Abstract
A functional single nucleotide polymorphism in the 5 genomic region of CD14 (CD14/−159) is one of the most widely tested genetic variations in relation to asthma and associated traits. The results of these studies have shown a remarkable, statistically significant heterogeneity, with some studies indicating the T-allele as a risk factor, others the C-allele, and others finding no association. Recent studies in which exposure to house-dust endotoxin or to domestic sources of microbial exposure were assessed concomitantly with CD14/−159 have shown a consistent, replicable gene-environment interaction. Specifically, results suggest that the C-allele is a risk factor for allergic phenotypes at low levels of exposure, whereas the T-allele is a risk factor at high levels of exposure. This finding seems to be explained by a genetically-determined heterogeneity for the protective effect of microbial exposure on allergic phenotypes, with homozygotes for the C-allele showing a much stronger negative association between exposure and allergic outcomes than carries of the other two genotypes. These results suggest that the often encountered, limited replicability of genetic associations may, at least in part, be due to complex interactions between genes and environment in determining asthma-related outcomes.
Keywords: asthma, CD14, gene–environment interaction
In August 1999 (1) our group* first reported the existence of a single nucleotide polymorphism (SNP) in the 5′ genomic region of CD14 at position −159 with respect to the transcription start site (−260 when counting from the translation start site). From the point of view of asthma and allergies, interest in this gene was twofold: first, CD14 is located in chromosome 5q31, a region that several genome-wide searches have suggested contains variations relevant to asthma (3); and second, CD14 is a receptor that has specificity for lipopolysaccharides (LPS) and other bacterial wall–derived components (4, 5), and has also been suggested as a potential response element for the respiratory syncytial virus (6). In the late 1990s, a new interpretation of the hygiene hypothesis (7) was emerging; as initially proposed, this hypothesis postulated that infections (and lack of “hygiene”) in early life could be protective against the development of allergies. However, results of studies in which allergic outcomes were assessed in children who had clinical infections in early life (measles, tuberculosis, etc.) did not reveal a consistent pattern. Martinez and Holt first suggested that, more than any specific infection, it was the global microbial burden in early life, including exposure to nonpathogenic microbes, which could deviate immune responses away from those associated with allergic responses (8). It thus made sense to surmise that polymorphisms in CD14, a major player in the biological process leading to the detection of and development of biological responses against environmental microbial products, could be involved in a mechanism that resulted in protection against asthma and allergies.
CD14 is constitutively expressed primarily on the surface of monocytes, macrophages, and neutrophils as membrane CD14 (mCD14). A soluble form of CD14, sCD14, is abundant in serum and is apparently derived both from secretion of CD14 and from enzymatically cleaved glycosyl-phosphatidylinositol–anchored mCD14 (9). We found that both alleles (C and T) for CD14/−159 were of similar frequency in the white population from Tucson, Arizona that we studied, and that TT homozygotes had significantly higher levels of circulating sCD14 than CC homozygotes (1). We also reported that atopic children who were homozygotes for the T allele had significantly smaller mean number of positive skin test to aeroallergens than CC homozygotes. These results suggested to us the hypothesis that TT homozygotes could be less prone to develop allergic responses because, having higher expression of CD14, they would be more sensitive to environmental microbial exposures that would deviate immune responses to allergens away from those of the T-helper-2 (Th2) type (10).
Since that first report, over 200 studies have been published that assess CD14/−159 in relation not only to asthma and associated traits, but also to a wide variety of other illnesses, from cardiovascular (11) to autoimmunity (12), from infections (13) to malignancies (14). The most consistent result emerging from this large body of evidence has been the association between CD14/−159 and circulating sCD14: most studies with minimally sufficient numbers of participants have replicated our original finding that homozygotes for the T allele have higher serum levels of sCD14 than carriers of the other two genotypes. Higher mCD14 expression in circulating mononuclear cells in CD14/ −159T as compared with CD14/−159C has also been reported (2). These findings are compatible with functional genomic studies by Le Van and coworkers (15) showing that constructs carrying the T allele for CD14/−159 show higher transcription rates than those carrying the C allele when transfected into a mononuclear cell line.
Based on the new interpretation that we had proposed for the hygiene hypothesis for asthma (8), we surmised that carriers of the T allele for CD14/−159, having higher constitutive CD14 expression in cell membranes and in circulation, could be more likely to respond to what we assumed were low levels of exposure to LPS (measured in environmental studies as endotoxin) in an urban community such as Tucson, Arizona.
REPLICATION AND NOT
Several groups attempted to replicate our findings with diverse success, as has happened for most genetic association studies of complex diseases. Koppelman and colleagues (16) studied subjects living in an urban community in The Netherlands and obtained very similar results to ours using a similar phenotype, namely, number of positive skin tests to allergens. Leung and coworkers (17) studied atopic and nonatopic Chinese children and also found higher sCD14 levels in CD15/−159 TT homozygotes than in carriers of the other two genotypes. They found no association between allergic sensitization and CD14/−159, but among atopic children, homozygotes for the T allele had higher total serum IgE than carriers of the other two genotypes. Buckova and colleagues (18) reported that, among Czech children, carriers of the C allele for CD14/−159 were more likely than their peers to be sensitized to molds. Takeuchi and coworkers (19) reported that TT homozygotes for CD14/−159 had significantly fewer positive CAP-radioallergosorbent tests than CC homozygotes and CT heterozygotes. Kabesch and colleagues (20), on the other hand, replicated the association between CD14/−159 and serum sCD14 levels, but found no association between the polymorphism and markers of allergy. Similarly, Sengler and coworkers (21) failed to find any association between allergies and the CD14/−159 polymorphism in a large study of German children. Kedda and colleagues (22) in Australia performed one of the largest published studies to date, which included over 1,000 subjects, 55% of whom had asthma. They found no association of CD14/−159 with asthma and a weak association with atopy, with the T allele showing increased risk. These authors also performed a meta-analysis of 12 studies conducted before 2005, and reported no association between either asthma or allergies and CD14/−159 but, interestingly, they did find a significant heterogeneity (p = 0.01) between studies.
At first glance, these results suggest a disappointing inconsistency. However, an indication that behind this inconsistency a far more complex picture could emerge came from studies by Ober and coworkers (23) among the Hutterites, an inbred population living in communal farms in Midwestern United States. These authors did report a positive association between allergic sensitization and CD14/−159 but, contrary to most other positive studies and in the same direction as that by Kedda and colleagues (22), it was the T allele that was over-transmitted within atopic families. Given the exceptional environment in which this Hutterite population lives, these findings suggested that there could be an antagonistic interaction between environmental context and CD14/−159 as determinants of allergic sensitization: the T allele could be either protective or a risk factor, depending on the degree of exposure to environmental microbial products, which we surmised could be present in different concentrations in urban and rural settings (24).
In support of this contention, Braun-Farlander and other investigators of the Asthma and Endotoxin (ALEX) study (25) observed that, in children living in rural areas of German-speaking Western Europe, risk of allergic sensitization was inversely associated with the concentration of endotoxin in house dust. Given the role of CD14 as a receptor for endotoxin/LPS, it was plausible to surmise that differential exposure to endotoxin could explain the heterogeneity of the results revealed by the meta-analysis by Kedda and coworkers (22).
A REPLICATED GENE–ENVIRONMENT INTERACTION
To test this hypothesis, and in collaboration with the ALEX investigators, we genotyped all subjects for whom DNA samples were available in the ALEX study for CD14/−159, and reassessed the previously reported inverse association of allergic sensitization to endotoxin exposure and to indoor pets and farm animals after stratifying the population by the polymorphism (26). We found that the previously-reported, very strong protective effect of endotoxin exposure was mainly observed among CC homozygotes for CD14/−159, whereas the association was much weaker among TT homozygotes and heterozygotes (Figure 1). An interesting consequence of this genetic heterogeneity in environmental sensitivity can be noticed in the “norms of reaction” (27) presented in Figure 1: at low levels of exposure, it is homozygotes for the C allele that are at the highest risk for allergic sensitization, whereas at high levels these same individuals are at the lowest risk for sensitization.
Figure 1.
Association between serum specific IgE against aeroallergens and CD14/−159 by exposure to endotoxin in house dust in the ALEX study (26). Sensitization was strongly and inversely associated with exposure among CC homozygotes (dark blue columns), whereas the association was much weaker among carriers of the other two genotypes (lighter blue columns). As a result, TT homozygotes were at risk for sensitization, as compared with the other two genotypes, whereas opposite (albeit not significant) trends were observed at lower levels of exposure. Reprinted by permission from Reference 26.
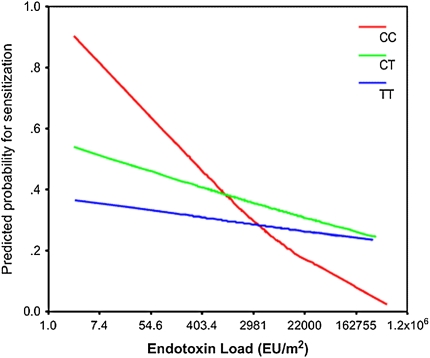
This pattern of antagonistic interaction between CD14 and endotoxin exposure is not exclusive of the somewhat exceptional environmental conditions present in the rural areas in which the ALEX study was conducted. Simpson and colleagues (28) performed very similar studies to those described above in Manchester (UK), and reported a strikingly similar pattern: the inverse relation between house dust endotoxin exposure and allergic sensitization was much stronger for CC homozygotes than for carriers of the other two genotypes (Figure 2). More recently, Keoki-Williams et al (29) assessed the same gene-environment interaction in a population of adults who were parents of children enrolled in a newborn cohort in Detroit and reported very similar results (Figure 3): allergic sensitization was inversely related to house dust endotoxin exposure, but only among CC homozygotes; no association was observed among carriers of the other two genotypes. In the studies of both Simpson and coworkers (28) and Williams and coworkers (29), as in that by Eder and colleagues (26), the association between CD14/−159 and allergic sensitization depended on level of exposure to endotoxin, with TT homozygotes being protected at low levels of exposure and at risk at high levels.
Figure 2.
Antagonistic interaction between house-dust endotoxin exposure and CD14/−159 genotypes as determinants of allergic sensitization in children enrolled in the Manchester Asthma and Allergy Study (28). As in Figure 1, the inverse relation between exposure and sensitization was strongest in CC homozygotes and less pronounced in the other two genotypes. Reprinted by permission from Reference 28.
Figure 3.
Association between total serum IgE and dust endotoxin exposure by CD14/−159 in adult participants in a Detroit cohort study (29). Gene–environment interactions are very similar to those described in Figures 1 and 2. Reprinted by permission from Reference 29.
The relevance for asthma of the findings of these three studies could not be adequately evaluated because all three were population-based and, therefore, subjects with asthma were too few to provide sufficient power to assess gene–environment interactions. Zambelli-Weiner and coworkers (30) studied nuclear families of African descent with one asthmatic proband in Barbados, West Indies. They reported a strong antagonistic interaction between level of endotoxin exposure in the home and CD14/−159 as determinants of both risk and severity of asthma. Among subjects exposed to low levels of house dust endotoxin, the TT genotype appeared protective for asthma (odds ratio [OR], 0.09; 95% confidence interval [95% CI], 0.03–0.27). However, TT individuals with high exposure to endotoxin were more than 11 (95% CI, 1.03–131.7) times more likely to have asthma than individuals with the CC genotype. Although these results were based on a small sample of TT homozygotes in the population, they revealed the same antagonistic interaction between endotoxin exposure and CD14/−159 observed for allergic sensitization in the three general population studies quoted earlier. Of particular interest was the finding that the relation between severity of asthma and CD14/−159 was also modified by endotoxin exposure: carriers of the TT genotype had significantly lower asthma severity scores than carriers of the other two genotypes, but only when house dust endotoxin exposure was also low. On the contrary, asthma severity scores were higher in TT homozygotes than in CC homozygotes among subjects with high exposures to house dust endotoxin, although numbers were small and the association did not reach statistical significance.
Thus, four studies provide consistently replicated evidence of a pattern of gene–environment interactions that, to our knowledge, has not been reported before in humans. It is important to stress here that important issues remain to be elucidated—for example, the biological mechanism that explains the marked differences in responsiveness to environmental endotoxin in carriers of different alleles for CD14/−159 has not been elucidated. One possible scenario could be that constitutive expression of CD14 may be higher in TT homozygotes than in carriers of the other two genotypes; this would make the former more susceptible to the protective effects of low levels of microbial products that interact with CD14. However, at higher doses of CD14 agonists, agonist-induced CD14 synthesis could be higher in carriers of the C allele than in carriers of the T allele.
It is important to stress here that there are several other polymorphisms in the CD14 gene (10) (Figure 4), and limited assessment of their role in these interactions is available. In addition, epistatic interactions with polymorphisms in other genes may also be important: Hong and colleagues (31) reported that, among Korean children with asthma, there was an interaction between CD14/−159 and a SNP in the tumor necrosis factor-α (TNF-α) gene, TNFα/−308, with only subjects with the risk alleles for each SNP (A for TNF-α/−08 and C for CD14/ −159) showing increased rates of bronchial hyperresponsiveness. How these epistatic effects may modify the gene–environment interactions we have described is unknown. Moreover, CD14 seems to be an adjuvant not only for responses to endotoxin/LPS but also for other microbial molecular patterns that signal through TLR2 (5) and even for viruses such as the respiratory syncytial virus (6). The role of these exposures in further modifying the association between CD14/−159 and asthma and allergies is unknown. In addition, studies in which this association was assessed after stratifying for exposures other than endotoxin suggest an added degree of complexity. Eder and coworkers (26) found no association between CD14/−159 and allergic sensitization among subjects with no exposure to animals, a protective effect of the T allele among those exposed to indoor domestic animals (cats or dogs) and a protective effects of the C allele among those exposed to farm animals. These associations were only partially explained by concomitant exposure to house dust endotoxin. Similarly, Gern and colleagues (32) found that exposure to indoor dogs was protective against the development of atopic dermatitis in early life but only among TT homozygotes; no association was observed among carriers of the other two genotypes. Again, exposure to house dust endotoxin did not explain the interaction. Finally, Choudhry and coworkers (33) reported that TT homozygote children with asthma who were exposed to environmental tobacco smoke had significantly lower levels of total serum IgE than CC homozygotes, but that total serum IgE was unrelated to CD14/−159 among unexposed children. The product present in tobacco smoke that may mediate these effects is unknown, but it is unlikely to be endotoxin because, in such case, the four studies reported earlier would suggest that CC homozygotes, and not TT homozygotes, should be protected when heavily exposed. Taken together, these studies suggest that other genetic variants and exposures may influence the interactions we have described in this paper. These caveats notwithstanding, these interactions appear robust and suggest mechanisms of disease in asthma and allergies that would have never been detected had genetic and environmental studies been performed separately.
Figure 4.
Schematic representation of the single nucleotide polymorphisms identified in the Innate Immunity Program for Genomic Applications (PGA) database (http://innateimmunity.net). The SNPs that tag for the four most frequent SNP bins among subjects of European origin are also included. Reprinted by permission from Reference 36.
GENE–ENVIRONMENT INTERACTIONS IN MICE
All the studies described earlier are subject to the restrictions imposed by ethical rules governing human subject research and, therefore, some sources of bias could still be present. Exposures were not randomized, and thus subjects may make environmental choices (voluntary or involuntary) based on their genetic susceptibility to certain exposures, thus creating gene–environment covariation that may confound or bias the results. We doubt that these sources of bias may be important in the studies that assessed endotoxin and animal exposure by CD14/ −159 interactions, because no association was reported between the SNP and exposures for any of these studies. For example, in the study by Eder and colleagues (26) there was no association between any of the exposures measured (indoor pets, farm animals, or endotoxin) and CD14/−159. Nevertheless, other more complex sources of confounding cannot be entirely excluded.
Very recent, pioneering work by Valdar and coworkers (34, 35) has provided unequivocal evidence that the interactions we have observed are not likely to be explained by confounding alone and are by no means exceptional among mammals. These authors studied almost 2,500 genetically heterogeneous mice and estimated the heritability of 88 phenotypes (anatomical, biochemical, behavioral, physiological, hematological, and pathological). Concomitantly, they assessed the impact on these phenotypes of a number of randomly assigned environmental factors (e.g., cage density, season, experimenter, etc.), and measured the size of gene by environment interactions. The results were apparently unexpected: gene–environment interactions were more frequent and accounted globally for a larger proportion of phenotypic variance than main effects measured separately. For example, half of the gene–environment interactions explained more than 20% of the variance of the phenotype under study, whereas main effects rarely explained more than 10% of the variance of any phenotype. The authors also noticed the existence of antagonistic interactions of the type described earlier between genetic and environmental factors and stressed that, in that case, the interaction may conceal the effect of both, even when both environmental factor and the genetic locus are included in a statistical model. Similar cases of genetic variation in susceptibility to environmental factors have been observed in Drosophila and in plants (27).
CONCLUSIONS
The prevailing paradigm for the study of the genetic and environmental studies of asthma and allergies has been that these two factors can be studied separately and independent of each other. This approach has indeed resulted in undeniable successes, and many exposures and genetic variants have been shown to consistently modify the risk for these conditions. However, with obvious exceptions such as, for example, smoking, the genetic and environmental effects observed are most often small and not consistently replicated in different populations. What the evidence that I have reviewed herein suggests is that the premise implicit in these studies of independent effects of genes and environment may not adequately address the etiologic processes at work in the determination of asthma and allergies. These phenotypes are rarely “genetic” or “environmental”; I propose that, most often, they are the result of genetically determined, inadequate responses to a complex array of environmental exposures. With rare exceptions, neither these exposures nor the genes they interact with are consistent and univocal determinants of disease: depending on the context in which they act, they may predispose for disease, be neutral, or protect from disease. The example of CD14 is particularly illustrative of this situation and suggests that a scientific strategy centered on the characterization of these diverse inherited patterns of environmental responsiveness provides the best hope for the development of new, individualized approaches to the prevention of asthma and allergies.
Supported by HL064307, HL080083, and HL056177.
Conflict of Interest Statement: F.D.M. in the last three years has served on a Merck Advisory Board and acted as a consultant for Genentech and Pfizer. He has received symposium reimbursement and honoraria from all three of the aforementioned companies. He has no additional relationships with commercial entities.
Footnotes
The same polymorphism was also reported by Hubacek and coworkers, 2 months later (2).
References
- 1.Baldini M, Lohman IC, Halonen M, Erickson RP, Holt P, Martinez FD. A polymorphism in the 5′-flanking region of the CD 14 gene is associated with circulating soluble CD14 levels and with total serum IgE. Am J Respir Cell Mol Biol 1999;20:976–983. [DOI] [PubMed] [Google Scholar]
- 2.Hubacek JA, Pit'ha J, Skodova Z, Stanek V, Poledne R. C(-260)→T polymorphism in the promoter of the CD14 monocyte receptor gene as a risk factor for myocardial infarction. Circulation 1999;99:3218–3220. [DOI] [PubMed] [Google Scholar]
- 3.Los H, Koppelman GH, Postma DS. The importance of genetic influences in asthma. Eur Respir J 1999;14:1210–1227. [DOI] [PubMed] [Google Scholar]
- 4.Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol 1995;13:437–457. [DOI] [PubMed] [Google Scholar]
- 5.Cleveland MG, Gorham JD, Murphy TL, Tuomanen E, Murphy KM. Lipoteichoic acid preparations of gram-positive bacteria induce interleukin-12 through a CD14-dependent pathway. Infect Immun 1996;64:1906–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, Walsh EE, Freeman MW, Golenbock DT, Anderson LJ, et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol 2000;1:398–401. [DOI] [PubMed] [Google Scholar]
- 7.Strachan DP. Hay fever, hygiene, and household size. BMJ 1989;299:1259–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez FD, Holt PG. Role of microbial burden in aetiology of allergy and asthma. Lancet 1999;354:SII12–SII15. [DOI] [PubMed] [Google Scholar]
- 9.Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol 1988;141:547–552. [PubMed] [Google Scholar]
- 10.Vercelli D, Baldini M, Martinez F. The monocyte/IgE connection: may polymorphisms in the CD14 gene teach us about IgE regulation? Int Arch Allergy Immunol 2001;124:20–24. [DOI] [PubMed] [Google Scholar]
- 11.Koenig W, Khuseyinova N, Hoffmann MM, Marz W, Frohlich M, Hoffmeister A, Brenner H, Rothenbacher D. CD14 C(-260)→T polymorphism, plasma levels of the soluble endotoxin receptor CD14, their association with chronic infections and risk of stable coronary artery disease. J Am Coll Cardiol 2002;40:34–42. [DOI] [PubMed] [Google Scholar]
- 12.de la Fontaine L, Schwarz M, Plischke H, Kleindienst N, Gruber R. Lack of association of the CD14/C-159T polymorphism with susceptibility and serological activity parameters of rheumatoid arthritis. Scand J Rheumatol 2006;35:20–22. [DOI] [PubMed] [Google Scholar]
- 13.Wiertsema SP, Khoo SK, Baynam G, Veenhoven RH, Laing IA, Zielhuis GA, Rijkers GT, Goldblatt J, Lesouef PN, Sanders EA. Association of CD14 promoter polymorphism with otitis media and pneumococcal vaccine responses. Clin Vaccine Immunol 2006;13:892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu MS, Cheng TY, Shun CT, Lin MT, Chen LC, Lin JT. Functional polymorphisms of CD14 and toll-like receptor 4 in Taiwanese Chinese with Helicobacter pylori-related gastric malignancies. Hepatogastroenterology 2006;53:807–810. [PubMed] [Google Scholar]
- 15.LeVan TD, Bloom JW, Bailey TJ, Karp CL, Halonen M, Martinez FD, Vercelli D. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol 2001;167:5838–5844. [DOI] [PubMed] [Google Scholar]
- 16.Koppelman GH, Reijmerink NE, Colin Stine O, Howard TD, Whittaker PA, Meyers DA, Postma DS, Bleecker ER. Association of a promoter polymorphism of the CD14 gene and atopy. Am J Respir Crit Care Med 2001;163:965–969. [DOI] [PubMed] [Google Scholar]
- 17.Leung TF, Tang NL, Sung YM, Li AM, Wong GW, Chan IH, Lam CW. The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol 2003;14:255–260. [DOI] [PubMed] [Google Scholar]
- 18.Buckova D, Holla LI, Schuller M, Znojil V, Vacha J. Two CD14 promoter polymorphisms and atopic phenotypes in Czech patients with IgE-mediated allergy. Allergy 2003;58:1023–1026. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi K, Suzuki S, Yagawa M, Yuta A, Majima YA. CD14 gene polymorphism is associated with the IgE level for Dermatophagoides pteronyssinus. Acta Otolaryngol 2005;125:966–971. [DOI] [PubMed] [Google Scholar]
- 20.Kabesch M, Hasemann K, Schickinger V, Tzotcheva I, Bohnert A, Carr D, Baldini M, Hackstein H, Leupold W, Weiland SK, et al. A promoter polymorphism in the CD14 gene is associated with elevated levels of soluble CD14 but not with IgE or atopic diseases. Allergy 2004;59:520–525. [DOI] [PubMed] [Google Scholar]
- 21.Sengler C, Haider A, Sommerfeld C, Lau S, Baldini M, Martinez F, Wahn U, Nickel R. Evaluation of the CD14 C-159 T polymorphism in the German Multicenter Allergy Study cohort. Clin Exp Allergy 2003;33:166–169. [DOI] [PubMed] [Google Scholar]
- 22.Kedda MA, Lose F, Duffy D, Bell E, Thompson PJ, Upham J. The CD14 C-159T polymorphism is not associated with asthma or asthma severity in an Australian adult population. Thorax 2005;60:211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet 2000;67:1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Von Mutius E, Braun-Fahrlander C, Schierl R, Riedler J, Ehlermann S, Maisch S, Waser M, Nowak D. Exposure to endotoxin or other bacterial components might protect against the development of atopy. Clin Exp Allergy 2000;30:1230–1234. [DOI] [PubMed] [Google Scholar]
- 25.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, Maisch S, Carr D, Gerlach F, Bufe A, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med 2002;347:869–877. [DOI] [PubMed] [Google Scholar]
- 26.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, Nowak D, Martinez FD, The A. Endotoxin Alex Study T. Opposite effects of CD14/-260 on serum IgE levels in children raised in different environments. J Allergy Clin Immunol 2005;116:601–607. [DOI] [PubMed] [Google Scholar]
- 27.Pigliucci M, Schlichting C. Phenotypic evolution: a reaction norm perspective. Sunderland, MA: Sinauer Associates, Inc.; 1996.
- 28.Simpson A, John SL, Jury F, Niven R, Woodcock A, Ollier WE, Custovic A. Endotoxin exposure, CD14 and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med 2006;174:386–392. [DOI] [PubMed] [Google Scholar]
- 29.Williams LK, McPhee RA, Ownby DR, Peterson EL, James M, Zoratti EM, Johnson CC. Gene-environment interactions with CD14 C-260T and their relationship to total serum IgE levels in adults. J Allergy Clin Immunol 2006;118:851–857. [DOI] [PubMed] [Google Scholar]
- 30.Zambelli-Weiner A, Ehrlich E, Stockton ML, Grant AV, Zhang S, Levett PN, Beaty TH, Barnes KC. Evaluation of the CD14/-260 polymorphism and house dust endotoxin exposure in the Barbados Asthma Genetics Study. J Allergy Clin Immunol 2005;115:1203–1209. [DOI] [PubMed] [Google Scholar]
- 31.Hong SJ, Kim HB, Kang MJ, Lee SY, Kim JH, Kim BS, Jang SO, Shin HD, Park CS. TNF-alpha (-308 G/A) and CD14 (-159T/C) polymorphisms in the bronchial responsiveness of Korean children with asthma. J Allergy Clin Immunol 2007;119:398–404. [DOI] [PubMed] [Google Scholar]
- 32.Gern JE, Reardon CL, Hoffjan S, Nicolae D, Li Z, Roberg KA, Neaville WA, Carlson-Dakes K, Adler K, Hamilton R, et al. Effects of dog ownership and genotype on immune development and atopy in infancy. J Allergy Clin Immunol 2004;113:307–314. [DOI] [PubMed] [Google Scholar]
- 33.Choudhry S, Avila PC, Nazario S, Ung N, Kho J, Rodriguez-Santana JR, Casal J, Tsai HJ, Torres A, Ziv E, et al. CD14 tobacco gene-environment interaction modifies asthma severity and immunoglobulin E levels in Latinos with asthma. Am J Respir Crit Care Med 2005;172:173–182. [DOI] [PubMed] [Google Scholar]
- 34.Valdar W, Solberg LC, Gauguier D, Cookson WO, Rawlins JN, Mott R, Flint J. Genetic and environmental effects on complex traits in mice. Genetics 2006;174:959–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 2006;38:879–887. [DOI] [PubMed] [Google Scholar]
- 36.LeVan TD, Guerra S, Klimecki W, Vasquez MM, Lohman IC, Martinez FD, Halonen M, Wright AL. The impact of CD14 polymorphisms on the development of soluble CD14 levels during infancy. Genes Immun 2006;7:77–80. [DOI] [PubMed] [Google Scholar]