Abstract
Caspases are a family of proteases that participate in the progression and execution of the apoptotic program. However, regulation of the caspase activation and their substrates has not yet been fully elucidated. Here we explore the effect of the ectopic expression of the human initiator caspases-8 and -10 in S. cerevisiae. Our results showed that the expression of human CASP10 and CASP8 triggers certain apoptotic markers such as a massive production of reactive oxygen species (ROS), chromatin condensation and phosphatidiylserine externalization, finally leading to cell death. In response to hydroxyurea (HU), yeast cells expressing caspase-10 did not reduce the replication of DNA and escaped to the intra-S checkpoint of the cell cycle. In addition, caspase-10 expression induced yeast vacuolization and a vacuole-associated phenotype resembling autophagy. Other intracellular alterations such as disorganization of the actin cytoskeleton, cell wall damage, and aberrations within the endoplasmic reticulum lumen were also associated with caspase-10 expression. Furthermore, caspase-induced cell death was completely dependent on the proteolytic activation of the enzyme but, in contrast, was not dependent on either of the endogenous yeast apoptotic proteins Aif1 and Mca1 or the mitochondria.
Keywords: caspase, yeast, oxidative stress, cell death, apoptosis, autophagy
Introduction
Caspases are a family of cysteine proteases that cleave a wide range of substrates at their aspartate residues. Caspases are involved both in inflammation and apoptosis, and their mechanisms of activation are fairly similar in both processes. Regarding their role during the course of apoptosis, caspases can be classified as initiators and effectors (scheme 1). Initiator caspases are responsible for eliciting the apoptotic pathways (extrinsic and intrinsic), acting as a link between the sensor/adaptor proteins and the effector caspases that function as the true executioners of the apoptotic program [1]. The extrinsic pathway is initiated by the activation of death receptors at the plasma membrane, whereas the intrinsic pathway is triggered by intracellular signals (scheme 1) [2].
scheme 1.
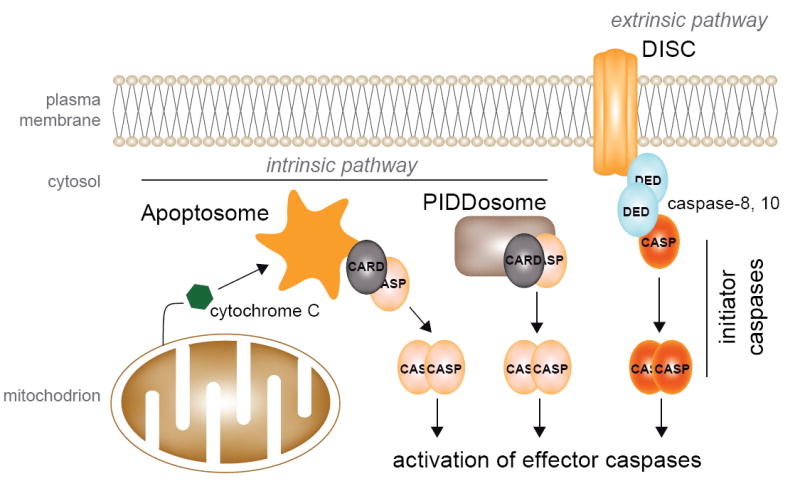
Apoptotic pathways in mammals.
Caspases are primarily expressed as inactive zymogens that need to be activated through proteolysis. Mammalian initiator caspases contain N-terminal protein-protein interaction domains, which belong to the death domain (DD) superfamily. The principal mechanism for the activation of initiatior caspases is their oligomerization and recruitment to death complexes designated as the PIDDosome, the DISC (death-inducing signaling complex) and the apoptosome (scheme 1), which allow the autocatalytic intrachain cleavage of the capases, dimerization of the catalytic subunits and activation [1, 2]. Caspase-8 and -10 are activated at the DISC, which includes death receptors at the plasma membrane (Fas), adaptor proteins (FADD) and caspases. Active caspases-8 and -10 are able to exert a proteolytic activation of downstream caspases-3, -6 and -7 that finally lead to the clearance of certain substrates [3, 4].
Apoptosis is a conserved physiological process that is also present in some unicellular organisms [5, 6]. Indeed, a number of groups have reported the existence of a suicide program in Saccharomyces cerevisiae as an adaptative mechanism to the changing environment [5, 7]. Several yeast orthologues of mammalian genes participating in apoptosis such as the metacaspase (MCA1), AIF1, Omi (NMA111) and EndoG (NUC1) have also been identified in S. cerevisiae [8-11]. In addition, the budding yeast S. cerevisiae has been successfully employed for the identification and characterization of several mammalian apoptotic proteins. For example, the mammalian Bax inhibitor-1 was first identified as a suppressor of Bax-induced cell death in S. cerevisiae [12]. Bax and Bak-induced lethality in yeast is also rescued by co-expression of anti-apoptotic Bcl-2 proteins [13]. Previous works have also reported that the heterologous expression of certain human caspases, including the initiator caspases-2 and -8, kills S. cerevisiae in a manner independent of the yeast MCA1 and AIF proteins; however, the mechanisms underlying this induced cell death have not been fully elucidated [14, 15]. Furthermore, it has also been reported that the isoform A of human caspase-10 does not induce cell death in S. cerevisiae [14].
Here we describe the effect of the heterologous expression of human initiator caspases-8 and -10 in S. cerevisiae. The full-length transcript variant of human CASP10 (encoding the longest isoform D), and two transcript variants of CASP8 (encoding the isoform C and a novel unknown isoform) induce cell death in S. cerevisiae. Several phenotypes associated with their expression are presented and discussed.
Materials and methods
S. cerevisiae strains and culture media
The S. cerevisiae strains used in this study are listed in Supplementary Data Table 1. Cells were routinely grown at 28°C in synthetic complete medium lacking either uracil or leucine (SC-Ura or SC-Leu) and containing either 2% glucose or 2% galactose plus 1% raffinose as carbon sources. Growth on liquid cultures was monitored spectrophotometrically at O.D.600 nm. The respiratory deficiency of the rho- strains was confirmed by complete lack of growth on obligatory respiratory media (2% glycerol as the only carbon source).
Cloning and expression of human CASP8 and CASP10 genes
The ORFs encoding human caspase-8 and -10 were amplified from a Jurkat cell cDNA library by PCR, using the primers listed in Supplementary Data Table 2. Human CASP8 and CASP10 ORFs were verified by DNA sequencing of the entire fragments. The ORFs corresponding to human CASP8 were cloned between the BamHI and SalI restriction sites of the yeast episomal expression vector pESC-LEU (Stratagene) under the control of the GAL1 promoter. Human CASP10 ORF was cloned into the NotI–SacI sites of the pESC-URA expression vector (Stratagene) under the control of the GAL10 promoter. The pESC-LEU/CASP8 and pESC-URA/CASP10 constructs were used to transform the S. cerevisiae strains BY4741, rho-, Δaif1 and Δmca1 following the TRAFO method [16]. The resulting transformants were selected for by either leucin or uracil prototrophy. Expression of the ORFs corresponding to human caspase-8 and caspase-10 was carried out in galactose/raffinose-containing medium.
Analysis of the DNA contents
Yeast cells were grown in synthetic complete media with either glucose or galactose/raffinose to an OD600 nm of 0.1. For HU treatment, the cultures were incubated with 0.2M HU for 2 hours. Aliquots were taken at the indicated times, centrifuged and fixed in 70% ethanol overnight at 4°C. The samples were resuspended in 400 μl of 50 mM sodium citrate, 0,2 μg/ml RNase and were incubated overnight at 37°C. Then, 200 μl of 50 mM sodium citrate, 55 mM HCl and 5 mg/ml pepsin were added to each sample and the samples were incubated for 15 minutes at 37°C. Finally, 400 μl of 50 mM sodium citrate containing 10 μg/ml propidium iodide was added and the samples were sonicated briefly to avoid agglutination. Samples were analyzed by flow cytometry using a FACS SORT (Becton Dickinson) and Cell Quest software.
Cell viability analyses
The cell viability of S. cerevisiae expressing human caspase-10 was assessed using three different methods: a clonogenic assay, FUN-1 staining (Molecular Probes) and the resazurin staining (Sigma). For the clonogenic assay, S. cerevisiae cells transformed with either the pESC-URA empty vector or the pESC-URA/CASP10 vector were grown in non-inducing conditions (glucose) to an OD600 nm of 0.1. The cultures were harvested, washed three times with sterile water and resuspended in SC-Ura media with either glucose (non-inducing conditions) or galactose/raffinose (inducing conditions) as carbon sources. After induction of caspase-10 expression at different time points the cells were plated onto SC-Ura media with glucose as a carbon source. The number of surviving colonies was counted and the results are shown as a percentage of the number of colonies in induced cultures with respect to the non-induced cultures. The FUN-1 stain allows metabolically active cells (living cells) to be determined, which are marked with red fluorescent intravacuolar structures; in contrast, dead cells exhibit a diffuse green citoplasmic fluorescence. Resazurin is a redox indicator that becomes fluorescent red in metabolically active cells. The FUN-1 staining was carried out as described [17] and the resazurin staining was performed as recommended by the manufacturer.
Site-directed mutagenesis of human CASP10
Amino acid substitutions in human caspase-10 were introduced by site-directed mutagenesis of the CASP10 gene through PCR techniques using the primers listed in Supplementary Data Table 2. The Cys401, which represents the active site, was changed to a glycine residue; also the Asp219 and Asp416, which are involved in the proteolytic activation of caspase-10, were replaced by alanine residues. The three mutant ORFs were checked by DNA sequencing of the entire fragments.
Tests for apoptotic markers in S. cerevisiae
Intracellular free radicals were detected with 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, Sigma) and dihydroethidium (DHE, Sigma). Phosphatidylserine externalization was detected with the Apoalert AnnexinV Apoptosis Kit (Clontech) as previously described [18]. Genomic DNA fragmentation was analyzed by pulsed-field gel electrophoresis using a CHEF system (Bio-Rad) to separate the yeast chromosomes as previously described [19].
Assessment of organelle integrity
SYTO18 yeast mitochondrial stain (Molecular Probes), MDY-64 membrane vacuole staining (Molecular Probes) and phalloidin (Molecular Probes) were used to analyze yeast mitochondria, vacuoles and the actin cytoskeleton, respectively.
Western-blot analysis of human caspase-10
Cells expressing human caspase-10 were resuspended in 5 ml of 20 mM Hepes ph 7,4; 80 mM KCl, 10 mM MgCl, 0,2 mM EDTA, 1mM DTT, 0,5% w/v CHAPS and disrupted using a French Press system at 1120 psi. Cell lysates were centrifuged at 13.000 rpm for 30 min and the soluble fraction was separated by SDS-PAGE. Proteins were blotted onto a PVDF membrane (Millipore) and human caspase-10 was detected with a polyclonal anti-caspase10 p10 (N-19) antibody (Santa Cruz Biotechnology).
Electron microscopy
Strain BY4741 carrying an empty vector or one expressing caspase-10 from the GAL1 promoter were grown overnight in SC-Ura medium and then diluted into SC-Ura containing 2% galactose and 1% raffinose as carbon sources. After 6 hours incubation at 30°C, cells were fixed by addition of glutaraldehyde to the growth medium, stained with potassium permanganate and prepared for electron microscopy as described previously [20]. Images were collected on an FEI BioTwin G2 microscope using an accelerating voltage of 80kV and an AMT XR-60 digital camera.
Results
The expression of human CASP8 and CASP10 is toxic in S. cerevisiae
The human CASP8 and CASP10 transcripts are subject to extensive alternative splicing that generates multiple isoforms within the cell. However, the isoform C of caspase-8 (also labeled as MACH-alpha-2 or MCH5-beta) and the isoform D of caspase-10 are the most common isoforms of caspase-8 and caspase-10 in human tissues [21, 22]. We therefore designed specific primers (see Supplementary Data Table 2) to amplify their corresponding ORFs from Jurkat cell cDNA by PCR. The sequences of the transcript variants C and D of CASP8 and CASP10, respectively, were amplified and verified by sequencing of the entire PCR products. We also isolated another splicing-variant of CASP8 (termed as variant_S) encoding for a novel unknown isoform (GenBank EU168332), which is shorter than the isoform C but also comprises two DED domains and a caspase domain (Fig. 1). The three ORFs were cloned into the yeast episomic expression vectors pESC-LEU (for CASP8 alleles) and pESC-URA (CASP10) and were introduced into the BY4741 haploid strain of S. cerevisiae. The corresponding empty vectors were also used to transform the BY4741 strain and were employed as negative controls in ensuing experiments.
Fig. 1.
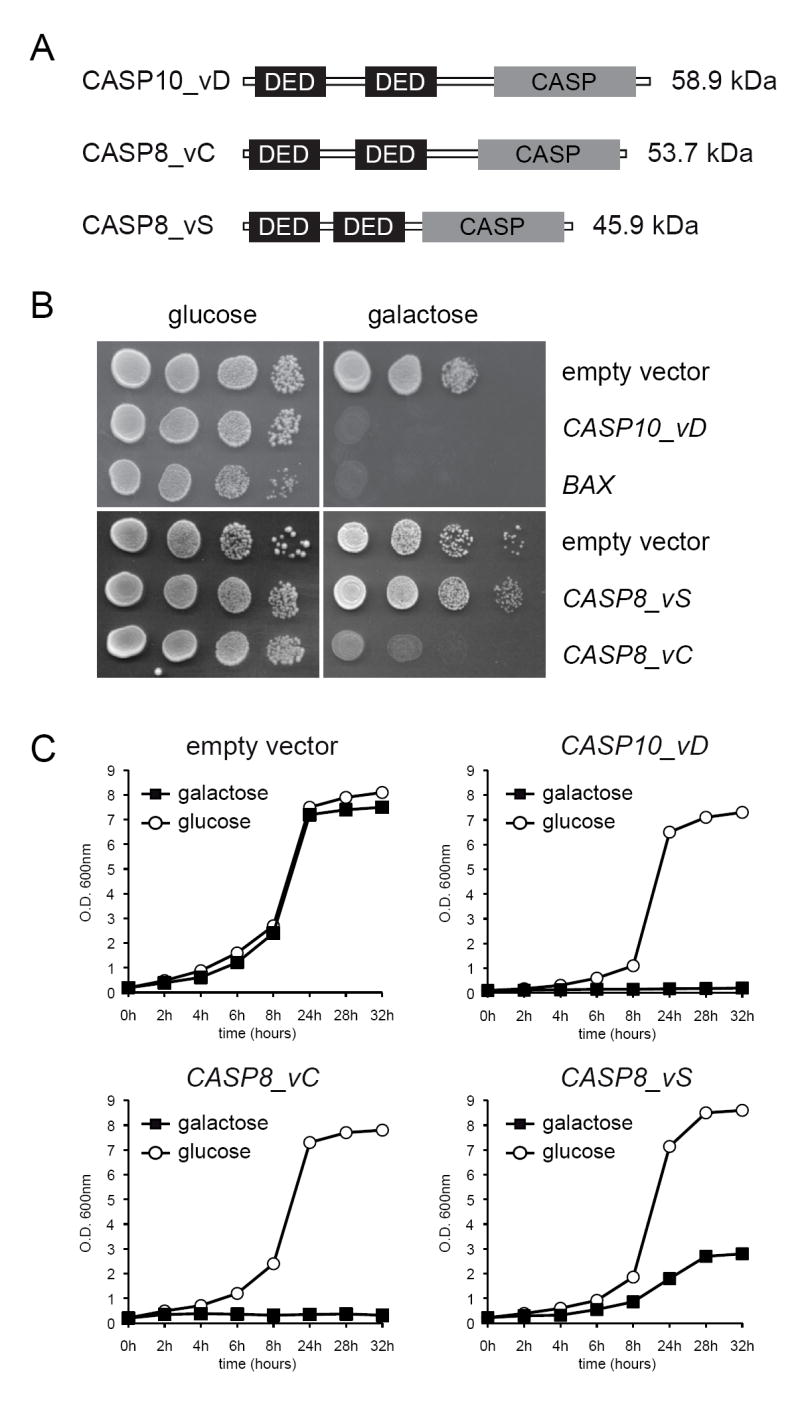
Heterologous expression of human caspase-8 and caspase-10 inhibits growth in S. cerevisiae. A, domain organization of the caspase-8 and -10 isoforms used in this study. B, plate assay of S. cerevisiae strains expressing CASP8 and CASP10 genes. The expression of BAX was used as a positive control for lethality and empty vectors were used as negative controls. C, growth curves of the caspase-expressing strains performed in either SC-Ura or SC-Leu and containing either 2% glucose or 2% galactose plus 1% raffinose as carbon sources. Data presented are means of three independent experiments.
The expression of caspases in the mutant strains was induced in galactose-containing media. As shown in figure 1, both the isoform C of caspase-8 and the isoform D of caspase-10 completely abolished the growth of S. cerevisiae. Indeed, their toxicity was comparable to that associated to the expression of Bax (Fig. 1B). However, the isoform S of caspase-8 was significantly less toxic than isoform C (Fig. 1C). For our next experiments we decided to use the isoform C of caspase-8 and the isoform D of caspase-10. Hereafter, these isoforms are referred to as caspase-8 and caspase-10.
Human CASP10-induced toxicity leads to cell death
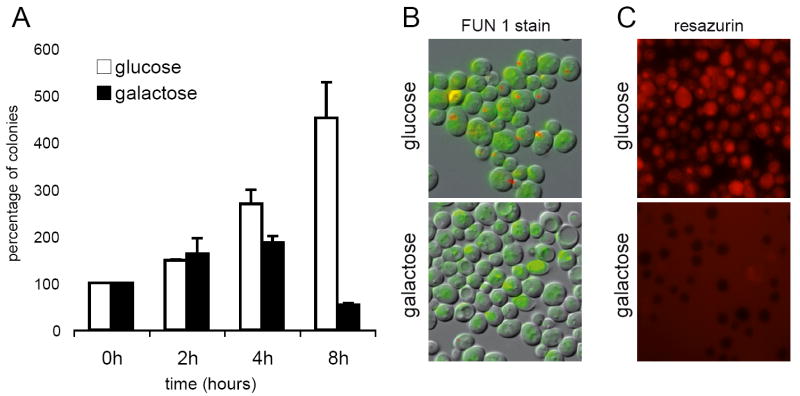
It has been reported that the expression of human caspase-8 induces cell death in S. cerevisiae [14, 15]. We therefore wished to know whether the toxicity associated with the expression of the isoform D of caspase-10 was also due to programmed cell death. We used three different methods to evaluate the CASP10-induced cell death. As shown in figure 2, the expression of caspase-10 strongly diminished the clonogenicity of S. cerevisiae after 4 hours of induction. Additionally, the vital dyes FUN1 and resazurin, which detect metabolically active cells, confirmed that caspase-10 killed most of the yeast cells after 6 hours of expression (Fig. 2). These results clearly revealed that human caspase-10 induces programmed cell death in S. cerevisiae.
Fig. 2.
Human caspase-10 induces S. cerevisiae cell death. A, clonogenic assay of the S. cerevisiae strain expressing human CASP10. Data are presented as percentages of surviving colonies as referred to non-inducing conditions at time 0h. B, FUN1 staining of S. cerevisiae cells expressing caspase-10. Metabolic active cells show red fluorescent intravacuolar structures. C, Resazurin staining of S. cerevisiae cells expressing caspase-10. Red fluorescence is only present in living cells.
It has been established that mitochondria and the proteins Aif1 and metacaspase are essential for both the life and death of S. cerevisiae [23]. Consequently, we investigated whether the cell death induced by caspase-8 and -10 was mediated through those cellular elicitors. We expressed both CASP8 and CASP10 genes in an aif1 mutant, an mca1 mutant and a rho- mutant, the latter of which lacks functional mitochondria. Our results showed that the expression of caspase-8 and caspase-10 also inhibited the growth of the mutants, indicating that neither the yeast apoptotic proteins Aif1 and metacaspase nor functional mitochondria are essential for human caspase-8 and -10 induced cell death (Fig. 3).
Fig. 3.
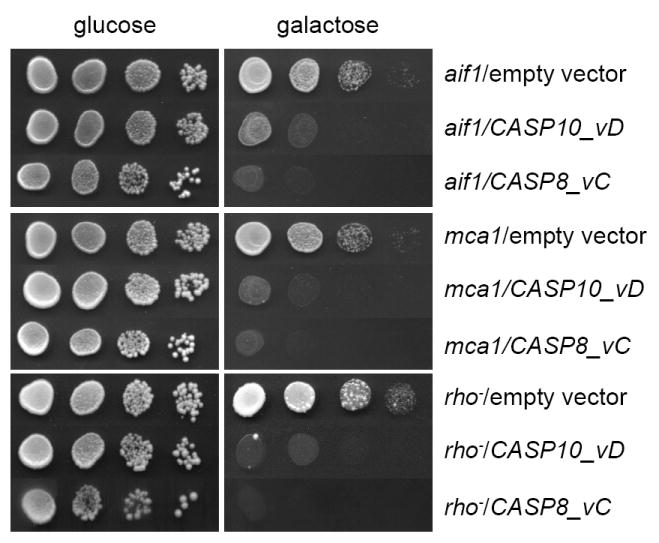
Caspase-8 and caspase-10 induce cell death independently of the yeast apoptotic machinery. Plate assay of Δaif1, Δmca1 and rho- strains expressing either human CASP8 or CASP10 genes. Empty vectors were used as controls.
Caspase-10 activation is essential for lethality in S. cerevisiae
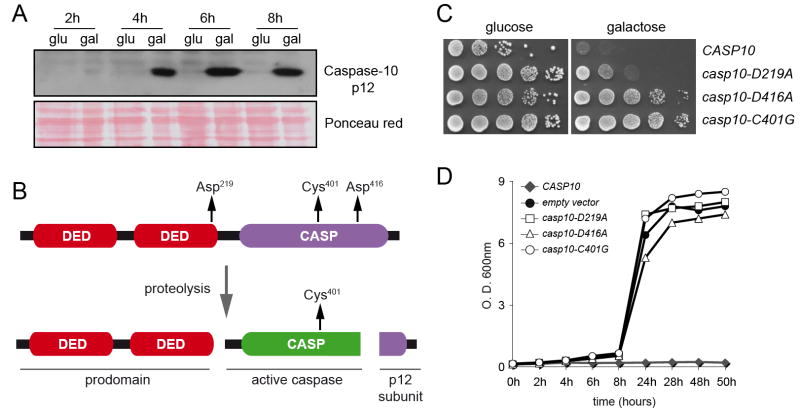
Previous studies have reported the enzymatic activity of caspases in protein extracts from human caspase-expressing yeast [14, 15]; however, there is no evidence about the proteolytic activation of human initiator caspases in S. cerevisiae. We carried out a western-blot analysis of protein extracts from the CASP10-expressing strain using an anti-caspase10 antibody to check whether human caspase-10 was efficiently cleaved in S. cerevisiae. As shown in figure 4A the antibody was able to detect the p10 small subunit (aprox. 12kDa) resulting from caspase-10 proteolysis demonstrating that human caspase-10 was efficiently activated in S. cerevisiae.
Fig. 4.
CASP10-mediated cell death requires proteolytic activation. A, western-blot analysis of protein crude extracts from a caspase-10 expressing strain. Ponceau red staining of the blot was used as protein loading control. B, schematic representation of the proteolytic activation of caspase-10. The residues involved in either activation or catalysis are indicated with arrows. C, plate assay of S. cerevisiae strains expressing different CASP10 alelles. D, growth curves of S. cerevisiae strains expressing different CASP10 alelles.
We next wished to investigate whether caspase-10 activation was necessary to induce cell death in S. cerevisiae. Accordingly, we designed mutagenic primers (see Supplementary Data Table 2) to create three mutant alleles of CASP10 deficient in either the catalytic active cysteine (Cys401) or the aspartate residues involved in activation (Asp219 and Asp416) (Fig. 4B). The substitutions carried out were as follows: Cys401 was changed to glycine (G); Asp219 was replaced by an alanine residue (A); and Asp416 was changed to alanine (A). Once we had amplified the three mutant ORFs by mutagenic PCR, the presence of the mutations was verified by DNA sequencing and the CASP10 mutant ORFs were cloned into the pESC-URA expression vector. Expression of the mutant alleles of CASP10 in S. cerevisiae was performed in galactose-containing media (Fig 4C-D). Our results showed that none of the three mutant isoforms (Casp10-D219A, Casp10-D416A or Casp10C401G) was lethal for S. cerevisiae, indicating that those replaced residues were essential for CASP10-induced cell death.
Human CASP8 and CASP10 trigger a massive production of ROS and cause phosphatidylserine externalization
The expression of human pro-apoptotic proteins such as Bax is accompanied by the generation of oxygen radicals [24]. We therefore examined the production of reactive oxygen species (ROS) associated with the expression of human caspase-8 and -10. Dihydroethidium (DHE) and 2,7-dichlorofluorescein-diacetate (H2-DCF-DA) were used to evaluate the production of superoxide and peroxide in the CASP8 and CASP10-expressing strains. ROS-associated fluorescence was evident after 2 and 4 hours of CASP8 and CASP10 expression, respectively (Fig. 5). Dead cells (ghosts) were also clearly visible after 4 hours of caspase expression. It is remarkable that after 4 hours of caspase expression almost all cells showed a high level of oxidative stress, as indicated by the visualization of the fluorescent dyes.
Fig. 5.
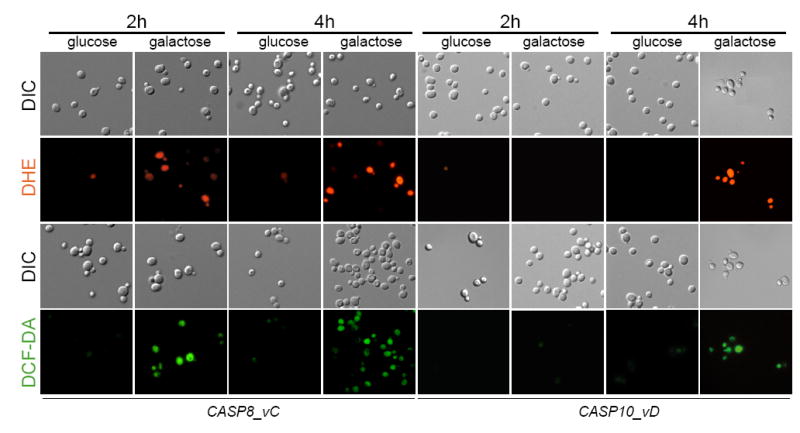
Human caspase-8 and caspase-10 elicit the accumulation of ROS in S. cerevisiae. CASP8 and CASP10-expressing strains were stained with either DHE or DCF-DA and yeast cells were visualized under fluorescence microscopy at the indicated times after caspase expression. DIC images are also included. Glucose cultures were used as negative controls.
We next investigated whether the expression of caspase-8 and -10 produced other apoptotic phenotypes such as phosphatidylserine (PS) exposure or DNA degradation. Zymolyase-treated cells expressing either caspase-8 or caspase-10 were annexin V-positive, but most of them were PI-negative (Fig. 6). The same results were obtained when intact whole cells were assayed (not shown). Additionally, we analyzed chromosomal DNA from yeast cells expressing either human caspase-8 or -10 by pulsed field gel electrophoresis. Strikingly, the chromosomes seem to be intact and DNA was not degraded as happened when cells were treated with hydrogen peroxide (Supplementary Data Fig. 1). Taken together, our results show that the expression of caspase-8 and caspase-10 induces a type of cell death accompanied by certain apoptotic phenotypes such as ROS production and PS externalization, but it does not show genomic DNA degradation.
Fig. 6.
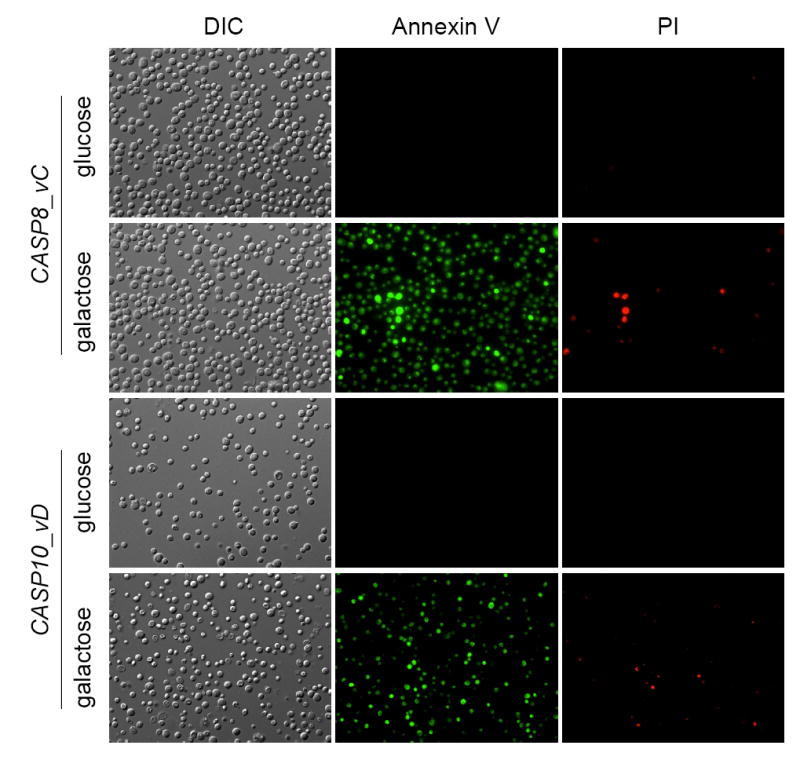
Expression of either CASP8 or CASP10 in S. cerevisiae causes phosphatidylserine externalization. Yeast cells harbouring either CASP8 or CASP10 were induced in galactose-containing media for 4 hours and the Apoalert AnnexinV Apoptosis Kit (Clontech) was used to observe the Annexin V- and the PI-associated fluorescence. Glucose cultures were used as negative controls.
Human initiator caspases abolish an HU-induced cell-cycle arrest
Apoptosis and cell-cycle progression are connected through the regulation of the checkpoints that control the activity of CDKs [25]. In our model, no alteration in the progression of the cell-cycle in asynchronous cultures was appreciated by FACS analysis after the expression of human initiator caspases (data not shown).
In response to DNA damage, cells activate the intra-S checkpoint and the replication of the DNA is reduced [26]. Therefore, we treated the yeast cells with HU, which is an inhibitor of the ribonucleotide reductase and induces a cell-cycle arrest in the S-phase, to evaluate the effect of caspase expression in the activation of the intra-S checkpoint in response to DNA damage during the replication of DNA. Yeast cells transformed with an empty vector showed a HU-dependent block of the cell-cycle as expected (Fig. 7); however, the cells expressing either caspase-8 or caspase-10 did not response to the HU treatment and were completely insensitive to the activation of the intra-S checkpoint (Fig. 7).
Fig. 7.
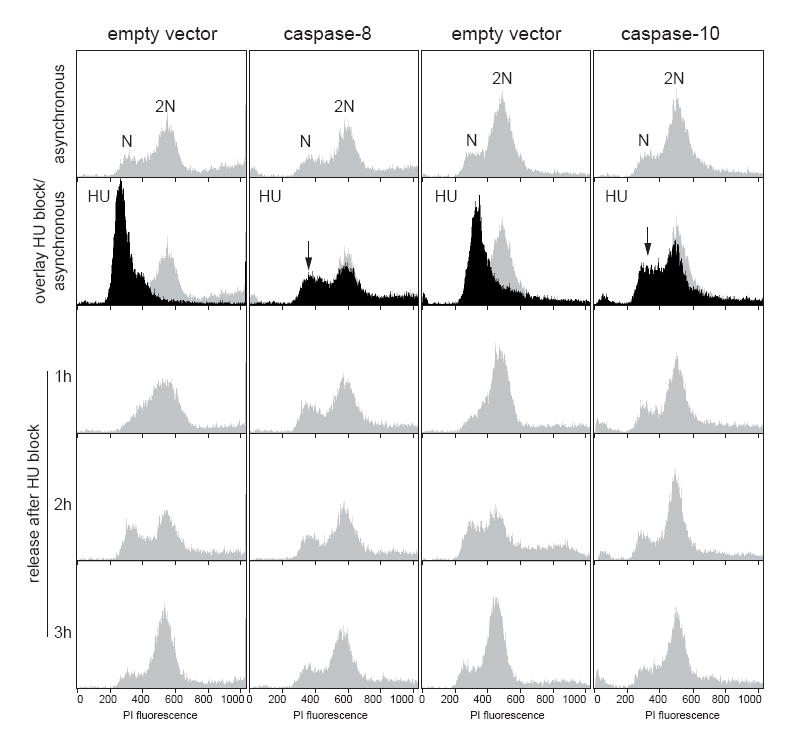
The expression of human initiator caspases prevents the HU-induced cell-cycle arrest. HU-treated cultures of yeast cells expressing either caspase-8 or caspase-10 were analyzed by FACS.
Actin disorganization follows human CASP10 expression in S. cerevisiae
Actin dynamics plays a central role in several signaling pathways and it is also important in the regulation of yeast cell death. Indeed, the organization of actin from discrete patches and cables to large clumps correlates with the levels of ROS [27, 28]. To determine whether the expression of human caspases might affect the organization of the actin cytoskeleton, we stained yeast cells with phalloidin after CASP10 induction. Large fluorescent actin clumps could be seen after 4 hours of caspase-10 expression (Fig. 8), when a ROS high level was also observed (see Fig. 5). In contrast, normal actin patches and cables were present in non-inducing conditions. Furthermore, large actin aggregates were associated within the bud necks of dividing cells after caspase-10 activation (Fig. 8F-G). Overall, our results indicate that caspase-10 expression leads to the disorganization of the actin cytoskeleton in S. cerevisiae.
Fig. 8.
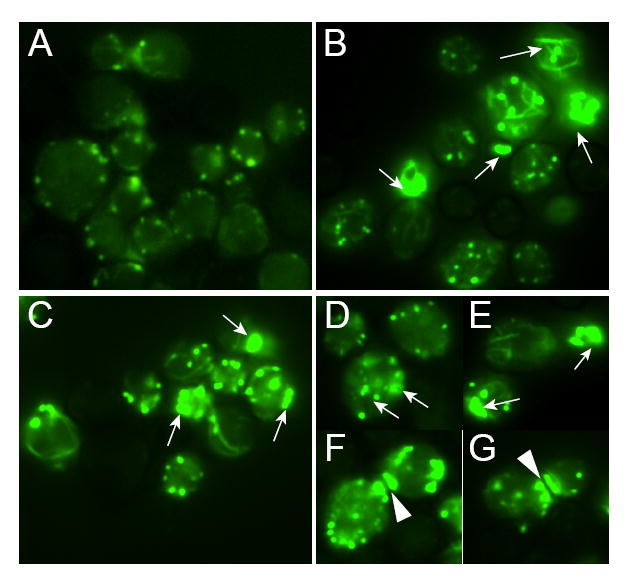
The actin cytoskeleton becomes disorganized after caspase-10 expression in S. cerevisiae. Yeast cells carrying human CASP10 were cultured for 4 hours in either glucose- (A) or galactose-containing media to induce caspase-10 expression (B-G). Cells were stained with phalloidin and were visualized under fluorescence microscopy. Arrows indicate large actin clumps. Arrowheads point to actin aggregation in the bud necks.
Subcellular phenotypes associated with the expression of Caspase-10
It has been described that apoptotic yeast cells show certain phenotypes at the subcellular level such as a high degree of vacuolation, chromatin condensation and alterations of the ER lumen [18, 29]. Also, mitochondrial disturbance has been reported upon expression of human caspase-3 and -8 in S. cerevisiae [15]. We therefore focused on the subcellular effects of the expression of caspase-10 in S. cerevisiae and analyzed mitochondria and vacuoles by fluorescence microscopy. As shown in figure 9A, highly vacuolated cells were visible after caspase-10 expression; however, the mitochondria remained unaltered (Fig. 9B). To gain more insight into the effect of caspase-10 expression on the organelles of S. cerevisiae, yeast cells expressing human CASP10 were examined by transmission electron microscopy. We observed that caspase-10 expression induced organelle degradation in the yeast cells (Fig. 10). Chromatin condensation and ER with an altered lumen could also be seen in some cells (Fig. 10B). In addition, alterations in the cell wall, such as abnormal bud necks and thickening of the cell wall, were frequent (Fig. 10). Consistent with the fluorescence microscopy results, many of the cells expressing CASP10 appeared highly vacuolated, with a proliferation of what appeared to be autophagic vesicles present in the vacuolar lumen. Some cells displayed nuclear material inside the vacuole, resembling piecemeal microautophagy of the nucleus that degrades nuclear material [30] (Fig. 10D). Taken together, our results indicate that vacuole-dependent organelle degradation, which is a typical marker of autophagy, occurs when caspase-10 is expressed in S. cerevisiae.
Fig.9.
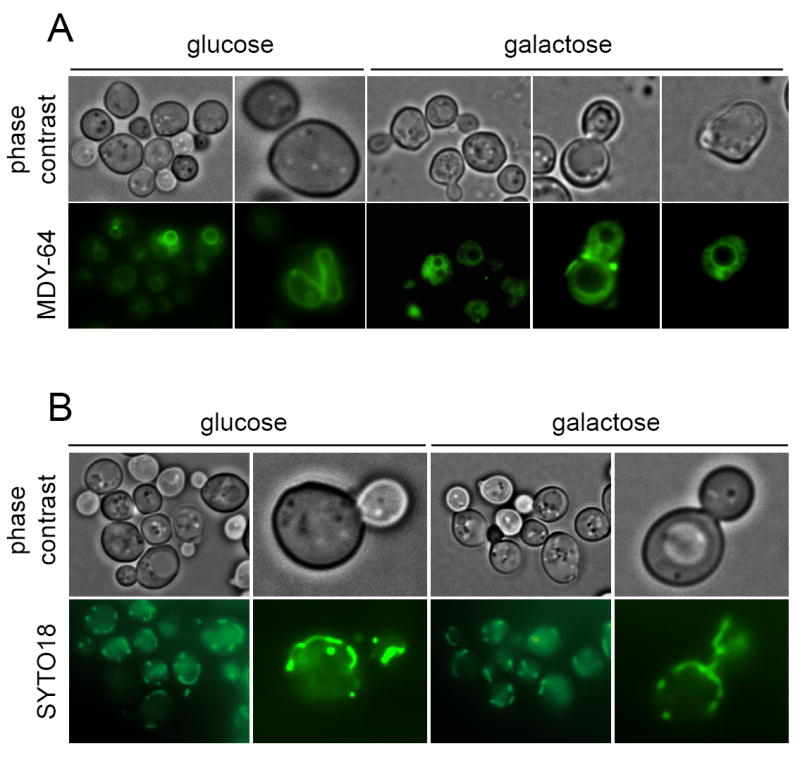
Caspase-10 expression induces vacuolization in S. cerevisiae. Yeast cells expressing CASP10 were stained with the vacuole marker MDY-64 and were visualized under fluorescence microscopy. Glucose cultures were used as negative controls.
Fig. 10.
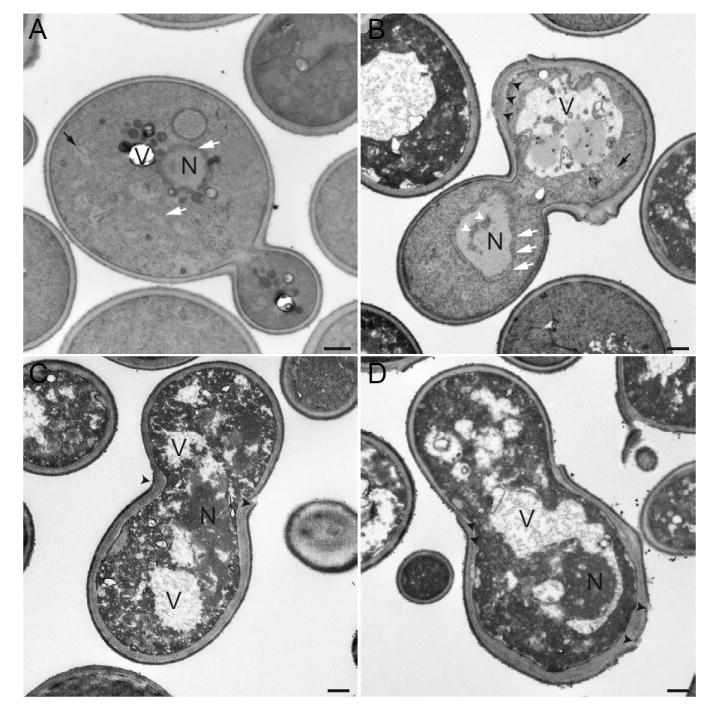
Electron micrographs of S. cerevisiae expressing human caspase-10. A, BY4741 transformed with the empty vector. B-D, BY4741/CASP10. N, nucleus; V, vacuole; white arrows, ER; black arrows, mitochondria; white arrowheads indicate chromatin condensation; black arrowheads point to cell wall abnormalities. Bars, 500 nm.
Discussion
Programmed cell death (PCD) is a term that includes different types of cell death that have been genetically programmed [31]. PCD has been described in multicellular organisms and, more recently, it has also been reported in unicellular organisms as an adaptative mechanism [5]. Apoptosis is a form of PCD that involves a number of morphological changes within the cell, including chromatin condensation and DNA degradation. The death executioners of the apoptotic program include caspases, which are responsible for eliciting the two main pathways of apoptosis: intrinsic and extrinsic [1, 32]. However, caspases and other apoptotic effectors also seem to be involved in cell death-unrelated functions [32, 33]. Yeast cells lacking the typical apoptotic pathways can serve to uncover yet unknown functions of human caspases. Thus, in order to develop a new tool for the investigation of unknown counterparts for human caspases, in the present work we have characterized the effect of the heterologous expression of the human initiator caspases-8 and -10 in S. cerevisiae.
The CASP8 and CASP10 genes map at the same region within human chromosome 2, suggesting that both genes would have arisen from a duplication event after the divergence of mice from humans in the course of evolution [1, 34]. Whether caspase-8 and -10 exert different functions in apoptosis is controversial; however, it seems to be accepted that these caspases play different roles in death receptor signaling and other cellular processes [34, 35].
Here we cloned and expressed the most common isoforms of caspase-8 (isoform C) and caspase-10 (isoform D) episomically in S. cerevisiae. Additionally, we examined the effect of a novel isoform of caspase-8 (isoform S). Cell death was observed after expression of human caspases-8 and -10 in S. cerevisiae, as indicated by the lack of clonogenic survival and staining with vital dyes; however, their lethality differed significantly between the caspase-8 isoforms, suggesting a differential function among the splicing variants of caspases. Indeed, a previous report has described that the isoform A of caspase-10 did not elicit cell death when expressed in yeast [14], confirming that the different isoforms of caspase-10 may also have diverse effects in S. cerevisiae.
We also observed that the proteolytic activation of caspase-10 was essential for its lethality in S. cerevisiae and we further observed that the expression of caspase-8 isoforms either lacking the caspase domain or with a truncated caspase domain did not cause yeast cell death (data not shown). Moreover, cell death associated with the expression of caspase-8 and -10 was not mediated, at least essentially, either by mitochondria or the yeast apoptotic proteins Aif1 and Mca1, indicating that other cellular elicitors must be involved in the lethal effect of caspases in S. cerevisiae. As previously described for the mammalian Bax inhibitor-1 [12], the identification of those caspase-interacting proteins in S. cerevisiae might allow the human orthologues involved in unidentified caspase-related functions to be discovered.
As mentioned, the expression of human initiator caspase-8 and -10 in S. cerevisiae encompasses some typical apoptotic phenotypes, such as chromatin condensation, ROS accumulation, and phosphatidylserine externalization. However, DNA degradation was not detected, as previously reported for caspase-3 and caspase-8 [15]; this result contrasts with our electronic microscopy observations regarding chromatin condensation. Additionally, it became clear that mitochondria, a central organelle in apoptosis [23], are not essential for the caspase-induced toxicity in S. cerevisiae since the cell death was not abolished in a rho- strain.
It has previously been reported that apoptosis and cell cycle regulation are linked, and that caspases, including human caspase-8, -10 and yeast Mca1, have non-death roles in the regulation of the cell cycle and proliferation [4, 36, 37]. Oxidative DNA damage causes a replication stress that activates the cell-cycle checkpoints to prevent the replication of damaged DNA and repair the defects [38]. In addition, it has been described that DNA replication defects cause apoptotic cell death in yeast [25]. The expression of human initiator caspases in S. cerevisiae triggers a massive production of ROS, which might eventually activate the G1 or intra-S checkpoints of the cell cycle in response to the oxidative damage to DNA. However, our results revealed that yeast cells expressing either caspase-8 or caspase-10 did not respond to HU treatment and continued dividing, thus indicating a possible role of those caspases in the regulation of the cell-cycle through the inactivation of DNA replication checkpoint proteins.
The actin cytoskeleton, the cell wall, and endomembranous organelles such as vacuoles and ER are somehow altered as a consequence of caspase-10 expression in S. cerevisiae. Actin aggregation in yeast caused by decreased actin turnover has been related to the release of ROS by mitochodria [27]. It has also been reported that the actin cytoskeleton is sensitive to changes in the levels of oxidative stress and that disulphide bonding in the cysteine residues of actin may function as an oxidative stress sensor [28, 39, 40]. Accordingly, it maybe speculated that the caspase-associated phenotype over the actin cytoskeleton would be an effect of elevated oxidative stress. Additionally, the observed alterations in the cell wall and bud necks could also be attributed to increased ROS levels through the induction of the cell integrity pathway, which has been shown to regulate the cell wall and actin dynamics in response to different stimuli, including oxidative stress [41, 42].
Yeast vacuoles are acidic organelles with storage and degradative functions that play a pivotal role under stress conditions [43]. Such vacuoles are responsible for the degradation of specific organelles through a process of autophagy [30, 44], which is also considered as an alternative form of PCD. We observed that caspase-10 expression induced a vacuole-associated response that led to organelle degradation, which might resemble microautophagy. Indeed, it has been reported that the heterologous expression of Bax also induces autophagic and mitophagic features [45, 46]. Moreover, both types of PCD (apoptosis and autophagy) must be related since it has been described that cells undergoing autophagy display apoptotic markers and that caspases also play a role in autophagy [47, 48].
The expression of caspase-10 in S. cerevisiae also induced an expansion of the ER lumen in some cells. This phenotype has been found in cdc48 yeast mutants, and was indeed identified as the first hint of yeast apoptosis [18], and it has been related to the dysfunction of the ER-associated degradation pathway (ERAD) [49]. ERAD dysfunction caused by mutations in CDC48 is, in turn, associated with the accumulation of ROS and caspase-dependent cell death [49]. However, a recent report has shown that induced ER stress is associated with non-apoptotic cell death, preceded by the accumulation of ROS [50]. As stated, mitochondria were not essential for caspase-induced cell death in S. cerevisiae. Therefore, the ER might be considered as an alternative source of the oxidative stress associated to the expression of human caspase-8 and -10 in yeast.
According to our results, all the identified phenotypes caused by human initiator caspase-10 in S. cerevisiae appeared immediately after the expression and activation of the caspase. Indeed, we were only able to detect the expression of caspase-10 after 4 hours of induction in galactose-containing media and we also observed cell death and other associated phenotypes after that time, indicating that the expression of human initiator caspases triggers an extremely fast lethal response. As mentioned above, the present yeast model might be considered as an appropriate tool to investigate as yet unknown functions of human initiator caspases.
Supplementary Material
Chromosomal DNA integrity after the expression of human initiator caspases in yeast. Chromosomes of BY4741 yeast cells transformed with either CASP8_vC or CASP10_vD were separated by pulse-field gel electrophoresis. Glu, glucose cultures; gal, galactose cultures. Yeast cells transformed with an empty vector were used as negative controls. Hydrogen peroxide (5mM) treatment of the strain transformed with an empty vector was used as a positive control for DNA degradation. Roman numerals indicate the number of the yeast chromosomes.
Yeast cells transformed with empty vectors were used as negative controls in all experiments performed. A, clonogenic analysis. B, ROS analysis. C, AnnexinV/PI staining. D, SYTO18 mitochondrial staining. E, MDY-64 vacuole staining. F, phalloidin staining.
Acknowledgments
This work was supported by grants from Ministerio de Ciencia e Innovación of Spain (AGL2005-07245-C03-03 to J. L. R., and SAF2008-02251 and RD06/0020/1037 - Red Temática de Investigación Cooperativa en Cáncer, Instituto de Salud Carlos III to F. M.), Fundación “la Caixa” (BM05-30-0) and NIH grant GM621284 to A. M. N.. P. L. S. and A. C. M. are recipients of FPU predoctoral fellowships from the Spanish Ministerio de Educación y Ciencia. We thank M. D. Sánchez for excellent technical help, A. González-Novo for help with the FACS experiments, Susan van Horn in the Stony Brook Center for Microscopy for assistance with sample preparations and N. Skinner for correcting the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ho PK, Hawkins CJ. Mammalian initiator apoptotic caspases. The FEBS journal. 2005;272:5436–5453. doi: 10.1111/j.1742-4658.2005.04966.x. [DOI] [PubMed] [Google Scholar]
- 2.Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell death and differentiation. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- 3.Festjens N, Cornelis S, Lamkanfi M, Vandenabeele P. Caspase-containing complexes in the regulation of cell death and inflammation. Biological chemistry. 2006;387:1005–1016. doi: 10.1515/BC.2006.124. [DOI] [PubMed] [Google Scholar]
- 4.Tibbetts MD, Zheng L, Lenardo MJ. The death effector domain protein family: regulators of cellular homeostasis. Nature immunology. 2003;4:404–409. doi: 10.1038/ni0503-404. [DOI] [PubMed] [Google Scholar]
- 5.Buttner S, Eisenberg T, Herker E, Carmona-Gutierrez D, Kroemer G, Madeo F. Why yeast cells can undergo apoptosis: death in times of peace, love, and war. J Cell Biol. 2006;175:521–525. doi: 10.1083/jcb.200608098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamann A, Brust D, Osiewacz HD. Apoptosis pathways in fungal growth, development and ageing. Trends in microbiology. 2008;16:276–283. doi: 10.1016/j.tim.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Frohlich KU, Fussi H, Ruckenstuhl C. Yeast apoptosis--from genes to pathways. Seminars in cancer biology. 2007;17:112–121. doi: 10.1016/j.semcancer.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, Sigrist S, Madeo F. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Fahrenkrog B, Sauder U, Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- 10.Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Frohlich KU. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 11.Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Frohlich KU, Manns J, Cande C, Sigrist SJ, Kroemer G, Madeo F. An AIF orthologue regulates apoptosis in yeast. J Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 13.Tao W, Kurschner C, Morgan JI. Modulation of cell death in yeast by the Bcl-2 family of proteins. The Journal of biological chemistry. 1997;272:15547–15552. doi: 10.1074/jbc.272.24.15547. [DOI] [PubMed] [Google Scholar]
- 14.Kang JJ, Schaber MD, Srinivasula SM, Alnemri ES, Litwack G, Hall DJ, Bjornsti MA. Cascades of mammalian caspase activation in the yeast Saccharomyces cerevisiae. The Journal of biological chemistry. 1999;274:3189–3198. doi: 10.1074/jbc.274.5.3189. [DOI] [PubMed] [Google Scholar]
- 15.Puryer MA, Hawkins CJ. Human, insect and nematode caspases kill Saccharomyces cerevisiae independently of YCA1 and Aif1p. Apoptosis. 2006;11:509–517. doi: 10.1007/s10495-006-5114-2. [DOI] [PubMed] [Google Scholar]
- 16.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in enzymology. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 17.Eggleston MD, Marshall PA. Saccharomyces cerevisiae samples stained with FUN-1 dye can be stored at -20 degrees C for later observation. Journal of microscopy. 2007;225:100–103. doi: 10.1111/j.1365-2818.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- 18.Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann SH, Mesner PW, Jr, Samejima K, Tone S, Earnshaw WC. Detection of DNA cleavage in apoptotic cells. Methods in enzymology. 2000;322:3–15. doi: 10.1016/s0076-6879(00)22003-x. [DOI] [PubMed] [Google Scholar]
- 20.Neiman AM. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng PW, Porter AG, Janicke RU. Molecular cloning and characterization of two novel pro-apoptotic isoforms of caspase-10. The Journal of biological chemistry. 1999;274:10301–10308. doi: 10.1074/jbc.274.15.10301. [DOI] [PubMed] [Google Scholar]
- 22.Scaffidi C, Medema JP, Krammer PH, Peter ME. FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. The Journal of biological chemistry. 1997;272:26953–26958. doi: 10.1074/jbc.272.43.26953. [DOI] [PubMed] [Google Scholar]
- 23.Eisenberg T, Buttner S, Kroemer G, Madeo F. The mitochondrial pathway in yeast apoptosis. Apoptosis. 2007;12:1011–1023. doi: 10.1007/s10495-007-0758-0. [DOI] [PubMed] [Google Scholar]
- 24.Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinberger M, Ramachandran L, Feng L, Sharma K, Sun X, Marchetti M, Huberman JA, Burhans WC. Apoptosis in budding yeast caused by defects in initiation of DNA replication. J Cell Sci. 2005;118:3543–3553. doi: 10.1242/jcs.02477. [DOI] [PubMed] [Google Scholar]
- 26.Grallert B, Boye E. The multiple facets of the intra-S checkpoint. Cell Cycle. 2008;7:2315–2320. doi: 10.4161/cc.6389. [DOI] [PubMed] [Google Scholar]
- 27.Gourlay CW, Ayscough KR. The actin cytoskeleton: a key regulator of apoptosis and ageing? Nature reviews. 2005;6:583–589. doi: 10.1038/nrm1682. [DOI] [PubMed] [Google Scholar]
- 28.Leadsham JE, Gourlay CW. Cytoskeletal induced apoptosis in yeast. Biochimica et biophysica acta. 2008;1783:1406–1412. doi: 10.1016/j.bbamcr.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Zheng K, Pan JW, Ye L, Fu Y, Peng HZ, Wan BY, Gu Q, Bian HW, Han N, Wang JH, Kang B, Pan JH, Shao HH, Wang WZ, Zhu MY. Programmed cell death-involved aluminum toxicity in yeast alleviated by antiapoptotic members with decreased calcium signals. Plant physiology. 2007;143:38–49. doi: 10.1104/pp.106.082495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroemer G, El-Deiry WS, Golstein P, Peter ME, Vaux D, Vandenabeele P, Zhivotovsky B, Blagosklonny MV, Malorni W, Knight RA, Piacentini M, Nagata S, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell death and differentiation. 2005;12(Suppl 2):1463–1467. doi: 10.1038/sj.cdd.4401724. [DOI] [PubMed] [Google Scholar]
- 32.Garrido C, Kroemer G. Life’s smile, death’s grin: vital functions of apoptosis-executing proteins. Current opinion in cell biology. 2004;16:639–646. doi: 10.1016/j.ceb.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Galluzzi L, Joza N, Tasdemir E, Maiuri MC, Hengartner M, Abrams JM, Tavernarakis N, Penninger J, Madeo F, Kroemer G. No death without life: vital functions of apoptotic effectors. Cell death and differentiation. 2008;15:1113–1123. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kischkel FC, Lawrence DA, Tinel A, LeBlanc H, Virmani A, Schow P, Gazdar A, Blenis J, Arnott D, Ashkenazi A. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. The Journal of biological chemistry. 2001;276:46639–46646. doi: 10.1074/jbc.M105102200. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Chun HJ, Wong W, Spencer DM, Lenardo MJ. Caspase-10 is an initiator caspase in death receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13884–13888. doi: 10.1073/pnas.241358198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilot D, Serandour AL, Ilyin GP, Lagadic-Gossmann D, Loyer P, Corlu A, Coutant A, Baffet G, Peter ME, Fardel O, Guguen-Guillouzo C. A role for caspase-8 and c-FLIPL in proliferation and cell-cycle progression of primary hepatocytes. Carcinogenesis. 2005;26:2086–2094. doi: 10.1093/carcin/bgi187. [DOI] [PubMed] [Google Scholar]
- 37.Lee RE, Puente LG, Kaern M, Megeney LA. A non-death role of the yeast metacaspase: Yca1p alters cell cycle dynamics. PLoS ONE. 2008;3:e2956. doi: 10.1371/journal.pone.0002956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nature reviews. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 39.Dalle-Donne I, Rossi R, Milzani A, Di Simplicio P, Colombo R. The actin cytoskeleton response to oxidants: from small heat shock protein phosphorylation to changes in the redox state of actin itself. Free radical biology & medicine. 2001;31:1624–1632. doi: 10.1016/s0891-5849(01)00749-3. [DOI] [PubMed] [Google Scholar]
- 40.Farah ME, Amberg DC. Conserved actin cysteine residues are oxidative stress sensors that can regulate cell death in yeast. Mol Biol Cell. 2007;18:1359–1365. doi: 10.1091/mbc.E06-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staleva L, Hall A, Orlow SJ. Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent Manner. Mol Biol Cell. 2004;15:5574–5582. doi: 10.1091/mbc.E04-02-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vilella F, Herrero E, Torres J, de la Torre-Ruiz MA. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress. The Journal of biological chemistry. 2005;280:9149–9159. doi: 10.1074/jbc.M411062200. [DOI] [PubMed] [Google Scholar]
- 43.Li SC, Kane PM. The yeast lysosome-like vacuole: Endpoint and crossroads. Biochimica et biophysica acta. 2008 doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khoury CM, Greenwood MT. The pleiotropic effects of heterologous Bax expression in yeast. Biochimica et biophysica acta. 2008;1783:1449–1465. doi: 10.1016/j.bbamcr.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 46.Kissova I, Plamondon LT, Brisson L, Priault M, Renouf V, Schaeffer J, Camougrand N, Manon S. Evaluation of the roles of apoptosis, autophagy, and mitophagy in the loss of plating efficiency induced by Bax expression in yeast. The Journal of biological chemistry. 2006;281:36187–36197. doi: 10.1074/jbc.M607444200. [DOI] [PubMed] [Google Scholar]
- 47.Gozuacik D, Kimchi A. Autophagy and cell death. Current topics in developmental biology. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- 48.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun RJ, Zischka H. Mechanisms of Cdc48/VCP-mediated cell death: from yeast apoptosis to human disease. Biochimica et biophysica acta. 2008;1783:1418–1435. doi: 10.1016/j.bbamcr.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Dudgeon DD, Zhang N, Ositelu OO, Kim H, Cunningham KW. Non-apoptotic death of Saccharomyces cerevisiae cells that is stimulated by Hsp90 and inhibited by calcineurin and Cmk2 in response to ER stresses. Eukaryotic cell. 2008 doi: 10.1128/EC.00291-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosomal DNA integrity after the expression of human initiator caspases in yeast. Chromosomes of BY4741 yeast cells transformed with either CASP8_vC or CASP10_vD were separated by pulse-field gel electrophoresis. Glu, glucose cultures; gal, galactose cultures. Yeast cells transformed with an empty vector were used as negative controls. Hydrogen peroxide (5mM) treatment of the strain transformed with an empty vector was used as a positive control for DNA degradation. Roman numerals indicate the number of the yeast chromosomes.
Yeast cells transformed with empty vectors were used as negative controls in all experiments performed. A, clonogenic analysis. B, ROS analysis. C, AnnexinV/PI staining. D, SYTO18 mitochondrial staining. E, MDY-64 vacuole staining. F, phalloidin staining.