Abstract
Proteomics is a rapidly developing field and it opens new horizons in many research areas of life sciences. In the field of medicine, proteomics promises to accelerate the discovery of new drug targets and protein disease markers useful for in vitro diagnosis. In this article, we review the current proteomics technologies for biomarker discovery and validation, which include two-dimensional gel electrophoresis, one- and two-dimensional liquid chromatography, and proteomic microarrays. We will also review proteomic strategies for protein–protein interactions and identification of post-translational modifications. Selection of the more effective technology or combination of technologies is required to maximize the interpretation and utility of the data.
Keywords: 2D electrophoresis, multiple dimensional chromatography, post-translational modification, proteomic microarrays, proteomics
The proteome is the temporal cell or tissue-specific protein complement of the genome, encompassing all proteins expressed at any given time, including various protein isoforms and their co- and post-translational modified forms (1–4). Proteomics is an emerging scientific field that involves the identification, characterization, and quantification of proteins in whole cells, tissues, or body fluids (1). Protein characterization, in an ideal situation, includes amino acid sequence analysis, post-translational modifications (PTMs), splice variants, and the identification of its binding partners and cellular localization (1).
The underlying reason for proteomic investigations is that proteins are often expressed in quantities and physical forms that can not be predicted from DNA and mRNA analysis. Thus expression analysis directly at the protein level is necessary to unravel the critical proteome changes that occur as part of disease pathogenesis. The proteome ultimately dictates the function of the cell, and therefore dictates phenotype. The proteome undergoes dynamic changes as it continuously responds to autocrine, paracrine, and endocrine factors, bloodborne mediators, temperature, drug treatment, and developing disease over time. This complex interplay results in a highly variable proteome, dynamically reflecting protein production, co- and post-translational modification, degradation, and secretion. Proteomic strategies are attracting increasing interest for identification of tissue markers and for providing data for analysis. In addition, proteomics gives the unique opportunity to develop blood-based biomarkers to be used for diagnosis, prognosis, and therapy modulation (5).
Proteomics is driven by the state-of-the-art analytical and biochemical technologies and it opens new horizons in many research areas of life sciences, particularly in the field of medicine. Clinical proteomics has been defined as application of proteomics specifically to the field of medicine, which promises to accelerate the discovery of new drug targets and protein disease markers useful for in vitro diagnosis (6). Useful biomarkers predict the extent and duration of organ damage, anticipate clinical outcomes, and evaluate the usefulness of therapeutic strategies. In theory, any protein change or protein modification that is tightly associated with a disease state has the potential to be a biomarker. The minimum criteria used to determine whether a particular protein is a potential biomarker (7–9) are listed in Table 1. Sample sources for biomarker could be tissues, body fluids, or cells.
TABLE 1.
MINIMUM CRITERIA OF A BIOMARKER
| 1. | Is specific to (or closely associated with) organ and/or to disease process |
| 2. | Can segregate the different clinical entities |
| 3. | Is not present (or increased) in diseases sporadically associated with milder forms of the disease |
| 4. | Preferably not present in healthy individuals regardless of age (novel expression with disease process with little or no biological variation [age]) |
| 5. | Is present in sufficient concentration in blood for reliable measurement |
| 6. | Is stable in blood (serum or plasma) and with storage over time |
Typically there are two strategies for biomarker discovery: tissue to serum or serum directly. In the first strategy, biomarkers are discovered in tissue and validated in serum. In the second strategy, biomarkers are discovered and validated in serum or plasma directly. The development of proteomics biomarkers should proceed in a systematic way and follow the phases of discovery and validation.
This review will discuss current proteomics technologies that are used for biomarker discovery and validation, and will also cover protein–protein interactions, and identification of PTMs, which are closely associated with biomarker and drug discovery.
PROTEOMICS PROFILING AND DISCOVERY OF BIOMARKERS
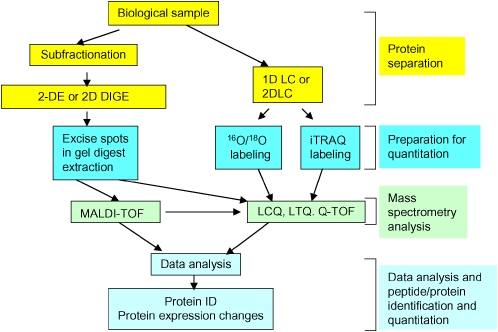
The first approach for proteomics-based biomarkers is global proteomics profiling. Owing to its diversity and complexity, the proteome cannot be resolved completely by a single technology. Proteomic studies have demonstrated that the most effective proteomic analysis of even a simple biological system requires combinations of protein separation and identification techniques. Listed in Table 2 are the major proteomics tools for quantitative analysis, and listed in Table 3 are proteomics tools for characterization (or nonquantitative) analysis. The use of combinations of complement technologies allows us to analyze a large spectrum of the proteome. However, the choice of which technologies to use must be driven by underlying clinical or biological questions. For an example of such a strategy see Figure 1.
TABLE 2.
PROTEOMIC TOOLS FOR QUANTITATIVE ANALYSIS
| Method* | Discovery or Validation | Peptide or Protein | Quantitative | Characterization (isoform + PTM) | Limitations | Cost |
|---|---|---|---|---|---|---|
| One-dimensional gel electrophoresis | Discovery | Protein | Very limited† (densitometry) | Very limited§ | Very limited resolution; limited PTM and isoform determination | Inexpensive |
| Two-dimensional gel electrophoresis | Discovery | Protein | Yes (densitometry/flourescence) | Yes | Limited solubility with loss of high and low Mw and hydrophobic proteins, complex image analysis | Reasonable |
| One-dimensional liquid chromatography (RP, SEC, CXC or AXC) | Discovery | Protein | Very limited† (absorbance or MS-based) | Limited§ | Solubility dependent extraction, complex protein mixtures per fraction, complex data analysis | Reasonable |
| Two-dimensional liquid chromatography | Discovery | Protein | Limited†‡ (absorbance or MS-based) | Yes | Solubility dependent extraction, complex protein mixtures per fraction, complex data analysis | Expensive |
| iTRAQ MS-based (or ICAT MS-based) | Discovery | Peptide | Yes (mole ratio) | No | Limited proteome coverage unless coupled with protein LC prefractionation | Expensive |
| Heavy atom labeling (SILAC) | Discovery | Peptide or protein | Yes (mole ratio) | No | Need to be coupled with peptide or protein separation | Expensive |
| 16O/18O labeling | Discovery | Peptide | Yes (mole ratio) | No | Need to be coupled to protein separation | Expensive |
| Antibody multiplex assays | Validation | Protein (limited with peptide) | Yes | No¶ | Detection by a secondary antibody increases specificity, Detection by MS limits quantification but allows potential detection of PTM | Expensive |
| ELISA (RIA) | Validation | Protein | Yes | No¶ | assessment of single protein | Inexpensive |
Definition of abbreviations: AXC, anion exchange chromatography; CXC, cation exchange chromatography; ELISA, enzyme-linked immunosorbent assay; iTRAQ, isobaric tagging for relative and absolute quantitation; LC, liquid chromatography; MS, mass spectrometry; PTM, post-translational modifications; RIA, radioimmune assay; RP, reversed-phase; SEC, size exclusion chromatography; SILAC, stable isotope labeling by amino acids in cell culture.
Common proteomic methods in practice today; however, other specialized methods have not been included.
Very limited due to complexity of protein/peptide per fraction (co-elution or co-migration); however, can be coupled to downstream MS-based quantitative method.
Limited, but if monitored at 210 nm or 280 nm can be semiquantitative.
Can detect PTM on one parameter that is used for the protein separation.
PTM can be detected if specific antibodies are used.
TABLE 3.
PROTEOMIC TOOLS FOR CHARACTERIZATION ANALYSIS
| Method* | Discovery or Validation | Peptide or Protein | Quantitative | Characterization (isoform + PTM) | Limitations | Cost |
|---|---|---|---|---|---|---|
| One-dimensional liquid chromatography (RP, SEC, CXC or AXC) | Discovery | Peptide | No† (absorbance or MS-based) | No | Solubility-dependent extraction, complex protein mixtures perfraction, complex data analysis | Expensive |
| IMAC MS | Discovery | Peptide | No | Phosphorylation | Normally needs to be coupled to protein separation method and LC/MS/MS | Expensive |
| Glycopeptide capture | Discovery | Protein | No | N-linked carbohydrates | Requires LC/MS/MS and appropriate software for analysis | Expensive |
| Antibody-based purification | Discovery | Protein | No | Specific to antibody | Nonspecific binding of antibody, incomplete binding of all possible proteins, normally needs additional protein separation | Expensive |
| Reversed arrays | Discovery/validation | Protein | No | Limited | Assessment of single protein per assay, potential denaturation of antigen, nonspecific binding | Reasonable |
Definition of abbreviations: AXC, anion exchange chromatography; CXC, cation exchange chromatography; IMAC, immobilized metal affinity chromatography; LC, liquid chromatography; MS, mass spectrometry; PTM, post-translational modifications; RP, reversed-phase; SEC, size exclusion chromatography.
Common proteomic methods, however, other specialized methods have not been included.
Very limited due to complexity of protein/peptide per fraction (co-elution or co-migration), however, can be coupled to downstream MS-based quantitative method.
Figure 1.
Schematic diagram of proteomic profiling analysis. The proteins are either separated using two-dimensional gel electrophoresis (2-DE) or one-/two-dimensional liquid chromatography (1D LC or 2D LC). The resulting spots are digested and identified by mass spectrometry (MS) in the 2-DE approach, and the quantitation data are obtained by gel image analysis. In the 2D LC approach, the resulting fractions are digested and labeled with isotopic reagents followed by MS analysis. The MS data are used for protein identification and quantitation.
Sample Preparation and Subcellular Fractionation
The technical difficulty in identifying and characterizing proteins is proportional to the complexity of the sample being analyzed. Essentially, there are two approaches to overcome this complexity: isolation of subproteomes (that reflects functional units such as organelles), or isolation of classes of proteins with similar chemical property (e.g., phosphorylation). The “best” subproteome preparations will have minimal contamination from other subproteomes or organelles, preserve any PTM induced during the biological intervention while not introducing any artificial PTM during processing, and be reproducible. Such subproteomes include mitochondria, membrane fractions, ER, and golgi, as well as individual protein networks. An example of a specific subproteome protocol that was developed for proteomics is IN-Sequence, which separates the cardiac muscle proteome into cytoplasmic proteins and myofilament/contractile proteins (10–13).
Protein Separation Methods
There are two approaches for proteome analysis: intact protein separation and peptide separation. Protein separation methods include one- and two-dimensional gel electrophoresis (1-DE and 2-DE), one- and two-dimensional liquid chromatography (1D LC and 2D LC), and affinity chromatography for selective isolation of a target protein or protein complex. Peptide separation is more limited and includes multidimensional liquid chromatography and selective enrichment of a subset of peptides which is highly representative of the parent proteins.
Two-dimensional gel electrophoresis.
A cornerstone of proteomic analysis and protein separation remains 2-DE. Two-dimensional gel electrophoresis separates intact proteins in the first dimension based on intrinsic pI (isoelectric focusing [IEF]), and in the second dimension by molecular weight (MW or mass). Two-dimensional gel electrophoresis is one of only a few methods that are able to routinely detect PTMs of proteins even in complex mixtures. However, 2-DE is limited by the solubility and mass of the proteins. Therefore, considerable effort is needed to optimize technical issues involved in maximizing protein solubilization at each stage of 2-DE (14, 15). This includes the optimization of sample solubilization, loading and running conditions for IEF which require testing various combinations of detergents, optimizing the ramping and duration of IEF (14, 15), and specialized methods to improve incorporation of high and low mass proteins (16) and basic proteins (17).
Gel image analysis.
Advanced gel image analysis is necessary to compare multiple gels for accurate protein quantification. This quantification can be accomplished using sophisticated comparison algorithms in software programs. Unfortunately, due to variation between gels, no two gel images are directly superimposable, and warping is required to overlay and compare. This limitation makes image comparison complex, particularly between samples with markedly different spot patterns or when subtle protein changes are under investigation. Currently two options are available to investigators: (1) software programs can be purchased and used in-house for gel image analysis with extensive manual editing, and (2) companies will provide image analysis on a contract basis.
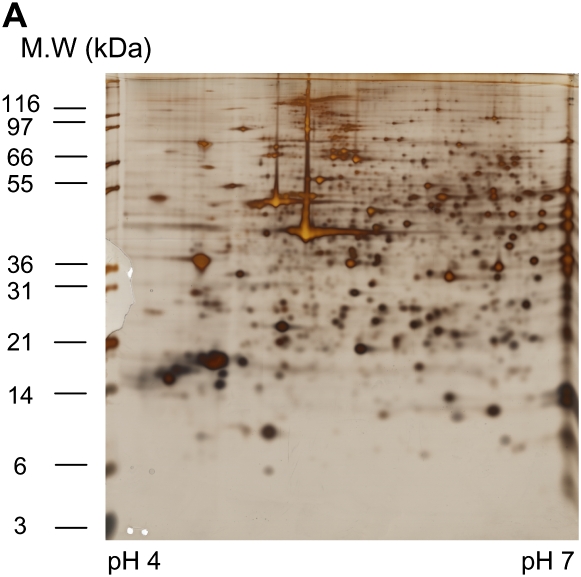
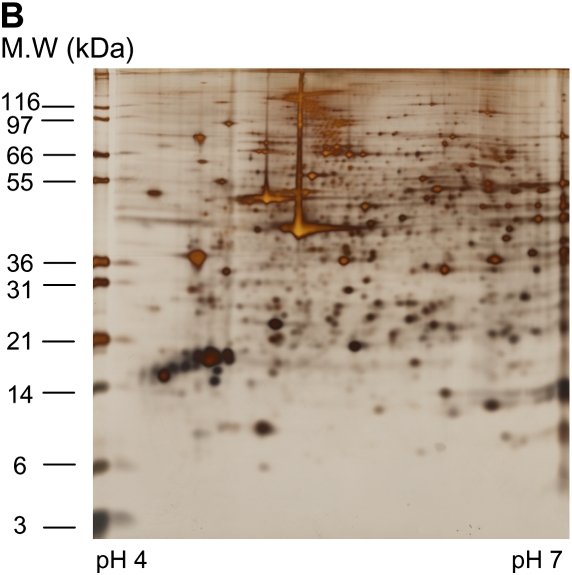
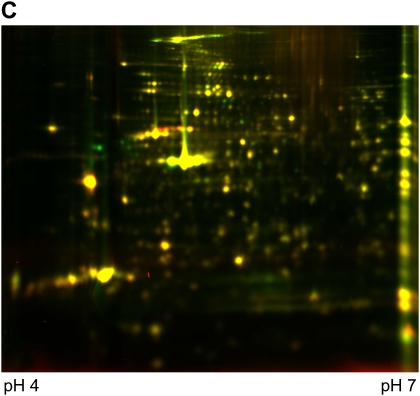
Differential in-gel electrophoresis (2D-DIGE) is a fairly recent improvement of the 2-DE technology. Before gel electrophoresis, the proteins from different disease states or experimental treatments are separately labeled with two different fluorescent dyes and an internal pooled standard labeled with the third dye. These three dyes are matched with mass and charge and each has a different emission wavelength. The labeled samples are then combined and subjected to 2-DE. The gel is scanned at different emission wavelengths and multiple images corresponding to a set of samples are generated and overlaid. Figure 2 shows an example of a pair of samples analyzed on two different gels stained with silver and also analyzed using 2D-DIGE. DIGE allows the differentially regulated proteins to be viewed as changed in color. Although 2D-DIGE significantly improves gel reproducibility and minimizes alignment issues (at least within the paired samples) (18, 19), some issues regarding quantification and translating gel maps to allow protein spot excising for downstream mass spectrometry (MS) identification remain.
Figure 2.
An example of two samples stained with silver (A and B) for comparison or analyzed using 2D DIGE (C). (A) Sino-atrial node cells without treatment (control); 150 μg was loaded onto 18 cm 10% Bis-Tris gels, silver stained. (B) Sino-atrial node cells treated with protein kinase inhibitor for 5 min; 150 μg was loaded onto 18 cm 10% Bis-Tris gels, silver stained. (C) 2D DIGE image with control labeled with Cy3 (green) and protein kinase inhibitor treated sample labeled with Cy5 (red).
Mass spectrometry analysis coupled to 2-DE analysis.
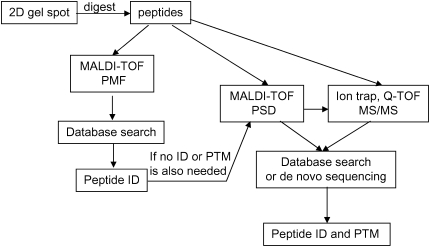
Proteomics and proteomics-based biomarker discovery have been greatly advanced with the development of MS methods. Spots from 2-DE are excised and the proteins are subjected to in-gel digestion; the resulting peptide fragments are then identified by MS. Typically there are three ways to identify the peptides by MS: peptide mass fingerprinting (PMF), tandem MS (commonly referred to as MS/MS), and post-source decay (PSD). With peptide mass fingerprinting (PMF), high accuracy mass-to-charge (m/z) ratios are usually obtained from a matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) or Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS). These m/z ratios for the observed peptides are used to compare with theoretical peptide masses (calculated from each amino acid sequence entry in protein database) to identify the parent protein. MS/MS spectra are usually generated from an ion trap or quadrupole time-of-flight mass spectrometer (Q-TOF MS), and PSD information is from MALDI-TOF. In MS/MS, the parent ion is isolated and fragmented to daughter ions, and the daughter ions are then detected. In PSD, the fragmentation occurs after the source, in the flight tube. MS/MS and PSD can be used for de novo sequencing and the exact sites of PTMs can be located. For identification of proteins from 2D gel spots, MALDI-TOF (for PMF) is generally used; however, if unambiguous results cannot be obtained or PTM information is needed, MS/MS information or PSD is required. Figure 3 shows the schematic diagram for the workflow of 2D gel spot analysis.
Figure 3.
Schematic diagram for the work flow of 2D gel spot. Typically the proteins are in situ digested to peptides and the resulting peptide mixture is then analyzed on MALDI-TOF MS for PMF. After database search, if the protein is not identified or PTM is also needed, the peptide mixture will be analyzed on MALDI-TOF in the PSD mode, or the peptide mixture will be analyzed on an ion trap instrument or Q-TOF to get MS/MS spectra. After database search or de novo sequencing, the peptide can be identified and the PTM can be identified and located.
Frischer and coworkers have applied 2-DE successfully to study cystic fibrosis (CF) (20). They generated a differential protein expression pattern in CF bronchial biopsies from eight control subjects and nine patients with CF. Biopsies (pools of three per patient) were taken; proteins were then extracted and run on 2-DE, with subsequent in-gel digestion and MS identification. Quantification of proteins was done using specific software. A total of 366 protein spots were identified and compared between groups. They found that the chaperone 75-kD glucose-regulated protein and ubiquinol–cytochrome c reductase complex core protein I and one form of nidogen, a pseudogene of aconitase 2, were increased in CF (p < 0.005). Aberrant protein levels may reflect molecular changes of CF as well as CF-linked inflammation, infection, and cellular stress response.
One- and two-dimensional liquid chromatography approaches to protein separation.
One particularly powerful methodology that has not been extensively used yet in proteomics is protein separation by one- and two- dimensional liquid chromatography (1D LC and 2D LC). One-dimensional liquid chromatography can be used to separate proteins based on their size (mass), pI (charge), or hydrophobicity—the three chemical characteristics that define any given protein. The most commonly used 1D LC is reversed phase chromatography, which separates proteins based on hydrophobicity. In proteomic studies, 1D LC has been used primarily for peptide separation before MS analysis, but it can be used for protein separation before protein enzymatic digestion and MS (21, 22). In 2D LC, proteins are separated in the first dimension by isoelectric chromatography (pI) (or strong cation exchange chromatography) and in the second dimension by hydrophobicity (23, 24), thereby increasing the extent of protein fractionation compared with 1D LC. As with 1D LC, this method has been used primarily in proteomics for peptide separation; however, it is increasingly being applied to separation of complex intact protein mixtures (22, 24–26). A combination of proteome separation by 2-DE, 1D LC, and 2D LC is synergistic and expands the observable proteome while allowing detection of protein isoforms and PTMs. In our laboratory, we compared 2-DE and 2D LC by creating an extensive database for serum (24) and isolated inner mitochondrial subproteome (22), and the findings were that only about 12% of the proteins observed were common between the two platforms.
When comparing the elution profiles of multiple samples obtained from different experiment conditions, it is essential to be able to both quantify (or obtain molar ratio) and identify the proteins present in the various fractions generated from the 2D LC separation. The reversed phase elution profile is monitored at 214 nm, the absorbance for peptide bonds. Hence, the elution peak volume is directly proportional to the number of peptide bonds present and thus reflects both the mass of the protein and its concentration. However, because in general multiple proteins are present in most of the 2D LC fractions, differential labeling is necessary at or after the time of tryptic digestion for the purpose of quantitation (see below). The standard strategy is to normalize and overlay elution profiles using sophisticated software and then quantify only the fractions that vary between samples. There are still needs for the development of more sophisticated quantitative and matching/alignment programs that would be similar to those used in 2-DE.
Isotopic labeling and mass spectrometry analysis.
For protein quantitation, several isotopic labeling techniques have been developed (27), and they can be divided into two main categories: globally adding labels in vitro before or after protein digestion, and metabolic labeling of proteins in vivo.
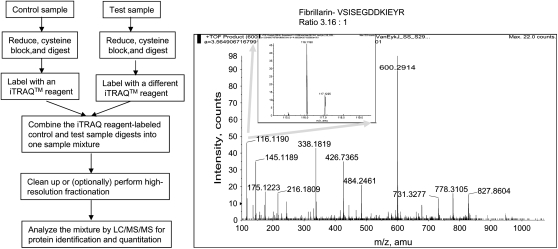
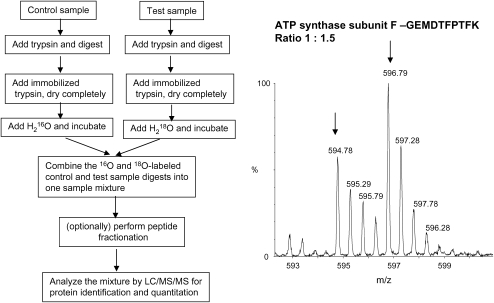
For the first category, two labeling techniques, isobaric tagging for relative and absolute quantitation (iTRAQ) (28) and 16O/18O labeling (29), are particularly useful. Briefly, iTRAQ reagents label free amines (N-termini of peptides and lysines). Up to four different samples with equivalent amounts are digested with trypsin separately, and each digest is then labeled with an iTRAQ reagent 114, 115, 116, or 117. Strong cation exchange is carried out to clear the peptide mixture from the reaction reagents, or to fractionate complex peptide mixture before LC-MS analysis. The intensity of the reporter ions in the tandem mass spectra (114.1, 115.1, 116.1, and 117.1) is used to quantify the peptides, and the average ratios of different peptides from the same proteins are used to quantify the protein amount. Second, the incorporation of the heavy or light oxygen isotopes can be accomplished during proteolysis—often referred to as 16O/18O labeling, which allows quantitative comparison between two samples. During proteolytic cleavage of proteins, two oxygen atoms from the solvent are incorporated universally into the carboxyl termini of all tryptic peptides. Thus, by incubating the peptides in either heavy or light oxygen water after the enzymatic digestion, selective incorporation of 18O or 16O will occur. The ratio of abundance between two samples can be obtained by the ratio of the two peaks separated by 4/z (z is the charge state of the peptide). This method may require reverse labeling to confirm the results, and it is restricted to two samples. However, the big cost difference between the two methods makes it reasonable to use the much more expensive iTRAQ method only when it is necessary to compare more than two samples. Figures 4 and 5 show the schematic diagrams of iTRAQ and 16O/18O labeling methodologies and example mass spectra. 16O/18O labeling has been applied for comparative proteomics analysis in adenovirus (29) and breast cancer (30, 31). iTRAQ has been successfully used to quantitate proteins from yeast whole cell lysates (28), murine cell cultures (32), and Escherichia coli (33).
Figure 4.
Overview of iTRAQ reagents methodology. The left schematic diagram shows the flow of iTRAQ methodology, and the spectra on the right shows an example MS/MS spectrum. The control was labeled with 116.1 and the test sample was labeled with 117.1 with a ratio of 3.16:1 as shown in the insert on the right. This peptide VSISEGDDKIEYR is from fibrillarin. The MS/MS spectrum is used courtesy of Shijun Sheng, Johns Hopkins University.
Figure 5.
Overview of 16O/18O labeling methodology. The left schematic diagram shows the flow of 16O/18O methodology, and the spectra on the right shows an example mass spectrum. The control was labeled with 16O and the test sample was labeled with18O with a ratio of 1:1.5, and this peptide GEMDTFPTFK is from ATP synthase subunit F. The MS spectrum is used courtesy of Shijun Sheng, Johns Hopkins University.
An alternative approach is to grow proteins already labeled (metabolic labeling). Lahm and Langen (34) and subsequently Oda and coworkers (35) used the 15N-substituted media for the purpose of quantifying differences between states of microorganisms. These cells labeled with 15N-proteins were then mixed with a second pool of cells grown in media containing naturally abundant nitrogen. The value of labeling the proteins in vivo is that labeled and unlabeled cells or tissues can be mixed together before introduction of any variability that will occur during the sample preparation process. Conrads and associates extended this approach to mammalian cells by using commercially available media containing 15N-labeled amino acids (36). A variety of 15N-, 2H-, and 13C-labeled amino acids have been used to label specific amino acids in cells grown in culture (37, 38). The use of labeled proteins is not only restricted to cell culture systems, it has been extended to tissue and even to entire multicellular organisms. Krijgsveld and colleagues labeled nematodes and fruit flies by feeding them 15N-enriched bacteria and yeast, respectively (39). Wu and coworkers have demonstrated that it is feasible to grow a rat entirely labeled with 15N (40).
Two-Dimensional Liquid Chromatography Approach to Peptide Separation
Analysis of complex peptide mixtures of digested proteomes is termed shot-gun or multidimensional protein identification technology (MudPIT) (41). This method involves tryptic digestion of protein mixture followed by multidimensional liquid chromatography (typically strong cation exchange followed by reverse phase chromatography) and MS analysis. Using MudPIT in combination with 1-DE, Guo and associates have identified 297 unique proteins from mouse bronchoalveolar lavage fluid (BALF) proteome, greatly expanding the BALF proteome by about threefold regardless of species (42).
Another peptide-based protein identification technique, different from MudPIT, is called combined fractional diagonal chromatography (COFRADIC). In COFRADIC, the proteins are first digested to peptides. A subset of peptides, which is highly representative of the parent proteins originally present in the lysate, are then selected. COFRADIC reduces the complexity of the peptide mixture. Theoretically, any peptide carrying a group that can be specifically and quantitatively modified can in principle be selected. It is sensitive and is characterized by a broad protein coverage, including abundant and rare, large and small, acidic and basic, and hydrophobic proteins. This concept has been applied to select methionine-containing peptides (43) and N-terminal peptides (44).
Whole Mass Monitoring of Intact Proteins
Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. In addition, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The intact proteins can also be ionized using electrospray ionization (ESI) or MALDI and introduced into a mass analyzer, primarily a TOF MS or FT-ICR MS. These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. MALDI produces predominately singly charged molecular ions (although larger proteins can produce multiply charged ions), making the analysis of mixtures very much straightforward. In ESI, multiply charged molecular species are formed from the analytes that contain more than one possible site of proton attachment. Proteins usually exhibit a characteristic series of multiple charged ions. The molecular mass of the protein can be easily calculated using the observed masses of any two adjacent ions in the series. Using whole mass monitoring, we have identified 24 proteins from the serum albuminome (a subset of proteins associated with albumin in serum) (45).
Proteomics Profiling Using Surface-enhanced Laser Desorption/Ionization TOF MS
Surface-enhanced laser desorption/ionization (SELDI)-TOF has led to a vast increase in the number of publications about new serum biomarkers since its introduction (46), and it has excellent potential for protein profiling. Technically, ProteinChip array-based SELDI-TOF MS is a variant of MALDI-TOF, but the on-chip purification is a great advantage. Twelve eight-spot chips are assembled in 96-well “bioprocessors,” which improves the expression analyses of many samples simultaneously. The subfraction of the proteome bound to the chips can be analyzed with MS on the same chip, resulting in a “pattern” of proteins characterized by mass-to-charge ratio (m/z). Furthermore, the technique is especially suitable for analyzing the low-molecular-weight proteome. The advantage of SELDI-TOF MS is that it does not rely on evidence of a gold-standard biomarker, but rather on combinations of peptide signals. However, substantial caveats remain, including limited biomarker discovery in complex biological samples, PTM analysis of proteins altered with diseases, identification of proteins/peptides, and precise protein/peptide quantitation (47). Platforms for direct on-chip sequencing of detected biomarkers by Q-TOF MS have become available, thus increasing the feasibility of the identification of discriminative proteins (48). SELDI-TOF MS has been applied to cancer tissue (49), plasma (50), serum (51), and nipple aspirate fluid (52). de Torre and coworkers have successfully used SELDI-TOF and 2-DE to assess markers of lung inflammation (53). They studied the changes in bronchoalveolar lavage (BAL) protein in 33 subjects challenged with local bronchial lung endotoxin and saline and in 11 patients with acute respiratory distress syndrome (ARDS). The temporal changes in acute inflammatory BAL (6, 24, and 48 h after endotoxin challenge) on hydrophobic binding chip surfaces revealed the differential presence of four proteins (all p < 0.001) in the inflammatory BAL. The differential pattern was also found in the ARDS BAL. They also analyzed the hydrophobic fraction of the inflammatory BAL using 2-DE and identified increased levels of apolipoprotein A1, and S100 calcium-binding proteins A8 and A9 in the inflammatory BAL. This pattern was also found in ARDS BAL after immunoblot analysis. These approaches will be useful to improve current methods of montoring lung inflammation and to identify new therapeutic targets.
PROTEIN–PROTEIN INTERACTIONS
The stimulation of cells from outside triggers cascades of signal transduction that results in cellular responses such as growth, differentiation, and movement. These signals are transduced by networks of interacting proteins (54). One method to probe protein–protein interactions is to use a bait protein labeled with an affinity tag, expressed in cell culture, and then isolated from lysed cells along with its associated partners by affinity chromatography. The resulting proteins either are separated on 1D gel, or are digested directly with trypsin and analyzed by LC/MS. For tissues, antibodies are in general used to pull down the bait protein and its binding partners. The protein–protein microarrays are the high-throughput platforms for study of such interactions. Proteomics, through protein–protein association studies, will eventually provide a detailed map of all protein interactions in the healthy and diseased cells and thus facilitate development of drugs that selectively target disease-associated pathways while minimizing unwanted side effects. Understanding how proteins function by interacting with relevant cellular partners will also make it possible to evaluate the consequences of gene mutations on the operation of the cell. This, in turn, should accelerate the advance of gene therapy and individualized medicine in general (1).
IDENTIFICATION OF PTMS
Protein structure and activity is often regulated by the enzymatic attachment and removal of covalent modifications. PTMs may occur at different stages of disease development, providing clues indicative of early or late events of transformation. Common PTMs include phosphorylation, acetylation, glycosylation, ubiquitination, farnesylation, methylation, and sialylation (55). There are more than 200 post-translational modifications known to affect a variety of protein functions such as protein–protein interaction and nucleic acid–protein interaction, stability, localization, and half-life (56). PTM-based biomarkers are proteins or peptides modified on a specific amino acid residue or residues and implicated in a specific pathway or biological network leading to initiation or promotion of a disease. For example, cardiac troponin I (cTnI) is specifically and selectively degraded at the C-terminus in the heart with myocardial ischemia. During acute myocardial infarction, the intact molecule and any degradation products are released from the heart cell into the blood where they can be detected (57). Although the detection of cTnI is now the gold standard for the diagnosis of myocardial infarction, increased quantities of degradation products have been proposed to increase risk and poor outcome (57).
Protein phosphorylation is one of the most important PTMs, and it is involved in most cellular signal pathways including cell cycling (58), signal transduction (59), DNA repair (60), and carcinogenesis (61). Nevertheless, despite phosphorylation being acknowledged as a crucial modification involved in many cellular events, determining the sites of phosphorylation on proteins is not a routine task. Recently, MS-based methods have emerged as powerful and preferred tools for the analysis of PTMs, including phosphorylation due to higher sensitivity, selectivity, and speed than most biochemical techniques (62–64). MS approaches for the analysis of phosphorylation sites have mostly relied on using an instrument capable of tandem MS experiments to determine the sequence and exact sites of phosphorylation on peptides. Because of the low stoichiometry of phosphopepitdes in a biological sample, they are in general enriched using affinity chromatography before MS analysis (65).
PROTEOMIC MICROARRAYS: VALIDATION AND DISCOVERY
Proteomics profiling is useful for biomarker discovery; however, it has inherent limitations. First, it is limited by sample complexity and has great difficulty in finding the low abundance biomarkers from hundreds to thousands of other proteins in the biological sample. Second, it does not provide (or provides only limited) information on specific proteins, such as biological role of the proteins, except in context of a particular subproteome. Third, proteomics is, in general, expensive and time-consuming, and not compatible with processing large amounts of clinical samples. Therefore, simultaneous efforts are being made to identify proteins and develop antibody assays for the candidate proteins identified from global proteomics profiling or other useful proteins (e.g., cytokines).
There has been fascinating growth in the field of large-scale and high-throughput biology, resulting in a new era of technology development and the collection and analysis of information. Elucidatiing the function of every encoded gene and protein, and understanding the cellular events mediating complex disease processes, are the challenges ahead. Miniaturized and parallel assay systems, especially microarray-based analysis, are crucial to high-throughput biological analysis. In a microarray format, capture molecules are immobilized in a very small area, and probed for various biochemical activities. There are two general types of protein microarrays: analytical microarrays and functional protein microarrays (66).
Analytical Microarrays: Quantitative Assessment of Potential Biomarkers and Useful Proteins
In antibody microarrays, antibodies, antibody mimics, or other proteins are arrayed and used to measure the presence and concentrations of proteins in complex mixtures. They are essentially the same as enzyme-linked immunosorbent assay (ELISA) except that multiple analytes are quantified simultaneously. They are desirable platforms for high-throughput and quantitative assessment of potential biomarkers resulting from proteomics profiling.
The most common form of analytical arrays are antibodies/antibody mimic arrays, in which antibodies (or similar reagents) that bind specific antigens are arrayed on a glass slide at high density. A lysate is passed over the array and the bound antigen is detected after washing. Detection is usually carried out by using labeled lysates or using a second antibody that recognizes the antigen of interest (multiplex sandwich immunoassay). Assay performance is evaluated based on (1) ability to measure analytes across broad dynamic range at sufficiently low coefficients of variation (CVs), (2) detection of proteins at levels requisite to capture biologically relevant expression difference, and (3) generation of standard curves to calculate analyte concentrations based on detected intensity data. Antibody microarrays require calibration standard curves to be included in the experiment so that the concentration of analytes in the samples can be calculated according to the assay signal. It is critical to obtain recombinant protein standards that mimic the native proteins in the particular biofluids to be analyzed. The stability and solubility of the recombinant proteins should also be evaluated, since insolubility of the recombinant protein can have a major impact on assay precision. Several factors make high quality multiplex sandwich immunoassay limited to date (67). Matched antibody pairs are often unavailable for novel proteins, and the cross-reactivity of the detection antibodies limits the degree of multiplexing. Also, with each additional analyte added, it can be time consuming and costly to select capture and detection antibodies with high sensitivity and low cross-reactivity for quantitative mulplexing analysis.
In addition to antibody microarrays, other analytical microarrays have been developed, including microarrays for profiling antibodies in the patient's serum and plasma, essentially the reciprocal of that described above. Joos and colleagues used 18 diagnostic markers for autoimmune diseases to form an autogen microarray and screened for antigen–antibody interactions (68).
Functional Protein Microarrays
Functional protein arrays are composed of sets or proteins or even an entire proteome that are arranged in an orderly fashion on a small surface. Unlike the antibody microarrays (mainly developed for diagnostics and profiling of expression), protein microarrays have enormous potential in assaying for a wide range of biochemical activities, as well as drug and drug target identification. Protein microarrays can be used to perform protein–protein, protein–ligand, protein–DNA, protein–RNA, protein–drug, and enzyme–substrate interactions. For example, Miksich has demonstrated the potential in using protein microarrays to conduct enzymatic assays to identify downstream targets of kinases (69). Protein microarrays will be the approach of choice to close the information gap between genomics and proteomics in the development of new markers for the early detection, diagnosis, and classification of diseases, as well as drug development and drug target identification.
The first great obstacle to overcome is the purification of large number of recombinant proteins in a high-throughput manner. Many research groups and companies have contributed tremendous effort in developing high-throughput protein purification methods, and recombinant proteins have been purified from E. coli, yeast, insects, and humans (70–74). Leuking and coworkers cloned cDNAs from human fetal brain tissues as C-terminal Hisx6-tagged fusions (75). In a later report, Braun and colleagues created a system (FLEXP) that performs from cDNA cloning to protein production from E. coli in a fully automated fashion (73). However, because eukaryotic proteins expressed in prokaryotic systems are not post-translationally modified, Zhu and coworkers has developed a high-throughput protein purification method from the budding yeast (71). For the same reason, Albala and associates chose 72 unique human cDNA clones to create an array of recombinant baculoviruses, from which 42% of the clones produced soluble fusion proteins in a 96-well format (74). Functional protein chips like traditional assays performed in microtiter plates (76) are suitable for a wide variety of biochemical analyses. Unlike microtiter plates, however, they are much more amenable to high-throughput studies and use small amounts of reagents. Studies analyzing large sets of proteins have recently been performed. Using a nanowell chip mounted on glass slides, Zhu and colleagues analyzed the activity of 119 yeast kinases for 17 different substrates (77).
FUTURE PERSPECTIVES AND CHALLENGES
Proteomics complements genomics-based approaches, providing additional information but presenting different technical challenges. Because no protein equivalent of PCR for amplification of low-abundant proteins is present, detection methods with large dynamic range are needed. The folded structures are important for the protein properties, but generic methods are difficult to design and apply. The analysis of PTMs provides another challenge. Certain technological processes, particularly protein separation and analysis, are difficult to automate.
Acknowledgments
The authors acknowledge Shijun Sheng for providing courtesy figures. They also thank Lesley A. Kane for her critical reading of the manuscript.
Funding sources: NHLBI Proteomics Initiative (content N01-HV-28180) (J.E.V.E., Y.G.).
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Hunt DF. Personal commentary on proteomics. J Proteome Res 2002;1:15–19. [DOI] [PubMed] [Google Scholar]
- 2.Graham DR, Elliott ST, Van Eyk JE. Broad-based proteomic strategies: a practical guide to proteomics and functional screening. J Physiol 2005;563:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dos Remedios CG, Liew CC, Allen PD, Winslow RL, Van Eyk JE, Dunn MJ. Genomics, proteomics and bioinformatics of human heart failure. J Muscle Res Cell Motil 2003;24:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilello JA. The agony and ecstasy of “OMIC” technologies in drug development. Curr Mol Med 2005;5:39–52. [DOI] [PubMed] [Google Scholar]
- 5.Alaiya A, Al-Mohanna M, Linder S. Clinical cancer proteomics: promises and pitfalls. J Proteome Res 2005;4:1213–1222. [DOI] [PubMed] [Google Scholar]
- 6.Vitzthum F, Behrens F, Anderson NL, Shaw JH. Proteomics: from basic research to diagnostic application. a review for requirements & needs. J Proteome Res 2005;4:1086–1097. [DOI] [PubMed] [Google Scholar]
- 7.Granger C, Mockrin S, Van Eyk JE, Anderson L. NHLBI Clinical Proteomics Working Group Report. Circulation 2005;109:1697–1703. [DOI] [PubMed] [Google Scholar]
- 8.Stanley B, Gundry RL, Cotter R, Van Eyk JE. Heart disease, clinical proteomics and mass spectrometry. Dis Markers 2005;20:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffe AS, Van Eyk JE. Troponin fragments: diagnostic potential in cardiac biomarkers in the management of cardiac disease. In: Morrow DA, editor. Contemporary cardiology series. Totowa, NJ: Humana Press. (In press)
- 10.Arrell DK, Neverova I, Fraser H, Marban E, Van Eyk JE. Proteomic analysis of pharmacologically preconditioned cardiomyocytes reveals novel phosphorylation of myosin light chain 1. Circ Res 2001;89:480–487. [DOI] [PubMed] [Google Scholar]
- 11.Neverova I, Van Eyk JE. Application of reversed phase high performance liquid chromatography for subproteomic analysis of cardiac muscle. Proteomics 2002;2:22–31. [PubMed] [Google Scholar]
- 12.Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, Robinson J, Murata M, Murphy AM, Marban E, et al. Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circ Res 2006;99:706–714. [DOI] [PubMed] [Google Scholar]
- 13.Kane LA, Neverova I, Van Eyk JE. Subfraction of heart tissue: the “IN Sequence” myofilament protein extraction of myocardial tissue. In: Vivanco F, editor. Methods in molecular biology series: cardiovascular proteomics. Totowa, NJ: The Humana Press, Inc. (In press) [DOI] [PubMed]
- 14.Stanley B, Neverova I, Van Eyk JE. Effect of detergents on two-dimensional gel electrophoresis of human cardiac tissue. Proteomics 2003;3:815–820. [DOI] [PubMed] [Google Scholar]
- 15.Stanley BA, Van Eyk JE. A solubility optimization protocol for 2-D gel electrophoresis of cardiac tissue. In: Vivanco F, editor. Methods in molecular biology series: cardiovascular proteomics. Totowa, NJ: The Humana Press, Inc. (In press) [DOI] [PubMed]
- 16.Graham DR, Garnham CP, Fu Q, Robbins J, Van Eyk JE. Improvements in two-dimensional gel electrophoresis by utilizing a low cost in-house neutral pH SDS-PAGE electrophoresis system. Proteomics 2005;5:2309–2314. [DOI] [PubMed] [Google Scholar]
- 17.Sabounchi-Schutt F, Astrom J, Olsson I, Eklund A, Grunewald J, Bjellqvist B. An immobiline DryStrip application method enabling high-capacity two-dimensional gel electrophoresis. Electrophoresis 2000;21:3649–3656. [DOI] [PubMed] [Google Scholar]
- 18.Von Eggeling F, Gawriljuk A, Fiedler W, Ernst G, Blaussen U, Klose J, Romer I. Fluorescent dual colour 2D-protein gel electrophoresis for rapid detection of differences in protein patter with standard image analysis software. Int J Mol Med 2001;8:373–377. [DOI] [PubMed] [Google Scholar]
- 19.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics techonology. Proteomics 2001;1:377–396. [DOI] [PubMed] [Google Scholar]
- 20.Frischer T, Myung JK, Maurer G, Eichler I, Szepfalusi Z, Lubec G. Possible regulation of chaperon and metabolic proteins in cystic fibrosis bronchial tissue. Proteomics 2006;6:3381–3388. [DOI] [PubMed] [Google Scholar]
- 21.Neverova I, Van Eyk JE. Role of chromatographic techniques in proteomic analysis. J Chromatogr B Analyt Technol Biomed Life Sci 2005;815:51–63. [DOI] [PubMed] [Google Scholar]
- 22.McDonald T, Sheng S, Stanley B, Chen D, Ko Y, Cole RN, Pederson P, Van Eyk JE. Expanding the subproteome of the inner mitochondria using protein separation technologies: one and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol Cell Proteomics (In press) [DOI] [PubMed]
- 23.Wall DB, Kachman MT, Gong S, Hinderer T, Parus S, Misek DE, Hanash SM, Lubman DM. Isoelectric focusing nonporous HPLC: a two-dimensional liquid-phase separation method for mapping of cellular proteins with identification using MALDI-TOF mass spectrometry. Anal Chem 2000;72:1099–1111. [DOI] [PubMed] [Google Scholar]
- 24.Sheng S, Chen D, Van Eyk JE. Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol Cell Proteomics 2006;5:26–34. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Wu R, Cho KR, Shedden KA, Barder TJ, Lubman DM. Classification of cancer cell lines using an automated two-dimensional liquid mapping method with hierarchical clustering techniques. Mol Cell Proteomics 2006;5:43–52. [DOI] [PubMed] [Google Scholar]
- 26.Chen EI, Hewel J, Felding-Habermann B, Yates JR III. Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT). Mol Cell Proteomics 2006;5:53–56. [DOI] [PubMed] [Google Scholar]
- 27.MacCoss MJ, Matthews DE. Quantitative MS for proteomics: teaching a new dog old tricks. Anal Chem 2005;77:294A–302A. [DOI] [PubMed] [Google Scholar]
- 28.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S et al. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004;3:1154–1169. [DOI] [PubMed] [Google Scholar]
- 29.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem 2001;73:2836–2842. [DOI] [PubMed] [Google Scholar]
- 30.Brown KJ, Fenselau C. Investigation of doxorubicin resistance in MCF-7 breast cancer cells using shot-gun comparative proteomics with proteolytic 18O labeling. J Proteome Res 2004;3:455–462. [DOI] [PubMed] [Google Scholar]
- 31.Zang L, Toy DP, Hancock WS, Sgroi DC, Karger BL. Proteomics analysis of ductal carcinoma of the breast using laser capture microdissection, LC-MS, and 16O/18O isotopic labeling. J Proteome Res 2004;3:604–612. [DOI] [PubMed] [Google Scholar]
- 32.Unwin RD, Pierce A, Watson RB, Sternberg DW, Whetton AD. Quantitative proteomic analysis using isobaric protein tags enables rapid comparison of changes in transcript and protein levels in transformed cells. Mol Cell Proteomics 2005;4:924–935. [DOI] [PubMed] [Google Scholar]
- 33.Aggarwal K, Choe LH, Lee KH. Quantitative analysis of protein expression using amine-specific isobaric tags in Escherichia coli cells expressing rhsA elements. Proteomics 2005;5:2297–2308. [DOI] [PubMed] [Google Scholar]
- 34.Lahm HW, Langen H. Mass spectrometry: a tool for the identification of proteins separated by gels. Electrophoresis 2000;21:2105–2114. [DOI] [PubMed] [Google Scholar]
- 35.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA 1999;96:6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conrads TP, Alving K, Veenstra TD, Belov ME, Anderson GA, Anderson, DJ, Lipton MS, Pasa-Tolic L, Udseth HR, Chrisler WB, et al. Quantitative analysis of bacterial and mammalian proteomes using a combination of cysteine affinity tags and 15N-metabolic labeling. Anal Chem 2001;73:2132–2139. [DOI] [PubMed] [Google Scholar]
- 37.Ong S, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002;1:376–386. [DOI] [PubMed] [Google Scholar]
- 38.Ibarrola N, Molina H, Iwahori A, Pandey A. A novel proteomic approach for specific Identification of tyrosine kinase substrates using [13C]tyrosine. J Biol Chem 2004;279:15805–15813. [DOI] [PubMed] [Google Scholar]
- 39.Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, Artal-Sanz M, Verrijzer CP, Plasterk RHA, Heck AJR. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat Biotechnol 2003; 21:927–931. [DOI] [PubMed] [Google Scholar]
- 40.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR. Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem 2004;76:4951–4959. [DOI] [PubMed] [Google Scholar]
- 41.Washbrun MP, Wolters D, Yates JR III. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 2001;19:242–247. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y, Ma S, Grigoryev D, Van Eyk J, Garcia JGN. One-dimensional polyacryamide gel electrophoresis mass spectrometry and two-dimensional liquid chromatography mass spectrometry analysis of the mouse bronchoalveolar lavage proteome. Proteomics 2005;5:4608–4624. [DOI] [PubMed] [Google Scholar]
- 43.Gevaert K, Van Damme J, Goethals M, Thomas GR, Hoorelbeke B, Demol H, Martens L, Puype M, Staes A, Vandekerckhove J. Chromatographic isolation of methionine-containing peptides for gel-free proteome analysis. Mol Cell Proteomics 2002;1:896–903. [DOI] [PubMed] [Google Scholar]
- 44.Gavaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol 2003;21:566–569. [DOI] [PubMed] [Google Scholar]
- 45.Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics-clinical applications (In press) [DOI] [PMC free article] [PubMed]
- 46.Hutchens TW, Yip TT. New desorption strategies for the mass spectrometric analysis of macromolecules. Rapid Commun Mass Spectrom 1993;7:576–580. [Google Scholar]
- 47.White MY, Gundry RL, Cordwell SJ. When does a fingerprint constitute a diagnostic? Lancet 2006;368:971–973. [DOI] [PubMed] [Google Scholar]
- 48.Caputo E, Moharram R, MartinBM. Methods for on-chip protein analysis. Anal Biochem 2003;321:116–124. [DOI] [PubMed] [Google Scholar]
- 49.Traub F, Feist H, Kreipe HH, Pich A. SELDI-MS-based expression profiling of ductal invasive and lobular invasive human breast carcinomas. Pathol Res Pract 2005;201:763–770. [DOI] [PubMed] [Google Scholar]
- 50.Caputo E, Lombardi ML, Luongo V, Moharram R, Tornatore P, Pirozzi G, Guardiola J, Martin BM. Peptide profiling in epithelial tumor plasma by the emerging proteomic techniques. J Chromatogr B Analyt Technol Biomed Life Sci 2005;819:59–66. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem 2002;48:1296–1304. [PubMed] [Google Scholar]
- 52.Sauter ER, Shan S, Hewett JE, Speckman P, Du Bois GC. Proteomic analysis of nipple aspirate fluid using SELDI-TOF-MS. Int J Cancer 2005;114:791–796. [DOI] [PubMed] [Google Scholar]
- 53.de Torre C, Ying SX, Munson PJ, Meduri GU, Suffredini AF. Proteomic analysis of inflammatory biomarkers in bronchoalveolar lavage. Proteomics 2006;6:3939–3957. [DOI] [PubMed] [Google Scholar]
- 54.Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev 2000;14:1027–1047. [PubMed] [Google Scholar]
- 55.Bode AM, Dong Z. Posttranslational modification of p53 in tumorigenesis. Nat Rev Cancer 2004;4:793–805. [DOI] [PubMed] [Google Scholar]
- 56.Banks RE, Dunn MJ, Hochstrasser DF, Sanchez J, Blackstock W, Pappin DJ, Selby PJ. Proteomics: new perspectives, new biochemical opportunities. Lancet 2000;356:1749–1756. [DOI] [PubMed] [Google Scholar]
- 57.McDonough JL, Van Eyk JE. Develiping the next generation of cardiac markers: disease-induced modifications of troponin I. Prog Cardiovasc Dis 2004;47:207–216. [DOI] [PubMed] [Google Scholar]
- 58.Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 1997;278:455–460. [DOI] [PubMed] [Google Scholar]
- 59.Downward J. The ins and outs of signaling. Nature 2001;411:759–762. [DOI] [PubMed] [Google Scholar]
- 60.Chao C, Saito S, Anderson CW, Appella E, Xu Y. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc Natl Acad Sci USA 2000;97:11936–11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.al-Obeidi FA, Wu JJ, Lam KS. Protein tyrosine kinases: structure, substrate specificity, and drug discovery. Biopolymers 1998;47:197–223. [DOI] [PubMed] [Google Scholar]
- 62.McLachlin DT, Chait BT. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr Opin Chem Biol 2001;5:591–602. [DOI] [PubMed] [Google Scholar]
- 63.Annan RS, Huddleston MJ, Verma R, Deshaies RJ, Carr SA. A Multidimensional electrospray MS-based approach to phosphopeptide mapping. Anal Chem 2001;73:393–404. [DOI] [PubMed] [Google Scholar]
- 64.Mann M, Ong SE, Grønborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol 2002;20:261–268. [DOI] [PubMed] [Google Scholar]
- 65.Garcia BA, Shabanowitz J, Hunt DF. Analysis of protein phosphorylation by mass smpectrometry. Methods 2005;35:256–264. [DOI] [PubMed] [Google Scholar]
- 66.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol 2003;7:55–63. [DOI] [PubMed] [Google Scholar]
- 67.Fu Q, Van Eyk JE. Proteomics and heart disease: identifying biomarkers of clinical utility. Expert Rev Proteomics 2006;3:237–249. [DOI] [PubMed] [Google Scholar]
- 68.Joos TO, Schrenk M, Hopfl P, Kroger K, Chowdhury U, Stoll D, Schorner D, Durr M, Herick K, Rupp S, et al. A microarray enzyme-linked immunosorbent assay for autoimmune diagnostics. Electrophoresis 2000;21:2641–2650. [DOI] [PubMed] [Google Scholar]
- 69.Houseman BT, Huh JH, Kron SJ, Mrksich M. Peptide chips for the quantitative evaluation of protein kinase activity. Nat Biotechnol 2002;20:270–274. [DOI] [PubMed] [Google Scholar]
- 70.Stoll D, Templin MF, Schrenk M, Traub PC, Vohringer CF, Joos TO. Protein microarray technology. Front Biosci 2002;7:c13–c32. [DOI] [PubMed] [Google Scholar]
- 71.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science 2001;293:2101–2105. [DOI] [PubMed] [Google Scholar]
- 72.Bussow K, Cahill D, Nietfeld W, Bancroft D, Scherzinger E, Lehrach H, Walter G. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res 1998;26:5007–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Braun P, Hu Y, Shen B, Halleck A, Koundinya M, Harlow E, LaBaer J. Proteome-scale purification of human proteins from bacteria. Proc Natl Acad Sci USA 2002;99:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albala JS, Franke K, McConnell IR, Pak KL, Folta PA, Rubinfeld B, Davies AH, Lennon GG, Clark R. From genes to proteins: high-throughput expression and purification of the human proteome. J Cell Biochem 2000;80:187–191. [DOI] [PubMed] [Google Scholar]
- 75.Lueking A, Horn M, Eickhoff H, Bussow K, Lehrach H, Walter G. Protein microarrays for gene expression and antibody screening. Anal Biochem 1999;270:103–111. [DOI] [PubMed] [Google Scholar]
- 76.Mendoza LG, McQuary P, Mongan A, Gangadharan R, Brignac S, Eggers M. High-throughput microarray-based enzyme-linked immunosorbent assay (ELISA). Biotechniques 1999;27:778–788. [DOI] [PubMed] [Google Scholar]
- 77.Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, Klemic KG, Smith D, Gerstein M, Reed MA, Snyder M. analysis of yeast protein kinases using protein chips. Nat Genet 2000;26:283–289. [DOI] [PubMed] [Google Scholar]