Abstract
It has been well established that genetic factors strongly affect susceptibility to asthma and its associated traits. It is less clear to what extent genetic variation contributes to the ethnic disparities observed for asthma morbidity and mortality. Individuals of African descent with asthma have more severe asthma, higher IgE levels, a higher degree of steroid dependency, and more severe clinical symptoms than individuals of European descent with asthma but relatively few studies have focused on this particularly vulnerable ethnic group. Similar underrepresentation exists for other minorities, including Hispanics. In this review, a summary of linkage and association studies in populations of African descent is presented, and the role of linkage disequilibrium in the dissection of a complex trait such as asthma is discussed. Consideration for the impact of population stratification in recently admixed populations (i.e., European, African) is essential in genetic association studies focusing on African ancestry groups. With the most recent update on the International HapMap Project, efficient selection of haplotype tagging single nucleotide polymorphisms (htSNPs) for African Americans has accelerated and efficiency of htSNPs chosen from one population to represent other continental groups (e.g., African) has been demonstrated. Cutting-edge approaches, such as genomewide association studies, admixture mapping, and phylogenetic analyses, offer new opportunities for dissecting the genetic basis for asthma in populations of African descent.
Keywords: genetic epidemiology, asthma, African descent, linkage disequilibrium
Asthma is a complex disease of unknown etiology characterized by intermittent inflammation of the airways, which, over time, may lead to irreversible airway remodeling and intractable airflow limitation. The prevalence of asthma has increased markedly since the latter half of the 20th century, doubling in the United States from 6.7 million in 1980 to 17 million in 1998, and now affects over 20 million people in the United States alone and 155 million individuals worldwide (1). Asthma can be described as an epidemic that disproportionately affects children and underserved minorities, and confers a substantial public health burden. Individuals of African descent with asthma have more severe asthma than those of European descent, but studies focusing on both the epidemiology and especially the genetic basis for asthma among this vulnerable ethnic group are limited. Although it is not known to what extent genetic susceptibility contributes to disparities in risk to asthma, the limited linkage and association studies performed in non-European populations have shown evidence of genetic control that differs among ethnic minority groups.
The huge efforts and expense that have been committed to asthma genetics over nearly two decades have already begun to modify the perception of the etiology of asthma and allergic disease. Not surprisingly, a number of target candidates that have been associated with asthma susceptibility are relevant to the mucosa and the bronchial epithelium (i.e., DPP10 [2], SPINK5 [3], GPRA [4]). Some of the earliest efforts in asthma genetics were directed—and efforts continue to be directed—toward candidate genes in pathways associated with treatment (i.e., ADRB2 [5, 6], ALOX5 [7, 8], CRHR1 [9]). As an outcome of what was, in the early 1990s, a relatively novel dichotomization of the cytokine milieu in the context of asthma, Th2 cytokines were a high priority set of priority candidate genes in some of the earliest association studies on asthma (i.e., IL4, IL13), and represent some of the most replicated associations to date (10–12). The biggest surprise is perhaps the preponderance of associations between asthma and polymorphisms in host defense genes, especially those in the pattern-recognition receptor signaling pathway (i.e., CD14 [13–23]). These findings not only suggest that the pathology associated with asthma is more complex than previously believed but also implicate complicated gene-by-environment interactions, as was recently demonstrated in the case of a functional variant in the CD14 gene and the role that domestic endotoxin levels played in conferring asthma risk versus protection (23).
Currently, there are essentially two major shortcomings in our understanding of the genetic epidemiology of asthma in African Americans. First, asthma genetics studies focused on sufficiently powered datasets of families and/or cases and control subjects of African descent are limited; therefore, the role that variants play in conferring risk of asthma among African Americans for most of the priority candidate genes described above (and the many other candidates published to date) is uncertain. Second, there is a plethora of environmental factors, which likely are relevant in gene–environment interactions associated with asthma, but for which exposure could differ according to ethnicity, independent of genetic background. Such differences might result from disparities in socioeconomic status, which in turn might lead to greater cockroach or mouse allergen exposure as a result of infestations in poor, inner-city housing (24–26). A higher proportion of residence in the urban environment can subject disenfranchised groups to more air pollutants, which also contribute to asthma (27). Cultural practices, including environmental tobacco smoke exposure dependent on smoking habits, may also differ (28). This review will focus on what is known to date regarding genetic studies of asthma in populations of African descent, the implications of the findings thus far, and, importantly, considerations for design of genetic epidemiology–based studies of complex traits among African Americans, and interpretation of the findings.
BIOLOGICAL RELEVANCE OF THE SOCIAL CONSTRUCT OF ETHNICITY AND RACE
After much recent debate in the biomedical and scientific literature on whether “race” is a biological or social construct, data emerging from human genome variation research leave little room for argument that race is an imperfect surrogate of biology (for in-depth reviews on this topic, see Nature Genetics supplement, volume 36, 2004), and one that carries with it certain social, cultural, educational, and economic variables that influence the epidemiology of minority health and health disparities as indicated in the data on African Americans with asthma. Consequently, the use of race as a surrogate for biology limits scientists in not only separating and identifying the real environmental and genetic determinants but also in determining the relationship between genome variation and population differences in asthma. More importantly, the social construct of ethnicity and race can and therefore must be used instructively and constructively to interrogate the biology of population-based gene–environment interactions in asthma susceptibility, severity, and response to medications. The reality of race as a social construct can be sanctioned when used beneficially to empower social identity in ways that positively impact disease prevention, health promotion, and the elimination of health disparities.
Consider, for example, the striking racial and ethnic disparities in disease prevalence for many of the common disorders characterized by inflammation and/or altered immunologic responses, including hypertension (29), non–insulin-dependent diabetes mellitus (30), and obesity (for review, see Reference 31). Although ethnic differences in disease incidence and prevalence have traditionally been dismissed as a mix of environmental, social, cultural, or economic factors in etiology, genetic factors cannot be ignored.
LINKAGE DISEQUILIBRIUM AND LESSONS LEARNED FROM THE HAPMAP
In recent years, the “Recent African Origin” model (RAO, or “Out of Africa” hypothesis [32]) has been adopted to explain evolution of modern human origins, purporting that Homo sapiens evolved in Africa some 100,000 to 200,000 years ago (probably from an East African gene pool), after which their anatomically modern members migrated and replaced archaic human groups throughout Europe and Asia. From this, we can estimate that biogeographical variation in phenotypes occurred within the past 100,000 years, and during varied points in time (60,000−3,000 years ago) there have been expansions across Europe, Asia, the Americas, and the Pacific, respectively, which are evident by patterns of allele frequency variation (33). Higher levels of genetic variation exist in the more ancient African populations, and less diversity is present in the younger, non-African populations (34).
Genetic diversity is the net result of naturally occurring recombination events. When an individual inherits a copy of genetic material from each parent during gamete formation, large portions of DNA travel together as a result of recombination that shuffles up chromosomal segments (35). Within these large segments are multiple polymorphic sites (alleles) that are physically linked. Combinations of these neighboring alleles, descended from single, ancestral chromosomes, comprise “haplotypes.” Linkage disequilibrium (LD) is the nonrandom association of these alleles (or haplotypes) at two linked loci. Measures of LD are based on the departure of observed haplotype frequencies from what is expected under random assortment of alleles at different loci (D′), or a function of the correlation coefficient between alleles at different markers (e.g., r2). Over time, our genetic diversity is enhanced because of the gradually eroding ancestral LD, which results in new combinations of alleles. Recombination events do not occur at a uniform rate and, as a result, LD is organized into a blocklike structure interspersed with recombination “hot spots” (36). In addition to naturally occurring recombination events, LD can also occur as a result of mutation, random genetic drift, migration, or selection in response to environmental forces, all of which may result in diverse patterns of LD across biogeographical groups. Natural selection can be a particularly powerful influence on LD patterns and lead to increasing frequency of a particular variant within a given population. A classic example is the LD observed between the FY locus on chromosome 1 and unlinked markers as much as 22 cM apart in African Americans (37); the FY locus confers a strong selective advantage against malaria (reviewed in Reference 38) and the “protective” variant occurs in the majority of African individuals (39). Thus, positive selection has created a block of LD extending far beyond the usual distance of several kilobases among Africans. Genetic epidemiologists can rely on the measure of LD as a useful tool for associating inheritance of a phenotype with genetic markers by asking whether one or more genetic markers occur at a frequency significantly higher or lower in an affected group compared with an unaffected group. If the difference is significant, one can assume that either the marker itself is associated with the trait, or the marker is in LD with the causal variant.
Our knowledge of LD patterns in human populations is rapidly expanding as a direct result of the International HapMap Project. The HapMap project is a collaboration between academic, public, and private institutions from the United States, Japan, China, Nigeria, the United Kingdom, and Canada, in which groups from these sites provide liaisons to populations willing to participate as subjects and provide blood samples, sample collection and storage support, genotyping, funding, data management oversight, and data analysis (40–42). The aim of the project is to gather data on similarities and variations in the human genome and the project is based on 269 DNA samples representing four biogeographical groups (whites from the United States with northern and western European ancestry; Yorubans from Ibadan, Nigeria [YRI]; Han Chinese from Beijing, China; and Japanese from Tokyo, Japan) to help scientists identify variants influencing disease and response to different environmental factors and medication. Genotypes are made publicly available for the purpose of developing new analytic methods and investigating patterns of variation and LD. The project began in 2002, and the latest set of data was released in January 2006. Genotypes on one million SNPs at a spacing of 5 kb or more were made available through the website in phase I of the project, and 5.8 million SNPs in each of the four populations were made available after completion of phase II (October 2006). The latest release provides build-35 chromosomal coordinates for phase I and II data (41, 42).
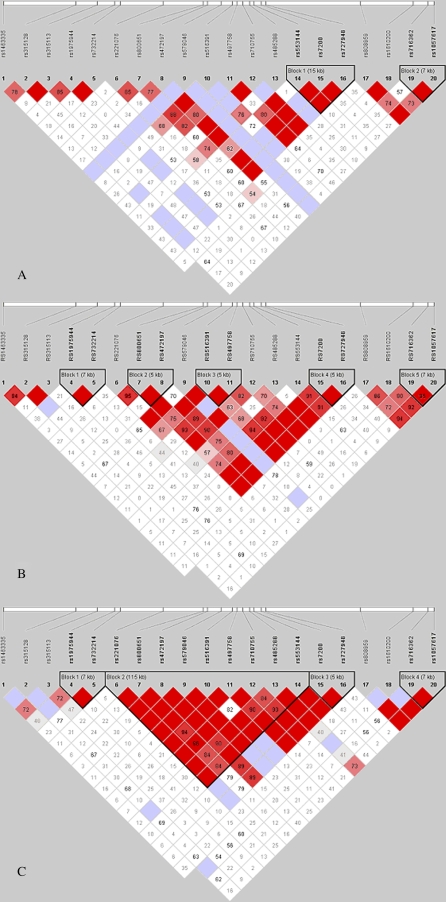
It is anticipated that a direct outcome of the HapMap will be the identification of genetic determinants for the most complex of diseases. The rationale behind this notion is the expectation that as of yet unobserved disease-causing variants will be in significant LD with surrounding markers that can be readily genotyped, and that they will be easily detected using association-based mapping approaches. In the context of studying complex diseases, such as asthma in African Americans, the HapMap has been invaluable. Consider, for example, that just as more ancient populations (e.g., African) will express greater heterogeneity than younger populations at the marker level, so the older populations will have smaller regions of LD, due to more recombination events and more time for alternative selective forces to exert influences at the genetic level. To demonstrate the impact of LD in illustrating our genetic diversity, consider the juxtaposition of LD between an “ancient” population, in this case founders from the YRI population participating in the HapMap project (www.hapmap.org), and an African-Caribbean group of founders from Barbados, characterized by European and West African admixture. The European admixture in Barbados is similar to that observed among African Americans, at around 21% (43), and the population is a result of essentially the same intermix of European colonists with West African slaves several hundred years ago, or about a dozen or so (25-year) generations. In Figure 1, a dense panel of single nucleotide polymorphisms (SNPs) in a large region on chromosome 12 is compared in both groups. In the “younger” Barbados founder population panel, several “blocks” of LD are relatively conserved among founders, compared with decay of LD for the same markers among founders from the YRI population.
Figure 1.
Comparison of pairwise linkage disequilibrium (LD) estimates (D′) in three populations. Pairwise LD estimated between each of 20 single nucleotide polymorphism (SNP) markers genotyped on chromosome 12q (67953342−68500427; [National Center for Biotechnology Information (NCBI) build 35; Illumina panel 42 (Illumina, Inc., San Diego, CA)] using Haploview (Julian Maller, developer, Massachusetts Institute of Technology). Haplotype blocks are shown according to the definition by Gabriel and colleagues (45). A represents the LD between 20 single nucleotide polymorphisms (SNPs) and founders from the Yoruban (YRI) population published by the HapMap project (www.hapmap.org; estimated from 120 chromosomes); B shows the same set of SNPs in an African-Caribbean population from Barbados (estimated from 900 chromosomes); and C shows the same set of SNPs in European Americans from Tangier Island, Virginia (estimated from 354 chromosomes). The 20 SNPs are aligned to the pairwise LD plot. Squares illustrate strong (red), little/no (white), and nonsignificant (blue) LD.
The differences in LD are even more striking, however, when we compare LD between a European-American, 12-generation population originally founded by three unrelated northern Europeans, with an average inbreeding coefficient of 0.018 for inbred individuals (44), and both the African-Caribbean group and the more ancient YRI group. In this case, a very large block of LD is conserved among founders in the relatively young European-American isolate, compared with some decay of LD for the same markers among the founder of African descent in Barbados, and even greater decay among the YRI founders.
Contrary to the assumptions derived from these examples, the HapMap project has demonstrated that LD is in fact broadly similar across populations (45); it is low near telomeres of each chromosome, and higher in duplicated regions of the genome, and within 5 Mb of centromeres (41). Regions of high and low LD when considering the genome in quartiles have a higher density of coding bases, with genes in high-LD regions involved in the cell cycle and genes in low-LD regions involved in the immune response, for example. Evolutionary hypotheses can be invoked to explain these observations. A measure of genetic variance among populations as a fraction of total genetic variance (FST) can be used to identify candidate loci likely to have undergone selection (46), and in the limited study populations to date, functional variation does not appear to be population specific. A study of FST values in the four HapMap populations and in the three populations studied by Perlegen Sciences (African American, European American, and Asian American) showed that values are too variable to be useful in detection of selection (47). New methods are being developed to detect regions of the genome where selection is likely to have had an effect (48).
AFRICAN AMERICANS AND DISPARITIES IN ASTHMA AND ALLERGIC DISEASE: A GENETIC BASIS?
The burden of asthma has increased over the past two decades; however, recent evidence suggests that asthma prevalence and death rates in the general population have been leveling off. Despite this, asthma morbidity and mortality are disproportionately high and continue to increase among African Americans (49–52). In 2002, African Americans had emergency department visit rates of 380%, hospitalization rates of 225%, and mortality rates of over 200% higher than non-Hispanic whites (CDC report, http://www.cdc.gov/nchs/products/pubs/pubd/hestats/asthma/asthma.htm). In addition, asthma appears to vary phenotypically according to ancestry, with African Americans showing different patterns of allergic sensitization, IgE levels, and bronchial hyperresponsiveness, when compared with Americans of European ancestry (52). These striking racial and ethnic disparities in disease characteristics and prevalence for common disorders such as allergic asthma cannot be explained entirely by environmental, social, cultural, or economic factors (53).
Although it is not known to what extent genetic susceptibility contributes to asthma-related disparities, differences according to ethnicity in linkage and association studies between asthma and associated traits and genetic markers have been observed. The NHLBI-funded Collaborative Study on the Genetics of Asthma (CSGA) was one of the first genomewide linkage screens for asthma and included families representing three United States' ethnic groups: European American, African American, and Hispanic American (54). In that study, evidence for linkage to six novel and several previously reported chromosomal regions and asthma was observed; curiously, the best evidence for African Americans (5p15, 17p11.1-q11.2), European Americans (11p15, 19q13), and Hispanics (2q33, 21q21) did not overlap, suggesting distinct genes may be acting in different groups, or that unique gene-by-environment interactions exist.
Another likely explanation is that the genes that confer risk are the same across ethnic groups, but the frequency of the polymorphisms associated with asthma is different according to ethnicity. There are many examples of substantial variation in frequencies of “high risk” variants in candidate genes associated with asthma and atopy according to self-reported ancestry, such as the following: (1) the 237G allele of the β chain of the high-affinity IgE receptor (FCER1B) (55); (2) the −589T allele of IL4 (56); (3) the Ile50 allele of the IL4RA α gene (57); (4) the P46L (c.224C>T) variant in the gene encoding member 1A of tumor necrosis factor receptor superfamily (TNFRSF1A) (58); (5) the −174 G/G genotype in the proinflammatory cytokine interleukin IL6 gene (59); and (6) the −401A allele of RANTES (regulated on activation, normal T-cell expressed and secreted) (60). As a further example, consider the comparisons in Table 1 of allele frequencies in SNPs representing the IL4 gene across four distinct populations: West African, African American, Brazilian African Caribbean, and European American. The biggest difference in IL4 variant allele frequencies is predominantly between the two populations most distinct from each other, the European Americans and Africans. Frequencies of variant alleles in the two African and European admixed populations are mostly between these two extremes, and in several cases most closely approximate frequencies in the African group. Thus, it can be concluded that, whereas IL4 is a strong candidate gene for asthma, differences according to ethnicity in frequencies of the SNPs tested in genetic association studies may lead to different conclusions.
TABLE 1.
ALLELIC FREQUENCIES OF SINGLE NUCLEOTIDE POLYMORPHISMS IN THE IL4 GENE IN FOUR ETHNIC GROUPS (WEST AFRICAN, AFRICAN AMERICAN, BRAZILIAN, EUROPEAN)
| Minor Allelic Frequencies (%)
|
|||||
|---|---|---|---|---|---|
| IL4 Marker | Allele | YRI* | GRAAD† | Brazil‡ | CEU§ |
| rs2070874 | C/T | 46.7 | 44.8 | 39.9 | 15.8 |
| rs2227284 | C/A | 99.2 | 44.4 | 40.6 | 27.1 |
| rs2243267 | G/C | 37.5 | 36.5 | 32.9 | 16.7 |
| rs2243270 | A/G | 77.5 | 37.0 | 37.7 | 15.5 |
| rs2243291 | C/G | 25.8 | 43.0 | 41.2 | 82.8 |
| rs734244 | A/G | 53.3 | 42.5 | 48.0 | 84.2 |
YRI, Yoruba people in Ibadan, Nigeria; allelic frequencies based on founders from 30 parent-and-adult-child trios participating in the International HapMap Project.
GRAAD, the consortium on Genomic Research on Asthma in the African Diaspora; allelic frequencies based on 306 healthy control subjects participating in an asthma genetics study in the Baltimore–Washington, DC, metropolitan area (unpublished data).
Allelic frequencies based on 205 founders participating in a family-based study on susceptibility to asthma and schistosomiasis in the Conde district, Bahia, Brazil (unpublished data).
CEU, European Americans from Utah, from the Centre d'Etude du Polymorphisme Humain collection; allelic frequencies based on founders from 30 parent-and-adult-child trios participating in the International HapMap Project.
UPDATE ON GENETIC STUDIES OF ASTHMA IN POPULATIONS OF AFRICAN DESCENT
Asthma and its associated trait of atopy were perhaps some of the first complex diseases for which a strong genetic basis was established (12, 61). To date, nearly a dozen genomewide screens have been performed on asthma and its associated phenotypes, for which 10 regions have been reproducible. From several of these screens, six novel genes have been identified by positional cloning (2, 4, 62–65). The Genetic Association Database (http://geneticassociationdb.nih.gov), a public archive of published gene-based genetic association studies, currently contains data from over 500 published genetic associations studies on asthma, and continues to grow (66). In one of the most thorough reviews on asthma genetics to date, Ober and Hoffjan (12) examined nearly 500 studies and identified 79 genes that had been associated with asthma or its associated phenotypes in two or more independent populations, 25 of which have been replicated in six or more populations, with an additional 54 genes associated in two to five populations (12).
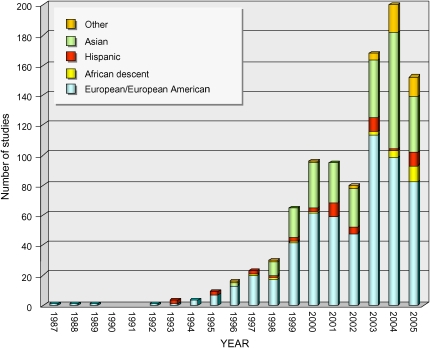
Regretfully, the voluminous data to date on asthma genetics are not representative of the diverse human populations afflicted with asthma, particularly those from groups that suffer disproportionately both in morbidity and mortality. In their review, Ober and Hoffjan (12) also characterized the populations in which these studies were performed and summarized the actual numbers of cases and control subjects used in each study. As illustrated in Figure 2, studies in white populations (i.e., of northern and western European ancestry) constitute the largest number of genetic associations since the earliest reported study (Reference 247 in the Ober and Hoffjan review [12]) through 2005, representing 60% of the associations summarized by Ober and Hoffjan. Reports on Asian populations are the second-most represented, comprising 28% of these studies, but there are only 25 studies (3%) based on populations of African ancestry (African American, African Caribbean, and two populations from continental Africa). In most of these studies, power is in question because the number of cases and/or control subjects is less than 100. Underrepresentation in asthma genetic studies is not limited to African Americans; only 41 studies in the Ober and Hoffjan review are based on Hispanic populations (Puerto Rican, Mexican American, Costa Rican, Venezuelan). This is also troubling, given that one of these groups, Puerto Ricans, reportedly has even higher asthma prevalence, morbidity, and mortality than do African Americans (67, 68).
Figure 2.
Number of published genetic association studies on asthma and associated phenotypes in different ethnic populations (European/European American, African descent, Hispanic, Asian, other) during 1987–2005. Summaries are derived from the data provided in the online supplement by Ober and Hoffjan (12), which include summaries of associations between specific variants in 120 HLA and non-HLA genes, phenotype, and the population studied, and are based on a compilation of 492 references. In studies that included more than one independent population, each population was counted. “African descent” includes African-American and African-Caribbean populations, and populations from continental Africa (Tunisia, South Africa). “Hispanic” includes Puerto Rican, Mexican American, Costa Rican, Mexican, and Venezuelan. “Other” includes ethnically mixed populations, and aboriginal Australian, Indonesian, and Hawaiian populations.
The NHLBI-funded CSGA was the first and only genomewide linkage study to date to include a population of African descent. As described above, there was evidence for linkage of asthma in African-American families to two novel regions: 5p15 (p = 0.0008) and 17p11.1-q11.2 (p = 0.0015) (54). Although novel at the time, linkage to the 17q locus was eventually replicated. Linkage to markers in 17q11.2-q21.2 was initially replicated in an independent population of African descent from Barbados (69), although significant evidence for linkage to 17q was only observed after conditioning on linkage to chromosome 12q markers. Elsewhere, linkage to the same region on 17q was replicated in a large panel of French families (70). Replication of linkage to the 5p locus has been more problematic; significant evidence for linkage to a region downstream of the 5p15 locus was previously observed in a European-American religious isolate for markers at 5p13 (71), and recent findings among both the Hutterites and a German population suggest the presence of at least two to five genes associated with asthma and bronchial hyperreactivity in this broad region (72). Therefore, and despite the many linkage replications to date, the 5p15 locus identified in the CSGA African Americans remains a unique locus. Conversely, the initial linkage observed in the African-Caribbean Barbados population on 12q13-24 has been one of the most replicated loci, predominantly in European populations (54, 70, 73–79), and therefore, like chromosome 17q, would not appear to be a locus unique to populations of African descent.
In a second-stage analysis among a subset of the CSGA African-American families, strong evidence for linkage has been observed at 11q21 (80). Follow-up fine-mapping studies have narrowed the critical linkage region to one containing several interesting candidates, including CD44, EHF, ELF5, and GSTP1 (81, 82). Another candidate gene in this general region is CRTH2 (83); two common SNPs in the 3′ untranslated region showed significant evidence of association, and functional analyses demonstrated that the two-SNP haplotype confers a significantly higher level of reporter mRNA stability when compared with a nontransmitted haplotype. Other studies in populations of African descent include significant associations between markers in innate immunity candidate genes and asthma in a family-based population living in Barbados, where a well-known (CD14) and a relatively unknown (AOAH) innate immunity gene have been shown to be associated with asthma (23, 84). The CD14 (−260) C/T variant has been shown to be negatively associated with asthma and associated phenotypes in a number of European ancestry and Asian populations (13–23), and most recently Hispanics (18); AOAH has not yet been tested. It still remains uncertain at this time whether there are asthma risk loci (polymorphisms) that are uniquely important to populations of African descent.
CONSIDERATIONS IN STUDY DESIGN AND TESTS FOR ASSOCIATION IN POPULATIONS OF AFRICAN DESCENT
SNP Selection in Populations of African Descent
The availability of high-density sets of well-characterized genetic markers for complete chromosomes or large contiguous regions has revolutionized approaches in selecting nonredundant markers, or haplotype-tagging SNPs (htSNPs), to both maximize information provided in dense marker maps and minimize the cost of genotyping. This approach involves genotyping only those SNPs necessary to predict the majority of observed haplotypes. HapMap data can be used to define “bins” of SNPs with all members sharing high pairwise LD (r2 ⩾ 0.8), and instead of selecting all SNPs in a bin for a genotyping project, one representative or tagging SNP can be chosen. A project by Perlegen Sciences (46) aims to capture genetic variation in Americans of European, African, and Asian ancestry, on a set of more than 1.5 million SNPs. An examination of bins from the Perlegen populations across the genome shows that the overall bin size among African Americans is lower than for European Americans. In examining the fraction of common SNPs that are highly redundant with other SNPs, there is a high degree of redundancy for the European-American and Han Chinese populations, and less for the African Americans. At a threshold of r2 = 0.8 for SNPs within bins, 73% common variation is captured for European Americans compared with 71% for African Americans. When selecting one tagging SNP from each bin across the genome (from a total set of ∼ 1 million SNPs), 30% are retained for European Americans, 28% for Han Chinese, and 50% for African Americans (46), once again highlighting the decay of LD in the population for whom the parental ancestry is more ancient, and the practical point that more SNPs need to be genotyped in African Americans than in other groups.
HapMap genotype data from the Utah and YRI populations can be used to select representative tagging SNPs for association studies in populations with African-European admixture. Several algorithms for selecting representative SNPs based on patterns of LD have been proposed, and the utility of five of these methods in terms of ability to retain associations observed in a fine-mapping project (713 SNPs) in an African-Caribbean, family-based population selected for asthma has recently been explored (85). These methods include both structured approaches, which take chromosomal position into account (i.e., Gabriel's and colleagues' block definition [45] and solid spline of LD [SSLD] [86]), and unstructured approaches (i.e., TagSNP [87], htSNP [88, 89], Tagger [90]), which only take correlations between alleles into consideration. It was determined that the greatest degree of information about association while achieving high efficiency is retained in htSNP, TagSNP, and Tagger (< 90% signals at an α of 0.05), with Tagger implemented through the HapMap website.
Figure 3 illustrates results from application of the Tagger algorithm on a set of 20 SNPs from chromosome 12q using an r2 = 0.8 in the same family-based population from Barbados as illustrated in Figure 1, in addition to individuals selected from different nuclear families in an isolated European-American population from Tangier Island, Virginia. In the Tangier population (Figure 3A), there is greater pairwise LD overall compared with founders from Barbados (Figure 3B), as would be expected. Also highlighted is the fact that the number of tagging SNPs selected is different: 12 of 20 (60%) SNPs should be retained in the Tangier sample to capture variation in this chromosomal region, compared with 18 of 20 (80%) SNPs for the population of African descent. SNPs retained are shown in blue for the two populations. In regions with greater local pairwise LD in both populations, a smaller number of tagging SNPs are necessary. This highlights the fact that genetic studies in populations of African descent require more SNPs than a European-American population.
Figure 3.
LD plots and tagging SNPs from a selected segment on chromosome 12q for two populations. Illustration of SNPs selected by the Tagger algorithm (90) on a set of 20 SNPs at r2 = 0.8. A represents the LD between 20 SNPs and European Americans from Tangier Island, Virginia (estimated from 354 chromosomes), and B represents the same set of SNPs in founders from the African-Caribbean population from Barbados (estimated 900 chromosomes). SNPs retained by Tagger are shown with blue marker identifiers for the two populations and include 12 of 20 SNPs for the Tangier sample and 18 of 20 (80%) SNPs for the Barbados sample.
Adjusting for Population Stratification
The unique population history of minorities in the United States, which has been characterized by a mixture of European ancestry with West Africans, indigenous groups (northern and southern Native Americans, as in the case of Mestizos in Central and South America), and a multitude of peoples from other distinct geographical regions, has diminished the role of geographical distance in defining populations according to their biogeographical ancestors. The impact of admixture is an especially important consideration in genetic epidemiologic studies of African Americans, because it is compounded when the disease of interest is more prevalent in a particular group (or groups) within a given population in that any alleles that are more common among the minority group(s) of interest will tend to be associated with the disease, even if completely unlinked to the disease-causing locus.
Various approaches to deal with the problem of stratification are available. One approach is to match the ethnic backgrounds of cases and control subjects, but this is problematic because there still may be considerable cryptic or hidden stratification remaining (91). Alternatively, unlinked genetic markers throughout the genome that show a high difference in allele frequencies across source populations relevant to the admixed population, or “Ancestry Informative Markers” (AIMs), can be typed in the study population to attribute an estimate of proportion of ancestry from the source population samples to each individual in the study population. This information can be used to adjust association test statistics and correct for hidden population stratification in case-control designs using self-reported ethnic membership (92–96). Simulation studies have demonstrated that upward of 100 AIMs are needed for optimal adjustment (97, 98). For example, in a reanalysis of previously published findings based on 35 AIMs (99), Halder and colleagues selected 177 AIMs for maximum information content. Although the original analysis produced significant associations for skin pigmentation and bone mineral density and the disease markers, the second analysis identified a suggestive association for fat-free mass and skin pigmentation, but not bone mineral density (Indrani Halder, Ph.D., Penn State University, personal communication, July 2006).
CUTTING-EDGE APPROACHES AND FUTURE DIRECTIONS
Genomewide Association Studies
Data readily available from the HapMap project, combined with more accurate approaches in selecting tagging markers sufficiently dense to capture most of the common variation in the human genome, have paved the way for the next step in gene hunting: genomewide association (GWA) studies. GWA studies offer a potential solution to the limitation in association studies to date: that each causal variant in each candidate gene will only make a modest contribution to overall heritability. By screening dense panels of markers representing the most common genetic variation across the genome, it is assumed that many more risk variants will be identified than what could be accomplished using conventional candidate gene–based studies. Candidate gene studies, after all, are only as good as the hypothesis on which they are based; if the biological hypothesis is inaccurate (e.g., the proposed pathway from which the candidates are selected is too broad or inaccurate), or if the physiologic defects are unknown, the candidate gene approach will not be successful (100).
The potential drawbacks have been extensively debated in the literature, even though to date there have been only a few published GWA studies. Issues related to multiple comparisons and the potential for multiple type I errors (i.e., false discovery rate) and ideal study designs (i.e., multistage vs. single stage) continue, and are unlikely to be settled until more empiric data are at hand. Conversely, solutions related to cost and feasibility, and the effects of bias due to population stratification, or admixture, are forthcoming. Costs are dropping rapidly and simultaneously; the size of the SNP panels is increasing (and consequential representation across the genome). Adjustments for the effect of admixture in an African-American population under study in a GWA analysis are similar to approaches described above, the main difference being that it is not necessary to genotype a specific panel of AIMs, because one can take advantage of the half million or so SNPs to be genotyped in the GWA panel, and evaluate admixture directly from those. Of particular relevance, newly released panels exceeding 500,000 SNPs have been selected on the basis of adequate average minor allele frequencies for the Centre d'Etude du Polymorphisme Humain (CEPH) project in Utah, Han Chinese of Beijing (CHB), individuals of Japanese ancestry from the Tokyo area (JPT), and YRI HapMap populations, and promise greater efficiency in coverage (i.e., tag SNP selection) than the earlier panels for which tagging SNPs were generated from European-ancestry data.
Admixture Mapping
Family-based linkage mapping has proved useful in interrogating the genome for disease genes in complex traits, especially the rare Mendelian diseases. As illustrated in this review and elsewhere (12), population-based association studies have been more useful in homing in on causative genes for a complex trait such as asthma, and the potential power in GWA studies to detect even more of these genes is described above. A major drawback to the genomewide approach is the expense of genotyping anywhere from 300,000 to 1,000,000 markers. Until very recently, success in mapping genes through GWA in populations of African descent has been hampered by the limited efficiency of haplotype tagging (101), due in part to the overall reduced levels of LD in African populations (33).
An approach that is gaining attention is admixture mapping, also referred to as mapping by admixture linkage disequilibrium (MALD). The approach is predicated on the assumption that when an admixed population is created from previously isolated populations (i.e., Africans and Europeans), and the ancestral population is disproportionately predisposed to a particular disease as a result of differences in risk allele frequencies, it will be possible to identify genomic regions where individuals with the trait of interest will have a higher proportion of ancestry from the parental population more likely to be affected by the trait. In other words, susceptibility genes can be identified by mapping distortions of ancestry (102). For example, a multiple sclerosis locus was recently identified on chromosome 1 where there is an exceptionally high level of European ancestry in a group of African-American cases and control subjects (103). Indeed, the discovery of the Duffy blood group locus was founded on this principal.
Admixture mapping is most successful when the differences in susceptibility allele frequencies and disease prevalence between the two parental populations are large, and when the populations have been recently admixed (104–106). Admixture LD diminishes over time, although the rate between linked markers is much slower than for unlinked markers (107). African Americans and African Caribbeans as a group were formed with the convergence of Europeans and West Africans and the colonial history of North and South America and the West Indies, just a dozen or so generations ago. As a result, the allelic associations in these groups created by admixture extend over large chromosomal distances (averaging 10–20 cM) (107), and contain far fewer genetic markers than GWA studies, as few as several thousand compared with upward of 1 million markers. A limitation until recently has been the availability of admixture panels, but these are increasingly available (108, 109), especially for African Americans, and new panels are on the horizon.
Phylogenetic Studies
Once a causal variant in a gene has been identified, the logical next step is to determine its effective significance. Predictions of the consequence of an amino acid change on protein function are most typically undertaken after significant genetic associations between the marker at the variant and the trait have been identified. Another approach involves phylogenetic analyses, whereby the degree of conservation of the modified amino acid is evaluated with the notion that the most functionally important residues will be the most conserved residues (110). In its simplest form, phylogenetics seeks to determine the rates and patterns of changes in our genome to reconstruct the evolutionary history of genes, and to identify the basis for positive selective pressures that confer functional changes, such as substitutions. Despite the more than 500 genetic associations of asthma to date, relatively few causal variants have been identified and validated. Given the remarkable diversity of the human adaptive immune system and the apparent and critical role that the more ancient and conserved host defense pathways play in the pathogenesis of and susceptibility to asthma, it is likely that studies focused on the evolutionary and selective basis for causal variants in asthma candidate genes will be forthcoming.
CONCLUSIONS
Despite the health disparities among underrepresented minorities for a number of complex diseases, our understanding of the role of the contribution of genetic variation in the mix of environmental, social, cultural, or economic factors involved in asthma is limited, due in large part to how relatively few well-designed and sufficiently powered genetic association studies have been conducted in nonwhite groups, such as African Americans, as illustrated in Figure 2. The striking and consistent differences in SNP frequencies across populations (i.e., Table 1) reflect variation in the human genome, which most typically results from naturally occurring recombination events. Patterns of LD reflect these events, and due in large part to the success of the International HapMap Project, these patterns are readily apparent (Figures 1 and 3). Advances made by the HapMap as well as other, similar endeavors (e.g., Perlegen Sciences) have specifically benefited studies in minority groups such as African Americans, enabling scientists to develop more efficient panels of markers for genetic association studies, and panels for adjustments to the confounding characteristic of admixed populations. Novel approaches including GWA studies and admixture mapping will likewise be dependent on the more efficient characterization of the human genome. It is hoped that these collective advances and insights will lead to a greater understanding of the pathogenetic basis of asthma and its associated traits.
Acknowledgments
The authors thank Pat Oldewurtel and William Shao for technical assistance in the preparation of this manuscript and Dr. H. J. Tsai for helpful comments.
Supported by an Asthma & Allergy Foundation of America New Investigator Award grant (K.C.B.), and NIH grants R01 AI50024, U01 HL72455, and U01 HL66615. K.C.B. was supported in part by the Mary Beryl Patch Turnbull Scholar Program.
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Centers for Disease Control and Prevention. Asthma Prevalence, health care use and mortality, 2000–2001. Atlanta, GA: National Center for Health Statistics; 2003.
- 2.Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, Bhattacharyya S, Tinsley J, Zhang Y, Holt R, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet 2003;35(3):258–263. [DOI] [PubMed] [Google Scholar]
- 3.Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B, Lawrence R, Wong K, Abecasis GR, Jones EY, Harper JI, et al. Gene polymorphism in Netherton and common atopic disease. Nat Genet 2001;29(2):175–178. [DOI] [PubMed] [Google Scholar]
- 4.Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, Makela S, Rehn M, Pirskanen A, Rautanen A, et al. Characterization of a common susceptibility locus for asthma-related traits. Science 2004;304:300–304. [DOI] [PubMed] [Google Scholar]
- 5.Martinez FD, Graves PE, Baldini M, Solomon S, Erickson R. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest 1997;100:3184–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lima JJ, Thomason DB, Mohamed MH, Eberle LV, Self TH, Johnson JA. Impact of genetic polymorphisms of the beta2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin Pharmacol Ther 1999;65:519–525. [DOI] [PubMed] [Google Scholar]
- 7.Drazen JM, Yandava CN, Dubé L, Szczerback N, Hippensteel R, Pillari A, Israel E, Schork N, Silverman ES, Katz DA, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet 1999;22(2):168–170. [DOI] [PubMed] [Google Scholar]
- 8.Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, Wang J, Sylvester J, Holbrook J, Wise R, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med 2006;173:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, Liggett SB, Gelfand EW, Rosenwasser LJ, Richter B, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet 2004;13:1353–1359. [DOI] [PubMed] [Google Scholar]
- 10.Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish LL. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy 1995;25:74–78. [DOI] [PubMed] [Google Scholar]
- 11.Walley AJ, Cookson WO. Investigation of an interleukin-4 promoter polymorphism for associations with asthma and atopy. J Med Genet 1996;33(8):689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun 2006;7:95–100. [DOI] [PubMed] [Google Scholar]
- 13.Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A polymorphism* in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol 1999;20(5):976–983. [DOI] [PubMed] [Google Scholar]
- 14.Gao PS, Mao XQ, Baldini M, Roberts MH, Adra CN, Shirakawa T, Holt PG, Martinez FD, Hopkin JM. Serum total IgE levels and CD14 on chromosome 5q31. Clin Genet 1999;56(2):164–165. [DOI] [PubMed] [Google Scholar]
- 15.Leung TF, Tang NL, Sung YM, Li AM, Wong GW, Chan IH, Lam CW. The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol 2003;14:255–260. [DOI] [PubMed] [Google Scholar]
- 16.Koppelman GH, Reijmerink NE, Colin Stine O, Howard TD, Whittaker PA, Meyers DA, Postma DS, Bleecker ER. Association of a promoter polymorphism of the CD14 gene and atopy. Am J Respir Crit Care Med 2001;163:965–969. [DOI] [PubMed] [Google Scholar]
- 17.Buckova D, Holla LI, Schuller M, Znojil V, Vacha J. Two CD14 promoter polymorphisms and atopic phenotypes in Czech patients with IgE-mediated allergy. Allergy 2003;58:1023–1026. [DOI] [PubMed] [Google Scholar]
- 18.Choudhry S, Avila PC, Nazario S, Ung N, Kho J, Rodriguez-Santana JR, Casal J, Tsai HJ, Torres A, Ziv E, et al. CD14 tobacco gene-environment interaction modifies asthma severity and immunoglobulin E levels in Latinos with asthma. Am J Respir Crit Care Med 2005; 172:173–182. [DOI] [PubMed] [Google Scholar]
- 19.Sackesen C, Karaaslan C, Keskin O, Tokol N, Tahan F, Civelek E, Soyer OU, Adalioglu G, Tuncer A, Birben E, et al. The effect of polymorphisms at the CD14 promoter and the TLR4 gene on asthma phenotypes in Turkish children with asthma. Allergy 2005;60:1485–1492. [DOI] [PubMed] [Google Scholar]
- 20.Woo JG, Assa'ad A, Heizer AB, Bernstein JA, Hershey GK. The –159 C→T polymorphism of CD14 is associated with nonatopic asthma and food allergy. J Allergy Clin Immunol 2003;112(2):438–444. [DOI] [PubMed] [Google Scholar]
- 21.Sharma M, Batra J, Mabalirajan U, Goswami S, Ganguly D, Lahkar B, Bhatia NK, Kumar A, Ghosh B. Suggestive evidence of association of C-159T functional polymorphism of the CD14 gene with atopic asthma in northern and northwestern Indian populations. Immunogenetics 2004;56:544–547. [DOI] [PubMed] [Google Scholar]
- 22.Martin AC, Laing IA, Khoo SK, Zhang G, Rueter K, Teoh L, Taheri S, Hayden CM, Geelhoed GC, Goldblatt J, Lesouef PN. Acute asthma in children: relationships between CD14 and CC16 genotypes, plasma levels and severity. Am J Respir Crit Care Med 2006;173:617–622. [DOI] [PubMed] [Google Scholar]
- 23.Zambelli-Weiner A, Ehrlich A, Stockton ML, Grant AV, Zhang S, Levett PN, Beaty TH, Barnes KC. Evaluation of the CD14/–260 polymorphism and house dust endotoxin exposure in the Barbados asthma genetics study. J Allergy Clin Immunol 2005;115:1203–1209. [DOI] [PubMed] [Google Scholar]
- 24.Rosenstreich DL, Eggleston P, Kattan M, Baker D, Slavin RG, Gergen P, Mitchell H, McNiff-Mortimer K, Lynn H, Ownby D, et al. The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthmam. N Engl J Med 1997;336:1356–1363. [DOI] [PubMed] [Google Scholar]
- 25.Eggleston PA, Rosenstreich D, Lynn H, Gergen P, Baker D, Kattan M, Mortimer KM, Mitchell H, Ownby D, Slavin R, et al. Relationship of indoor allergen exposure to skin test sensitivity in inner-city children with asthma. J Allergy Clin Immunol 1998;102(4 Pt 1):563–570. [DOI] [PubMed] [Google Scholar]
- 26.Matsui EC, Simons E, Rand C, Butz A, Buckley TJ, Breysse P, Eggleston PA. Airborne mouse allergen in the homes of inner-city children with asthma. J Allergy Clin Immunol 2005;115:358–363. [DOI] [PubMed] [Google Scholar]
- 27.Breysse PN, Buckley TJ, Williams D, Beck CM, Jo SJ, Merriman B, Kanchanaraksa S, Swartz LJ, Callahan KA, Butz AM, et al. Indoor exposures to air pollutants and allergens in the homes of asthmatic children in inner-city Baltimore. Environ Res 2005;98:167–176. [DOI] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. Cigarette smoking among adults: United States. MMWR Morb Mortal Wkly Rep 1993;42: 230–233. [PubMed] [Google Scholar]
- 29.Jamerson KA. The disproportionate impact of hypertensive cardiovascular disease in African Americans: getting to the heart of the issue. J Clin Hypertens (Greenwich) 2004;6(4, Suppl 1): 4–10. [DOI] [PMC free article] [PubMed]
- 30.Cowie CC, Harris MI, Silverman RE, Johnson EW, Rust KF. Effect of multiple risk factors on differences between blacks and whites in the prevalence of non-insulin-dependent diabetes mellitus in the United States. Am J Epidemiol 1993;137:719–732. [DOI] [PubMed] [Google Scholar]
- 31.Cossrow N, Falkner B. Race/ethnic issues in obesity and obesity-related comorbidities. J Clin Endocrinol Metab 2004;89:2590–2594. [DOI] [PubMed] [Google Scholar]
- 32.Stringer CB, Andrews P. Genetic and fossil evidence for the origin of modern humans. Science 1988;239:1263–1264. [DOI] [PubMed] [Google Scholar]
- 33.Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton, NJ: Princeton University Press; 1994.
- 34.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet 2003;4:293–340. [DOI] [PubMed] [Google Scholar]
- 35.Bertranpetit J, Calafell F, Comas D, Gonzalez-Neira A, Navarro A. Structure of linkage disequilibrium in humans: genome factors and population stratification. Cold Spring Harb Symp Quant Biol 2003; 68:79–88. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein DB. Islands of linkage disequilibrium. Nat Genet 2001;29:109–111. [DOI] [PubMed] [Google Scholar]
- 37.Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol 2001;114(1):18–29. [DOI] [PubMed] [Google Scholar]
- 38.Hadley TJ, Peiper SC. From malaria to the chemokine receptor: the emerging physiological role of the Duffy blood group antigen. Blood 1997;89:3077–3091. [PubMed] [Google Scholar]
- 39.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 1995;10:224–228. [DOI] [PubMed] [Google Scholar]
- 40.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 41.International HapMap Consortium. A haplotype map of the human genome. Nature 2005;437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project web site. Genome Res 2005;15:1592–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickel RG, Willadsen SA, Freidhoff LR, Huang S-K, Caraballo L, Naidu RP, Levett PN, Bumenthal M, Banks-Schlegel S, Bleecker E, et al. Determination of Duffy-genotypes in three populations of African descent using PCR and sequence specific oligonucleotides. Hum Immunol 1999;60:738–742. [DOI] [PubMed] [Google Scholar]
- 44.Mathias RA, Bickel CA, Beaty TH, Petersen GM, Hetmanski JB, Liang K-Y, Barnes KC. A genealogical analysis of the Tangier Island, Virginia, population. J Phys Anthropol 2000;112:29–38. [DOI] [PubMed] [Google Scholar]
- 45.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–2229. [DOI] [PubMed] [Google Scholar]
- 46.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science 2005;307:1072–1079. [DOI] [PubMed] [Google Scholar]
- 47.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res 2005;15:1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res 2005;15:1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans R. Recent observations reflecting increases in mortality from asthma. J Allergy Clin Immunol 1987;80:377–379. [DOI] [PubMed] [Google Scholar]
- 50.Wissow LS, Gittelsohn AM, Szyko M, Starfield B, Mussman M. Poverty, race, and hospitalization for childhood asthma. Am J Public Health 1988;78:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss KB, Wagener DK. Changing patterns of asthma mortality: identifying target populations at high risk. JAMA 1990;264(13):1683–1687. [PubMed] [Google Scholar]
- 52.Joseph CL, Williams LK, Ownby DR, Saltzgaber J, Johnson CC. Applying epidemiologic concepts of primary, secondary, and tertiary prevention to the elimination of racial disparities in asthma. J Allergy Clin Immunol 2006;117:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins FS. What we do and don't know about “race,” “ethnicity,” genetics and health at the dawn of the genome era. Nat Genet 2004; 36(11 Suppl):S13–S15. [DOI] [PubMed] [Google Scholar]
- 54.The Collaborative Study on the Genetics of Asthma. A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 1997;15:389–392. [DOI] [PubMed] [Google Scholar]
- 55.Unkelbach K, Gardemann A, Kostrzewa M, Philipp M, Tillmanns H, Haberbosch W. A new promoter polymorphism in the gene of lipopolysaccharide receptor CD14 is associated with expired myocardial infarction in patients with low atherosclerotic risk profile. Arterioscler Thromb Vasc Biol 1999;19(4):932–938. [DOI] [PubMed] [Google Scholar]
- 56.Hubacek JA, Stuber F, Frohlich D, Book M, Wetegrove S, Rothe G, Schmitz G. The common functional C(-159)T polymorphism within the promoter region of the lipopolysaccharide receptor CD14 is not associated with sepsis development or mortality. Genes Immun 2000;1(6):405–407. [DOI] [PubMed] [Google Scholar]
- 57.Caggana M, Walker K, Reilly AA, Conroy JM, Duva S, Walsh AC. Population-based studies reveal differences in the allelic frequencies of two functionally significant human interleukin-4 receptor polymorphisms in several ethnic groups. Genet Med 1999;1(6):267–271. [DOI] [PubMed] [Google Scholar]
- 58.Tchernitchko D, Chiminqgi M, Galacteros F, Prehu C, Segbena Y, Coulibaly H, Rebaya N, Loric S. Unexpected high frequency of P46L TNFRSF1A allele in sub-Saharan West African populations. Eur J Hum Genet 2005;13:513–515. [DOI] [PubMed] [Google Scholar]
- 59.Ness RB, Haggerty CL, Harger G, Ferrell R. Differential distribution of allelic variants in cytokine genes among African Americans and White Americans. Am J Epidemiol 2004;160:1033–1038. [DOI] [PubMed] [Google Scholar]
- 60.Nickel R, Casolaro V, Wahn U, Beyer K, Barnes KC, Plunkett B, Freidhoff LR, Sengler C, Plitt J, Schleimer RP, et al. Atopic dermatitis is associated with a functional mutation in the promoter of the CC chemokine RANTES. J Immunol 2000;164:1612–1616. [DOI] [PubMed] [Google Scholar]
- 61.Barnes KC. Genetics and epidemiology. Curr Opin Allergy Clin Immunol 2001;1:383–385. [DOI] [PubMed] [Google Scholar]
- 62.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, Kuldanek S, Donfack J, Kogut P, Patel NM, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet 2005;76:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noguchi E, Yokouchi Y, Zhang J, Shibuya K, Shibuya A, Bannai M, Tokunaga K, Doi H, Tamari M, Shimizu M, et al. Positional identification of an asthma susceptibility gene on human chromosome 5q33. Am J Respir Crit Care Med 2005;172:183–188. [DOI] [PubMed] [Google Scholar]
- 64.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 2002;418(6896):426–430. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, Edser P, Bhattacharyya S, Dunham A, Adcock IM, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet 2003;34(2):181–186. [DOI] [PubMed] [Google Scholar]
- 66.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet 2004;36(5):431–432. [DOI] [PubMed] [Google Scholar]
- 67.Carter-Pokras OD, Gergen PJ. Reported asthma among Puerto Rican, Mexican-American, and Cuban children, 1982 through 1984. Am J Public Health 1993;83:580–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Homa DM, Mannino DM, Lara M. Asthma Mortality in U.S. Hispanics of Mexican, Puerto Rican, and Cuban heritage, 1990–1995. Am J Respir Crit Care Med 2000;161(2):504–509. [DOI] [PubMed] [Google Scholar]
- 69.Barnes KC, Nickel R, Mathias RA, Freidhoff LR, Casolaro V, Stockton ML, Xue X, Ehrlich E, Naidu RP, Levett PN, et al. Interaction between loci on chromosomes 12q and 17q increases susceptibility to elevated total IgE in two distinct populations. J Allergy Clin Immunol 2000;104(1, Pt. 2):S370. [Google Scholar]
- 70.Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, Annesi-Maesano I, Boussaha M, Bousquet J, Charpin D, Degioanni A, Gormand F, Grimfeld A, et al. Genome screen for asthma and related phenotypes in the French EGEA study. Am J Respir Crit Care Med 2000;162: 1812–1818. [DOI] [PubMed] [Google Scholar]
- 71.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet 2000;67(5):1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurz T, Hoffjan S, Hayes MG, Schneider D, Nicolae R, Heinzmann A, Jerkic SP, Parry R, Cox NJ, Deichmann KA, et al. Fine mapping and positional candidate studies on chromosome 5p13 identify multiple asthma susceptibility loci. J Allergy Clin Immunol 2006;118:396–402. [DOI] [PubMed] [Google Scholar]
- 73.Nickel R, Wahn U, Hizawa N, Maestri N, Duffy DL, Barnes KC, Beyer K, Forster J, Bergmann R, Zepp F, et al. Evidence for linkage of chromosome 12q15-q24.1 markers to high total serum IgE concentrations in children of the German Multicenter Allergy Study (MAS '90). Genomics 1997;46:159–162. [DOI] [PubMed] [Google Scholar]
- 74.Wilkinson J, Grimley S, Collins A, Thomas NS, Holgate ST, Morton N. Linkage of asthma to markers on chromosome 12 in a sample of 240 families using quantitative phenotype scores. Genomics 1998; 53(3):251–259. [DOI] [PubMed] [Google Scholar]
- 75.Ober C, Cox NJ, Abney M, Di Rienzo A, Lander ES, Changyaleket B, Gidley H, Kurtz B, Lee J, Nance M, et al. Genome-wide search for asthma susceptibility loci in a founder population. The Collaborative Study on the Genetics of Asthma. Human Molecular Genetics 1998; 7(9):1393–1398. [DOI] [PubMed]
- 76.Wjst M, Fischer G, Immervoll T, Jung M, Saar K, Rueschendorf F, Reis A, Ulbrecht M, Gomolka M, Weiss EH, et al. A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics 1999;58(1):1–18. [DOI] [PubMed] [Google Scholar]
- 77.Malerba G, Lauciello MC, Scherpbier T, Trabetti E, Galavotti R, Cusin V, Pescollderungg L, Zanoni G, Martinati LC, Boner AL, et al. Linkage analysis of chromosome 12 markers in italian families with atopic asthmatic children. Am J Respir Crit Care Med 2000;162(4 Pt 1):1587–1590. [DOI] [PubMed] [Google Scholar]
- 78.Xu J, Bleecker ER, Jongepier H, Howard TD, Koppelman GH, Postma DS, Meyers DA. Major recessive gene(s) with considerable residual polygenic effect regulating adult height: confirmation of genomewide scan results for chromosomes 6, 9, and 12. Am J Hum Genet 2002; 71:646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shao C, Suzuki Y, Kamada F, Kanno K, Tamari M, Hasegawa K, Aoki Y, Kure S, Yang X, Endo H, et al. Linkage and association of childhood asthma with the chromosome 12 genes. J Hum Genet 2004;49:115–122. [DOI] [PubMed] [Google Scholar]
- 80.Xu X, Fang Z, Wang B, Chen C, Guang W, Jin Y, Yang J, Lewitzky S, Aelony A, Parker A, et al. A genomewide search for quantitative-trait loci underlying asthma. Am J Hum Genet 2001;69(6):1271–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang SK, Mathias RA, Ehrlich E, Plunkett B, Liu X, Cutting GR, Wang XJ, Li XD, Togias A, Barnes KC, et al. Evidence for asthma susceptibility genes on chromosome 11 in an African-American population. Hum Genet 2003;113:71–75. [DOI] [PubMed] [Google Scholar]
- 82.Mathias RA, Gao P, Goldstein JL, Wilson AF, Pugh EW, Furbert-Harris P, Dunston GM, Malveaux FJ, Togias A, Barnes KC, et al. A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet 2006;7:38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang JL, Gao PS, Mathias RA, Yao TC, Chen LC, Kuo ML, Hsu SC, Plunkett B, Togias A, Barnes KC, et al. Sequence variants of the gene encoding chemoattractant receptor expressed on Th2 cells (CRTH2) are associated with asthma and differentially influence mRNA stability. Hum Mol Genet 2004;13:2691–2697. [DOI] [PubMed] [Google Scholar]
- 84.Barnes KC, Grant A, Gao P, Baltadjieva D, Berg T, Chi P, Zhang S, Zambelli-Weiner A, Ehrlich E, Zardkoohi O, et al. Polymorphisms in the novel gene acyloxyacyl hydroxylase (AOAH) are associated with asthma and associated phenotypes. J Allergy Clin Immunol 2006;118:70–77. [DOI] [PubMed] [Google Scholar]
- 85.Chi PB, Duggal P, Kao WH, Mathias RA, Grant AV, Stockton ML, Garcia JG, Ingersoll RG, Scott AF, Beaty TH, et al. Comparison of SNP tagging methods using empirical data: association study of 713 SNPs on chromosome 12q14.3–12q24.21 for asthma and total serum IgE in an African Caribbean population. Genet Epidemiol 2006;30: 609–619. [DOI] [PubMed] [Google Scholar]
- 86.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 87.Stram DO, Haiman CA, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Pike MC. Choosing haplotype-tagging SNPs based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the multiethnic cohort study. Hum Hered 2002;55:27–36. [DOI] [PubMed] [Google Scholar]
- 88.Clayton D. Choosing a set of haplotype tagging SNPs from a larger set of diallelic loci. Available from: www.nature.com/ng/journal/v29/n2/extref/ng1001-233-S10.pdf. 2001 (Accessed July 12, 2006).
- 89.Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet 2005;37:1243–1246. [DOI] [PubMed] [Google Scholar]
- 90.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005;37:1217–1223. [DOI] [PubMed] [Google Scholar]
- 91.Ewens WJ, Spielman RS. The transmission/disequilibrium test: history, subdivision, and admixture. Am J Hum Genet 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- 92.Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999;55(4):997–1004. [DOI] [PubMed] [Google Scholar]
- 93.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet 2000;64(2):171–186. [DOI] [PubMed] [Google Scholar]
- 94.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000;155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol 2001; 20(1):4–16. [DOI] [PubMed] [Google Scholar]
- 96.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet 2003;72:1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol 2005;28: 289–301. [DOI] [PubMed] [Google Scholar]
- 98.Tsai HJ, Choudhry S, Naqvi M, Rodriguez-Cintron W, Burchard EG, Ziv E. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum Genet 2005;118:424–433. [DOI] [PubMed] [Google Scholar]
- 99.Bonilla C, Shriver MD, Parra EJ, Jones A, Fernandez JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York City. Human Genetics 2004;115(1):57–68. [DOI] [PubMed] [Google Scholar]
- 100.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005;6:95–108. [DOI] [PubMed] [Google Scholar]
- 101.Ke X, Durrant C, Morris AP, Hunt S, Bentley DR, Deloukas P, Cardon LR. Efficiency and consistency of haplotype tagging of dense SNP maps in multiple samples. Hum Mol Genet 2004;13:2557–2565. [DOI] [PubMed] [Google Scholar]
- 102.Sawcer S. A new era in the genetic analysis of multiple sclerosis. Curr Opin Neurol 2006;19:237–241. [DOI] [PubMed] [Google Scholar]
- 103.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet 2005;37:1113–1118. [DOI] [PubMed] [Google Scholar]
- 104.McKeigue PM. Prospects for admixture mapping of complex traits. Am J Hum Genet 2005;76:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reich D, Patterson N. Will admixture mapping work to find disease genes? Philos Trans R Soc Lond B Biol Sci 2005;360:1605–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet 2005;6:623–632. [DOI] [PubMed] [Google Scholar]
- 107.Halder I, Shriver MD. Measuring and using admixture to study the genetics of complex diseases. Hum Genom 2003;1:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet 2004;74(5):1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, Schurr TG, Zhadanov SI, Osipova LP, Brutsaert TD, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genom 2005;2:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Santibanez Koref MF, Gangeswaran R, Santibanez Koref IP, Shanahan N, Hancock JM. A phylogenetic approach to assessing the significance of missense mutations in disease genes. Hum Mutat 2003;22:51–58. [DOI] [PubMed] [Google Scholar]