Abstract
The integrated effect of multiple pathways, molecules, genetic polymorphisms, environmental stimuli, and possible infection determines the lung phenotype in idiopathic pulmonary fibrosis (IPF), a chronic progressive and often lethal lung disease. Systems biology approaches aim to provide a systemwide view of biological process using computational tools and high-throughput technologies. Although much of the analysis of genome-level transcriptional high-resolution profiles of IPF was reductionist, usually focusing on a single factor in the disease process, there are some studies that implement systems approaches. We discuss these analyses and provide examples of the global analysis of IPF, hypersensitivity pneumonitis, and nonspecific interstitial pneumonia. Detailed quantitative phenotyping and correlation with microarray results as well as high-throughput genotyping should provide us with the datasets to implement systems biology approaches in fibrosis research. Interdisciplinary teams and training of junior investigators in the vocabulary of systems biology should allow us to use these datasets integratively and generate a global model of human pulmonary fibrosis.
Keywords: Cancer Biomedical Informatics Grid, caBIG; idiopathic pulmonary fibrosis; interstitial lung disease; microarrays
Idiopathic pulmonary fibrosis (IPF) is a progressive and relatively poorly understood fibrotic lung disease whose median survival (2.5–3 yr) is unaffected by currently available medical therapies (1). In the last two decades, we have experienced an unprecedented increase in our understanding of lung fibrosis in general. Studies using advanced molecular biology approaches, genetically modified animals, virally administered genes, and high-throughput transcriptional profiling approaches provided evidence for multiple pathways, molecules, and systems that may be involved in fibrosis. On the basis of these studies, it seems that pulmonary fibrosis, at least the form induced by bleomycin in the mouse lung, is in part dependent on intact tumor necrosis factor (TNF) pathways (2), intact transforming growth factor (TGF)-β activation and signaling pathways (3), angiogenesis, cell trafficking and recruitment (4), coagulation cascades (5), apoptosis (6), lipid mediator metabolisms (7), and expression of multiple regulatory molecules by the alveolar epithelium (8). We have data to support the role of alveolar epithelial cells, myofibroblasts, circulating mesenchymal pleuripotent cells, T cells, macrophages, and endothelial cells in lung fibrosis (8–10). The success of these studies has presented us with a biological Rashômon. Like the viewers of the film by the legendary Japanese filmmaker Akira Kurosawa (11), which provides multiple versions to a crime observed by four witnesses, we observe multiple, mismatched, often conflicting versions of the same event that leave us wondering whether we could resolve the picture, or more importantly in this case, can we really understand human pulmonary fibrosis in a way that will allow us to significantly impact the disease?
The answer, we believe, lies in several major developments that happened in the last decade. The availability of complete sequences of the human and other genomes and the introduction of high-throughput technologies for gene and protein expression provide at least part of the answer. It is relatively easy to invert the experimental design now and, instead of identifying a molecule in a mouse model of disease, one can profile or mine publicly available transcriptional profiles of human disease, identify a gene or protein of interest using genetically modified animals or other methods for gene knockdown, and study its function. In addition, instead of studying the expected phenotypic outcomes of experiments that manipulate the expression or function of a gene of interest in animal model of disease, we can now look at the global impact of these perturbations. The advent of high-throughput methodologies to study genetic background variability, epigenetic regulation, and transcription factor–based gene expression regulation should allow us to provide an additional part of the answer by explaining how individual variability explains disease susceptibility in mice and humans. Furthermore, these technologies allow us to query how biological information gets dynamically translated into context- and cell-specific gene and protein expression patterns, which, in turn, serve to change the cellular context. The rapid increase in computing power and connectivity translates into more rapid and efficient data manipulation and sharing; this releases us from the need to query multiple articles to generate a hypothesis because much of the data we require to “connect and project” are now readily available in relatively easy-to-use and widely accessible databases. Indeed, within a relatively brief period of time, the use of high-throughput techniques for gene expression profiling and of computerized databases has become a mainstay of biomedical research.
Although all of these exciting technological advances that exponentially increase the levels of knowledge about every disease and model serve as facilitators of integration, they do not inherently provide integrative models of disease. For this to occur, a shift in thinking is required. Instead of a single-factor/reductionist approach, which is highly effective in the lab, we need to think “globally.” We need to shift from an approach that tries to explain lung fibrosis using “one molecule at a time, one cell type at a time” to an approach that looks at the network of interactions between multiple molecules, pathways, and cells, and characteristics of the organism, as they converge to determine the lung phenotype in pulmonary fibrosis. Systems biology is the field of biology that aims to provide this “holistic” view. In this review, we will discuss and define systems biology and its application as well as relevance to the study of IPF. We will provide a brief description of microarray studies of IPF (recently reviewed by us [12]) as well as examples of systems biology analysis of these data. We will also describe the requirements and challenges in implementing systems biology to IPF research.
SYSTEMS BIOLOGY
Systems biology is the field that addresses the need to shift from a component-based reductionist view of biology to a systemwide perspective. It can be described as a global quantitative analysis of how all components in a biological system interact to determine its phenotype. Although the definitions may vary, systems biology can usually be characterized as interdisciplinary, iterative, computationally intensive, and information greedy. An explanation of these terms is beneficial to set the stage for the remainder of this discussion:
Interdisciplinary: Although it is sometimes possible to answer a reductionist question by a collaboration of scientists from the same discipline, a global view requires a multidisciplinary team. Scientists from multiple quantitative disciplines (mathematics, computer science, statistics), medical specialties (clinicians, radiologists, epidemiologists), and life sciences disciplines (molecular and cell biologists, geneticists, chemists, physiologists) need to work together for a real systemwide model to be developed.
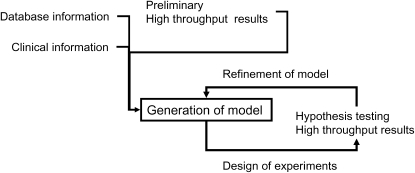
Iterative: An intriguing aspect of systems approaches is the iterative nature of the process, in which experiments are designed to provide information required for modeling; based on these experiments, a model is created and tested by further experiments. The results of these experiments are in turn used to refine the model (Figure 1).
Computationally intensive: The integration of multiple levels of information, multiple datasets from high-throughput technologies (25,000 genes on hundreds of patients, millions of single nucleotide polymorphisms), the use of advanced mathematical and computational tools, and the iterative nature of the analysis require high-powered and parallel computing.
Information greedy: In contrast to reductionist approaches, where only recognized, directly relevant information is used, in systems biology all information is relevant; therefore, the use of high-throughput technologies for profiling genotypes and phenotypes is critically important, as it may be the combination of several elements that best describe the phenotype. Also important is the use of digitalizable methods for clinical phenotyping. The integrative nature of such experiments requires rapid access to multiple levels of information from public databases that summarize the results of millions of reductionist experiments into pathways, gene annotations, protein interactions, and so forth.
Figure 1.
The iterative nature of systems biology research. Note that every refinement of the model needs additional data generation for retesting of specific hypotheses.
In a recent review, Alan Aderem (13) described three concepts that are also worth introducing in any discussion of systems biology, as follows:
Emergence: The most basic principle in systems biology. The popular equivalent of this concept is “the whole is bigger than the sum of its parts.” In systems biology, it means that biological systems will express emergent characteristics that cannot be predicted from only knowing their parts. As an example, the presence of collagen I and III and matrix metalloproteases (MMPs) in the lung does not necessarily result in fibrosis; it is the combination of multiple factors and their deregulation that leads to the abnormal phenotype.
Robustness: This concept describes how stable the system is in response to environmental stresses and genetic variation. This includes understanding of internal regulatory loops and positive and negative feedback mechanisms. In general, negative feedback loops increase system robustness and positive feedback loops reduce it. It is worth considering that similar organ phenotypes may differ in their robustness. For example, bleomycin-induced fibrosis in the mouse is relatively easily preventable and most often resolves spontaneously, whereas the pulmonary fibrosis in IPF is persistent despite active repair mechanisms and may be stable for long periods of time, until a shift happens and the system becomes unstable (14). Understanding the system's robustness and the factors that increase its sensitivity may be important in design of therapeutic interventions.
Modularity: This means that complex biological systems are organized by functional modules, where multiple components are coregulated by a process and, when activated, lead to a similar outcome. A module can be a set of genes located in a similar genomic region that are activated by the same transcription factors, or a set of chemokines that are bound to a similar glycoprotein; an increase in the peptidase that degrades this protein will cause predictable response through all of the associated molecules. The benefit of modular analysis is that it reduces dependence on the relative variability in levels of single molecules. Modularity often helps resolving conflicting pieces of information by allowing for spatial, temporal, and contextual organization of the information.
HOW ARE MICROARRAYS RELEVANT TO SYSTEMS BIOLOGY?
Microarrays allow for the simultaneous profiling of the complete genome. By their nature, they provide global views of the mRNAs expressed in a tissue or a cell. Therefore, the data generated by microarray experiments are highly amenable to analyses that implement systems biology approaches. Although, in general, microarray data do not provide us information regarding the robustness of the system, they do provide us information regarding the emergent characteristics of the system studied and much about its modularity. In a recent review, we contrasted two approaches to analysis of microarray experiments: the reductionist, or “cherry-picking,” approach and the global “systems” approach. Generally, most microarray studies belong to the former, partly because reductionist approaches are still easier to complete, and most often because reviewers are more familiar and comfortable with articles and grants that describe reductionist approaches, thus making such studies easier to publish. Unfortunately, this is also the case with funding. Naturally, the full extent of the information available in a microarray experiment is used only by applying a systems approach to the data. Such an approach does not try to identify the single most interesting gene but instead tries to understand the general themes, characteristics, and functional elements that are present in the data. A critical element here is the “information greediness” of the process. In the systems approach, every piece of information is obtained, downloaded, and used to create gene attribute files that allow functional characterization of the biological question at hand. As previously described by us (15), it is not only the biological information that is relevant but also all the additional information about the experiments, from demographics, to dates of experiments, and every other characteristic. In addition to revealing the systematic characteristics of IPF lungs, and their regulatory mechanisms, the wealth of information may provide unexpected observations and confounders that may not be uncovered in any other way. Although it is obvious that protein–protein interactions and post-translational modifications determine many phenotypic manifestations, the footprint of such events may also be evident in transcriptional data and provide additional insights into regulatory mechanisms,
MICROARRAY ANALYSIS OF PULMONARY FIBROSIS
We have recently reviewed most of the work that implemented microarrays in lung fibrosis (12, 16); these included studies of animal models of fibrosis, analysis of human lungs, and analysis of cells obtained from lungs of patients with the disease. These articles and analytic approaches are summarized in Table 1. A brief synopsis of these areas follows.
TABLE 1.
ARTICLES USING MICROARRAYS IN PULMONARY FIBROSIS
| First Author | Organism | Main Follow-up | Approach | Reference |
|---|---|---|---|---|
| Kaminski | Mouse | RT-PCR, in silico | Combined | 18 |
| Katsuma | Mouse | RT-PCR | Combined | 39 |
| Zuo | Human | Knockout mice | Reductionist | 26 |
| Du | Mouse | Transgenic mice | Reductionist | 19 |
| Liu | Rat | Protein and mRNA validation | Reductionist | 17 |
| Functional in vitro | ||||
| Cosgrove | Human | Immunohistochemistry | Reductionist | 27 |
| Functional in vitro | ||||
| Lemay | Mouse | RT-PCR | Combined | 22 |
| Haston | Mouse | RT-PCR | Combined | 21 |
| Pardo | Human | Immunohistochemistry and protein levels in BAL | Reductionist | 30 |
| Functional in vitro | ||||
| Selman | Human | Immunohistochemistry | Combined | 31 |
| In silico |
Definition of abbreviations: BAL = bronchoalveolar lavage; RT-PCR = reverse transcriptase–polymerase chain reaction.
Microarray Analysis of Animal Models of Disease
We have previously described the earliest articles that applied microarrays in bleomycin-induced fibrosis, including the work of Liu and colleagues, which analyzed lung gene expression profiles after bleomycin exposure in the rat and identified FIZZ1 as a potential regulator of fibrosis (12, 16, 17). Interestingly, much of the work using arrays in mouse lung fibrosis focused on comparing gene expression patterns in mice susceptible and resistant to fibrosis. In our first microarray article (18), we applied a clustering-based approach to obtain global insights about bleomycin-induced fibrosis using mice homozygous for a null mutation of the integrin β6 subunit gene (β6−/−), which develop inflammation but not fibrosis in response to bleomycin, and wild-type mice. Our analysis allowed us to dissect the transcriptional programs associated with inflammation or fibrosis in bleomycin-induced lung injury. Similarly, Du and coworkers (19) compared gene expression patterns of a fibrosis-susceptible C3.SW-H2b strain that differs from the fibrosis-resistant C3H strain by only 6 to 8 cM within the major histocompatibility (MHC) region on chromosome 17, previously suggested to be associated with susceptibility to bleomycin-induced lung fibrosis (20). They identified 70 differentially expressed genes and chose to focus on H2-EA, which encodes the α chain of MHC class II antigen E. This antigen is one of two class II antigens that are expressed on the cell surface of antigen-presenting cells, including macrophages, B cells, and dendritic cells. They then demonstrated that the fibrosis susceptibility was associated with loss-of-function deletion of 600 bp in the promoter and first exon of the H2-Ea gene. H2-EA transgenics had a better survival in response to bleomycin, suggesting that it does confer protection from bleomycin-induced lung injury and fibrosis. Following the same vein, Haston and colleagues (21) used microarrays to identify genes differentially expressed between resistant and susceptible strains. Sixty-seven of the genes mapped to the previously mapped bleomycin susceptibility loci Blmpf1 and Blmpf2 on chromosomes 17 and 11 (20). Applying a similar approach, they also compared resistant (A/J) and susceptible (C57BL/6J) strains and tried to distinguish a list of fibrosis susceptibility genes (22). Although such approaches have the potential to identify sets of genes and genomic regions associated with fibrosis, it is not completely obvious that susceptibility to bleomycin is indeed the best criterion, and it is not completely clear from these experiments whether these mouse strains exhibit a resistance to bleomycin-induced acute lung injury or to fibrosis. Interestingly, searching the Gene Expression Omnibus (GEO), we found at least two more publicly available but unpublished datasets (GSE452-453 and GSE485) that compare genetically susceptible and resistant mouse strains.
Microarray Analysis of Cells Isolated from the Lung
Considering that multiple groups have isolated primary fibroblast lung cell lines, it is quite surprising that there are only a few articles that have been published. One possibility is that the global differences between IPF and normal lung fibroblasts are not dramatic, therefore requiring additional experiments. Choi and colleagues (23) found increased expression of CCL7 in fibroblasts isolated from lungs of patients with IPF, nonspecific interstitial pneumonia, and respiratory bronchiolitis–interstitial lung disease compared with controls without interstitial lung disease using inflammatory gene expression arrays. Renzoni and colleagues analyzed gene expression patterns in normal control fibroblasts and in IPF fibroblasts using oligonucleotide arrays (24) but did not find any dramatic differences between IPF and normal lung fibroblasts, and therefore focused on the effects of TGF-β. Although this observation may sound disappointing, it highlights one of the strengths of global observations: the ability to determine that populations are relatively similar, as was the case with their fibroblast cell lines (24).
Microarray Analysis of Whole Human IPF Lungs
Microarrays have been implemented in analysis of IPF lungs and have led to the identification of MMP7 as a potential regulator in IPF (25, 26). These articles have previously been reviewed by us (16). We (26) also provided a qualitative global analysis and identified genes encoding for proteins expressed in smooth muscle differentiation and muscle contractile machinery, genes that encoded extracellular matrix proteins and a coordinated increase in the levels of several MMPs (MMP1, MMP2, MMP7, and MMP9); however, this analysis was intuitive and quantitative. In a recent review (12), we used the data from the previous article to look for enrichment of Gene Ontology (GO) annotations in the genes up-regulated in IPF. We used NIH DAVID, a Web-based tool that provides a comprehensive set of functional annotation tools allowing exploration of biological meaning in large lists of genes (http://david.abcc.ncifcrf.gov/). IPF lungs were characterized by an increased catabolic state, which was also associated with an increase in glucose metabolism, potentially reflecting the active remodeling that occurs in IPF lungs. Analysis of cellular localization revealed enrichment for genes associated with protein biosynthesis and extracellular matrix, and enrichment in molecular functions reflected by enrichment for both structural constituents and genes with endopeptidase activity; the functional themes contained some unexpected groups like antigen binding and hydrogen ion transporter activity. This simple analysis of the first human IPF dataset demonstrates the power of the global approach and its discovery potential.
Cosgrove and colleagues (27) identified pigment epithelium-derived factor (PEDF), a protein with angiostatic properties in IPF lungs, using microarrays, and we focused on osteopontin, a phosphoprotein previously shown to be required for development of bleomycin-induced fibrosis in murine lungs (28, 29), which was highly up-regulated in IPF lungs (30). Recently, we compared gene expression patterns among samples from patients with IPF, hypersensitivity pneumonitis (HP), and nonspecific interstitial pneumonitis (NSIP) (31). We identified multiple genes that distinguished IPF from HP. We used NIH DAVID to look for enrichment of GO annotations in genes overexpressed in IPF and genes overexpressed in HP. The GO analysis was highly successful: genes that characterized HP were enriched with multiple inflammatory annotations, including T-cell activation, immune response, and defense response, whereas the genes that characterize IPF were enriched with ectoderm development, metalloendopeptidase activity, and extracellular matrix. This functional gene expression signature fit well the clinical observation that patients with IPF rarely respond to antiinflammatory treatment (1). It also suggested that we might be able to identify functional modules that will guide therapy.
Encouraged by these results, and especially for this article, we reanalyzed the data using Genomica (http://genomica.weizmann.ac.il/index.html), a more advanced analysis and visualization tool, which evolved from the GeneXpress and allows integration of multiple levels of information (15, 32) in analysis of high-throughput data. For each gene, we generated attribute files that included its GO annotation and its involvement in known pathways. For pathway information, we used several resources (reviewed in Reference 33), including Kyoto's Encyclopedia of Genes and Genomes (KEGG; http://www.genome. jp/kegg/) for pathways, GENMapp pathways (http://www. genmapp.org/), and others, as well literature information. One of the advantages of Genomica is the simplicity by which information files can be generated (32). We then looked for enrichment of these annotations in the genes that distinguished IPF, HP, and NSIP (normal controls were not included in this analysis). Figure 2 provides the functional map of these interstitial lung diseases. As expected from our previous article and analysis, IPF is dramatically functionally different from HP. Although HP is characterized by cytokine activation, T-cell activation, humoral immune response, and multiple other inflammatory annotations, IPF is characterized by cell adhesion, extracellular matrix, smooth muscle differentiations, and genes associated with lung development, heparin binding, enzyme inhibitor activity, and insulin growth factor binding (Figure 2). Although it was very difficult to find individual genes that distinguished NSIP from IPF or HP (partly because the small number of NSIP samples limited the significance of the results), the global analysis revealed that, compared with IPF, NSIP tissues were enriched with genes associated with cellular defense response and IPF tissues were enriched with genes associated with cellular regulation of cell cycle (Figure 2). Although preliminary, this analysis, taken together with our previous data, suggests that using global approaches will enable us to generate mechanistic hypotheses and modeling of relevant information.
Figure 2.
Functional analysis of genes that distinguish idiopathic pulmonary fibrosis (IPF), nonspecific interstitial pneumonia (NSIP), and hypersensitivity pneumonitis (HP). Filled boxes = enriched annotation. All annotations were statistically significant at a false discovery rate of 5%.
TOWARD SYSTEMS BIOLOGY OF PULMONARY FIBROSIS
The need for systems biology approaches in IPF is obvious. The complexity and multitude of pathways, cells, biological processes, and molecular families involved in generating the lung phenotype in the disease demand an integrative approach. Although our examples suggest that microarrays provide data that can be used for systems analyses, it is also obvious that to generate integrative models of pulmonary fibrosis, additional layers of information and education are needed:
Transcriptional Profiles
It is obvious that many more data are required. Cross-sectional profiles do not provide the dynamic information required for modeling; dynamic profiles, in which gene expression patterns change in time or after intervention, are required for modeling. For humans, this would translate into obtaining samples at multiple stages of the disease, because patients who undergo two lung biopsies are very rare. Unfortunately, most IPF tissues are obtained relatively late in the disease and when the disease is stable. To address progressive and exacerbating disease, we have initiated a warm autopsy program in our institute that allows us to harvest lungs for research from patients who die of IPF (34). This program should allow us to identify the dynamic processes that characterize acute exacerbations of IPF. We also try to capture all lung explants and biopsies.
An additional and critical component required for modeling is intrapulmonary transcriptional profiling. We have recently completed analysis of epithelial cells adjacent to fibrotic regions and in normal regions. Transcriptional profiles of lung microenvironments are critical to understanding of the transcriptional networks within the lung and will be valuable for the modeling experiments. Limitations of human samples can somewhat be mitigated by using data from animal models of disease. Multiple models of fibrosis (e.g., asbestos, adenoviral TGF-β), detailed time courses, and multiple strains and genetically modified animals need to be profiled. Naturally, all of these data should be widely available and this should be a request before funding or publication.
High-Throughput Haplotype Maps of Patients with IPF
Recently, Lawson and colleagues reviewed the genetics of familial and sporadic IPF (35). Although some polymorphisms are associated with sporadic IPF, it is clear that the relationship between the genetic background and the disease may be complex. It is possible, for instance, that a set of modifier genes will be associated with disease progression without being causative for the disease. In our view, to implement a systems approach, it is necessary to investigate the global genetic background of different populations with IPF and analyze the data in conjunction with results of high-throughput gene expression data. In addition, systematic and targeted genotyping of disease-relevant pathways could also provide valuable information.
Detailed and Digital Phenotyping Efforts
This is the place to stress that genomic and high-throughput profiles are only part of the modeling effort. The other critical component is the clinical phenotyping of the patient and the relationship among the molecular phenotype (gene expression profile, proteomic profile), the global genetic background, and the patient's clinical characteristics. One of the advantages of pulmonary medicine and research is that many of the phenotypic characteristics of patients are easily digitalizable. Patients have physiologic evaluations and the imaging systems are highly amenable to quantitative analyses (36). In fact, many consider physiology as the original systems biology because it always contained mathematical representation of biological functions; however, despite this natural alliance, only a few physiologists engage in systems biology. A strong effort is required to add physiologists to multidisciplinary teams to enrich the models with physiologic parameters (37). Efforts like the NIH-sponsored Lung Tissue Resource Consortium, in which in addition to careful collection of samples, a detailed phenotype that includes physiologic parameters, a symptoms questionnaire, and a high-resolution computed tomography scan is collected for every patient and made available to the scientific community, may facilitate the implementation of systems biology approaches in pulmonary research.
Information Sharing
Our knowledge and understanding of the basic mechanism underlying lung fibrosis has greatly increased and will continue to increase in the near future. A major challenge is the development of standards for meaningful information sharing among investigators. For example, even the most basic model, intratracheal bleomycin, is performed and interpreted differently by different groups, making sharing of detailed quantitative information regarding relative strain resistance to bleomycin nearly impossible. To address and integrate information, we require a framework for sharing of information generated by any kind of experiment (clinical, molecular biological, genomic, in silico). The model for such a framework was created in 2003 by the National Cancer Institute and is called the Cancer Biomedical Informatics Grid (caBIG; https://cabig.nci.nih.gov/) (38). This is a voluntary grid of individuals and institutions that collaborate to enable sharing of data and tools in cancer research. Among the resources generated are caArray, a tool for sharing and publication of microarray data; caMOD, an open-source data management system for development and sharing of data of animal models; caDSR, a cancer data standards repository; aCMAP, a cancer molecular analysis tool; and CRIX, a clinical research information exchange that implements a common, secure standards-based electronic infrastructure to support the sharing of clinical research data. Of particular interest to pulmonary researchers is the Lung Imaging Database Consortium at the National Cancer Imaging Archive. This database contains lung images from low-dose helical computed tomography scans of adults screened for lung cancer. It is expected that, by generating standards and tools for data sharing using these standards, caBIG will transform information sharing, collaboration, and, most important, data integration. The availability of caBIG greatly reduces the need for generation of new tools for the pulmonary fibrosis community. In fact, these open-source tools can relatively easily be translated to address the need of the pulmonary research community and create a pulBIG (pulmonary bioinformatics grid). The availability of such tools and resources will greatly facilitate the implementation of systems biology approaches in pulmonary research.
A Cadre of Investigators That Can Lead and Participate in Systems Biology Research
Systems biology offers the perfect marriage between clinical and basic science—the real translation. However, this marriage does require unique training. The vocabulary of systems biology is different than that of molecular biology or clinical research. The collaborative effort is more interdisciplinary and the analytic systems are still evolving. However, the biggest challenge is the shift in thinking. Traditionally, the molecular biology instruction of trainees that enter the lab is highly reductionist. They are trained to deconstruct systems, to develop specific hypotheses, and to devotedly follow them up. Although this training is invaluable and critical for scientific thinking, an additional layer to scientific education is required: a layer that encourages integrative thinking, generates expertise in diverse information management and retrieval, and promotes familiarity with computational and genomic vocabularies and resources. It is important to mention that currently there is only a small number of pulmonary investigators who could serve as mentors in systems biology. Therefore, creating the multidisciplinary infrastructure through design of specialized training programs for translational systems biology of lung fibrosis is required. Such programs will immerse fellows and students in systems biology approaches early on in their career and provide them with the tools and skills to become leaders in this exciting field.
CONCLUSIONS
In this review, we provided a brief introduction to systems biology and made the case why it is critical for our ability to translate our bench understanding of pulmonary fibrosis to the benefit of our patients with IPF. We described microarray experiments relevant to IPF and how they related to systems biology and provided an example of how we can globally understand interstitial lung disease. The addition of multiple datasets and the collection of digital patient information should provide us with the information required to implement systems biology approaches in IPF. Interdisciplinary teams and young trainees well versed in computational vocabularies should enhance our ability to use these datasets to develop a predictive model of pulmonary fibrosis.
Supported by NIH grant HL073745 (N.K.) and by a generous donation from the Simmons family.
Conflict of Interest Statement: S.M.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. N.K. received $5,000 for serving on a Biogen IDEC advisory board in November 2005. He is also a recipient of an investigator-initiated grant from Biogen IDEC in August 2006 ($674,800 for 2 yr).
References
- 1.Selman M, Thannickal VJ, Pardo A, Zisman DA, Martinez FJ, Lynch JP. Idiopathic pulmonary fibrosis: pathogenesis and therapeutic approaches. Drugs 2004;64:405–430. [DOI] [PubMed] [Google Scholar]
- 2.Kelly M, Kolb M, Bonniaud P, Gauldie J. Re-evaluation of fibrogenic cytokines in lung fibrosis. Curr Pharm Des 2003;9:39–49. [DOI] [PubMed] [Google Scholar]
- 3.Sheppard D. Transforming growth factor β: a central modulator of pulmonary and airway inflammation and fibrosis. Proc Am Thorac Soc 2006;3:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agostini C, Gurrieri C. Chemokine/cytokine cocktail in idiopathic pulmonary fibrosis. Proc Am Thorac Soc 2006;3:357–363. [DOI] [PubMed] [Google Scholar]
- 5.Chambers RC. Role of coagulation cascade proteases in lung repair and fibrosis. Eur Respir J Suppl 2003;44:33s–35s. [DOI] [PubMed] [Google Scholar]
- 6.Thannickal VJ, Horowitz JC. Evolving concepts of apoptosis in idiopathic pulmonary fibrosis. Proc Am Thorac Soc 2006;3:350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charbeneau RP, Peters-Golden M. Eicosanoids: mediators and therapeutic targets in fibrotic lung disease. Clin Sci (Lond) 2005;108:479–491. [DOI] [PubMed] [Google Scholar]
- 8.Selman M, Pardo A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc Am Thorac Soc 2006;3:364–372. [DOI] [PubMed] [Google Scholar]
- 9.Huaux F, Liu T, McGarry B, Ullenbruch M, Xing Z, Phan SH. Eosinophils and T lymphocytes possess distinct roles in bleomycin-induced lung injury and fibrosis. J Immunol 2003;171:5470–5481. [DOI] [PubMed] [Google Scholar]
- 10.Lama VN, Phan SH. The extrapulmonary origin of fibroblasts: stem/progenitor cells and beyond. Proc Am Thorac Soc 2006;3:373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurosawa A. Rashômon. Japan: Janus Films; 1950.
- 12.Kaminski N, Rosas IO. Gene expression profiling as a window into idiopathic pulmonary fibrosis pathogenesis: can we identify the right target genes? Proc Am Thorac Soc 2006;3:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aderem A. Systems biology: its practice and challenges. Cell 2005;121: 511–513. [DOI] [PubMed] [Google Scholar]
- 14.Martinez FJ, Safrin S, Weycker D, Starko KM, Bradford WZ, King TE Jr, Flaherty KR, Schwartz DA, Noble PW, Raghu G, et al. The clinical course of patients with idiopathic pulmonary fibrosis. Ann Intern Med 2005;142:963–967. [DOI] [PubMed] [Google Scholar]
- 15.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat Genet 2005; 37:S38–S45. [DOI] [PubMed] [Google Scholar]
- 16.Kaminski N. Microarray analysis of idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 2003;29:S32–S36. [PubMed] [Google Scholar]
- 17.Liu T, Dhanasekaran SM, Jin H, Hu B, Tomlins SA, Chinnaiyan AM, Phan SH. FIZZ1 stimulation of myofibroblast differentiation. Am J Pathol 2004;164:1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski N, Allard JD, Pittet JF, Zuo F, Griffiths MJ, Morris D, Huang X, Sheppard D, Heller RA. Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis. Proc Natl Acad Sci USA 2000;97:1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du M, Irani RA, Stivers DN, Lee SJ, Travis EL. H2-Ea deficiency is a risk factor for bleomycin-induced lung fibrosis in mice. Cancer Res 2004;64:6835–6839. [DOI] [PubMed] [Google Scholar]
- 20.Haston CK, Wang M, Dejournett RE, Zhou X, Ni D, Gu X, King TM, Weil MM, Newman RA, Amos CI, et al. Bleomycin hydrolase and a genetic locus within the MHC affect risk for pulmonary fibrosis in mice. Hum Mol Genet 2002;11:1855–1863. [DOI] [PubMed] [Google Scholar]
- 21.Haston CK, Tomko TG, Godin N, Kerckhoff L, Hallett MT. Murine candidate bleomycin induced pulmonary fibrosis susceptibility genes identified by gene expression and sequence analysis of linkage regions. J Med Genet 2005;42:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemay AM, Haston CK. Bleomycin-induced pulmonary fibrosis susceptibility genes in AcB/BcA recombinant congenic mice. Physiol Genomics 2005;23:54–61. [DOI] [PubMed] [Google Scholar]
- 23.Choi ES, Jakubzick C, Carpenter KJ, Kunkel SL, Evanoff H, Martinez FJ, Flaherty KR, Toews GB, Colby TV, Kazerooni EA, et al. Enhanced monocyte chemoattractant protein-3/CC chemokine ligand-7 in usual interstitial pneumonia. Am J Respir Crit Care Med 2004;170:508–515. [DOI] [PubMed] [Google Scholar]
- 24.Renzoni EA, Abraham DJ, Howat S, Shi-Wen X, Sestini P, Bou-Gharios G, Wells AU, Veeraraghavan S, Nicholson AG, Denton CP, et al. Gene expression profiling reveals novel TGFbeta targets in adult lung fibroblasts. Respir Res 2004;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgrove GP, Schwarz MI, Geraci MW, Brown KK, Worthen GS. Overexpression of matrix metalloproteinase-7 in pulmonary fibrosis. Chest 2002;121:25S–26S. [PubMed] [Google Scholar]
- 26.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben-Dor A, Lollini L, Morris D, Kim Y, DeLustro B, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci USA 2002;99:6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosgrove GP, Brown KK, Schiemann WP, Serls AE, Parr JE, Geraci MW, Schwarz MI, Cool CD, Worthen GS. Pigment epithelium-derived factor in idiopathic pulmonary fibrosis: a role in aberrant angiogenesis. Am J Respir Crit Care Med 2004;170:242–251. [DOI] [PubMed] [Google Scholar]
- 28.Berman JS, Serlin D, Li X, Whitley G, Hayes J, Rishikof DC, Ricupero DA, Liaw L, Goetschkes M, O'Regan AW. Altered bleomycin-induced lung fibrosis in osteopontin-deficient mice. Am J Physiol Lung Cell Mol Physiol 2004;286:L1311–L1318. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi T, Ebihara Y, Manabe A, Tsuji K, Nakamura T, Nakahata T, Iwamoto A. Surfactant protein D and KL-6 as serologic indicators of Pneumocystis carinii pneumonia in a child with acute lymphoblastic leukemia. J Med 2001;32:41–51. [PubMed] [Google Scholar]
- 30.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med 2006;173:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dave NB, Kaminski N. Analysis of microarray experiments for pulmonary fibrosis. Methods Mol Med 2005;117:333–358. [DOI] [PubMed] [Google Scholar]
- 33.Cary MP, Bader GD, Sander C. Pathway information for systems biology. FEBS Lett 2005;579:1815–1820. [DOI] [PubMed] [Google Scholar]
- 34.Lindell KO, Erlen JA, Kaminski N. Lessons from our patients: development of a warm autopsy program. PLoS Med 2006;3:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawson WE, Loyd JE. The genetic approach in pulmonary fibrosis: can it provide clues to this complex disease? Proc Am Thorac Soc 2006; 3:345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez AT, Coxson HO, Hogg JC, Gibson K, Thompson PF, Rogers RM. Use of CT morphometry to detect changes in lung weight and gas volume. Chest 2005;128(4):2471–2477. [DOI] [PubMed] [Google Scholar]
- 37.Strange K. The end of “naive reductionism”: rise of systems biology or renaissance of physiology? Am J Physiol Cell Physiol 2005;288:C968–C974. [DOI] [PubMed] [Google Scholar]
- 38.Saltz J, Oster S, Hastings S, Langella S, Kurc T, Sanchez W, Kher M, Manisundaram A, Shanbhag K, Covitz P. caGrid: design and implementation of the core architecture of the cancer Biomedical Informatics Grid. Bioinformatics 2006;22:1910–1916. Epub 2006 Jun 9. [DOI] [PubMed]
- 39.Katsuma S, Nishi K, Tanigawara K, Ikawa H, Shiojima S, Takagaki K, Kaminishi Y, Suzuki Y, Hirasawa A, Ohgi T, et al. Molecular monitoring of bleomycin-induced pulmonary fibrosis by cDNA microarray-based gene expression profiling. Biochem Biophys Res Commun 2001;288:747–751. [DOI] [PubMed] [Google Scholar]