Abstract
Abortive initiation, when first discovered, was an enigmatic phenomenon, but fully three decades hence, it has been shown to be an integral step in the transcript initiation process intimately tied to the promoter escape reaction undergone by RNA polymerase at the initiation-elongation transition. A detailed understanding of abortive initiation-promoter escape has brought within reach a full description of the transcription initiation mechanism. This enormous progress was the result of convergent biochemical, genetic, and biophysical investigations propelled by parallel advances in quantitation technology. This chapter discusses the knowledge gained through the biochemical approach and a high-resolution method that yields quantitative and qualitative information regarding abortive initiation-promoter escape at a promoter.
Keywords: abortive initiation, promoter escape, steady-state transcription reactions, quantitative initiation parameters, abortive RNA sequencing
1. Introduction
Abortive initiation refers to the repetitive synthesis and release of short nascent RNAs by RNA polymerase. It was first observed in transcription reactions containing only the first two nucleoside triphosphate (NTP) substrates [1]. Missing the full complement of NTPs clearly prevented elongation by RNA polymerase, but the release of the nascent RNA was unexpected. This finding gave way to a bigger surprise when abortive initiation was shown to occur in the presence of all four NTPs to yield a mix of abortive RNAs ranging from 2–8 nucleotides (nt) [2]. This study showed that abortive initiation normally accompanies steady-state transcription, that there is a finite potential to release the RNA after each phosphodiester bond incorporation event (during the initiation phase), and that abortive initiation may be linked to the role of sigma subunit in the transcription cycle [3]. These studies marked a promising beginning to an emerging field of research.
Research to clarify abortive initiation came in two waves. First wave led to the characterization of lacUV5 as the first example of a promoter whose transcription initiation kinetics is rate-limited at the promoter escape step, due to the insightful work of Gralla and colleagues [2, 4–6]. They defined the term promoter escape to indicate that productive synthesis represents an escape from abortive cycling. Second wave came up with the arrival of the phosphorimaging technology that greatly facilitated the quantitative measurement of transcription reaction products. The new technology was applied to the in vitro transcription analysis of three phage promoters T7 A1, T5 N25 and N25anti, and has helped put the current initiation research on a rigorous quantitative footing [7, 8].
In this chapter, I shall briefly review the important understandings regarding abortive initiation, but focus on the quantitative protocol involved. Many studies and approaches that have contributed to achieving a mechanism of abortive initiation and promoter escape have recently been reviewed [9, 10].
1.1 Steps after open complex formation influence promoter strength
Promoter-specified transcription is foremost regulated at the initiation stage when RNA polymerase binds to a promoter DNA to form an open complex with varying potential at de novo initiation. Based on the abortive initiation phenomenon, McClure and coworkers designed a steady-state assay for measuring the initiation frequencies of RNA polymerase at various promoters [11]. Using the abortive initiation assay, it was possible to derive KB, the equilibrium binding constant for closed complex formation, and k2, the forward isomerization rate constant for open complex formation, assuming that k2 is the rate-limiting step and k−2 (the rate constant for open complex collapse) is negligible [12]. The product KB k2 reflects the apparent second order rate constant for open complex formation, and a high value indicates a strong efficacy of forming an active and stable open complex that displays high initiation frequency [13]. KB k2 for ~30 E. coli Eσ70 promoters that differed by four orders, in turn, showed a direct correlation with the homology score, which measures the degree of agreement of a promoter sequence to the consensus [14], allowing these parameters to be considered as indicators of promoter strength in vitro.
These parameters, however, failed to predict promoter strength in vivo. Bujard and coworkers compared the in vivo level of mRNA measured from a set of well-studied strong promoters with their in vitro determined kON (the pseudo-first-order rate constant for complex formation) [15] and homology score. Whereas kON and homology score correlated well with each other, no correlation was found between the in vivo promoter strength (measured as mRNA levels relative to that of an internal standard promoter) and homology score [16]. This discrepancy points out that, in considering promoter function and activity, early emphasis was placed on promoter features essential for efficient recognition and binding by the polymerase based on in vitro measured kON and homology score. Promoter activity in vivo must involve steps after binding and de novo initiation. During the initiation phase, these steps were shown subsequently to correspond to the steps of abortive initiation and promoter escape.
1.2 lacUV5 is the first escape rate-limited promoter
In investigating the kinetics of transcription initiation, lacUV5 promoter was shown to be a much slower initiator than T7 A1 and λ PR and PL promoters, but its slowest step in initiation comes after open complex formation [4, 5]. Using a gel assay, Carpousis and Gralla [2] monitored simultaneously the synthesis of short and long RNAs and found that both abortive and productive initiations were supported in the presence of all four NTPs and that from lacUV5 promoter, short RNAs of hexanucleotides were detected. The formation of both abortive and productive RNAs suggests that, of all de novo initiated chains, only a fraction would reach the productive length. In the same study, abortive initiation was found to persist in the presence of heparin, which selectively inhibits free rather than complexed RNA polymerase. Coupling this observation to the fact that abortive RNAs are made in large molar excess over the amount of promoter DNA present led to the suggestion that RNAP, upon releasing its short RNA, does not dissociate from the promoter but cycles back to the open complex state and repeatedly initiate to give the high abortive yield [2]. Footprinting analysis later confirmed that the default conformation for RNAP during abortive cycling is the open complex conformation [17].
Abortive cycling implies the existence of a step limiting the conversion of nascent transcripts to the full-length RNA. Because productive synthesis could be viewed as an escape from abortive cycling by the polymerase, this rate-limiting step was called promoter escape now known to correspond with the initiation-elongation transition [2]. By measuring the ratio of abortive to productive RNA synthesized, both lacUV5 and Tn5 promoters were found to engage in more abortive synthesis indicating more difficulty at escape than the T7 A1 promoter [18].
Extending their early insightful work, Carpousis et al. [6] compared the relative abortive to productive yield of four lac promoter variants that differ in −35 and −10 elements and showed that the closer the promoter sequence is to the consensus, the poorer is the escape. This result was the first evidence that the strength of interaction of the core promoter elements with RNA polymerase is involved in determining the relative extent of abortive initiation and promoter escape.
1.3 Studies of T5 N25 promoter shed new light on abortive initiation and promoter escape
In their study of the phage T5 N25 promoter, Bujard and coworkers sought to uncover the promoter elements responsible for its high activity [19, 20]. They subdivided the promoter fragment (spanning −50 to +20 corresponding to the DNase I protected region of an open complex footprint) into three regions: the upstream region (USR), from −50 to −36; the core region, −35 to −1, and the downstream region (DSR), from +1 to +20, and investigated changes in the USR and DSR on N25 activity. While USR was uniformly required for the high activity of the T5 phage early promoters, an “anti” version of the DSR (involving A↔C and G↔T changes) rendered the N25 promoter ~10 fold weaker in mRNA synthesis without affecting its kON [16, 20]. These authors concluded that DSR affects a step after open complex formation, namely the promoter clearance step, to reduce productive mRNA synthesis. This finding prompted our participation to examine the abortive-productive RNA synthesis by the two promoters, N25 and N25antiDSR. (Note that we have abbreviated the N25antiDSR designation to N25anti [8].)
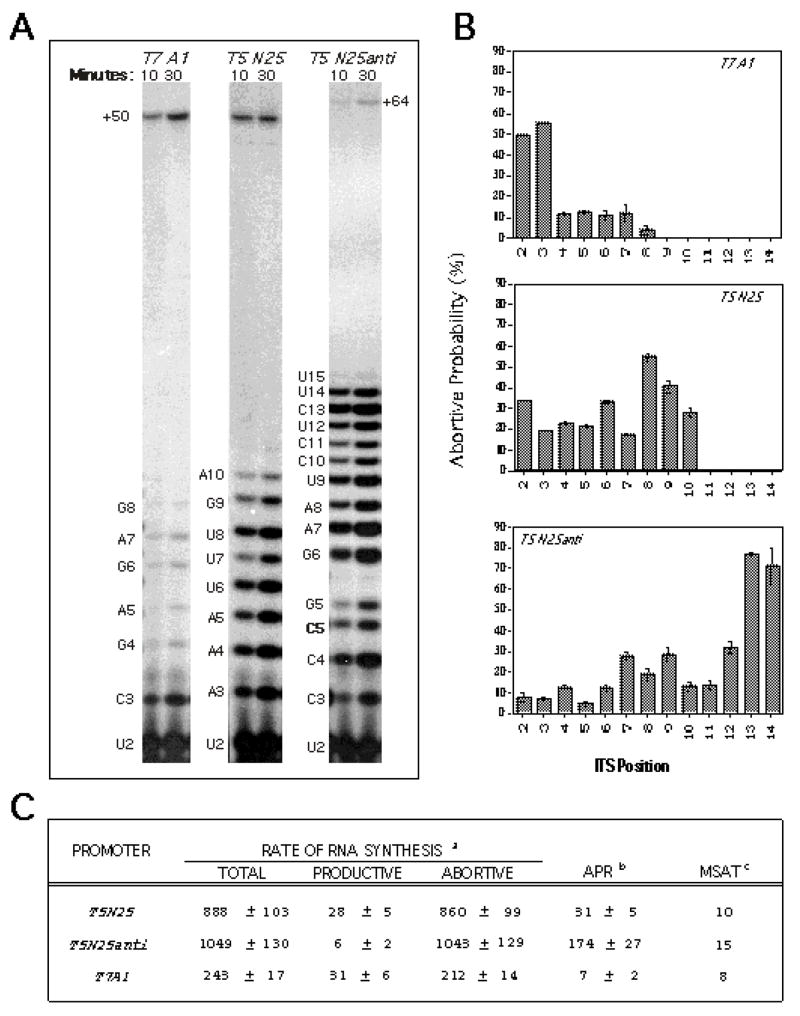
For this analysis, we coupled the steady-state in vitro transcription reaction and high percentage gel fractionation to quantitative phosphorimaging analysis [7]. When applied to the analysis of T7 A1, T5 N25, T5 N25anti promoters, we found several striking results [8]. One, these promoters displayed very different abortive RNA ladders, indicating different positions of the escape transition on individual promoters. Two, with T5 N25anti, the abortive RNA ladder ranged 2–15 nt in length; this was the first example that abortive synthesis can proceed beyond the +10 position [3]. (Here, it should be pointed out that all of the abortive RNAs were not convertible into longer RNA during high [NTP] chase and shown to be released from the enzyme complex by molecular sieve chromatography [8].) Three, these three promoters differ drastically in their abortive-productive ratio, and therefore, their productive transcription efficiency. In this study, we devised quantitative initiation parameters to describe the abortive initiation-promoter escape process at each promoter. Figure 1 shows the qualitative pattern of abortive-productive transcription from these three promoters (Figure 1A), their abortive probability profile (Figure 1B), and tabulated quantitative parameters (Figure 1C). This study led to the conclusion that the abortive initiation-promoter escape process is controlled by the promoter sequence – both the promoter recognition region (PRR; the untranscribed region spanning −60 to −1) and initial transcribed sequence (ITS; the transcribed region from +1 to +20) elements. This quantitative methodology will be discussed in greater detail in the section on Description of Method.
Figure 1.
A typical result of the quantitative and qualitative analysis of transcription. The figure shown is taken from Ref. 8 where three phage promoters –T7 A1, T5 N25, and N25anti -- were analyzed. A. A time-course investigation involving two time points (10-minute and 30-minute) was performed to obtain the abortive and productive rates. B. Abortive probability was calculated as described [7]. Plotting the abortive probability of an initial transcribing complex to its abortive product length yields the abortive probability profile, which shows up the location of high abortive barriers on each promoter. Abortive probability profiles for all three promoters are displayed. C. Tabulation of the various quantitative parameters. aProductive and abortive rates measured as (femto)moles of RNA per (femto)mole of RNA polymerase per hour. bAPR: the abortive:productive ratio. cMSAT: the maximal size of abortive transcript.
1.4 Strength of PRR interactions establishes the rate-limiting step
Applying the quantitative analysis to a comprehensive set of core promoter variants that deviated from the consensus features by 1 bp revealed an inverse correlation of consensus agreement of a promoter with its ability to support escape by RNA polymerase [21]. This result supports a conclusion reached earlier using the lac promoters [6]. Thus, a consensus promoter binds very tightly to RNA polymerase and interferes with the escape process. Having maximized KB and k2, a consensus promoter becomes rate-limited at the next step in its initiation program, i.e., the promoter escape step. A polymerase that cannot escape undergoes abortive initiation instead, resulting in higher abortive-productive RNA ratio. Another consequence of converting a promoter to a consensus version leads to lengthening of the abortive ladder, as shown with the T7 A1 promoter, from 8 to 16 nt; thus, delaying the escape transition [22].
1.5 ITS directs the abortive initiation-promoter escape program
When transcribed under the same conditions, N25 and N25anti promoters show comparable levels of total initiation, but very different levels of abortive vs. productive initiation. Because N25 and N25anti share identical PRR but differ in ITS, the above results suggest that PRR determines the level of initiation at a promoter, but ITS governs the extent of abortive versus productive synthesis [8]. To further investigate the role of ITS, we constructed ~40 N25 promoter-random ITS variants and found that the randomized ITS conferred a 25-fold modulation of productive synthesis from the same N25 promoter [23]. An inverse correlation exists between the productive yield and total initiation among the N25-random ITS promoter variants, suggesting that a failure at escape relegates the polymerase to repetitive abortive initiation. Each ITS variant further shows a distinct abortive probability profile, indicating that the ITS sequence directly controls the promoter escape process – in terms of how long an abortive program is carried out and where the high abortive barriers are during the initial transcription program.
Of PRR and ITS elements that both exert some degree of influence on promoter escape, our extensive investigation with the N25 promoter and its numerous (PRR and ITS) variants led to the conclusion that PRR is the primary determinant of escape involved in setting the rate-limiting step of transcription initiation. ITS plays a secondary role in that it affects the pattern of escape only when it is affiliated with an escape-rate-limited promoter [23].
1.6 GreA/B diminishes abortive synthesis to enhance productive expression
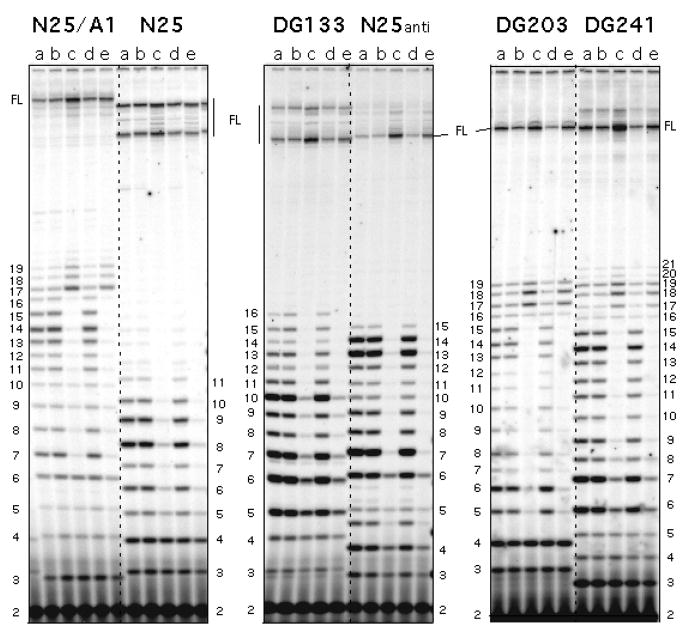
The level of abortive initiation at a promoter – to the extent that it reflects an impairment at promoter escape – can be dramatically reduced in the presence of Gre factors, GreA or GreB [24, 25]. Gre are transcription factors that rescue arrested complexes by stimulating the intrinsic cleavage activity of RNAP [26]. Transcript cleavage removes 2–3 nt (GreA) or up to 10 nt (GreB) from 3′-end of the backtracked RNA to generate a new 3′-OH group at the active site, thus allowing re-extension of the 5′-piece of nascent transcript to reach the full length. Gre-mediated rescue of abortive synthesis suggests that the abortive RNAs, before their release from the enzyme, were transiently associated with backtracked initial transcribing complexes [24, 25]. Of the two factors, GreA was deemed the more important cleavage stimulatory factor in vivo [27, 28], while GreB was found to be more effective at cleavage-rescue of long-abortive promoters [23]. Figure 2 shows the effect of GreB on a set of N25 promoter variants. With each promoter, GreB brought about a sharp reduction in abortive RNA levels and a concomitant increase in productive RNA level [23].
Figure 2.
Effect of GreB and NusA on promoter escape from N25-random ITS promoter variants. The figure shown was published in Ref. 10. Six N25 promoter variants (N25/A1, N25, DG133, N25anti, DG203, DG241) differ only in their initial transcribed sequence (ITS), which dictated the appearance of distinct abortive ladder for each promoter. Each promoter was transcribed under five different enzymatic conditions for 10 min at 37 °C. The transcripts were labeled with [γ-32P]-ATP. Enzymatic conditions were: (a) GreA−GreB− RNAP; (b) wt RNAP; (c) wt RNAP:GreB (1:10); (d) wt RNAP:NusA (1:10); and (e) wt RNAP:GreB:NusA (1:10:10). RNAP was preincubated for 10 min at room temperature with the accessory protein(s) prior to its addition to the reaction. This gel showed that NusA has no effect on abortive initiation and promoter escape. GreB, however, decreased abortive synthesis and stimulated productive synthesis (compare lane c in each panel to the neighboring lanes). Numbers on the border refer to the length of abortive RNA bands located next to it. FL: full-length RNA.
1.7 Couching abortive initiation-promoter escape in the context of a transcription initiation rate diagram
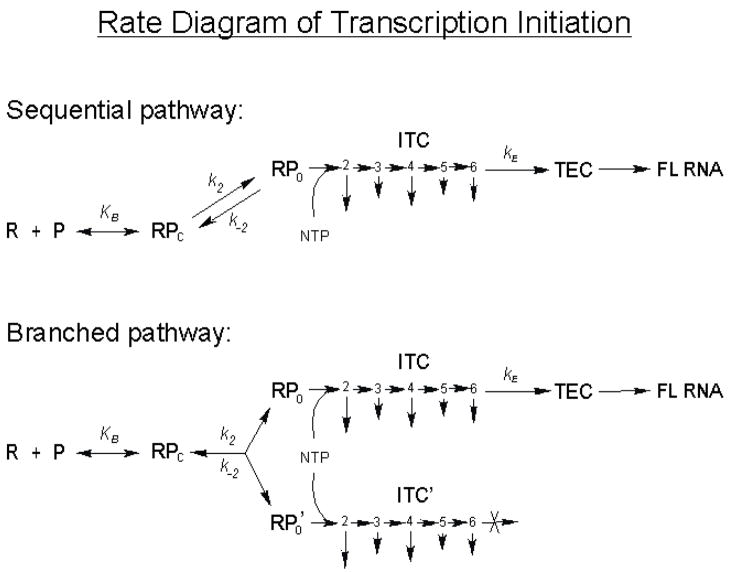
To understand the implications of the abortive level at a promoter, it is necessary to place the abortive initiation-promoter escape process into the sequential multi-step rate diagram of transcription initiation (Figure 3). According to this diagram, the abortive level at a promoter is determined by the entry into and exit from initial transcription by RNA polymerase, respectively governed by the kinetic constants − KB k2/k−2, which sets the level of total initiation (during a specified time period) and kE, the composite rate constant of escape, which determines the fraction of initiated events that successfully undergoes escape to become full-length (FL) transcripts [9]. Knowing the various constants associated with a promoter, one can account for the abortive level observed at that promoter. For example, T5 N25 has a very high KB k2 value and a negligible k−2 [16], and it is rate-limited at the promoter escape step (presumably due to a low kE); this combination of rate constants gives rise to high yields of abortive RNA [8]. Compared to T5 N25, the T7 A1 promoter has a lower KB k2 value but a higher k−2 [16], so it supports a lower level of total initiation. However, it is less rate-limited at escape. Under similar transcription conditions, the combination of rate constants for T7 A1 led to a slightly higher level of productive transcripts than that obtained with T5 N25 [8]. For rrnB P1 promoter, its KB k2 value is slightly lower than that of T7 A1 [14], but it has a high k−2 such that open complexes are extremely short-lived [29]. Initiation occurs only in the presence of high [NTP] which, by mass effect, pulls the isomerization equilibrium in favor of k2. Because rrnB P1 is rate-limited at open complex half-life and stability, and not at the promoter escape step, all initiated transcripts from rRNA promoters – under the conditions that have been tested – appear to be readily elongated to the full length without being released as abortive RNA [30; T. Gaal and R. L. Gourse, personal communication]. Note in the above discussions that the comparisons cannot be very exact. This is because many kinetic constants (especially kE) have not been measured; and of the measured constants, many were determined under different reaction conditions. These issues preclude a rigorous direct comparison between the various promoters.
Figure 3.
Rate diagram of transcription initiation. This figure was previously described in Ref. 9. The sequential pathway of transcription initiation shows RNA polymerase (R) first binds the promoter (P) to form a closed complex (RPc) with an equilibrium constant KB. RPc next isomerizes to form the open complex (RPo) with a forward rate constant k2 and a reverse rate constant k−2. The open complexes are competent at de novo initiation, and in the presence of NTP, forms initial transcribing complexes (designated by the numbers 2, 3, 4, 5, 6 to indicate the position of the ITC) which abortively release their nascent RNAs to different degrees (as indicated by the length of the downward arrow). After some rounds of abortive initiation, all ITCs eventually escape with a rate constant kE to form ternary elongation complex (TEC) which can be processively elongated to form the full-length (FL) RNA. Here, kE, the rate constant of escape, is measured from pre-formed open complexes under single-cycle transcription condition as the rate of (short) FL RNA synthesis. As such, kE represents a composite constant which encompasses the rates of all of the abortive initiation steps before escape, the escape step itself, and the rates of all of the elongation steps (~20 steps) after escape. The sequential pathway predicts that every RNA polymerase molecule eventually escapes to synthesize the full-length RNA. The branched pathway model of transcription initiation shows the formation of productive and unproductive open complexes (RPo and RPo’, respectively) at a promoter. In the presence of NTP, RPo forms productive ITC which is capable of escape after some rounds of obligate abortive initiation. RPo’, however, forms unproductive ITCs (ITC’) that cannot escape and are trapped to perform continual abortive synthesis.
For highly abortive promoters, a branched initiation diagram (Figure 3) is necessary to account for the existence of productive and unproductive initial transcribing complexes (unproductive ITCs) detected by an inverse pulse-chase assay [31, 32]. The productive ITCs are capable of escape to produce full-length RNA, but only after going through obligate rounds of abortive initiation. The unproductive ITCs are trapped in abortive transcription continuously and cannot undergo promoter escape. Thus, the abortive RNAs made at such a promoter are derived from both the productive and unproductive ITCs. Because the unproductive complexes contribute disproportionately to the abortive yield, their existence greatly complicates the quantitative analysis of abortive initiation. Interested readers are referred to a recent review [10] for further discussion of this topic.
1.8 Regulatory consequences at abortive initiation
The level of transcription initiation is often used to gauge the regulatory consequences at a promoter [Busby, this volume]. Here, it is not sufficient to monitor only the level of abortive initiation, but also productive initiation that yields full-length transcripts. Looking at the abortive initiation levels alone, there is not a straightforward correlation of its fluctuation with repression or activation. This is because, according to the transcription initiation rate diagram (Figure 3), the abortive initiation level exhibited by a promoter is subject to change at two points: at the total initiation level, due to regulation exerted at KB k2, and at the promoter escape process, due to regulation exerted at kE. Accordingly, a decrease in the abortive level can result from a repression of KB k2 without changing kE, or an activation of kE without affecting KB k2. The activating effect of the cAMP-CRP complex at the malT promoter is an example of the latter where reduced abortive level is accompanied by elevated full-length RNA synthesis [33]. However, these constants rarely change in isolation. When the KB k2 value at a promoter is elevated (which leads to a further stabilization of the polymerase-promoter open complex contacts), the escape by RNA polymerase (which requires the disruption of its open complex contacts) is automatically compromised, leading to a decrease in kE. These concurrent but opposite changes in the two sets of rate constants can be brought about by promoter sequence mutations [21] or by regulatory proteins [34]. The latter is showcased by B. subtilis phage Φ29 protein p4 and its effect on the early A2c promoter [34]. The reverse situation where a regulatory protein slows open complex formation (thus decreasing KB k2) and accelerates promoter clearance (presumably by enhancing kE) is exemplified by phage P22 Arc-SL35 protein on a consensus version of the Pant promoter [35].
The steady-state quantitative transcription method [7] is well suited for the purpose of deducing the targets of regulation. One can obtain, from one gel run, the abortive initiation and productive initiation levels. These levels sum up to give the total initiation level, which reflects the KB k2 associated with a promoter. The abortive to productive ratio (APR) of initiated transcripts gives an indication of the escape efficiency. By comparing the transcriptional outcome of these two parameters – total initiation and APR – in reactions containing or lacking a regulatory factor, one can readily pinpoint the target of regulation during the initiation phase.
1.9 Regulation at transcript slippage vs. abortive initiation
Although abortive initiation is a non-productive form of transcription, the abortive products and their synthesis process are specified by the template sequence and have been shown to be obligatory steps towards productive transcription [32, 36]. Abortive initiation is therefore an integral part of the transcription initiation mechanism.
Another form of non-productive transcription during the initiation stage involves slippage synthesis at homopolymeric triplet sequences, especially near the transcriptional start site, to produce slippage transcripts that contain reiterative addition of the triplet repeat nucleotide [37]. The slippage transcripts are usually released immediately from the enzyme and cannot be re-elongated in a template sequence-specified manner (with one recent exception to be discussed below). Therefore, the slippage transcripts may be considered abortive products. Many bacteria have appropriated the slippage transcription design for regulating a group of UTP biosynthetic pathway genes in a [UTP] dependent manner. In these genes, a TTT (non-template strand) repeat near the start site of transcription steers the enzyme towards the synthesis of polyU-containing slippage transcripts at high [UTP], which prevents transcriptional read-through to the downstream structural gene sequence, repressing its expression. At low [UTP], RNA polymerase carries out templated transcription, producing a normal set of abortive transcripts on its way to the productive expression of the structural gene sequence. Thus, two types of abortive initiation pathways exist at these promoters to govern the expression of the downstream structural gene in a [UTP]-dependent manner [37].
The one exception mentioned above is concerned with the fact that the G-slippage transcript at the B. subtilus pyrG start site, after incorporating 1 to ~10 additional G residues, can re-enter normal transcription. pyrG encodes CTP synthase whose transcriptional expression is feedback regulated by the cellular [CTP]. Low [CTP] triggers the G-slippage. The polyG stretch at the 5′-end of the resultant transcript subsequently pairs with a pyrimidine-rich stretch downstream to form an antiterminator hairpin that allows transcriptional read-through to the structural gene [37, 38].
1.10 In vivo evidence of promoter escape rate limitation
The prominence of abortive initiation in vitro has raised questions regarding its relevance in vivo. While no one has demonstrated the existence of abortive RNAs in vivo, transcription with supercoiled promoter templates suggests that abortive initiation can occur in the cellular context [29, 39]. This possibility is strengthened by numerous evidences of promoter escape rate-limitation – the root cause of abortive initiation – in vivo. A direct evidence was obtained with permanganate footprinting in vivo which caught RNA polymerase stalling at the early transcribed region (i.e., at +6 to +12) of a consensus promoter [40]. A comprehensive investigation of GreA’s role in vivo led to the conclusion that GreA mainly stimulates transcript cleavage during early stages of initiation to facilitate promote escape and productive target gene expression [27]. The involvement of GreA’s cleavage stimulatory activity implies the existence of arrested transcripts. Lately, the ChIP-chip technique has become a powerful tool in probing the genome location and distribution of a target enzyme in vivo [see Rhodius and Wade, this volume]. With E. coli Eσ70 RNA polymerase, 23% of those bound at the promoter region were not associated with a transcript. It was suggested that these “poised” RNA polymerase molecules may be rate-limited at promoter escape in vivo [41]. The prevalence of RNA polymerase stalling at promoter-proximal sites in vivo was confirmed by KMnO4 footprinting on chromosomal DNA [28]. The half-lives of these stalled complexes are subject to GreA modulation [28], indicating that stalling is the result of escape rate-limitation. The abundant examples of escape rate-limitation above suggest abortive initiation can occur in vivo. A question remains regarding its degree of occurrence.
2. Description of Method
The arrival of the phosphorimaging technology in the early 1990s made possible the ready quantitation in situ of a large number of 32P-labeled transcript bands fractionated and displayed on polyacrylamide gels. At the time, we wished to apply this technology to the quantitative determination of abortive and productive transcriptions by E. coli RNA polymerase and had to design an in vitro transcription reaction protocol for the purpose of obtaining large amounts of quantifiable information on individual promoters so that the transcription initiation process can be described by quantitative parameters. Such a protocol was achieved by incorporating several constraints; these have been discussed in detail [7]. The quantitative assay gave us a handle for comparing the effect of promoter sequence mutations, RNA polymerase mutations, as well as other intrinsic and extrinsic elements that affect abortive/productive transcription. Below, I briefly review the important considerations involved and describe an updated version of the protocol with its various modifications and utilities.
2.1 Important considerations in protocol design
2.1.1 Transcription templates
To study the transcriptional expression from a single promoter, we decided to use PCR-amplified single-promoter fragments as templates for transcription. PCR amplification allows us to specify the length of the run-off (productive) RNA such that both the abortive and productive RNAs could be fractionated and resolved in one gel. For this purpose, we usually design the promoter fragments to yield run-off RNAs at ~50–60 nucleotides (nt) in length; however, under our modified gel protocol (see below), we now can fractionate RNAs from 2 to ~100 nt in the same gel. PCR-amplification also allows one to specify the upstream boundary of the template fragment, which can vary depending on whether a promoter is subject to regulation by upstream-binding protein factors. For factor-independent promoters, upstream boundary to −60 (relative to the +1 start site) is usually sufficient, although sequences up to −100 were shown to facilitate steps towards open complex formation [42, 43].
Abortive initiation is not limited to transcription from fragment templates. Supercoiled DNA minicircles containing a single promoter (gal P1 or gal P2) support abortive initiation and the activation-repression pattern of P1 and P2 expression by cAMP-CRP resembles the in vivo situation [40]. Abortive initiation has further been studied from promoters cloned into a supercoiled plasmid vector that contains many endogenous promoters [29]. When transcribing from supercoiled plasmid templates, one has to ensure that the reaction conditions only allow (abortive) initiation from the target promoter.
2.1.2 Steady-state vs. single-cycle transcription
Of the two ways of performing in vitro transcription reactions – single-cycle or steady-state – we chose the latter so that we can normalize the multiple-cycle nature of abortive initiation (whether in single-cycle or steady-state condition) to multiple rounds of productive transcription. The recycling synthesis of abortive products during single-cycle reactions leads to an overestimation of the abortive yield. The over-representation of abortive transcripts occurs even with the steady-state conditions, due to the existence of productive ITCs and unproductive ITCs, both of which give rise to abortive RNAs but only productive ITC synthesizes the long RNA [31, 32].
2.1.3 [γ-32P]- vs. [α-32P]-NTP labeling
Of the two means of labeling the transcripts through nucleotide incorporation, whether by single end-labeling using [γ-32P]-NTP or by “body-labeling” using [α-32P]-NTP, we opted for the former for the following reasons. With single labeling at the 5′-end of each transcript, the visual intensity of the transcript bands directly reflects their molar ratio. This feature greatly facilitates the subsequent quantitation and analysis. In contrast, α-32P labeling may present a number of problems depending on the template involved. One is the appearance of intrinsic or (contaminating) Gre-stimulated cleavage products; they form distinct bands and may interfere with RNA identification and band assignment. Two, alpha-labeling proportionately increases the signal incorporated into the longer RNAs. If a long RNA is a contaminant species of similar size to the run-off RNA, quantitation of the run-off RNA would be greatly exaggerated. With a contaminant RNA, because one does not know its composition, additional error would be incorporated when correcting its signal to obtain the molar quantity. Figure 4 presents a transcript profile obtained with γ-32P vs. α-32P labeling that illustrates the differences mentioned above. Finally, if the single promoter being studied undergoes initiation at more than one start site, [α-32P]-labeling would result in the synthesis of multiple sets of abortive products all superimposed within the same gel lane. This presents a highly complicated picture for quantitative analysis. (Of course, if the alternate site starts with the same nucleotide, the advantage of using [γ-32P]-labeling for resolving overlapping abortive ladders would be lost.)
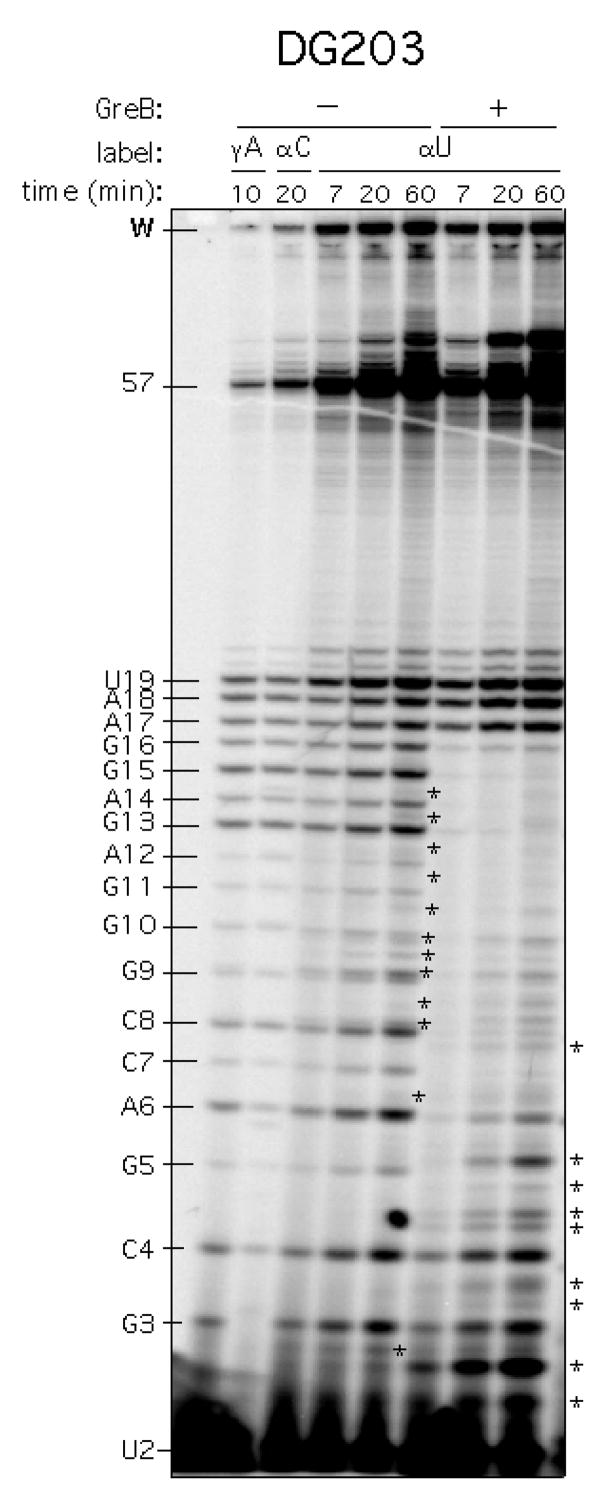
Figure 4.
Comparison of gamma- vs. alpha-32P labeling of DG203 promoter. The DG203 promoter was transcribed for 7, 20 or 60 min at 37 °C in 200 mM KCl, 1X transcription buffer (see text), and 100 μM NTP with [γ-32P]-ATP labeling (leftmost lane), [α-32P]-CTP labeling (second lane from left), or [α-32P]-UTP labeling (in all other lanes). The abortive ladder is indicated on the left border. The abortive RNAs are each designated by a letter-number combination where the number represents the length of the abortive RNA and the letter indicates the identity of the 3′-most nucleotide. Full-length RNA is 57-nt in length. W indicates the bottom edge of the sample wells. This gel shows two sharp differences between α-U and γ-A labeling. One is the large intensity difference associated with the full-length band, which impacts accurate quantitation. The other is the appearance of 3′-cleavage products indicated by asterisks. (Asterisks on the gel indicate intrinsic cleavage products. Those on the border indicate additional cleavage products derived from GreB stimulation.) Cleavage products are 5′-monophosphorylated RNAs. As such, they migrate slower on the gel than their 5′-triphosphorylated abortive RNAs of the same size (L. Hsu, unpublished results).
2.1.4 Optimizing reaction conditions
Two solution conditions – [KCl] and [NTP] --need to be optimized for each promoter. The [KCl] dependence of productive transcription exhibits a broad bell-shaped curve, with the optimum ranging from 150 to 250 mM KCl for the N25-random ITS promoter variants [Sara Barnes, senior thesis]. At the optimal [KCl], one obtains the highest ratio of productive versus abortive initiation. Our recent analysis showed that the [KCl] dependence is crucial for the recycling of RNA polymerase during the steady-state reactions [Barnes, senior thesis]. This observation may be related to the fact that RNA polymerase tends to arrest at the ends of fragment templates during in vitro transcription reactions [27]. A second reaction condition requiring optimization is [NTP] which controls the absolute level of initiation -- the higher the [NTP], the higher the initiation frequency. This dependence does reach saturation, and for the N25 promoter variants, saturation is reached at 100 μM NTP [8]. This parameter differs from the 20 μM we used in the original protocol [7].
2.1.5 Ethanol precipitation recovery using glycogen carrier
For quantitation purposes, it is ideal to measure all of the transcription products, from dinucleoside tetraphosphate (product of the first phosphodiester bond) to the full-length RNA. One can ensure the presence of all transcripts by directly loading the reaction mixture (after mixing with an equal volume of a loading dye). Due to the high level of [γ-32P]-NTP used, this strategy leaves a high radioactive background, especially around the di-, or tri-nucleotide region of the gel, and compromises the accuracy of quanitating the 2- and 3-nt RNA. Given that the 2-nt product usually comprises ~50% of total abortive RNAs [2, 8], a large error here impacts the subsequent calculation of the abortive yield, and as well, the abortive probability of each RNA [7]. To remove the radioactive background, we perform an ethanol precipitation step after the transcription reaction, using glycogen as the precipitation carrier, to remove much of the labeling NTP, and ~15% of the 2-nt product [7], but complete recovery of the 3-nt product and up is achieved. This trade-off produces a gel with “clean” background so that the subsequent quantitation is both easier and more accurate. RNase-free glycogen can be obtained either commercially or prepared by repeated phenol extraction [7].
2.1.6 Gel electrophoresis conditions
In the original protocol, reaction products were fractionated in 15–17% (19:1) polyacrylamide gel containing 7 M urea. Electrophoresis was carried out in electrolyte gradient buffer of 0.5X TBE in the top buffer reservoir and 0.67X TBE containing 1 M NaAc in the bottom buffer reservoir [44]. We have since switched to using a higher crosslinked gel (23% [10:1] polyacrylamide gel in 7 M urea) and a shallower salt gradient buffer (1X TBE in the top reservoir, and 1X TBE with 0.3 M NaAc in the buttome reservoir) to achieve the high resolution of abortive and productive RNAs. Here, the steeper the salt gradient, the more “packed” are the abortive RNAs, and the longer can be the productive RNA that enters the gel. Thus, one can vary the salt gradient buffer conditions to make sure all sizes of the desired reaction products become included in the gel. To assess whether gel electrophoresis has proceeded long enough (i.e. having run off the mononucleotides from the bottom of the gel), we have replaced the bromophenol blue (BPB) dye with amaranth, which migrates with a 2-nt product and reliably indicates when electrophoresis can be terminated (i.e. when the amaranth dye reaches within 1 cm from the bottom of the gel).
2.2 Steady-state transcription reaction protocol
2.2.1 In vitro transcription reaction
The analytical transcription reactions are routinely set up in 10 μL volume containing:
1 μL 300 nM promoter DNA
1 μL 10X transcription buffer (0.5 M Tris-HCl, pH 8, 0.1 M MgCl2, 0.1 M β-mercaptoethanol, 0.1 mg/mL acetylated BSA)
1 μL 2 M KCl
1 μL 1 mM NTP
0.5 μL [γ-32P]-ATP (Perkin Elmer NEG002Z; 6000 Ci/mmol supplied at 10μCi/μL on the reference date; final specific activity of ~10 cpm/fmol)
4.5 μL DEPC-treated H2O
9 μL; mix and quick spin
Add 1 μL of RNA polymerase (freshly diluted to 500 nM with diluent buffer [45]: 10 mM Tris-HCl, pH 8, 10 mM β-ME, 10 mM KCl, 5% v/v glycerol, 0.1 mM Na2EDTA, 0.4 mg/mL BSA, 0.1% v/v Triton X-100)
Quickly mix, and incubate at 37 °C for 10 minutes
Terminate reaction by adding 100 μL of a GES mix (1 mg/mL glycogen, 10 mM Na2EDTA, 0.3 M NaAc); carefully mix by pipetting with a yellow tip.
Add 330 μL ethanol; again, mix by pipetting with a yellow tip.
Leave at −20 °C freezer overnight.
Spin 15 minutes at 13,000 rpm (at 4 °C) to collect the precipitate.
Pipet off the ethanol supernatant completely and discard.
Dry the pellet in a Speed Vac rotary desiccator for 15 minutes.
Redissolve the pellet in 10 μL of a formamide loading buffer (FLB: 80% deionized formamide, 1X TBE, 10 mM Na2EDTA, 0.08% xylene cyanol, 0.08% amaranth; made fresh weekly)
Vortex the sample vigorously and spin; repeat three times.
Load 4 μL per sample and electrophorese at 35 watts until the amaranth dye reaches within 1 cm from the bottom of the gel. (For short transcripts that have little secondary structures, it is not necessary to pre-heat the samples to 90 °C and plunge on ice before loading.)
2.2.1.1 Technical comment on reaction set up
In setting up a reaction, we pipet in the various ingredients in the following order, first the water, then transcription buffer, salt, DNA, and the nucleotide mixes. After mixing and a brief spin, RNA polymerase is added to start the reaction. Realistically, however, a transcription experiment usually involves many samples. To reduce pipetting error and increase between-sample consistency during the reaction set up, we make up master mixes containing several common ingredients so that they can be added in one volume. As an example, to set up a reaction as above for comparing the transcription from 8 different promoter templates, we would set up 9 reactions -- one for each of the 8 templates, plus a 9th one that is a minus-enzyme control for background radioactivity in the sample. To set up these reactions, we would make up two master mixes: Mix 1 contains the water, transcription buffer and salt, and Mix 2 contains the labeled and unlabeled nucleotides (see comments in section 2.2.1.2 below). Each master mix would be 10X in volume to allow for pipetting errors incurred in sampling the 9 aliquots. With the master mixes at hand, setting up the 9 reaction tubes simply involves pipetting 6.5 μL each of Mix 1 into all tubes, add 1 μL of (different) promoter DNA into each tube, followed by pipetting 1.5 μL each of Mix 2 into all tubes. Mix and spin the samples, and one is ready to add enzyme to start the reaction. In a multi-sample experiment, the enzyme is added in a 30-second staggered fashion. Thus, for a reaction that is to receive a 10-minute incubation, one can carry out an experiment containing 20 samples. At the end of the 10-minute incubation, the samples are terminated in a staggered fashion by the addition of a stop mix.
2.2.1.2 Technical comment on the nucleotide mixture
The nucleotide mixture can be made up in various ways to achieve the intended goals of your reaction. For obtaining quantitative parameters on A-starting promoters, we generate a nucleotide mixture by mixing a 10X NTP solution with enough [γ-32P]-ATP stock to obtain a calculated final radioactive specificity of ~5–10 cpm/fmol for ATP. Because the quantitative parameters we desire are all fractional numbers (see comments in section 2.2.3 below), it is not necessary to determine the accurate cpm/fmol for this analysis.
However, if one wishes to determine the exact (femto)moles of a transcript made during a reaction, the nucleotide mix is made up differently by mixing a 10X G/C/UTP stock and a separately prepared 10X stock of ATP/[γ-32P]-ATP at the desired specificity. With the 32P-ATP stock, one can determine the actual cpm/fmol value on the day of gel exposure from two numbers: the cpm/μL number (from scintillation counting) and the fmol/μL (i.e. nM) number (from A260 absorbance). Dividing cpm/μL by fmol/μL yields the radioactive specificity as cpm/fmol. From the nM number, one can adjust the volume of 32P-ATP stock to be taken for the reaction.
For quantitating radioactive counts embedded in a gel, it is necessary to spot on standards containing known amount of counts (in cpm) on Whatman 3 MM filter paper (3 mm × 10 mm) [we use several amounts for obtaining an average later], tape them on the Saran Wrap covering the gel (in an area of the gel that does not contain transcript bands) for exposure to the phosphorimager along with the gel. The phosphorimager counts (measured as ImageQuant Volume units, or IQV) divided by its associated cpm yields the conversion factor IQV/cpm, which along with cpm/fmol, allows the conversion of IQV counts of each transcript band into femtomole quantities.
2.2.1.3 Technical comment on different ways of initiating steady-state transcription reactions
By adding RNA polymerase to start the reaction, the incubation time includes time for polymerase binding and open complex formation before initiation and elongation. For promoters that form open complex quickly, this process does not detract from the reaction time significantly. However, for promoters that form open complex slowly, one might need to lengthen the reaction time appropriately to capture many rounds of transcription. There are alternate ways of initiating transcription reactions. One approach involves pre-forming the open complexes in the absence of NTP, and adding the NTP mix to start the reaction; this is usually how single-cycle kinetic reactions are carried out [46]. Another approach involves pre-forming the open complexes in the absence of Mg++, and adding Mg++ to start the reaction [47]. These strategies eliminate the open complex formation time for the first cycle only, and do not alter the requirements for steady-state transcription in subsequent cycles.
2.2.2 Denaturing PAGE
To pour a 23% (10:1) 7 M urea polyacrylamide gel, we prepare an 80-mL gel mixture for pouring a sequencing-sized gel (37 cm × 43 cm × 0.4 mm). The gel recipe is as follows:
33.6 g ultrapure urea
8 mL 10X TBE (0.89 M Tris Base, 0.89 M Boric Acid, 25 mM Na2EDTA)
46 mL 40% (10:1) acrylamide-bisacrylamide stock1
Dissolve to yield 80 mL
Polymerize with 400 μL 10% APS (ammonium persulfate) and 40 μL TEMED (N,N,N′,N′-tetramethylethylenediamine)
Pour into a sequencing gel mold2, set the comb, and allow polymerization for ~1 hour at room temperature. (After inserting the comb, apply three large clamps across the comb. This prevents the formation of a thin gel layer above and below each tooth which interferes with sample loading.)
Electrophoresis was carried out in a salt gradient buffer (top reservoir buffer = 1X TBE; bottom reservoir buffer = 0.3 M NaAc in 1X TBE) that allows the short RNA bands to collect at the bottom (before they run off the gel) and the long RNA (60–100 nt in length) to enter the gel. The saltier the bottom buffer, the more closely packed would be the abortive RNAs, and the longer the run-off RNA can be.
Electrophoresis is usually carried out at 35 watts for ~5 hours until the amaranth dye (which migrates with the 2-nt RNA) is within 1 cm from the bottom edge of the gel. After the electrophoresis, pry apart the gel plates. Dab dry the gel with Kimwipe, before covering with Saran Wrap for exposure to a phosphorimager screen3. (Without dab drying, the wet spots – especially around the wells – get smeared throughout the gel when the Saran Wrap is applied. This allows radioactivity to seep out of the thin gel, resulting in dark smears on the phosphorimage.)
2.2.3 Quantitation using ImageQuant
To obtain the quantitative initiation parameters for a promoter, we quantitate all of the abortive and productive RNA bands, from 2-nt RNA to the full length, within each lane. The ImageQuant Volume (IQV) counts obtained are then corrected by subtracting the IQV values associated with a blank rectangle from an equivalent position in the no-enzyme control lane run on the same gel (see Figure 5). The background subtraction is especially important for the 2- and 3-nt spots. From the corrected IQV values, we can obtain the sum of abortive IQVs (i.e., abortive initiation), the sum of productive IQVs (i.e., productive initiation), and the total IQVs (i.e., total initiation). From these three values, we can derive all of the quantitative initiation parameters that include the abortive yield (abortive initiation as a percentage of total initiation), the productive yield (productive initiation as a percentage of total initiation), and the abortive-productive ratio (APR; the ratio of abortive yield over productive yield). For each abortive transcript, the abortive yield value is further divided by the fraction of RNA polymerase that reaches each position [see formula in Ref. 7] to yield a parameter called abortive probability. Abortive probability for a given ITS position reflects the tendency that an ITC at that template position might release its nascent RNA. A qualitative parameter derived from the gel profile is MSAT (maximum size of abortive transcript). MSAT marks the juncture where the initiation-elongation transition occurs to lead to the abrupt cessation of repetitive abortive cycling. As shown in Figure 5, this transition can occur at different positions on different promoters. (For the purpose of this discussion, we consider the MSAT to be 11-nt for N25, 15-nt for N25anti, and 19-nt for DG203.) However, the assignment of MSAT is somewhat arbitrary, due to the trailing presence of several faint bands after the escape transition point (see Figure 5). These bands result from the fact that the escape transition is accomplished over a 4–5 nucleotide span (L. Hsu, unpublished results). Their intensity varies from experiment to experiment (compare the abortive ladders for N25, N25anti, and DG203 in Figures 1, 2, 4, 5, and 6) depending on whether one is using fresh or aged radioactive stock, [α-32P]- or [γ-32P]-NTP label, and the length of reaction time. We assign the most consistently observed longest abortive transcript from a promoter as its MSAT. The assignment of the 11-nt RNA as MSAT for N25 was confirmed in the DNA scrunching investigation [36].
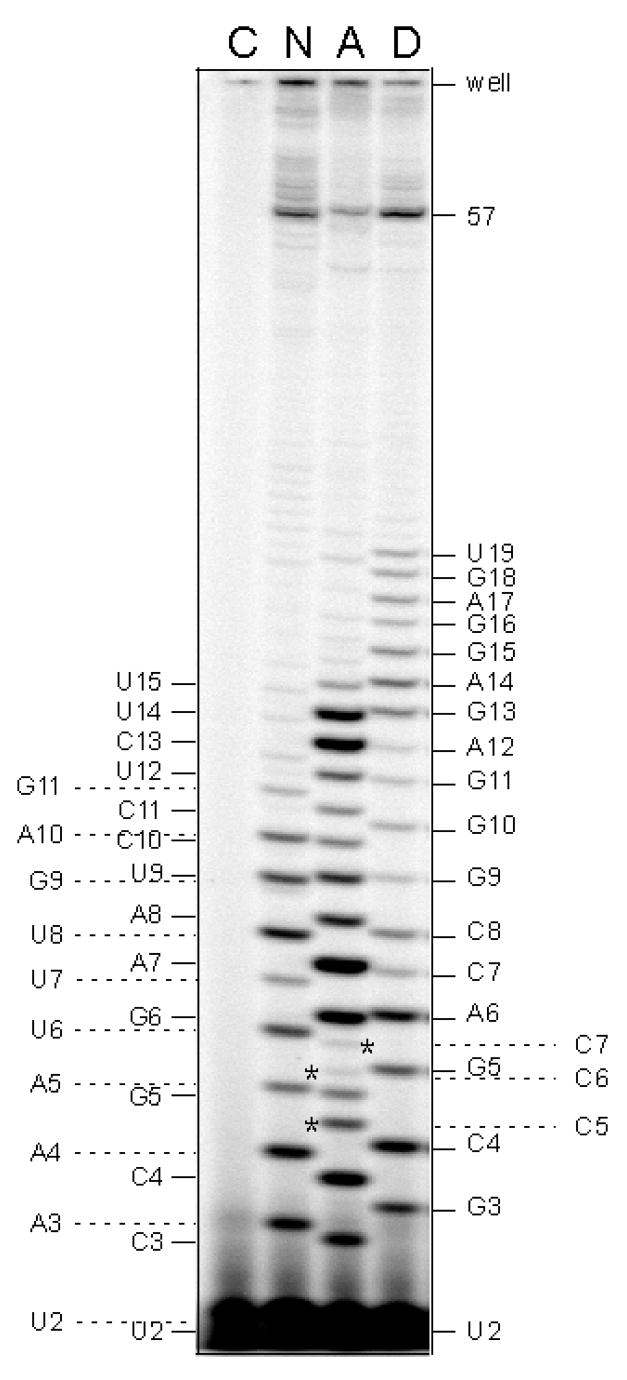
Figure 5.
A gel showing the importance of background subtraction during phosphorimager quantitation. Three promoters (designated N, A, and D) were transcribed under the steady-state reaction conditions with [γ-32P]-ATP labeling. N: N25; A: N25anti; D: DG203. The abortive ladders for each promoter are indicated on the border: N, the far left column; A: the nearby left column; D: the nearby right column. W: the bottom edge of the sample wells. 57: the length of run-off RNA. For quantitation purposes, a no-enzyme control reaction (designated as C) was included. (The control reaction contained N25 promoter, was incubated with enzyme-dilution buffer, and treated through all of the subsequent recovery steps.) One can see the substantial background contribution to the 2- and 3-nt spots. In lane A, N25anti transcription yields slippage products C5, C6, and C7, marked by asterisks. The identity of C5 (AUCCC) was established by nearest-neighbor analysis [8]. The identity of C6 (AUCCCC) and C7 (AUCCCCC) was inferred from steady-state transcription reactions carried out in the presence of limited NTPs, either the combination of [γ-32P]-ATP, UTP, and CTP, or the combination of ATP, UTP, and [α-32P]-CTP (results not shown).
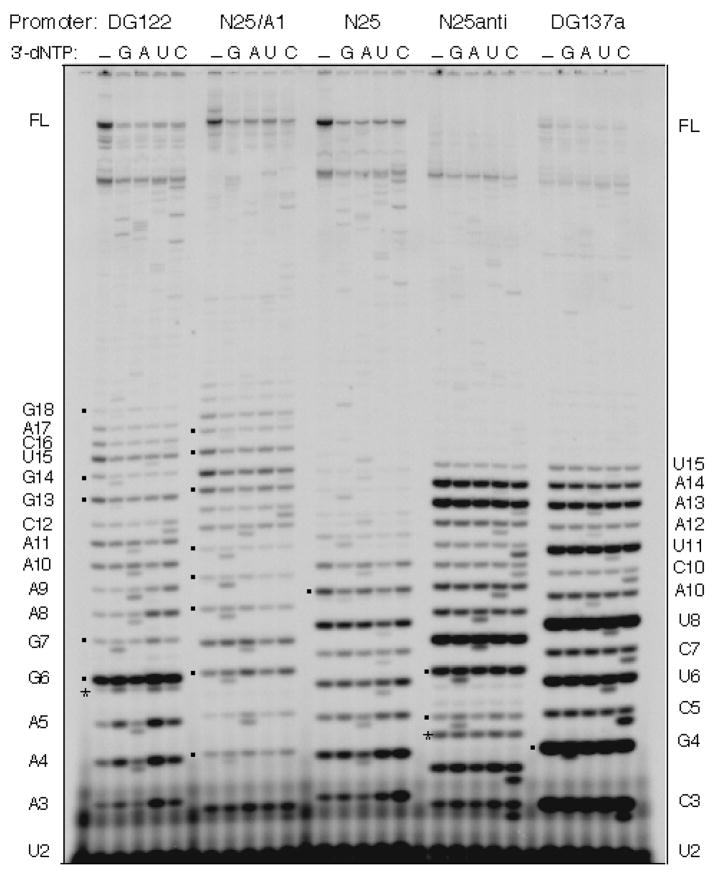
Figure 6.
Sequencing the abortive RNAs by chain termination. The figure shown is taken from ref. 23. The abortive RNA ladders from five promoters are displayed on a denaturing high percentage polyacrylamide gel. Each promoter is reacted under steady-state transcription conditions (containing 100 μM NTP) for 10 min at 37 °C under five conditions -- in the absence or presence of 100 μM of the specified 3′-deoxy-NTP (indicated by G, A, U, or C). In the lanes containing a 3′-dNTP, the 3′-dNMP-terminated abortive RNAs migrate just ahead of their regular abortive RNA counterparts, forming doublet bands. Square dots indicate the position of G-terminating bands, which always migrate to yield a larger gap from its precursor RNA than A, U-, or C-terminating bands. The abortive RNA sequence obtained from these five promoters agree with the DNA sequence of their ITS. Under the nucleotide concentrations used, slippage transcripts – for example, C5 in N25anti and A6 of DG122 (indicated by asterisks) -- do not undergo chain termination by incorporating the corresponding 3′dNMP. Abortive RNA ladder on the left border is that of DG122; on the right, that of DG137a.
The quantitative initiation parameters for each promoter are reasonably reproducible values determined under a specific set of reaction conditions [see Table 1 in Ref. 23]. Several factors in conducting the transcription reactions and phosphorimager quantitation are important in ensuring the level of reproducibility. First, the aforementioned use of master mixes in making up the reaction volume reduces the variability between samples on a gel. Second, the two primary parameters, abortive yield and productive yield, are internally normalized to the total initiation level, making their determination independent of the variable counts applied to each lane. Finally, a large variability is associated with phosphorimager quantitation in the size of an object (i.e., a rectangle or an oval) one draws around a band. To minimize this variability, we have resorted to drawing just a few objects that can be copied to capture all of the bands on a gel. (We routinely draw an oval for the 2-nt RNA, a rectangle around the most intense abortive RNA band for quantitating all other abortive RNAs, and another rectangle for the productive RNA bands.) Adopting regularized practices greatly reduces the variability between samples and experiments.
2.3 Verifying the sequence of an abortive RNA ladder
Although abortive initiation occurs in a template-specified manner, the identity of abortive RNAs bands obtained from a promoter, particularly from one that has not been previously characterized for its abortive pattern, may not be easily inferred from the template DNA sequence if there are slippage or misincorporation products mixed in [23]. Earlier strategies to establish the identities of abortive RNAs involved nearest-neighbor analysis [8] or the selective use of NTPs, dinucleotide primer as initiators, and the alpha-labeling nucleotide to synthesize specific abortive bands. These strategies are tedious and time –consuming, and soon lose their ability to resolve longer RNAs.
Faced with the challenge of identifying hundreds of abortive RNA bands [see Ref. 23], we modified the steady-state transcription reaction (described above in section 2.2.1) for sequencing the abortive RNAs using RNA chain terminators. By supplementing a 3′-deoxy NTP, say 3′-deoxy-ATP, to the reaction mixture containing 100 μM of all four regular NTPs, an abortive RNA that terminates in A now would be represented by two bands – the normal A-terminating band and the 3′-dA-terminating band. These two products migrate as a doublet with the 3′-deoxy-A-terminated band migrating faster in the gel due to its lower molecular weight. Thus, to sequence the abortive RNA ladder from one promoter, five reactions are set up – one without 3′-dNTP serves as the control, and the others are individually supplemented with one of the four 3′-dNTPs (Trilink Biotechnology, Inc.) to 100 μM. A typical sequencing gel result is shown in Figure 6.
This sequencing method, however, does not resolve the slippage or misincorporation bands [23]. In N25anti, we found a set of slippage RNA (designated C5, C6, and C7 in Figure 5) stemming from a CC dinucleotide in the initial transcribed region and mixed in with the normal abortive ladder [8]. The identity of C5 was established with nearest-neighbor analysis; that of C6 and C7, by transcription in limited NTPs [results not shown].
We also rely on the distinct gel mobility of G-terminating RNA as a confirmation of the sequence assignment [23]. In the gel system described above (section 2.2.2), G-terminating RNAs always mark a bigger gap during gel electrophoresis from its precursor one nucleotide shorter (see Figure 6). In this context, C-terminating RNAs mark the smallest gap, and A- or U-terminating RNAs show migration mobility somewhere in-between.
3. Concluding Remarks
The analysis of abortive RNAs has come a long way, aided by the high resolution gel protocol coupled with phosphorimaging technology that allow us to track changes both quantitatively and qualitatively. This methodology gave rise to a large body of information linking the extent and pattern of abortive initiation to its control by the promoter sequence. The biochemical knowledge gained, joined by biophysical evidence and structural understanding of RNA polymerase, has brought within reach a plausible mechanism of abortive initiation and promoter escape. However, there is much yet to be learned about this process. For example, there is evidence that the efficiency of escape on N25 promoter is modulated by the initial transcribed sequence (ITS) [23]. Two targets of modulation can be postulated based on the branched pathway transcription initiation diagram (see Figure 3). One is kE and the other, the partitioning of RNA polymerase at the promoter to form productive versus unproductive complexes. To solve whether either factor is involved in determining the extent of productive transcription will require extensive use of the quantitative transcription methods.
Acknowledgments
I thank Dr. Michael Chamberlin for suggesting that I work on the T5 N25 and N25anti promoters during a sabbatical leave in his lab in 1992–1993 supported by NSF grant (HRD-9252956) funded through the Visiting Professorships for Women Program. Our work cited in these pages was supported by NIH grant (R15 GM55907) and NSF grant (RUI-0077941). The work published in Ref. 23 was completed during a two-year sabbatical stay in Dr. Caroline Kane’s lab. I am extremely grateful for her hospitality, generosity, intellectual input, and scientific advice.
Footnotes
The 40% (10:1) acrylamide-bisacrylamide stock is prepared as follows. Place a 4-L plastic beaker containing ~1.5 L MilliQ water on a magnetic stirrer, first dissolve100 g of N,N′-methylene-bis-acrylamide (USB #75821). Then, mix in 1 kg of acrylamide (USB #75820) and add water to ~2.5 L. Let stir overnight and bring the solution to 2.75 L. After additional mixing, filter the solution through Whatman No. 1 filter (90 mm diameter) on a porcelain Buchner funnel (with fixed perforated plate; Fisher #10–356D). Change the filter every 500 mL. Store the stock in 1-L Corning bottles.
We treat the sequencing gel plates every time, on both inside surfaces, with a thin smear of Rain-X (obtained from the automotive department in a retail store) for 10 minutes at room temperature. Afterwards, the plates are wiped clean with water and ethanol before being assembled to form the gel mold. This treatment regime ensures smooth pouring of the gel mixture, and as well, ready separation of the gel with one of the plates after electrophoresis.
The exposure cassette used with the older models of Molecular Dynamics phosphorimager can accommodate a gel left on a thick glass plate, so direct exposure of the gel is possible. With the newer models, the glass plate is too thick for the gel cassette. For exposure, one might need to first transfer the gel to a thin support (such as a sheet of Whatman 3MM paper, or an old piece of X-ray film). However, this transfer does not work well with the high % polyacrylamide gels we use. Alternatively, one can simply set up the exposure in a dark drawer, making sure that the phosphorimager screen is pressed tightly and securely to the gel.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnston DE, McClure WR. In: RNA Polymerase, Cold Spring Harbor Laboratory. Losick R, Chamberlin M, editors. Cold Spring Harbor; N. Y: 1976. pp. 413–428. [Google Scholar]
- 2.Carpousis AJ, Gralla JD. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 3.Hansen UM, McClure WR. J Biol Chem. 1980;255:9564–9570. [PubMed] [Google Scholar]
- 4.Gralla JD, Carpousis AJ, Stefano JE. Biochemistry. 1980;19:5864–5869. doi: 10.1021/bi00566a031. [DOI] [PubMed] [Google Scholar]
- 5.Stefano JE, Gralla JD. J Biol Chem. 1980;255:10423–10430. [PubMed] [Google Scholar]
- 6.Carpousis AJ, Stefano JE, Gralla JD. J Mol Biol. 1982;157:619–633. doi: 10.1016/0022-2836(82)90502-2. [DOI] [PubMed] [Google Scholar]
- 7.Hsu LM. In: Methods in Enzymology. Adhya S, editor. Vol. 273. Academic Press; New York: 2002. pp. 59–71. [Google Scholar]
- 8.Hsu LM, Vo NV, Kane CM, Chamberlin MJ. Biochemistry. 2003;42:3777–3786. doi: 10.1021/bi026954e. [DOI] [PubMed] [Google Scholar]
- 9.Hsu LM. Biochim Biophys Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 10.Hsu LM. In: EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, editors. ASM Press; Washington, D.C: http://www.ecosal.org. 12 February 2008, posting date. [Google Scholar]
- 11.McClure WR, Cech CL, Johnston DE. J Biol Chem. 1978;253:8941–8948. [PubMed] [Google Scholar]
- 12.McClure WR. Proc Natl Acad Sci USA. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClure WR. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 14.Mulligan ME, Hawley DK, Entriken R, McClure WR. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunner M, Bujard H. EMBO J. 1987;6:3139–3144. doi: 10.1002/j.1460-2075.1987.tb02624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knaus R, Bujard H. In: Nucleic Acids and Molecular Biology. Eckstein F, Lilley DM, editors. Vol. 4. Springer-Verlag; Heidelberg, Germany: 1990. pp. 110–122. [Google Scholar]
- 17.Carpousis AJ, Gralla JD. J Mol Biol. 1985;183:165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- 18.Munson LM, Reznikoff WS. Biochemistry. 1981;20:2081–2085. doi: 10.1021/bi00511a003. [DOI] [PubMed] [Google Scholar]
- 19.Deuschle U, Kammerer W, Gentz R, Bujard H. EMBO J. 1986;5:2987–2994. doi: 10.1002/j.1460-2075.1986.tb04596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kammerer W, Deuschle U, Gentz R, Bujard H. EMBO J. 1986;5:2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. Biochemistry. 2003;42:3798–3811. doi: 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
- 22.Kulbachinskiy A, Mustaev A. J Biol Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- 23.Hsu LM, Cobb IM, Ozmore JR, Khoo M, Nahm G, Xia L, Bao Y, Ahn C. Biochemistry. 2006;45:8841–8854. doi: 10.1021/bi060247u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng G, Lee DN, Wang D, Chan CL, Landick R. J Biol Chem. 1994;269:22282–22294. [PubMed] [Google Scholar]
- 25.Hsu LM, Vo NV, Chamberlin MJ. Proc Natl Acad Sci USA. 1995;92:11588–11592. doi: 10.1073/pnas.92.25.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Proc Natl Acad Sci USA. 1005;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stepanova E, Lee J, Ozerova M, Semenova E, Datsenko K, Wanner BL, Severinov K, Borukhov S. J Bacteriol. 2007;189:8772–8785. doi: 10.1128/JB.00911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatoum A, Roberts J. Mol Microbiol. 2008;68:17–28. doi: 10.1111/j.1365-2958.2008.06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker MM, Gaal T, Josaitis CA, Gourse RL. J Mol Biol. 2001;305:675–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 30.Gourse RL. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubori T, Shimamoto N. J Mol Biol. 1996;256:449–457. doi: 10.1006/jmbi.1996.0100. [DOI] [PubMed] [Google Scholar]
- 32.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. Biochemistry. 2003;42:3787–3797. doi: 10.1021/bi0269613. [DOI] [PubMed] [Google Scholar]
- 33.Menedez M, Kolb A, Buc H. EMBO J. 1987;6:4227–4234. doi: 10.1002/j.1460-2075.1987.tb02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monsalve M, Calles B, Mencia M, Salas M, Rojo F. Mol Cell. 1997;1:99–107. doi: 10.1016/s1097-2765(00)80011-8. [DOI] [PubMed] [Google Scholar]
- 35.Smith TL, Sauer RT. Proc Natl Acad Sci USA. 1996;93:8868–8872. doi: 10.1073/pnas.93.17.8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Revyakin A, Liu C, Ebright RH, Strick TR. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turnbough CL, Jr, Switzer RL. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng Q, Turnbough CL, Jr, Switzer RL. Proc Natl Acad Sci USA. 2004;101:10943–10948. doi: 10.1073/pnas.0403755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choy HE, Adhya S. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellinger T, Behnke D, Bujard H, Gralla JD. J Mol Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- 41.Reppas N, Wade JT, Church GM, Struhl K. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 42.Ross W, Gourse RL. Proc Natl Acad Sci USA. 2005;102:291–296. doi: 10.1073/pnas.0405814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis CA, Bingman CA, Landick R, Record MT, Jr, Saecker RM. Proc Natl Acad Sci USA. 2007;104:7833–7838. doi: 10.1073/pnas.0609888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheen JY, Seed B. Biotechniques. 1988;6:942–944. [PubMed] [Google Scholar]
- 45.Gonzalez N, Wiggs J, Chamberlin MJ. Arch Biochem Biophys. 1977;182:404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- 46.Rutherford ST, Lemke J, Vrentas CE, Gaal T, Ross W, Gourse RL. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park JS, Roberts JW. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]