and the corresponding committees at the Texas Children's Hospital and The Methodist Hospital, Houston.
Unambiguous distinctions between affected and normal male subjects were based on clinical criteria including head circumference, external ear length, the position of the pinnae, the shape of the helices, hand and finger lengths, dental anomalies, and ocular characteristics (Table 1). Blood samples were obtained by a single observer with no knowledge of linkage information. Detailed examination findings of individuals and family pedigrees have been published elsewhere [1,6,16,17].
Table 1. Clinical characteristics of NHS patients.
| Family |
Congenital cataract |
Nystagmus |
Microcornea |
Microphthalmia |
Dental anomalies |
Mental retardation |
Brachymetacarpalia |
Dysmorphic facial features |
| XL-11 |
+ |
+ |
+ |
+ |
+ |
No |
+ |
+ |
| XL-56 |
+ |
+/- |
+ |
+ |
+ |
No |
+ |
+ |
| XL-51 |
+ |
+ |
+ |
+ |
+ |
No |
+ |
+ |
| XL-116 |
+ |
+ |
+ |
+ |
+ |
No |
+ |
+ |
| XL-39 | + | + | + | + | + | No | + | + |
Mutation screening and sequence analysis
Venous blood from family members established permanent EBV-transformed lymphoblastoid cultured cell lines with standard techniques [18]. Genomic DNA was isolated from EBV transformed cells using Qiagen kits, following the manufacturer's recommendations. Exons from the NHS gene were amplified from genomic DNA (100 ng) with primers that span intron-exon junctions and primers within single large exons (primer sequences listed in Table 2). This analysis included exon 3a which is variably expressed in human NHS transcripts [19]. PCR was performed with 0.5 U AmpliTaq gold polymerase (ABI, Foster City, CA), 10 mM Tris HCl, pH 8.3, 1.5 mM MgCl2, and 50 mu M each dATP, dTTP, dGTP, and dCTP in 50 mu l volume. Amplification was performed following these conditions: 94 °C for 5 min, followed by 35 cycles at 95 °C for 1 min, 55-60 °C for 30 s, and 72 °C for 1 min with a final extension at 72 °C for 7 min. PCR products were separated by gel electrophoresis, purified over gel filtration columns (Novagen, Madison, WI), and cloned into TOPO II vectors (Invitrogen, Carlsbad, CA), or sequenced directly as necessary on a 3900 DNA Sequencer (Applied Biosystems, Foster City, CA). For each mutation found in families XL116, XL11, and XL39, the presence of the mutation was also analyzed in two unaffected family members.
Table 2. Primer sequences used in PCR of genomic DNA.
|
NHS1 exon |
Primer sequence |
| 1 |
1F TATCCGGACTGCCAGATCGC |
| 1R GAGTAGTAAGGTGCAAGCTGC |
|
| 2 |
2F GTTGGCCAAAAGCACAACTT |
| 2R GGTGTGTTGGGGCTGATG |
|
| 3 |
3F ACTCCCAAGGGGAAAAGAGA |
| 3R TTCCTCAGCAGCAAGCATAG |
|
| 3aF ATACACTGTGTTGTGTGCACG |
|
| 3aR TCTGGACAGAGTGGGATAGG |
|
| 4 |
4F TTCCTTTGTCCTAAGGGCCTA |
| 4R TGGTATTCTTAGCAGCACAGA |
|
| 5 |
5F TGAGACCTATTTGTGGGTTGC |
| 5R TCTGTACTAGGCGGAGGAATG |
|
| 6 |
6.1 F TCACTGTGCTTTCCATGTGC |
| 6.1 R ATGTGGCTGCTAAGGAGGAC |
|
| 6.15F GGCCTGCTCTCAACATCTTC |
|
| 6.2F GCCACATGGACCAGAAAGAT |
|
| 6.2R CAAGAGGCAGCTTCATTTCC |
|
| 6.3F CAGCACCTGCCTCACAGTT |
|
| 6.3R TCTTCAGACTTGTTGATGGACCT |
|
| 6.4 FAAGCAG AAC ACAGTAGG AG AAACA |
|
| 6.4R TCCTTCTGTGGGAAAAGCAC |
|
| 6.5F TCTCCCTTATTTAGAGGAAAGCA |
|
| 6.5R TACCAGGCACTTTGTCATGG |
|
| 6.6F GCAGTTGAGATGGGACCAGA |
|
| 6.6R ATTCCAGGAAGTGCCATGAG |
|
| 7 |
7F TAGCGTGCTGGGTAACTTCC |
| 7R GGGGCAAAACCTTTGTTGTA |
|
| 8 |
8F GTGAGATGTTTGCCCCATTT |
| 8R GTAAGGGTTTTGGCCTTTGC |
|
| 8.1 F TGAGATGTTTGCCCCATTTT |
|
| 8.1 R TGGCAGACATGCGGTAACTA |
|
| 8.2F AAGAAAGGCAGTCGCTCAGA |
|
| 8.2R GTAAGGGTTTTGGCCTTTG |
Results & Discussion
In family XL-116, we identified a C >T nonsense mutation in all tested affected individuals, at nucleotide 4129 of the 4893 bp human NHS cDNA. This mutation introduces a putative premature stop codon at the end of exon 6 and would result in a truncated protein that lacks a small part of exon 6 and all of exons 7 and 8. Affected individuals in family XL-39 carry a C >A nonsense mutation that introduces a putative premature stop codon at nucleotide 3624. This mutation falls in the middle of exon 6 and results in a truncated protein lacking part of exon 6 and exons 7 and 8. Family XL-11 displays a C >T nonsense mutation at nucleotide 1108. This mutation occurs in exon 5 and predicts a truncated protein lacking part of exon 5 and all of exons 6, 7, and 8. None of these mutations was observed in any unaffected family members. No coding region or splice site alterations were identified in families XL-56 and XL-51. These mutation results are presented as Figure 1 and Table 3.
Figure 1.
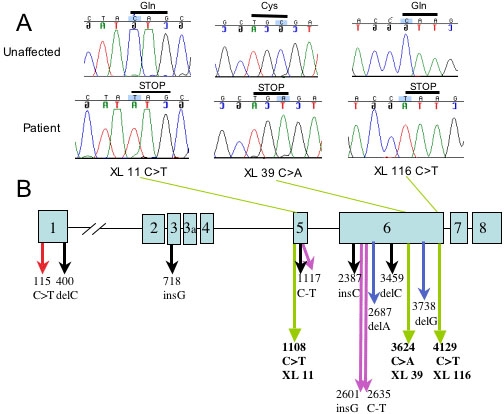
NHS mutation detection and localization. A: The identification of NHS nonsense mutations in three NHS families. Chromatograms from one affected and one unaffected individual are shown for each family. Above each tracing the base change is highlighted and the predicted stop codon is indicated. B: The localization of the fourteen reported NHS mutations within the NHS gene: red arrow [14]; black arrows [12]; green arrows (this study); purple arrows [15]; and blue arrows [13].
Table 3. Summary of mutation in the NHS gene.
|
Reference |
Family |
Exon |
Genomic mutation |
Predicted protein change |
| [12] |
1 |
6 |
c.2387insC |
p.S797 fsX |
| 2 |
6 |
c.3459delC |
p.L1154fsX28 |
|
| 3* |
5 |
c.1117C>T |
P.R378X |
|
| 4 |
3 |
c.718insG |
p.E240fsX36 |
|
| 5** |
1 |
c.400delC |
p.R134fsX61 |
|
| 6 |
- |
No mutation |
||
| [13] |
1 |
6 |
c.3738-3739delTG |
p.C1246-A1247fsX15 |
| 2** |
1 |
c.400delC |
p.R134fsX61 |
|
| 3 |
6 |
c.2687delA |
p.Q896fsX110 |
|
| CRX |
- |
No mutation |
||
| [14] |
1 |
1 |
C.1150T |
p.Q39X |
| [15] |
P8598 |
IVS 3-2 |
c.853-2 A>G |
Splice site change |
| P20079 |
6 |
c.2601insG |
p.K868E fsX5 |
|
| P21540* |
5 |
c.1117C>T |
P.R378X |
|
|
P24486 |
6 |
c.2635C>T |
p.R879X |
|
| This study | XL 39 |
6 |
c.3624C>A |
P.C1208X |
| XL 116 |
6 |
c.4129C>T |
P.Q1358X |
|
| XL 11 |
5 |
c.1108C>T |
P.Q370X |
|
| XL 51 |
- |
No mutation |
||
| XL 56 | - | No mutation |
Our analyses are consistent with previous reports that showed no correlation between the NHS genotype and the severity of NHS signs among affected family members [12,15]. For example, patients carrying the C >A mutation at bp 3624 present with typical NHS ocular and dental features [15]. In contrast, patients with a TG deletion at the neighboring positions 3738-3739 have bilateral cleft palate and the classical NHS signs [13] (family 3). Similarly, Ramprasad et al. [14] and Burdon et al. [12] identified two different NHS mutations within exon 1. However, the reported phenotype in patients bearing a mutation at nucleotide 400 is more severe than those with a mutation at nucleotide 115. A detailed structure-function analysis of the NHS protein should reconcile the phenotypic differences in the several NHS families.
Our study includes two families with no identified NHS mutations in the coding region. This is consistent with previous studies [12,13] and suggests the possibility of regulatory or intronic mutations in these families. Interestingly, the equivalent mouse Xcat mutant, which is the model for human NHS, does not display coding region mutations but does carry a large insertion mutation within the first intron of the mouse homolog of NHS (Nhs1) [20]. This insertion alters the expression of the Nhs1 transcript and the Nhs protein in Xcat mice. Perhaps the NHS families who do not display NHS coding region mutations carry alterations in intron 1 structure. It will be interesting to explore this possibility further through the use of cytogenetic and gene expression studies on the NHS transformed cell lines. This study identifies additional NHS gene mutations in NHS families and validates the model that the NHS gene is required for proper eye development in humans. In addition, this work is consistent with the role of mouse Nhs1 in cataract formation. Based on the phenotype of NHS patients, the NHS gene has many specialized functions in several different tissues. Functional studies on the NHS protein should help illuminate its role in eye, ear, tooth, and digit development.
Acknowledgements
Sincere appreciation is extended to the families described here for their willing and continuing cooperation in these investigations. These studies were supported in part by the U.S. Public Health National Eye Institute grant R01 EY13615 (K.M.H.) and unrestricted funds from Research to Prevent Blindness (RPB), New York, New York and Wellcome Trust UK (077477/Z/05/Z). Dr. Lewis is a Senior Scientific Investigator of RPB.
References
- 1.Nance WE, Warburg M, Bixler D, Helveston EM. Congenital X-linked cataract, dental anomalies and brachymetacarpalia. Birth Defects Orig Artic Ser. 1974;10:285–91. [PubMed] [Google Scholar]
- 2.Horan M, Billson F. X-linked cataract and hutchinsonian teeth. Aust Paediatr J. 1974;10:98–102. [Google Scholar]
- 3.Walpole IR, Hockey A, Nicoll A. The Nance-Horan syndrome. J Med Genet. 1990;27:632–4. doi: 10.1136/jmg.27.10.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toutain A, Ayrault AD, Moraine C. Mental retardation in Nance-Horan syndrome: clinical and neuropsychological assessment in four families. Am J Med Genet. 1997;71:305–14. doi: 10.1002/(sici)1096-8628(19970822)71:3<305::aid-ajmg11>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 5.Stambolian D, Lewis RA, Buetow K, Bond A, Nussbaum R. Nance-Horan syndrome: localization within the region Xp21.1-Xp22.3 by linkage analysis. Am J Hum Genet. 1990;47:13–9. [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis RA. Mapping the gene for X-linked cataracts and microcornea with facial, dental, and skeletal features to Xp22: an appraisal of the Nance-Horan syndrome. Trans Am Ophthalmol Soc. 1989;87:658–728. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu D, Alcorn DM, Antonarakis SE, Levin LS, Huang PC, Mitchell TN, Warren AC, Maumenee IH. Assignment of the Nance-Horan syndrome to the distal short arm of the X chromosome. Hum Genet. 1990;86:54–8. doi: 10.1007/BF00205172. [DOI] [PubMed] [Google Scholar]
- 8.Stambolian D, Favor J, Silvers W, Avner P, Chapman V, Zhou E. Mapping of the X-linked cataract (Xcat) mutation, the gene implicated in the Nance Horan syndrome, on the mouse X chromosome. Genomics. 1994;22:377–80. doi: 10.1006/geno.1994.1398. [DOI] [PubMed] [Google Scholar]
- 9.Toutain A, Ronce N, Dessay B, Robb L, Francannet C, Le Merrer M, Briard ML, Kaplan J, Moraine C. Nance-Horan syndrome: linkage analysis in 4 families refines localization in Xp22.31-p22.13 region. Hum Genet. 1997;99:256–61. doi: 10.1007/s004390050349. [DOI] [PubMed] [Google Scholar]
- 10.Toutain A, Dessay B, Ronce N, Ferrante MI, Tranchemontagne J, Newbury-Ecob R, Wallgren-Pettersson C, Burn J, Kaplan J, Rossi A, Russo S, Walpole I, Hartsfield JK, Oyen N, Nemeth A, Bitoun P, Trump D, Moraine C, Franco B. Refinement of the NHS locus on chromosome Xp22.13 and analysis of five candidate genes. Eur J Hum Genet. 2002;10:516–20. doi: 10.1038/sj.ejhg.5200846. [DOI] [PubMed] [Google Scholar]
- 11.Francis PJ, Berry V, Hardcastle AJ, Maher ER, Moore AT, Bhattacharya SS. A locus for isolated cataract on human Xp. J Med Genet. 2002;39:105–9. doi: 10.1136/jmg.39.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burdon KP, McKay JD, Sale MM, Russell-Eggitt IM, Mackey DA, Wirth MG, Elder JE, Nicoll A, Clarke MP, FitzGerald LM, Stankovich JM, Shaw MA, Sharma S, Gajovic S, Gruss P, Ross S, Thomas P, Voss AK, Thomas T, Gecz J, Craig JE. Mutations in a novel gene, NHS, cause the pleiotropic effects of Nance-Horan syndrome, including severe congenital cataract, dental anomalies, and mental retardation. Am J Hum Genet. 2003;73:1120–30. doi: 10.1086/379381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks SP, Ebenezer ND, Poopalasundaram S, Lehmann OJ, Moore AT, Hardcastle AJ. Identification of the gene for Nance-Horan syndrome (NHS). J Med Genet. 2004;41:768–71. doi: 10.1136/jmg.2004.022517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramprasad VL, Thool A, Murugan S, Nancarrow D, Vyas P, Rao SK, Vidhya A, Ravishankar K, Kumaramanickavel G. Truncating mutation in the NHS gene: phenotypic heterogeneity of Nance-Horan syndrome in an asian Indian family. Invest Ophthalmol Vis Sci. 2005;46:17–23. doi: 10.1167/iovs.04-0477. [DOI] [PubMed] [Google Scholar]
- 15.Florijn RJ, Loves W, Maillette de Buy Wenniger-Prick LJ, Mannens MM, Tijmes N, Brooks SP, Hardcastle AJ, Bergen AA. New mutations in the NHS gene in Nance-Horan Syndrome families from the Netherlands. Eur J Hum Genet. 2006;14:986–90. doi: 10.1038/sj.ejhg.5201671. [DOI] [PubMed] [Google Scholar]
- 16.Lewis RA, Nussbaum RL, Stambolian D.Mapping X-linked ophthalmic diseases. IV. Provisional assignment of the locus for X-linked congenital cataracts and microcornea (the Nance-Horan syndrome) to Xp22.2-p22.3. Ophthalmology 199097110–20.discussion120-1 [DOI] [PubMed] [Google Scholar]
- 17.Krill AE, Woodbury G, Bowman JE. X-chromosomal-linked sutural cataracts. Am J Ophthalmol. 1969;68:867–72. doi: 10.1016/0002-9394(69)94582-6. [DOI] [PubMed] [Google Scholar]
- 18.Wilson JM, Baugher BW, Mattes PM, Daddona PE, Kelley WN. Human hypoxanthine-guanine phosphoribosyltransferase. Demonstration of structural variants in lymphoblastoid cells derived from patients with a deficiency of the enzyme. J Clin Invest. 1982;69:706–15. doi: 10.1172/JCI110499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S, Ang SL, Shaw M, Mackey DA, Gecz J, McAvoy JW, Craig JE. Nance-Horan syndrome protein, NHS, associates with epithelial cell junctions. Hum Mol Genet. 2006;15:1972–83. doi: 10.1093/hmg/ddl120. [DOI] [PubMed] [Google Scholar]
- 20.Huang KM, Wu J, Duncan MK, Moy C, Dutra A, Favor J, Da T, Stambolian D. Xcat, a novel mouse model for Nance-Horan syndrome inhibits expression of the cytoplasmic-targeted Nhs1 isoform. Hum Mol Genet. 2006;15:319–27. doi: 10.1093/hmg/ddi449. [DOI] [PubMed] [Google Scholar]


