Abstract
The etiology of sarcoidosis remains uncertain. The hallmark of sarcoidosis is the epithelioid granuloma, which serves as a necessary starting point for considering disease etiology. Any etiologic agent of sarcoidosis must also explain the typical clinical behaviors and characteristic immunopathologic features of the disease. One clinical observation that serves as a bridge to the etiology of sarcoidosis is the Kveim reaction. In this reaction, local epithelioid granulomas develop several weeks after the intradermal injection of homogenates of sarcoidosis tissue. Our group capitalized on the known properties of the Kveim reagent to search for candidate pathogenic tissue antigens in sarcoidosis without other a priori hypotheses regarding possible microbial or autoimmune etiologies. Using a limited proteomics approach based on the physicochemical properties of Kveim reagent, we detected a limited number of poorly soluble antigenic proteins in sarcoidosis tissues by protein immunoblotting, using sarcoidosis sera. Matrix-associated laser desorption/ionization-time of flight mass spectrometry identified one of these antigens to be the Mycobacterium tuberculosis catalase–peroxidase protein (mKatG). We found IgG responses to recombinant mKatG in more than 50% of patients with sarcoidosis but rarely in purified protein derivative (PPD)-negative control subjects. These findings support the conclusion that mKatG is a tissue antigen and target of the adaptive immune response in sarcoidosis, providing further evidence of a mycobacterial etiology in a subset of sarcoidosis. More generally, the approach used in these studies might be employed to discover and validate other candidate pathogenic antigens in sarcoidosis or other granulomatous disorders.
Keywords: sarcoidosis, etiology, granuloma, proteomics, mycobacteria
Since sarcoidosis was first described in about 1900, the cause of the disease has been the subject of much speculation and study. Space–time clusters and familial aggregation lend support to an environmental trigger or triggers for sarcoidosis but have not defined a specific cause. Microbial etiologies of sarcoidosis have always been considered on the basis of clinical similarities to infectious granulomatous diseases (1). Mycobacterial and propionibacterial organisms have been most commonly implicated as potential etiologic agents by studies using polymerase chain reaction–based approaches that report the detection of microbial DNA from these organisms in tissues from patients with sarcoidosis from around the world (2, 3). However, results from different studies have varied considerably, with reports of microbial DNA in 0–80% of sarcoidosis tissues as well as 0 to more than 30% of control tissues (4, 5). The completed ACCESS (A Case Control Etiologic Study of Sarcoidosis) study provides data that support possible disease associations with select microbially rich environments (6). The overall disease risk from these associations was modest (odds ratios, about 1.5), and false positive associations from multiple comparisons could not be excluded. Taken together, these studies have failed to provide a consensus on the role specific microbial agents play as a cause of sarcoidosis. Given this uncertainty, a role for autoimmunity remains a postulated mechanism of sarcoidosis pathogenesis, although this mechanism could also be linked to inciting microbial triggers. However, to date, no autoantigens linked specifically to sarcoidosis have been discovered. Evidence that genetic factors, particularly genes from the major histocompatibility locus, play a role in susceptibility to sarcoidosis is consistent with either a microbial or autoimmune causation of sarcoidosis, but provides no further insight into specific etiologies (7, 8).
FUNDAMENTAL FEATURES OF SARCOIDOSIS
With an uncertain etiology, a review of the fundamental features of sarcoidosis may be a useful starting point for further studies. For sarcoidosis, this begins and ends with the granuloma. The formation of granulomas, an ancient and preserved pathologic response to (usually) foreign material, occurs around a nidus of poorly soluble or insoluble material that cannot be simply removed by a single cell. It is an organized structure involving multiple cells that develops in an orchestrated manner over days or weeks. The immunogenic amplification that occurs from antigenic stimulation at sites of granuloma formation can involve either polarized helper T-cell type 1 (Th1) or helper T-cell type 2 (Th2) immunologic responses (9). In sarcoidosis, the granulomatous inflammation interferes with local tissue homeostasis, resulting in organ impairment that is dependent on its location. Given that the epithelioid granuloma is the pathologic hallmark of sarcoidosis, any etiologic agent must be capable of inducing this pattern of inflammation.
Any etiologic agent of sarcoidosis must also explain the characteristic clinical behavior of the disease (10). Fundamental clinical features found in most patients include the following: a monophasic, progressive clinical course of the underlying granulomatous inflammation unless there is remission (exceptions include ocular or neurologic involvement or erythema nodosum); either a remitting or chronic clinical course; rare recurrence after remission; multiple organ involvement; variable rate of progression of granulomatous inflammation in different individuals and in different organs; suppression of granulomatous inflammation by antiinflammatory therapies that are not curative; and rare opportunistic infections implying no systemic impairment of host defense. Although there are some patients with sarcoidosis with clinical courses that fall outside these typical parameters, it remains necessary that any etiologic agent of sarcoidosis be capable of inducing a pathobiological process compatible with these basic clinical features.
In addition, any etiologic agent(s) of sarcoidosis must be capable of inducing characteristic immunopathologic features of the disease (11). These features include the following: dominant CD4+ T cells at sites of granulomatous inflammation; oligoclonal expansions of T cells expressing specific αβ+ or γδ+ T-cell receptor genes consistent with an antigen-driven response; polarized Th1 cytokine expression at the time of diagnosis (uncertain cytokine polarization in progressive, fibrotic sarcoidosis); “classically activated” macrophage phenotype at the time of diagnosis; and heightened expression of tumor necrosis factor and other cytokines and chemokines known to be involved in experimental granulomatous inflammation. These immunopathologic features of sarcoidosis provide another critical checklist for testing the compatibility of candidate etiologic agents or antigens.
THE KVEIM REACTION
Within this clinical and immunopathogenic framework, there is a clinical observation that may serve as a bridge to the etiology of sarcoidosis—the Kveim or Kveim–Siltzbach reaction. The Kveim reaction was initially observed as a local, nodular eruption several weeks after the intradermal injection of homogenates of syngeneic or allogeneic sarcoidosis lymph node or spleen tissue. Ansgar Kveim in the 1940s was the first to report that biopsy of these nodules demonstrated the presence of epithelioid granulomas that were histologically identical to granulomas seen in other sarcoidosis tissues (12). In contrast to a delayed-type hypersensitivity reaction, the development of epithelioid granulomas in this reaction occurs after many days, with well-formed granulomas seen at 2–4 weeks or more. Louis Siltzbach and others demonstrated that about 80% of patients with sarcoidosis worldwide have a positive reaction (granulomas) to validated Kveim reagent, with fewer than 1% false positive reactions in control subjects (13). More recent studies have demonstrated that this Kveim reaction is characterized by an influx of CD4+ T cells and histiocytes, with oligoclonal Vβ-specific T-cell expansions consistent with an antigen-specific immune response (14). Importantly, the physicochemical properties of the granuloma-inducing component of the Kveim reagent have been defined by in vivo studies of patients with sarcoidosis. These properties include the following: relative resistance to neutral detergents, heat, acidity, organic solvents, nucleases, and proteases (15, 16). The fact that Kveim reactivity is abrogated by potent denaturants suggests a protein component is responsible for the granuloma-inducing activity (17). These physicochemical properties presumably reflect the same characteristics of the factors that cause granulomas in patients with systemic sarcoidosis.
SEARCH FOR KVEIM-LIKE ANTIGENS
An approach for identifying pathobiologically relevant antigens in sarcoidosis can be constructed by capitalizing on the physicochemical properties of the granuloma-inducing component of the Kveim reagent, and by inference, native sarcoidosis granulomas. In this context, our group employed a limited proteomic approach to restrict our search for sarcoidosis tissue antigens to those antigens with biochemical properties similar to those of the Kveim reagent (18). We hypothesized that sarcoidosis is caused by linked T- and B-cell responses to poorly soluble protein aggregates of microbial and/or endogenous origins with physicochemical properties similar to those of the Kveim reagent. This hypothesis and approach had the advantage of avoiding possible prejudicial a priori hypotheses regarding specific microbial or environmental agents or autoantigens.
An outline of this approach is as follows:
Step 1: Limit the proteome set of tissue proteins on the basis of the physicochemical properties of the Kveim reagent.
Step 2: Assess for the presence of tissue antigens by protein immunoblotting, using sarcoidosis and control sera.
Step 3: Identify candidate antigens by matrix-associated laser desorption/ionization-time of flight mass spectrometry.
Step 4: Confirm the presence of candidate antigens in tissues by protein immunoblotting, using specific antibody reagents to the candidate protein.
Step 5: Evaluate B- and T-cell responses to recombinant candidate proteins and derived peptides identified by the initial studies.
To concentrate tissue antigens with properties similar to those of the Kveim reagent, we used two different approaches. First, an extraction procedure was developed to concentrate protease-resistant tissue proteins that were insoluble in neutral detergent. Second, hypothesizing that sarcoidosis antigens would form poorly soluble aggregates, we adapted methods used to concentrate prion or amyloid proteins (19). Both of these methods resulted in a loss of more than 90–99% of the starting frozen tissue. After extractions were made from sarcoidosis and control tissues, samples were run on protein immunoblots and sarcoidosis or control sera were used to detect potential tissue antigens. With both methods, we found discrete antigenic bands on protein immunoblots detected with circulating IgG or Fab fragments from patients with sarcoidosis but not control sera (18). These results indicated that poorly soluble antigens were present in sarcoidosis tissues consistent with possible pathogenic sarcoidosis antigens.
Mycobacterium tuberculosis CATALASE–PEROXIDASE AS A CANDIDATE PATHOGENIC ANTIGEN
To identify these unknown antigens, we employed matrix-associated laser desorption/ionization-time of flight mass spectrometry and peptide fingerprinting methods (18). Gel slices containing the antigenic bands were cut out, digested with trypsin, and subjected to mass spectrometry. The mass spectra was analyzed by peptide fingerprinting against established protein databases, using Web-accessible peptide fingerprinting algorithms. Our first attempts led to a highly statistically significant match of the peptide mass spectra to Mycobacterium tuberculosis catalase–peroxidase (mKatG) (18). Analysis of a sarcoidosis lung sample led to a potential match (nonstatistically significant) with Mycobacterium smegmatis catalase–peroxidase. To confirm the presence of mKatG protein in sarcoidosis tissues, we used protein immunoblotting and a specific anti-mKatG monoclonal antibody (IT42, provided by Colorado State University [Fort Collins, CO] under NIH contract AI-75320). With this approach we found evidence of mKatG protein in 55% of sarcoidosis tissues but in no control tissues. We also confirmed the presence of mycobacterial katG DNA in sarcoidosis tissues, using in situ hybridization with tyramide signal amplification (18). Because a role for this specific mycobacterial protein had not been considered previously in any prior published reports on sarcoidosis, these findings appeared to validate the use of this approach for the discovery of novel candidate pathogenic antigens in sarcoidosis.
Mycobacterial KatG is perhaps best known to clinicians because approximately 80% of isoniazid resistance results from deleted, inactive, or mutant katG (20). Isoniazid is a prodrug that must be converted to form an active mycobactericidal compound, an action accomplished by functional KatG. Mycobacterial KatG is a broad-spectrum heme-containing bifunctional enzyme with both catalase and peroxidase activity. This protein is a virulence factor allowing prolonged survival inside macrophages. In many mycobacterial species, KatG is a homodimer with subunits of about 80 kD. Immune responses to mKatG are found in patients with tuberculosis, although the role of this protein in host defense against this organism has remained undefined.
The finding that mKatG is a tissue antigen and target of the adaptive immune response in sarcoidosis has both specific and general implications for possible sarcoidosis etiologies. First, the presence of mycobacterial KatG and katG DNA in sarcoidosis tissues is further evidence of a mycobacterial etiology in at least a subset of patients with sarcoidosis. More generally, our findings substantiate that the sarcoidosis granuloma contains pathobiologically relevant material that can recapitulate the etiology of sarcoidosis. These findings do not exclude the possibility that other microbial organisms such as Propionibacterium acnes may be etiologic agents in other subgroups of patients with sarcoidosis (4). In this context, the presence of mycobacterial or other microbial DNA in sarcoidosis tissues is only one form of etiologic marker. To validate a potential etiologic role for any specific microbial agent, it remains necessary to identify those remnant microbial antigens in sarcoidosis tissues that are responsible for inducing T- and B-cell responses that take part in the local granulomatous inflammation.
The identification of mKatG as a candidate pathogenic antigen in sarcoidosis raises several obvious questions. Are mKatG responses pathogenic or only another marker of disease inflammation? Does mKatG have a dominant role as a pathogenic antigen or is it one of many mycobacterial antigens involved in sarcoidosis? Do mKatG-specific T cells have effector or regulatory phenotypes? Do these phenotypes differ in remitting versus chronic sarcoidosis? And finally, given that the presence of mKatG protein indicates a prior mycobacterial infection, what is the difference between sarcoidosis and active/latent mycobacterial infection?
To begin to answer these questions, studies are needed to characterize mKatG-responsive T cells and to correlate results from T- and B-cell assays with clinical phenotypes in sarcoidosis. Comparison studies involving purified protein derivative–negative (PPD−) healthy control subjects and PPD+ subjects, from BCG vaccination or M. tuberculosis infection, may provide clues to differences in immune responses in sarcoidosis and mycobacterial infection. Whether mKatG has special properties relevant to granuloma formation can be investigated in rodent models, in an attempt to develop a relevant preclinical model. Depending on the results of these studies, mKatG could be envisioned as part of a diagnostic test or as part of a vaccine strategy for the treatment of sarcoidosis. Although other tissue antigens are likely to be involved in sarcoidosis, at the very least the discovery of a specific candidate pathogenic antigen provides an opportunity to explore downstream pathobiological process that may define immunologic responses in sarcoidosis.
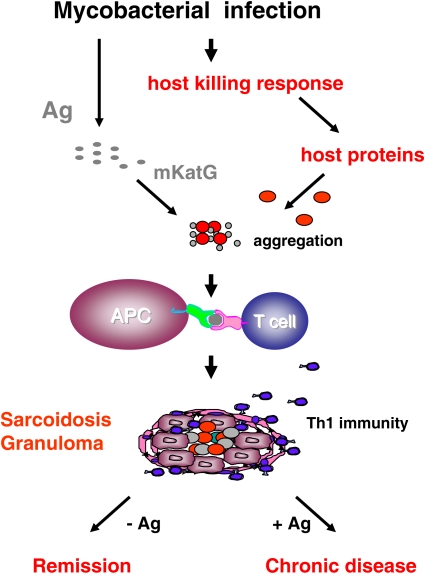
Although it is not known what specific properties contribute to the persistence of mKatG in sarcoidosis tissues, it is likely that mKatG is not the sole component of any nidus for epithelioid granuloma development. The nidi for sarcoidosis granulomas are likely to involve other host proteins and might also include other microbial proteins. We propose that these components form, together with mKatG, poorly soluble protein aggregates that provide both a nidus for granuloma formation and a depot of antigenic material (Figure 1). Within this conceptual framework, an updated hypothesis is proposed for one potential etiologic agent of sarcoidosis:
Figure 1.
Hypothetical model of a mycobacterial etiology of sarcoidosis. An occult mycobacterial infection results in the release of mycobacterial antigens (Ag) that induce an effective, mycobacteriocidal host response. In sarcoidosis, host proteins induced as part of this response aggregate with remnant mKatG proteins to form a poorly soluble nidus for granuloma formation and depots of microbial antigens. Macrophages and dendritic cells (antigen-presenting cells [APC]), activated directly by mKatG-host protein aggregates, produce the helper T-cell type 1 (Th1)–promoting cytokines IL-12, IL-18, and tumor necrosis factor. Antigen-specific Th1 responses orchestrate the complex process of epithelioid granuloma formation and accompanying inflammation. Removal of the inciting antigenic aggregates combined with immunosuppressive effects from transforming growth factor-β results in granuloma regression and disease remission. Failure to remove mKatG–host protein aggregates, possibly with the induction of autoimmunity, results in persistent inflammation and chronic disease.
Hypothesis: Sarcoidosis is caused by an effective antimicrobial host response to a mycobacterial infection that results in deposition of select mycobacterial antigens associated with focal aggregation of host proteins, which together form a nidus for granuloma formation that drives a pathogenic Th1 response and undergoes remission only with effective removal of stimulating antigens.
CONCLUSIONS
There remains a lack of consensus on the role of specific etiologic agents in sarcoidosis. Prior studies have largely relied on preconceived hypotheses regarding specific microbial, environmental, or autoantigenic factors. Using a limited proteomic approach based on the physicochemical properties of the Kveim reagent, our group identified a novel candidate pathogenic antigen of mycobacterial origin, the mKatG protein. This mycobacterial protein was detected in sarcoidosis tissues and was the target of an adaptive immune response in a subgroup of patients with sarcoidosis, supporting a mycobacterial etiology of sarcoidosis. Convincing an inherently skeptical research community that mKatG is pathogenic in sarcoidosis will require confirmatory studies by other groups. In addition, there is a need to identify other potential microbial antigens that may be pathogenic in different subgroups of patients with sarcoidosis. The limited proteomic approach used to discover mKatG in sarcoidosis tissues might be employed to validate other candidate pathogenic antigens in sarcoidosis or to discover novel pathogenic antigens in other granulomatous disorders.
Acknowledgments
The author thanks his collaborators and coworkers on this project and the patients with sarcoidosis and other subjects who participated in these studies. The author thanks Ed Chen for helpful comments on the manuscript and Figure 1.
Supported in part by grants HL68019, HL77732, and HL54658 from the National Heart, Lung, and Blood Institute, the Life and Breath Foundation, and the Hospital for the Consumptives of Maryland (Eudowood) Foundation.
Conflict of Interest Statement: D.R.M. and Johns Hopkins University have filed a Report of Invention entitled: Mycobacterial Catalase–peroxidase in the Diagnosis and Treatment of Sarcoidosis.
References
- 1.American Thoracic Society; European Respiratory Society; World Association of Sarcoidosis and Other Granulomatous Disorders. Statement on sarcoidosis. Am J Respir Crit Care Med 1999;160:736–755. [DOI] [PubMed] [Google Scholar]
- 2.Hance AJ. The role of mycobacteria in the pathogenesis of sarcoidosis. Semin Respir Infect 1998;13:197–205. [PubMed] [Google Scholar]
- 3.du Bois RM, Goh N, McGrath D, Cullinan P. Is there a role for microorganisms in the pathogenesis of sarcoidosis? J Intern Med 2003;253:4–17. [DOI] [PubMed] [Google Scholar]
- 4.Eishi Y, Suga M, Ishige I, Kobayashi D, Yamada T, Takemura T, Takizawa T, Koike M, Kudoh S, Costabel U, et al. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J Clin Microbiol 2002;40:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drake WP, Pei Z, Pride DT, Collins RD, Cover TL, Blaser MJ. Molecular analysis of sarcoidosis tissues for Mycobacterium species DNA. Emerg Infect Dis 2002;8:1334–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman LS, Rose CS, Bresnitz EA, Rossman MD, Barnard J, Frederick M, Terrin ML, Weinberger SE, Moller DR, McLennan G, et al.; ACCESS Research Group. A case control etiologic study of sarcoidosis: environmental and occupational risk factors. Am J Respir Crit Care Med 2004;170:1324–1330. [DOI] [PubMed] [Google Scholar]
- 7.Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA, Pandey JP, Newman LS, Magira E, Beznik-Cizman B, et al.; ACCESS Group. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet 2003;73:720–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schurmann M, Lympany PA, Reichel P, Muller-Myhsok B, Wurm K, Schlaak M, Muller-Quernheim J, du Bois RM, Schwinger E. Familial sarcoidosis is linked to the major histocompatibility complex region. Am J Respir Crit Care Med 2000;162:861–864. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel SL, Lukacs NW, Strieter RM, Chensue SW. Th1 and Th2 responses regulate experimental lung granuloma development. Sarcoidosis Vasc Diffuse Lung Dis 1996;13:120–128. [PubMed] [Google Scholar]
- 10.Moller DR. Rare manifestations of sarcoidosis. In: Drent M, Costabel U, editors: Sarcoidosis, Vol. 10, European Respiratory Society Monograph 32. Wakefield, UK: Charlesworth Group; 2005. pp. xx–xc.
- 11.Muller-Quernheim J. Sarcoidosis: immunopathogenetic concepts and their clinical application. Eur Respir J 1998;12:716–738. [DOI] [PubMed] [Google Scholar]
- 12.Kveim A. Em ny og specifikk kutans-reaksjon ved Boecks sarcoid, en forelobig meddelse. Nord Med 1941;9:169–172. Norwegian. [Google Scholar]
- 13.Siltzbach LE. The Kveim test in sarcoidosis: a study of 750 patients. JAMA 1961;178:476–482. [DOI] [PubMed] [Google Scholar]
- 14.Klein JT, Horn TD, Forman JD, Silver RF, Teirstein AS, Moller DR. Selection of oligoclonal Vβ-specific T-cells in the intradermal response to Kveim-Siltzbach reagent in individuals with sarcoidosis. J Immunol 1994;154:1450–1460. [PubMed] [Google Scholar]
- 15.Chase MW, Siltzbach LE. Concentration of the active principle responsible for the Kveim reaction. In: Turiaf J, Chabot J, editors. La sarcoïdose. Paris: Masson et Cie; 1967. pp. 150–160.
- 16.Munro CS, Mitchell DN. The Kveim response: still useful, still a puzzle. Thorax 1987;42:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons D, Donald S, Mitchell D, Asherson G. Chemical inactivation of the Kveim reagent. Respiration (Herrlisheim) 1992;59:22–26. [DOI] [PubMed] [Google Scholar]
- 18.Song Z, Marzilli L, Greenlee BM, Chen ES, Silver RF, Askin FB, Teirstein AS, Zhang Y, Cotter RJ, Moller DR. Mycobacterial catalase–peroxidase is a tissue antigen and target of the adaptive immune response in systemic sarcoidosis. J Exp Med 2005;201:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilmert H, Diringer H. A rapid and efficient method to enrich SAF-protein from scrapie brains of hamsters. Biosci Rep 1984;4:165–170. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase–peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 1992;358:591–593. [DOI] [PubMed] [Google Scholar]