Abstract
The recent completion of the Human Genome Project has made possible a high-throughput “systems approach” for accelerating the elucidation of molecular underpinnings of human diseases, and subsequent derivation of molecular-based strategies to more effectively prevent, diagnose, and treat these diseases. Although altered phenotypes are among the most reliable manifestations of altered gene functions, research using systematic analysis of phenotype relationships to study human biology is still in its infancy. This article focuses on the emerging field of high-throughput phenotyping (HTP) phenomics research, which aims to capitalize on novel high-throughput computation and informatics technology developments to derive genomewide molecular networks of genotype–phenotype associations, or “phenomic associations.” The HTP phenomics research field faces the challenge of technological research and development to generate novel tools in computation and informatics that will allow researchers to amass, access, integrate, organize, and manage phenotypic databases across species and enable genomewide analysis to associate phenotypic information with genomic data at different scales of biology. Key state-of-the-art technological advancements critical for HTP phenomics research are covered in this review. In particular, we highlight the power of computational approaches to conduct large-scale phenomics studies.
Keywords: computational genomics, gene–disease associations, phenomics, phenotype
GLOSSARY
Directed acyclic graph: a directed network structure with no loops
“Granularity” of phenotypic descriptors: level of detail provided to define a phenotype (class level of nomenclature level)
Ontology: structured hierarchy of classifications for entities
Pedigree: a familial history of ancestors
Phenomics: the genome-scale study of the relation of phenotypes to their molecular underpinnings in genetics, protein interactions, and so forth
Text mining: techniques used for identifying and extracting key concepts and terms and their relationships, often on a large scale
Since the first observation by Gregor Mendel that phenotypic traits of pea plants are faithful manifestations of their genetic inheritance, altered phenotypes, which are readily observed, described, and quantified, have been central to the discovery of gene functions and molecular relationships among genes in the field of genetics. Thomas Hunt Morgan's demonstration that genes linked in a chromosome (gene loci) may determine observable hereditary traits further illustrated the close relationship between genetic contents and phenotypic expression. The recent completion of the Human Genome Project has made possible a high-throughput “systems approach” for accelerating the elucidation of molecular underpinnings of human diseases, and subsequent derivation of molecular-based strategies to more effectively prevent, diagnose, and treat these diseases. Although the platform of molecular networks primarily derived from gene profiling under homeostatic or disease conditions has been intensively explored as a gateway to “systems medicine,” this molecular approach to analyzing genomic data is often complicated by genetic heterogeneity and the lack of cellular, tissue, organ, anatomic, or environmental context to accurately interpret the gene functions, which are highly context dependent. Furthermore, because mutations in different genes may yield identical or related phenotypes, a molecular characterization solely based on genes may neglect important relationships among molecularly distinct diseases at the phenotypic level. Although altered phenotypes are among the most reliable manifestations of altered gene functions, research using systematic analysis of phenotype relationships to study human biology is still in its infancy. The lack of high-throughput technologies to access well-networked and integrated phenotypes from heterogeneous sources and across multiple scales of biology under homeostasis or disease conditions has prevented the effective use of phenotypic information. As a result, development of phenotypic databases dramatically lags behind the rapid advance in genomic databases. A greater integration of medicine and biology calls for innovative computational and informatics tools and high-throughput discovery technologies for phenotypic research that aims to unlock gene–disease relationships, a key step for better understanding the genetic basis of human diseases and more effective gene-based disease management.
This article focuses on the emerging field of phenomics, which aims to capitalize on novel high-throughput computation and informatics technologies to derive genomewide molecular networks of genotype–phenotype associations, or “phenomic associations.” Currently, such large-scale high-throughput phenotyping (HTP) phenomic studies are limited due to our lack of knowledge about the relationships between molecular-level genotypes and their organism-level phenotypic manifestations.
To address this challenge in HTP phenomics research, several technological advancements will be critical in enabling the collection, organization, and computable encoding of large-scale, high-throughput phenotypes, and will be discussed in this review. In this article, we first provide a detailed analysis of the challenges facing HTP phenomics research, followed by an introduction of the current state of high-throughput phenotypic data collection (1), representation and encoding of phenotypes for computation, development of phenomic databases, and genomewide HTP phenomic analyses. In this last section, Whole Genome HTP Phenomic Analyses, we will also explore the feasibility of using computational phenomics approaches to enhance our understanding of genotype–phenotype relations and networks across different biological scales, from molecular biology to systems medicine.
CURRENT CHALLENGES FOR HTP PHENOMICS
One of the main factors hindering the progress of phenotypic discovery research is the limited accurate and timely access to comprehensive gene–phenotype networks associated with knowledge about biology and diseases. There are several obstacles restricting such access, as discussed in the following sections.
Lack of Understanding of Gene–Phenotype Relationships
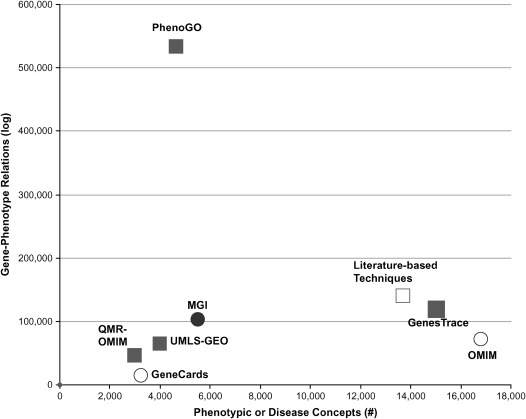
In the emerging field of phenomics, the pace of developing computable phenotypic databases and deriving networks of relationships among phenotypes and genes for use in constructing genotype–phenotype databases trails behind the rapid evolution of genomic databases. Currently, although many genomic databases of model organisms contain some phenotypic information, phenotypes are often coded at different levels of granularity, in different formats, and with different aims. In this case, we refer to “granularity” as the level of detail by which phenotypes are defined (e.g., “chronic obstructive lung disease” is less detailed than “centriacinar emphysema”). For example, PhenomicDB (2) allows only comparative genomic studies containing limited queries of textual (uncoded) phenotypic information associated with genes of interest. In contrast, state-of-the-art phenome-oriented methods require organization and encoding of phenotypes to genes before conducting combined genotypic/phenotypic analyses. However, most of such phenotypic databases are manually curated, and are thus limited in their breadth for high-throughput computing. Although high-throughput genotype–phenotype analyses were permitted via mining the wealth of scientific literature, such efforts yielded limited success due to the lack of expressiveness and granularity of text mining technology. To overcome these obstacles in developing phenotypic databases, our research group developed PhenoGO, a large-scale, ontology-anchored gene–phenotype network that we engineered and optimized for integration, classification, and analysis of well-encoded phenotypes. As shown in Figure 1, PhenoGO currently has the largest collection of relationship networks among phenotypes, genes, and the Gene Ontology (GO).
Figure 1.
Comparison of the number of distinct phenotypes and the number of gene–phenotype relationships in gene–phenotype databases and networks, showing that the PhenoGO is the largest network and that both literature text-mining techniques and OMIM provide the broadest annotations of distinct phenotypes with genes. Solid circle, ontology-anchored database; open circles, database with unstructured phenotypes; solid squares, ontology-anchored high-throughput phenotyping (HTP) phenomics; open square, HTP phenomics with unstructured phenotypes. MGI = Mouse Genome Informatics; OMIM = Online Mendelian Inheritance in Man; QMR = Quick Medical Reference; UMLS-GEO = Unified Medical Language System–Gene Expression Omnibus.
Phenotypes Are Poorly Integrated across the Model Organism Databases, Literature, and Human Disease Databases
Representation of phenotypic information is more complicated than biological data, and consequently there are few data standards and models for managing phenotypes across species and within human repositories. In addition, the granularity of phenotypic data varies from database to database, and current methods for accessing phenotypic information across databases are insufficient. Thus, there is an urgent need for the development of technologies to encode and organize phenotypic information for high-throughput computing. For example, although the Online Mendelian Inheritance in Man (OMIM) database has the largest collection of human diseases (3), the unstructured narrative content of its phenotypes makes it unsuitable for computational analysis. In contrast, the phenotypic concepts organized in our PhenoGO database are structured under standard ontology codes, allowing computation through networks of phenotypes.
Scarcity of Phenotypic Discovery Methods, Theories, and Predictions
There is a scarcity of phenotypic discovery methods, theories, and predictions to exploit the rich and untapped phenotypic data repositories in current genetic model organism databases and, soon, the databases of the National Institutes of Health (NIH) “Whole Genome Association” studies.
HIGH-THROUGHPUT COLLECTION OF PHENOTYPES
Over the past few years, several advances using experimental or imaging methods have made it possible to gather phenotypic information from different organisms in a high-throughput fashion. However, gene–phenotype analyses are currently limited to quantitative trait loci (QTL) studies requiring carefully curated pedigrees of individuals. For example, to map large-scale QTL to phenotypes, Solberg and colleagues (4) developed a protocol to collect multiple phenotypic measurements for high-throughput parallel phenotyping in populations of mice, and significantly reduced the high cost of genotyping in relation to the amount of information that can be derived from each phenotypic measurement. This protocol led to the detection of statistically significant variations among several inbred strains of mice from a population of over 2,500. However, because this method relies heavily on pedigree, it cannot be readily applied to clinical records and genetic databases because the pedigree associated with phenotypes is often absent. In other arenas, advances in imaging technologies, such as preclinical magnetic resonance imaging, have facilitated high-throughput phenotype imaging and reduced both the financial cost and time to characterize each individual animal (5). Similarly, advances in micro–computed tomographic scanning technology have brought down the expense of high-precision imaging. This technology has been applied to “virtual histology,” saving both the time and cost of phenotyping murine embryos while retaining image fidelity (6). In addition, genome-scale RNAi screens have been widely used in invertebrate systems for cellular-level phenotyping, and are now increasingly applied to more complex organisms (7).
REPRESENTATION OF PHENOTYPES FOR HIGH-THROUGHPUT ANALYSES
Although technological advancements have accelerated the pace of collecting phenotypic data, the task of coding and interpreting the output of high-throughput data collection is still left largely to humans, a labor-intensive and rate-limiting process in establishing phenotypic databases. Image-processing technologies, such as those used to automatically analyze imaging data from zebrafish (8), will play an increasingly important role in automating the evaluation and quantification of the massive amounts of phenotypic data. However, automated and accurate encoding and integration of heterogeneous phenotypic data remains challenging.
Applications of ontologies are now becoming a prevalent topic in the biomedical informatics field, largely due to the successful launch of GO. Scientists have invested considerable effort in establishing standards for the integration of phenotypes using ontologies. Since the launch of GO, a number of other ontology-based databases have been developed and have demonstrated the power of ontologies as the best standards for accelerating the data integration and analysis processes of biological and genomic data, which generally use unconstrained text and are too complicated to interpret. GO (9), which has been very successful in annotating genes with molecular functions, processes, and cellular locations, provides a good resource for the association of genes with cellular phenotypes. The Cell Type Ontology (CTO) (10) includes over 680 cell types covering the prokaryotic, fungal, animal, and plant worlds (11). The Mouse Genome Informatics (MGI) databases (12) and Rat Genome Database (13) contain genes, phenotypes coded in the Mammalian Phenotype Ontology (MPO) (12), unstructured phenotypic narratives, and references to PubMED. In the clinical domain, the Systematized Nomenclature of Medicine (SNOMED) (14), which is part of the Unified Medical Language System (UMLS), contains over a half million clinical concepts, such as disease, anatomy, morphology, functions, drugs, procedures, and treatments. To provide a unified framework for representing attributes of phenotypes requiring the composition of more than one code in any given ontology, the GO consortium has also initiated the development of the Phenotype Attribute Ontology (15) to reduce the structural barriers that limit the reuse of phenotypic databases. GO, SNOMED, CTO, and MPO are arranged as directed acyclic graphs (16), a data structure that allows standardized computational methods to process data in high throughput. These foundational ontology initiatives in both the biological and medical communities have set the stage for increasing the productivity of phenotypic research. However, many phenotypes stored in model organism databases remain buried in narratives or coded in terminologies specific to a community that are not cross-indexed with widespread standards.
In addition to the challenges associated with experimental methods for gathering phenotypic information are those associated with automatically encoding phenotypes collected in heterogeneous, unstructured forms. Although there has been a recent growth in text-mining research geared toward capturing gene–phenotype relationships from the literature (1, 17–21), it has failed to provide deep semantic and nested levels of associations from which ternary or higher order relations (e.g., a cell-type–dependent specific gene function) across concepts can be derived. Alternatively, some natural language processing (NLP) techniques can provide a deeper level of semantic relationship and a nested level of associations across concepts, allowing for more sophisticated computational studies. The Medical Language Extraction and Encoding NLP system (MedLEE), developed by Friedman and colleagues (22), was the first and most general NLP system, shown to be as accurate as clinicians in extracting phenotypic information from clinical reports (24). It has been evaluated in many different fields of clinical medicine, as evidenced by results of numerous independent evaluations (23–28). NLP systems are generally designed to extract phenotypes, but not to encode them in ontologies. Friedman and colleagues (29) and Tulipano and colleagues (30) have extended the capabilities of MedLEE to accurately encode phenotypes from clinical and imaging reports in comprehensive terminologies, such as the UMLS and SNOMED. Other NLP systems have also been shown to be robust but restricted in the specific task of extracting phenotypes from medical records (30–32). Although a few commercial NLP systems are currently available, to our knowledge they are incapable of encoding concepts from narratives in clinical reports. Rather, they classify concepts into simple classifications such as International Classification of Diseases Clinical Modification (ICD-9-CM), containing about 15,000 diseases (33). In contrast to clinical and imaging narratives, co-occurrence–based text-mining systems abound for mining the scientific literature, as reviewed by Jensen (34). However, they do not encode in terminologies, and thus generally are useful only for specifically designed studies and are not reusable in more general settings. Lussier and colleagues (35) and Friedman and colleagues (36) have recently completed BioMedLEE, the first NLP system for coding phenotypes in the scientific literature, which also allows for mining semantic relationships between genes and phenotypes that could not be captured by co-occurrence–based or statistics-based text-mining systems. BioMedLEE was successfully applied in high throughput over thousands of scientific abstracts and amassed the largest collection of gene–phenotype associations in PhenoGO (35), which will be described further in the following section.
Representing and encoding phenotypes in ontologies is an essential, yet insufficient step for automating the integration of coded phenotypes across heterogeneous databases. Indeed, many different terminologies and ontologies offer overlapping representations, sometimes at different levels of granularity. Cimino and Barnett first conceived lexical methods for creating translation tables across heterogeneous medical terminologies (37). Others thereafter have incrementally improved these techniques (37–49). For example, the UMLS has an extensive number of related tools such as MetaMap (MMTx), which can map terms to concepts in the UMLS Metathesaurus (38). Lussier and Li (50) and Sarkar and colleagues (51) pioneered the automated translation between heterogeneous phenotypic terminologies. An alternative approach to integrating phenotypes across terminologies is to rely on large-scale metathesauri designed specifically for that purpose, such as the NIH UMLS (52), or the National Cancer Institute's Metathesaurus (53), which includes hundreds of distinct biomedical terminologies that have been semiautomatically mapped to one another. Although the automated terminology integration approaches are limited in accuracy, they are scalable to any pair of terminologies and can be conducted in real time. In contrast, the metathesauri are more accurate but are rate-limited due to the many terminologies that have not yet been integrated, and perhaps more important, because the mappings may be out of synchronization with newer versions of the source terminologies.
In summary, it is noteworthy that automated coding and harmonization technologies are not widely available and remain the panacea of bioinformatics networks and research groups, whereas the metathesauri are freely available. Comprehensive dissemination of technologies and training will be required in the future for phenotypic datasets to be computer processable in real time (54).
DEVELOPMENT OF PHENOTYPIC DATABASES
In the process of associating phenotypes with genes, data integration plays a key role in correlating heterogeneous phenotypic data with genomic data at different scales. The current efforts to organize phenotypic information for high-throughput phenomics studies focus on both manual and computational methods for gathering phenotypes and their related genomic information. Both methods have their distinctive advantages and disadvantages. Although manual methods provide more accurate gene–phenotype relations, they are more time and labor consuming. In contrast, computational methods are able to generate large networks of gene–phenotype relationships in a relatively short amount of time, but generally lack accuracy when compared with the results of manual methods.
Manually Curated Databases
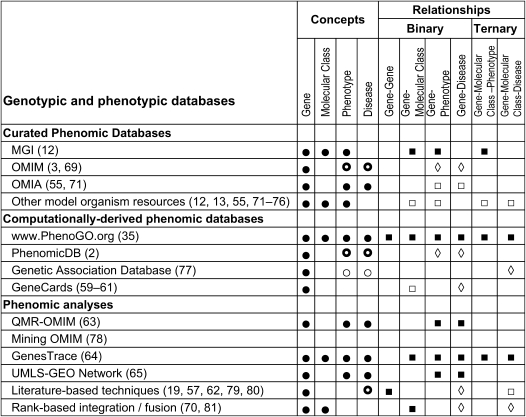
There are several databases that contain manually curated phenotypic information, including OMIM (3), the Online Mendelian Inheritance in Animals (OMIA) (55), and all model organism databases. The OMIM and OMIA databases contain unstructured phenotypic narratives and references to PubMED, from which it is computationally difficult to extract coded phenotypic data. Similarly, although many genomic databases of model organisms contain some phenotypic information, phenotypes are often coded at different levels of granularity, in different formats, and with different aims (56). Realizing the difficulties of using phenotypic narratives in organizing phenotypic information, the MGI database (12) chose to use coded and computable phenotypes in the MPO, as described above, to organize phenotypes in different mouse strains (12). Although phenotypic narratives can be more nuanced and detailed, coded phenotypes are classified in the MPO and are readily computable. The contents of these different databases are summarized in Figure 1, in which we present the quantity of distinct phenotypes (breadth) and the quantity of gene–phenotype associations (depth) for OMIM and MGI. Of the curated databases, MGI remains the best-organized database with the most variety of coded phenotypes and coded binary and ternary relationships (Figure 2).
Figure 2.
Descriptions of the conceptual content and expressiveness of relationships in gene-phenotype databases. The figure shows that the current gene-phenotype databases are limited in expressiveness because binary and higher-order relationships are scarce and available only in unstructured form. Solid circle = coded in terminology; thick circle = semistructured text; thin circle = unstructured (free) text; solid square = automated (structured concepts); open square = rate-limiting curation (structured concepts); open diamond = semistructured concepts.
Computationally derived Databases
To overcome the limitations of manual annotation for creating phenotypic datasets, scientists use computational techniques for identifying phenotype–gene relations. These approaches are generally based on high-throughput methods, such as text mining the scientific literature for phenotype–genotype co-occurrences (57, 58). In addition, some efforts have been made to integrate and standardize phenotypic data for the purposes of sharing. For example, the PhenomicDB (2) database provides a single portal for heterogeneous phenotypic information from a number of different model organisms, including humans. It contains over 15,000 distinct uncoded textual phenotypic terms and 120,000 genotypes for the mouse and human species. Similarly, GeneCards provides a single portal for integrating human genetic data with their related genomic information and textual disease relationships (59–61). The Genetic Association Database (16) provides a collection of standardized genetic association datasets, in which associated diseases are classified and structured. In addition, Gene2Disease was constructed over OMIM using text-mining methods coupled with analysis of the chromosomal locations of diseases (62). Although these resources allow scientists to browse phenotypes and their associated genes, and to conduct comparative genomics studies among different organisms, their analyses are limited to functional genomics datasets organized according to textual terms containing phenotypic information.
In contrast, Lussier and colleagues used NLP over the scientific literature combined with the GO database to amass and encode phenotypes in high throughput (35). The resulting database, PhenoGO (http://www.PhenoGO.org), contains the largest number of gene–phenotype associations (Figure 1), and provides the broadest variety of binary and ternary relationships between genes, GO concepts, and phenotypes (Figure 2 and Table 1). The PhenoGO database also differs from other gene–phenotype databases in that it also provides ternary relationships, such as biological process of a specific gene in a particular phenotypic context. For example, the PhenoGO database refines GO concepts though the assignment of phenotypic information, such as the cell type, tissue, and organ to GO–gene annotations. The addition of such phenotypic context to gene expression information could be a crucial step for understanding the development and the molecular underpinnings of the pathophysiology of diseases, as not all potential biological processes associated to a gene are possible in every cell type. Currently, PhenoGO consists of 532,406 phenotype–GO relations, with 33,224 distinct genes in 10 species, 5,680 unique GO concepts and 4,650 unique phenotypes coded in SNOMED, MPO, CTO, and UMLS. Manual evaluation of a random sample of gene–GO–(phenotype or disease) relationships revealed a precision (positive predictive value) of 85% (95% confidence interval [CI], 82–89%) and a recall of 76% (95% CI, 69–83%). To our knowledge, this is the first system that offers a level of precision not too far from that of manual curation.
TABLE 1.
DESCRIPTION OF GENOTYPIC AND PHENOTYPIC DATABASES
| Phenomic Databases | Description |
|---|---|
| MGI | Mouse Genome Informatics (MGI) provides integrated access to data on the genetics, genomics, phenomics, and biology of the laboratory mouse. |
| OMIM | The Online Mendelian Inheritance in Man (OMIM) is a database that catalogs relationships between human genes and genetic disorders. |
| OMIA | The Online Mendelian Inheritance in Animals (OMIA) is a database that catalogs genes, inherited disorders, and traits in more than 135 animal species (other than human and mouse). |
| PhenoGO | The PhenoGO is a computed database that provides phenotypic contexts and their associated GO terms for multiple organisms, including human, mouse, and rat. |
| PhenomicDB | The PhenomicDB is a multiorganism phenotype–genotype database, which is built by integrating data from several model organism databases. |
| Genetic Association Database | The Genetic Association Database archives human genetic association studies on complex diseases and disorders. |
| GeneCards | The GeneCards is a database of human genes with their associated genomic, proteomic, single nucleotide polymorphism, and disease information. |
| QMR-OMIM | This database integrates clinical knowledge and genomic data to define human trait–disease–gene relationships. |
| Mining OMIM | This study used OMIM to study relationships between human disease and genes. |
| GenesTrace | The GenesTrace defines ontology-anchored phenotypes from the UMLS and their statistical and semantic relationships to GO and model organism databases. |
| UMLS-GEO network | This study defines highly related phenotypic concepts and gene expressions by integrating phenotypically related concepts in UMLS and the microarray gene expression data from the NCBI's Gene Expression Omnibus (GEO). |
| Literature-based techniques | These methods mine literature by high-throughput computational method to identify relations between genes and unconstrained phenotypic contexts. |
Definition of abbreviations: NCBI = National Center for Biotechnology Information; QMR = Quick Medical Reference; UMLS = Unified Medical Language System.
In summary, given the size of current phenomic databases, computational approaches certainly have the edge over manual methods for quickly collecting and integrating large amounts of phenotypic information. However, computed techniques generally have a lower precision than manual curation (∼ 95%). Term co-occurrence and statistical NLP generally produce up to 75% precision, whereas semantic NLP, such as BioMedLEE and MedLEE, can reach above 85% precision. To illustrate the differences and similarity between these genome–phenome networks, Table 2 provides an example of the subset of a manually curated network (MGI) and additional computed phenotypes (found in the PhenoGO database).
TABLE 2.
SUBSETS OF GENE–PHENOTYPE NETWORK SHOWING MANUALLY CURATED KNOWLEDGE AND COMPUTED KNOWLEDGE
| Gene and Reference Found in GO and in PhenoGO | Biological Process Curated in GO | Phenotypic Context Computed in PhenoGO |
|---|---|---|
| Nerve growth factor β (Ngfb;MGI: 97321) | Perception of pain (GO: 0019233) | Afferent neuron (UMLS: C0027883) |
| Vascular endothelial growth factor C (Vegfc; MGI: 109124) (82) | Morphogenesis of embryonic epithelium (GO: 0016331) | Lymphatic vessel (UMLS: C0229889) |
Definition of abbreviations: GO = Gene Ontology; MGI = Mouse Genome Informatics; UMLS = Unified Medical Language System.
Curated relationships of biological processes are found in GO. Manual curation, a rate-limiting process, is generally considered more accurate than knowledge that can be computed in high throughput. The GO Consortium manually curated over 1,617,028 annotations of genes to Gene Ontology code in the last 5 yr and intercurator reliability of curated relationships in GO was estimated at about 93% (83). In contrast, it took about 3 yr to develop BioMedLEE and PhenoGO, a natural language processing system and a text-mining tool, together capable of encoding gene–GO–phenotypes in high throughput with a precision of 85% (35). The PhenoGO system can now process vast quantities of text within a reasonable time. The PhenoGO database, which contains about 550,000 gene–GO–phenotype annotations, can substantially facilitate whole genome association research by providing a well-organized and ontology-anchored genome–phenome network mined from massive amounts of information found in biomedical journal articles. These annotations, refined with phenotypic context, such as the cell type, tissue, and organ in which a gene is expressed and has a function, often specific to the cell type, provide a crucial step for understanding the development and the molecular underpinning of embryogenesis and possibly the pathophysiology of diseases. In the table, the biological process associated with each gene via curation is further refined with phenotypic context via computations.
WHOLE GENOME HTP PHENOMIC ANALYSES
Text-based HTP phenomics is designed to predict gene–disease associations; however, its methods vary broadly. To overcome the limitations of manual annotation to create phenotypic datasets, others in the field conducted high-throughput phenotype–genotype analyses by mining text on phenotype–genotype relationships from the scientific literature (57, 58), with limitations of text mining as described above. Korbel and colleagues conducted an analysis that combined data mining of the MEDLINE abstracts to extract terms of prokaryotic traits, and comparative genome analysis to identify association of phenotype to genotype relationships (57). Approximately 2,700 significant gene–trait associations were identified. Gene2Disease was constructed over OMIM using text-mining methods coupled with analysis of the chromosomal locations of diseases (62). However, in these two systems, the integration of phenotypes relies on the juxtaposition of the original lexical string of text in the same field across species. Thus, a textual search for a concept may miss synonyms, as well as related or subsumed concepts. Although these literature-based approaches allow scientists to browse phenotypes and their associated genes and to conduct comparative genomics analyses among different organisms, their analyses are merely functional genomics studies constrained to datasets organized according to textual terms containing phenotypic information. In addition, the resultant binary textual relationships lack context.
Lussier and coworkers pioneered ontology-anchored HTP phenomics in clinical databases. They integrated the Quick Medical Reference (QMR) with OMIM, from which relationships among genes, diseases, and traits of diseases were generated. Clustering of genes with traits of diseases demonstrated classification of diseases according to genes (63) and enabled association studies of environmental factors, such as drug intake and smoking, found in QMR with genes found in OMIM. This study was followed up with the GenesTrace method, a large-scale integrative study of ontology-anchored phenotypes from the UMLS and their statistical and semantic relationships to GO and model organism databases (64). We were able to infer approximately 3 million phenotype–gene associations among 22,040 phenotypic concepts in the UMLS and 16,894 gene products annotated using GO and its associated databases (64). Inferences were validated by comparing them to known gene–disease relationships, as defined in OMIM's Morbidmap. Approximately 30% of the predictions could be found in OMIM, and conversely, 9% of OMIM's relationships were found in Genestrace (64). In addition, our methods provided direct links to clinically significant diseases through established terminologies or ontologies. These observations demonstrate the significance of exploiting the existing manually curated relationships in biomedical resources as a tool for the discovery of potentially valuable new gene–disease relationships. Recently, Butte and Kohane (65) conducted the first ontology-anchored HTP phenomics study with phenotypically related concepts in UMLS (66) and microarray gene expression data from the NCBI's Gene Expression Omnibus (67) using a term presence/absence method. Significantly expressed genes above a threshold were correlated with UMLS phenotypic concepts via a resampling-based multiple testing simulation generating 64,003 relations among 281 biomedical concepts and 7,466 genes (65). This study provided an HTP phenomic method for identifying genes related to phenotype and environment.
Although HTP phenomics is in its early stages, there is sufficient evidence through validations that it is promising. In 2001, Jimenez-Sanchez and colleagues established a proof-of-concept study for HTP phenomics by manually relating about 1,000 disease-related genes to their molecular functions and observed that the frequency distribution of lethality of genes according to their molecular function recapitulates current knowledge about these molecular families (68). Since that proof of concept, GenesTrace provided additional evidence that integrating and systematically analyzing genome–phenome networks can accurately predict disease genes. More precisely, the GenesTrace study was based on patterns of GO annotations of genes (9) with the UMLS clinical knowledge base (66). Using the 1,407 single gene diseases of the OMIM (69) dataset as a control, GenesTrace predicted 124 distinct genes in the context of being related to their specific disease concept, and 290 distinct genes were erroneously associated with concepts, for a precision of 30% and recall of 8.8% (64). Kohane and Butte also merged the UMLS knowledge base, this time with microarray datasets, and accurately predicted novel findings corroborated in a new microarray study (65). Recently, Aertz and colleagues predicted gene phenotypes through a fusion of a large amount of heterogeneous genetic and clinical knowledge bases, including text mining of the literature (70). This technique, called “Endeavor” data fusion, identified a novel gene involved in craniofacial development and likely with DiGeorge-like birth defects. This prediction was further corroborated in zebrafish embryos that showed an underdeveloped lower jaw. The properties of these studies are summarized in Table 1 and their respective dataset size in Figure 1.
FUTURE CHALLENGES
HTP phenomics research faces the challenge of technological research and development to generate novel tools in computation and informatics to amass, access, integrate, organize, and manage phenotypic databases across species and enable genomewide analysis to associate phenotypic information with genomic data at different scales of biology. Currently, the lack of high-throughput technologies to access well-networked and integrated phenotypes from heterogeneous sources and across multiple scales of biology has prevented the effective usage of phenotypic information. Therefore, HTP phenomics research that aims to unlock gene–disease relationships will play a key role in a “systems approach” to molecular medicine and individualized medicine. In this review, we highlighted the state of the art in computational approaches to conduct large-scale phenomics studies. Among various strategies that could facilitate computational phenomics, ontologies have proved to be particularly effective at integrating and organizing a large number of phenotypic concepts on a computable platform. The success of GO underscores the importance of ontologies in phenotypic research. Similarly, the NLP techniques have increasingly shown their unique and efficient capacity to associate genes with the narrative phenotypic descriptions in the literature, which are often unconstrained and unstructured and could not be otherwise handled by other technologies. Although there are novel computational approaches proposed for conducting high-throughput association analysis, they generally lack a common benchmark for comparison, thus often yielding results that are difficult to compare. Because the NIH recently recognized the urgent need for a well-organized resource of human phenotypes and diseases, it launched Whole Genome Association studies. The Whole Genome Association will link genetic data with the rich phenotypic datasets of large-scale clinical studies accumulated over several generations of patients to generate large-scale common sharable datasets. Such unified efforts will accelerate the process of identifying the genetic and environmental factors associated with human disease. It will also provide a framework to use HTP phenomics methods in conjunction with methods based on quantitative trait loci methods. The emerging field of HTP phenomics is likely to have a focus on therapeutic predictions and delineate gene–disease associations.
Acknowledgments
The authors thank Lee Sam for his advice on improving the manuscript.
Supported in part by NIH/NLM grants 1K22 LM008308-01, R01 LM007659, and by the National Center for the Multiscale Analysis of Genomic and Cellular Networks (U54CA121852-01A1).
Conflict of Interest Statement: Y.A.L., as a member of the Columbia University Center for Advanced Technology, was mandated by his Department chairman to provide scientific advice to the executive board of John Wiley and Sons. He has received no stipends or payments; however, he did receive a sponsored research contract described below. Wiley will not benefit directly from this paper; indirectly, however, this research is related to its interests since mining the scientific literature is part of Wiley's potential future markets. In 2004–2005, Y.A.L. received a research contract from John Wiley and Sons to conduct studies on “Ontology-anchored Methods for Computable Biomedical Excerpts.” In the section of the manuscript pertaining to the Natural Language Processing and the text mining, he has not mentioned any publication that pertains to this contract, as he has not published our results yet. The research contract is completed and follow-up studies will help display clinical phenotypes in large clinical warehouses. Y.A.L. has a patent pending for computational terminology tools that the Columbia Center for Advanced Technology and the Columbia Science Venture groups are marketing. He is unaware of any companies currently interested; however, in the past, companies related to the mining of phenomic data were approached. He has received no money for these patents. Y.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Yakushiji A, Tateisi Y, Miyao Y, Tsujii J. Event extraction from biomedical papers using a full parser. Pac Symp Biocomput 2001;6:408–419. [DOI] [PubMed] [Google Scholar]
- 2.Kahraman A, Avramov A, Nashev LG, Popov D, Ternes R, Pohlenz HD, Weiss B. PhenomicDB: a multi-species genotype/phenotype database for comparative phenomics. Bioinformatics 2005;21:418–420. [DOI] [PubMed] [Google Scholar]
- 3.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM). Hum Mutat 2000;15:57–61. [DOI] [PubMed] [Google Scholar]
- 4.Solberg LC, Valdar W, Gauguier D, Nunez G, Taylor A, Burnett S, Arboledas-Hita C, Hernandez-Pliego P, Davidson S, Burns P, et al. A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm Genome 2006;17:129–146. [DOI] [PubMed] [Google Scholar]
- 5.McConville P, Moody JB, Moffat BA. High-throughput magnetic resonance imaging in mice for phenotyping and therapeutic evaluation. Curr Opin Chem Biol 2005;9:413–420. [DOI] [PubMed] [Google Scholar]
- 6.Johnson JT, Hansen MS, Wu I, Healy LJ, Johnson CR, Jones GM, Capecchi MR, Keller C. Virtual histology of transgenic mouse embryos for high-throughput phenotyping. PLoS Genet 2006;2:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods 2004;1:127–132. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, Lu J, Wang Y, Campbell WA, Huang L, Zhu J, Xia W, Wong ST. Computerized image analysis for quantitative neuronal phenotyping in zebrafish. J Neurosci Methods 2005;153:190–202. [DOI] [PubMed] [Google Scholar]
- 9.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rector AL, Rogers J, Roberts A, Wroe C. Scale and context: issues in ontologies to link health- and bio-informatics. Proc AMIA Symp 2002:642–646. [PMC free article] [PubMed]
- 11.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol 2005;6:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blake JA, Eppig JT, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): updates and enhancements. Nucleic Acids Res 2006;34:D562–D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Cruz N, Bromberg S, Pasko D, Shimoyama M, Twigger S, Chen J, Chen CF, Fan C, Foote C, Gopinath GR, et al. The Rat Genome Database (RGD): developments towards a phenome database. Nucleic Acids Res 2005;33:D485–D491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spackman KA, Campbell KE, Cote RA. SNOMED RT: a reference terminology for health care. Proc AMIA Annu Fall Symp 1997: 640–644. [PMC free article] [PubMed]
- 15.(PaTO), P.A.O., Open Global Ontologies (OBO). Available from: http://obo.sourceforge.net/. For PaTO, see ftp://ftp.geneontology.org/pub/go/gobo/phenotype.ontology/phenotype.txt (accessed August, 2003).
- 16.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 2004;428:493–521. [DOI] [PubMed] [Google Scholar]
- 17.Hafner CD, Baclawski K, Futrelle RP, Fridman N, Sampath S. Creating a knowledge base of biological research papers. Proc Int Conf Intell Syst Mol Biol 1994;2:147–155. [PubMed] [Google Scholar]
- 18.Bajdik CD, Kuo B, Rusaw S, Jones S, Brooks-Wilson A. CGMIM: automated text-mining of Online Mendelian Inheritance in Man (OMIM) to identify genetically-associated cancers and candidate genes. BMC Bioinformatics 2005;6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Iratxeta C, Bork P, Andrade MA. Association of genes to genetically inherited diseases using data mining. Nat Genet 2002;31:316–319. [DOI] [PubMed] [Google Scholar]
- 20.Raychaudhuri S, Altman RB. A literature-based method for assessing the functional coherence of a gene group. Bioinformatics 2003;19:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raychaudhuri S, Chang JT, Sutphin PD, Altman RB. Associating genes with gene ontology codes using a maximum entropy analysis of biomedical literature. Genome Res 2002;12:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman C, Hripcsak G, Shagina L, Liu HF. Representing information in patient reports using natural language processing and the extensible markup language. J Am Med Inform Assoc 1999;6:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman C, Knirsch C, Shagina L, Hripcsak G. Automating a severity score guideline for community-acquired pneumonia employing medical language processing of discharge summaries. Proc AMIA Symp 1999:256–260. [PMC free article] [PubMed]
- 24.Hripcsak G, Friedman C, Alderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med 1995;122:681–688. [DOI] [PubMed] [Google Scholar]
- 25.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med 1998;37:1–7. [PubMed] [Google Scholar]
- 26.Jain NL, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp 1997:829–833. [PMC free article] [PubMed]
- 27.Knirsch CA, Jain NL, Pablos-Mendez A, Friedman C, Hripcsak G. Respiratory isolation of tuberculosis patients using clinical guidelines and an automated clinical decision support system. Infect Control Hosp Epidemiol 1998;19:94–100. [DOI] [PubMed] [Google Scholar]
- 28.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics 2001;17:S74–S82. [DOI] [PubMed] [Google Scholar]
- 29.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tulipano PK, Tao Y, Zanzonico P, Kolbert K, Lussier Y, Friedman C. Natural language processing in the molecular imaging domain. AMIA Annu Symp Proc 2005:1143. [PMC free article] [PubMed]
- 31.Zweigenbaum P, Bachimont B, Bouaud J, Charlet J, Boisvieux JF. Issues in the structuring and acquisition of an ontology for medical language understanding. Methods Inf Med 1995;34:15–24. [PubMed] [Google Scholar]
- 32.Masseroli M, Kilicoglu H, Lang FM, Rindflesch TC. Argument-predicate distance as a filter for enhancing precision in extracting predications on the genetic etiology of disease. BMC Bioinformatics 2006;7:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mamlin BW, Heinze DT, McDonald CJ. Automated extraction and normalization of findings from cancer-related free-text radiology reports. AMIA Annu Symp Proc 2003:420–424. [PMC free article] [PubMed]
- 34.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet 2006;7:119–129. [DOI] [PubMed] [Google Scholar]
- 35.Lussier YA, Borlawsky T, Rappaport D, Friedman C. PhenoGO: a multistrategy language processing system assigning phenotypic context to gene ontology annotations. Pac Symp Biocomput 2006: 64–75. [PMC free article] [PubMed]
- 36.Friedman C, Borlawsky T, Shagina L, Xing H, Lussier Y. Bio-ontology and text: bridging the modeling gap. Bioinformatics 2006;22:2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cimino JJ, Barnett GO. Automated translation between medical terminologies using semantic definitions. MD Comput 1990;7:104–109. [PubMed] [Google Scholar]
- 38.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001:17–21. [PMC free article] [PubMed]
- 39.Hill DP, Blake JA, Richardson JE, Ringwald M. Extension and integration of the gene ontology (GO): combining GO vocabularies with external vocabularies. Genome Res 2002;12:1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodenreider O, Mitchell JA, McCray AT. Evaluation of the UMLS as a terminology and knowledge resource for biomedical informatics. Proc AMIA Symp 2002:61–65. [PMC free article] [PubMed]
- 41.Cimino JJ, Johnson SB, Peng P, Aguirre A. From ICD9-CM to MeSH using the UMLS: a how-to guide. Proc Annu Symp Comput Appl Med Care 1993:730–734. [PMC free article] [PubMed]
- 42.Zeng Q, Cimino JJ. Mapping medical vocabularies to the Unified Medical Language System. Proc AMIA Annu Fall Symp 1996:105–109. [PMC free article] [PubMed]
- 43.Tuttle MS, Suarez-Munist ON, Olson NE, Sherertz DD, Sperzel WD, Erlbaum MS, Fuller LF, Hole WT, Nelson SJ, Cole WG, et al. Merging terminologies. Medinfo 1995;8:162–166. [PubMed] [Google Scholar]
- 44.Tuttle MS, Cole WG, Sheretz DD, Nelson SJ. Navigating to knowledge. Methods Inf Med 1995;34:214–231. [PubMed] [Google Scholar]
- 45.Tuttle MS, Sherertz DD, Erlbaum MS, Sperzel WD, Fuller LF, Olson NE, Nelson SJ, Cimino JJ, Chute CG. Adding your terms and relationships to the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care 1991:219–223. [PMC free article] [PubMed]
- 46.Masarie FE Jr, Miller RA, Bouhaddou O, Giuse NB, Warner HR. An interlingua for electronic interchange of medical information: using frames to map between clinical vocabularies. Comput Biomed Res 1991;24:379–400. [DOI] [PubMed] [Google Scholar]
- 47.Lussier YA, Shagina L, Friedman C. Automating SNOMED coding using medical language understanding: a feasibility study. Proc AMIA Symp 2001:418–422. [PMC free article] [PubMed]
- 48.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care 1994:235–239. [PMC free article] [PubMed]
- 49.Rocha RA, Rocha BH, Huff SM. Automated translation between medical vocabularies using a frame-based interlingua. Proc Annu Symp Comput Appl Med Care 1993:690–694. [PMC free article] [PubMed]
- 50.Lussier YA, Li J. Terminological mapping for high throughput comparative biology of phenotypes. Pac Symp Biocomput 2004:202–213. [DOI] [PMC free article] [PubMed]
- 51.Sarkar IN, Cantor MN, Gelman R, Hartel F, Lussier YA. Linking biomedical language information and knowledge resources: GO and UMLS. Pac Symp Biocomput 2003:439–450. [DOI] [PMC free article] [PubMed]
- 52.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med 1993;32:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Covitz PA, Hartel F, Schaefer C, De Coronado S, Fragoso G, Sahni H, Gustafson S, Buetow KH. caCORE: a common infrastructure for cancer informatics. Bioinformatics 2003;19:2404–2412. [DOI] [PubMed] [Google Scholar]
- 54.Soldatova LN, King RD. Are the current ontologies in biology good ontologies? Nat Biotechnol 2005;23:1095–1098. [DOI] [PubMed] [Google Scholar]
- 55.Lenffer J, Nicholas FW, Castle K, Rao A, Gregory S, Poidinger M, Mailman MD, Ranganathan S. OMIA (Online Mendelian Inheritance in Animals): an enhanced platform and integration into the Entrez search interface at NCBI. Nucleic Acids Res 2006;34:D599–D601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biesecker LG. Mapping phenotypes to language: a proposal to organize and standardize the clinical descriptions of malformations. Clin Genet 2005;68:320–326. [DOI] [PubMed] [Google Scholar]
- 57.Korbel JO, Doerks T, Jensen LJ, Perez-Iratxeta C, Kaczanowski S, Hooper SD, Andrade MA, Bork P. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol 2005;3:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haft DH, Selengut JD, Brinkac LM, Zafar N, White O. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics 2005;21:293–306. [DOI] [PubMed] [Google Scholar]
- 59.Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, Ben-Dor U, Esterman N, Rosen N, Peter I, et al. GeneCards 2002: towards a complete, object-oriented, human gene compendium. Bioinformatics 2002;18:1542–1543. [DOI] [PubMed] [Google Scholar]
- 60.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 1998;14:656–664. [DOI] [PubMed] [Google Scholar]
- 61.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet 1997;13:163. [DOI] [PubMed] [Google Scholar]
- 62.Perez-Iratxeta C, Wjst M, Bork P, Andrade MA. G2D: a tool for mining genes associated with disease. BMC Genet 2005;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lussier YA, Sarkar IN, Cantor M. An integrative model for in-silico clinical-genomics discovery science. Proc AMIA Symp 2002:469–473. [PMC free article] [PubMed]
- 64.Cantor MN, Sarkar IN, Bodenreider O, Lussier YA. GenesTrace: phenomic knowledge discovery via structured terminology. Pac Symp Biocomput 2005:103–114. [PMC free article] [PubMed]
- 65.Butte AJ, Kohane IS. Creation and implications of a phenome-genome network. Nat Biotechnol 2006;24:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.National Library of Medicine. Unified Medical Language System® fact sheet. Available from: http://www.nlm.nih.gov/pubs/factsheets/umls.html (accessed March 23, 2006).
- 67.Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res 2004;32:D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jimenez-Sanchez G, Childs B, Valle D. Human disease genes. Nature 2001;409:853–855. [DOI] [PubMed] [Google Scholar]
- 69.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM): a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005;33:D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, et al. Gene prioritization through genomic data fusion. Nat Biotechnol 2006;24:537–544. [DOI] [PubMed] [Google Scholar]
- 71.Nicholas FW. Online Mendelian Inheritance in Animals (OMIA): a comparative knowledgebase of genetic disorders and other familial traits in non-laboratory animals. Nucleic Acids Res 2003;31:275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bogue M. Mouse Phenome Project: understanding human biology through mouse genetics and genomics. J Appl Physiol 2003;95:1335–1337. [DOI] [PubMed] [Google Scholar]
- 73.Bogue MA, Grubb SC. The Mouse Phenome Project. Genetica 2004;122:71–74. [DOI] [PubMed] [Google Scholar]
- 74.Bono H, Kasukawa T, Furuno M, Hayashizaki Y, Okazaki Y. FANTOM DB: database of Functional Annotation of RIKEN Mouse cDNA Clones. Nucleic Acids Res 2002;30:116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res 2002;30:69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mashimo T, Voigt B, Kuramoto T, Serikawa T. Rat Phenome Project: the untapped potential of existing rat strains. J Appl Physiol 2005;98:371–379. [DOI] [PubMed] [Google Scholar]
- 77.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet 2004;36:431–432. [DOI] [PubMed] [Google Scholar]
- 78.Cantor MN, Lussier YA. Mining OMIM for insight into complex diseases. Medinfo 2004;11:753–757. [PMC free article] [PubMed] [Google Scholar]
- 79.Krauthammer M, Kaufmann CA, Gilliam TC, Rzhetsky A. Molecular triangulation: bridging linkage and molecular-network information for identifying candidate genes in Alzheimer's disease. Proc Natl Acad Sci USA 2004;101:15148–15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet 2001;28:21–28. [DOI] [PubMed] [Google Scholar]
- 81.Tiffin N, Kelso JF, Powell AR, Pan H, Bajic VB, Hide WA. Integration of text- and data-mining using ontologies successfully selects disease gene candidates. Nucleic Acids Res 2005;33:1544–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karkkainen MJ, Haiko P, Sainio K, Partanen J, Taipale J, Petrova TV, Jeltsch M, Jackson DG, Talikka M, Rauvala H, et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat Immunol 2004;5:74–80. [DOI] [PubMed] [Google Scholar]
- 83.Lee V, Camon E, Dimmer E, Barrell D, Apweiler R. Who tangos with GOA?-Use of Gene Ontology Annotation (GOA) for biological interpretation of ‘-omics’ data and for validation of automatic annotation tools. In Silico Biol. 2005;5:5–8. [PubMed] [Google Scholar]