Abstract
Asthma is a common but complex respiratory ailment; current data indicate that interaction of genetic and environmental factors lead to its clinical expression. In the United States, asthma prevalence, morbidity, and mortality vary widely among different Latino ethnic groups. The prevalence of asthma is highest in Puerto Ricans, intermediate in Dominicans and Cubans, and lowest in Mexicans and Central Americans. Independently, known socioeconomic, environmental, and genetic differences do not fully account for this observation. One potential explanation is that there may be unique and ethnic-specific gene–environment interactions that can differentially modify risk for asthma in Latino ethnic groups. These gene–environment interactions can be tested using genetic ancestry as a surrogate for genetic risk factors. Latinos are admixed and share varying proportions of African, Native American, and European ancestry. Most Latinos are unaware of their precise ancestry and report their ancestry based on the national origin of their family and their physical appearance. The unavailability of precise ancestry and the genetic complexity among Latinos may complicate asthma research studies in this population. On the other hand, precisely because of this rich mixture of ancestry, Latinos present a unique opportunity to disentangle the clinical, social, environmental, and genetic underpinnings of population differences in asthma prevalence, severity, and bronchodilator drug responsiveness.
Keywords: genes, environments, Latinos, Hispanics, asthma
LATINO (HISPANIC) POPULATIONS
Latinos are the largest, youngest, and fastest growing minority population in the United States, accounting for 14% of the nation's total population. Currently, there are 42.6 million Latinos in the United States, not including residents of Puerto Rico. By 2050, it is predicted that 25% of the U.S. population will be Latino (2, 3). Furthermore, among all children in the United States, Latinos represent the largest demographic group (4). Latinos are not a homogeneous ethnic group, because there is great genetic, socioeconomic, educational, and demographic variation both between and within Latino ethnic groups. The diversity of the Latino population provides the intrinsic variability necessary to study the interactions of race, genetics, culture, and environment, and their impact on asthma. Some of the discussion of these studies has been previously reported in the form of original research (5–13).
LATINO POPULATIONS AND ANCESTRY
The term “Hispanic” or “Latino” describes a population with a shared cultural heritage and most often a universal language, but does not refer to race or a common ancestry. Race is usually considered as a fixed characteristic of the individual, linked to their genetic makeup, whereas ethnicity represents a broader construct based on cultural tradition, common history, language, religion, and often a shared genetic heritage (14). Although Latinos have been considered to be an ethnic group, they represent a heterogeneous mix of Native American, European, and African ancestries (8, 10, 11). Therefore, Latino individuals can self-identify as any race or of mixed race as defined by the 2000 U.S. Census. For example, in the 2000 U.S. Census, 97.9% of the non-Latino U.S. population self-identified as one of the five major racial categories. However, 48% of Latinos self-identified as white, 2% as African/African American, 1% as American Indian, and 42% as “some other race” (4). This demonstrates the complexity of self-identification of Latinos for epidemiologic studies.
U.S. DEMOGRAPHIC SHIFTS
Recently, there have been dramatic shifts in the “source country” of immigrants to the United States, with more than half coming from Latin America (15). The top 10 Latino sources of immigration to the United States are Mexico, El Salvador, the Dominican Republic, Colombia, Guatemala, Peru, Cuba, Ecuador, Brazil, and Honduras (4). Puerto Ricans are U.S. citizens by birth and not considered immigrants. This demographic shift has dramatically altered the Latino population within major cities in the United States. For example, in the Bronx, New York, in the 2000 U.S. Census, 48% of the population was Latino/Hispanic and, of this, the major Latino ethnic groups were as follows: Puerto Ricans (48%), Dominicans (21%), Mexicans (5%), Central Americans (3%), South Americans (3%), Cubans (1%), and other Latinos (17%).
ASTHMA AMONG LATINO AMERICANS
Asthma is a common respiratory disease that is caused by genetic and environmental factors. U.S. vital statistics for asthma demonstrate that asthma prevalence, morbidity, and mortality are highest in Puerto Ricans, intermediate in Dominicans and Cubans, and lowest in Mexicans and Central Americans (1, 16, 17). This is paradoxical since all groups are considered “Hispanic.” Socioeconomic and environmental differences measured to date have not fully explained the discrepancy of asthma among Latino Americans. The discrepancy in asthma burden, as well as the paucity of studies of asthma in Latinos and especially among different Latino ethnic groups, has led the American Academy of Pediatrics to identify asthma among Latinos as an urgent priority for further research (18).
ENVIRONMENTS AND ASTHMA
Freeman and colleagues administered an asthma symptom and household exposure factor questionnaire to 4,634 schoolchildren in Passaic, New Jersey (19). Passaic is a unique community in that it has a wide range of Latino ethnic groups representing the Caribbean, Mexico, and Central and South America. This diversity offers a unique opportunity to compare asthma and environmental exposures for a variety of Latino ethnic groups within the same urban and predominantly poor community. Asthma diagnosis was highest in Puerto Ricans (26%), intermediate for Dominicans (14%), and lowest for Mexicans (7%). Although environmental exposures were similar for all groups, environmental factors associated with asthma differed by ethnic group. For example, damp/moldy conditions were associated with asthma in Puerto Ricans but not Mexicans and Dominicans. These results suggest that there are ethnicity-specific gene–environment interactions for asthma and asthma-related phenotypes.
LATINO POPULATIONS IN THE UNITED STATES PROVIDE A UNIQUE OPPORTUNITY FOR GENETIC EPIDEMIOLOGY STUDIES
One of the primary limitations in dissecting the etiology of differences in health and disease experiences among racial/ethnic groups is confounding. Puerto Ricans, Dominicans, and Mexican Americans differ from one another in terms of culture, socioeconomic status, and levels of discrimination in work and housing, as well as genetic ancestry. The variation in ancestral proportions between Latino subgroups can be exploited by using a variety of epidemiologic study designs and modern genetic techniques, to potentially unravel some of the differences in disease incidence and outcomes.
Despite the large size of the Latino population and the extensive asthma research documenting a high asthma prevalence, morbidity, and mortality for specific Latino ethnic groups, Latinos have, for the most part, not been included in asthma clinical, genetic, and/or epidemiologic research. Although the difficulty in recruitment of minority populations is partly to blame for the paucity of research in these populations, the genetic complexity of admixed populations further complicates studies simply because of genetic confounding. However, the wide variation in admixture and environmental exposure in Latino ethnic groups provides the intrinsic variability needed to untangle complex gene–gene and gene–environment interactions in asthma susceptibility and severity. To this end, we have initiated the Genetics of Asthma in Latino Americans (GALA) study.
THE GALA STUDY
GALA is a multicenter, international effort designed to identify and directly compare clinical, genetic, and environmental risk factors associated with asthma, asthma severity, and drug responsiveness among Latino ethnic groups. In our first analysis of GALA participants (5), we compared asthma-related clinical characteristics between 300 Mexican and 386 Puerto Rican individuals with asthma recruited from San Francisco, New York City, Puerto Rico, and Mexico City. We found that Puerto Ricans with asthma had a higher risk of an emergency department visit in the previous year (odds ratio [OR], 2.63; 95% confidence interval [CI], 1.6–4.3; p < 0.001), and of previous hospitalization for asthma (OR, 1.94; 95% CI, 1.2–3.2; p = 0.009), than Mexicans. We also tested participants for responsiveness to albuterol, a β2-adrenergic receptor agonist and bronchodilator drug, by measuring the percentage change from baseline FEV1. Worldwide, albuterol is the most commonly prescribed treatment for asthma. Interestingly, Puerto Ricans with asthma had, on average, 7.3% (95% CI, 4.6–9.9; p < 0.001) lower bronchodilator responsiveness than Mexicans with asthma. This finding suggests that there may be subgroups of subjects with asthma that may not respond well to commonly prescribed asthma therapies. If replicated, this finding could have important clinical and public health implications.
Despite the ubiquitous use of albuterol in the treatment of asthma, there is significant variation in drug efficacy (20), as suggested by our finding in Puerto Ricans and Mexicans. Understanding the genetic basis of variability in drug response (pharmacogenetics) will help physicians to optimize their diagnosis and treatment of individual patients or patient groups. Among GALA participants, we demonstrated that there are ethnic-specific genetic factors that contribute to observed differences in physiologic response to albuterol. Specifically, we demonstrated that the Arg16 allele of the β2AR gene is associated with greater bronchodilator responsiveness in Puerto Ricans but not in Mexicans with asthma (9). In contrast, we demonstrated that the Arg-19Cys polymorphism in the β upstream peptide of the β2AR gene may play an important role in bronchodilator drug responsiveness in African-American subjects (21). Potential causes of this variation include differences in gene–gene and gene–environment interactions, and ethnic-specific differences in patterns of linkage disequilibrium (LD).
GENETIC ESTIMATION OF INDIVIDUAL ANCESTRY
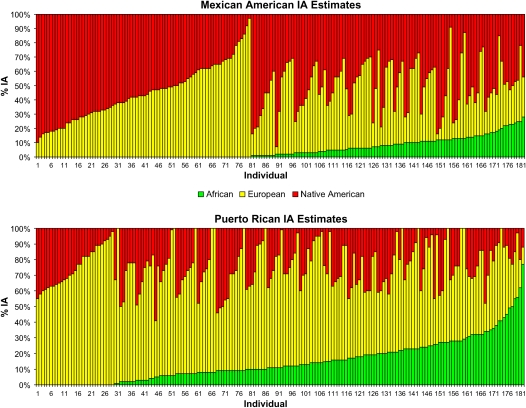
Self-reported ancestry among Latinos is inaccurate for determination of individual ancestry (IA) (8, 10, 11). However, genetic markers that are informative for ancestry and newly developed statistical methods are making the genetic estimation of ancestry increasingly more accurate (22–25). We genotyped 44 ancestry informative markers (AIMs) to determine IA estimates in a sample of 181 Mexican subjects with asthma and 181 Mexican control subjects from the San Francisco Bay Area and 181 Puerto Rican subjects with asthma and 178 Puerto Rican control subjects from Puerto Rico, collected as part of the GALA study. Although all participants, their biological parents, and all grandparents, self-identified as either Mexican or Puerto Rican, respective to their ethnicity, we still observed substantial variation in ancestry among individuals within these two Latino ethnic groups. In Figure 1, we show the distribution of IA estimates for Mexican American and Puerto Rican cases estimated using individual biogeographical ancestry (IBGA) which implements a maximum likelihood–based method described by Hanis and coworkers (22). As can be seen from this figure, there is dramatic heterogeneity in admixture levels among individuals within each Latino ethnic group despite stringent recruitment by ethnicity. For example, some Puerto Rican subjects had less than 10% African ancestry, whereas others had over 50% African ancestry. Moreover, European ancestry among Puerto Ricans ranged from under 20% to over 80%, whereas Native American ancestry showed less fluctuation, generally hovering between 5 and 20%.
Figure 1.
Individual ancestry (IA) estimates. Estimates for 181 Mexican Americans with asthma (top) and 181 Puerto Ricans with asthma (bottom) are shown, clustered by admixture level. The distribution of IA estimates in Mexican Americans covers the range of European and Native American proportions, whereas African ancestry contributes very little to this population. However, in Puerto Ricans, African and European ancestries show a high degree of variability, whereas Native American ancestry exhibits a more restricted pattern of variation. Note that some Mexican and Puerto Rican individuals overlap in terms of ancestry.
It is also clear that two Latino ethnic groups, Mexicans and Puerto Ricans, are, on average, different in terms of ancestry. Although Puerto Ricans have 66% European ancestry, Mexicans have 45%; Puerto Ricans have 16% African ancestry compared with 3% for Mexicans; Puerto Ricans have only 18% Native American ancestry compared with 52% for Mexicans.
POPULATION STRATIFICATION AND GENETIC CONFOUNDING
Population stratification in an admixed case-control cohort is the over- or underrepresentation of a particular ancestral proportion in cases versus control subjects. A primary cause of population stratification in admixed cohorts is the unequal distribution of disease risk between ancestral groups, which results in oversampling of the subjects with an excess of the “high risk” ancestry in the case group (8, 10–13, 26, 27). A stratified cohort can result in an excess of false-positive and/or false-negative results in genetic studies if the alleles tested are relatively specific to the differentially distributed ancestry. Ideally, investigators would match cases and control subjects with respect to ancestry and prevent this type of genetic confounding (13, 28). However, we have demonstrated that an approach based on self-reported ancestry cannot be reliably used in studies of asthma in genetically complex tri-hybrid Latino populations (8).
We have previously demonstrated that population stratification confounds genetic association studies of asthma (8, 11, 27). We used AIMs to identify and correct for population stratification among Mexican and Puerto Rican subjects participating in case-control studies of asthma. Three hundred and sixty-two subjects with asthma (Mexican, 181; Puerto Rican, 181) and 359 ethnically matched control subjects (Mexican, 181; Puerto Rican, 178) were genotyped for 44 AIMs. We observed a greater than expected degree of allelic association between pairs of AIMs on different chromosomes in Mexicans (p < 0.00001) and Puerto Ricans (p < 0.00002), providing evidence for population substructure and/or recent admixture. To assess the effect of population stratification on association studies of asthma, we measured differences in genetic background of cases and control subjects by comparing allele frequencies of the 44 AIMs. Among Puerto Ricans but not Mexicans, we observed a significant overall difference in allele frequencies between cases and control subjects (p = 0.0002); of 44 AIMs tested, 8 (18%) were significantly associated with asthma. However, after adjustment for IA, only two of these markers remained significantly associated with the disease. Our findings suggest that empirical assessment of the effects of stratification is critical to appropriately interpret the results of case-control studies in admixed populations.
The former study supports the argument that population stratification can likely not be controlled for in Latino populations despite meticulous matching based on self-reported ancestry. Rather, this example shows the potential power of using “genetically determined” individual ancestral proportions to correct for this type of confounding. In the next section, we detail different statistical methods and the number and type of AIMs required for adequate IA estimates and subsequent correction for population stratification.
CORRECTING FOR POPULATION STRATIFICATION USING IA ESTIMATES
Several methods have been proposed to estimate individual admixture proportions in admixed populations, which are then used to adjust for population stratification in genetic association tests. Most of these methods have been developed and implemented in two-way admixed populations, such as African Americans. We investigated the common case of three-way population admixture, which is relevant to Latino populations. Specifically, we evaluated and compared the performance of three different methods for estimating individual admixture and for eliminating excess type I error rate due to population stratification using various simulated datasets (12). The three methods are as follows: maximum likelihood estimation, ADMIXMAP (version 0.6.4), and STRUCTURE (version 2.1). According to our results, the method selected has a relatively small impact on the accuracy of individual admixture estimates (23–25) or on the type I error rate. By far the most important factor in determining accuracy of the admixture estimate and in minimizing the type I error rate appears to be the number of markers used to estimate admixture. We demonstrate that, for markers with a mean delta value (marker informativeness) of 0.4 (range from 0.1 to 0.8), approximately 100 markers are required to obtain estimates of admixture that correlate strongly (r > 0.9) with the true admixture estimates. In addition, after accounting for admixture information in association tests, the excess type I error rate is controlled at the 5% level when 100 markers are used to estimate admixture. These simulations provide some practical guidelines for investigators conducting association studies in admixed populations.
ADMIXTURE VARIATION AND QUANTITATIVE TRAITS OF ASTHMA
The extent of interindividual variation in ancestral proportions among Latinos presents the opportunity to correlate global ancestry with quantitative traits of asthma, such as FEV1, a surrogate for asthma severity, and bronchodilator response to albuterol. Differences in ancestry proportion may partly explain the dramatic differential in these traits between Puerto Ricans and Mexicans. To address this hypothesis, we used regression models to test for association between individual admixture (IA) estimates and the quantitative distributions of asthma severity, as defined by FEV1 and bronchodilator responsiveness (ΔFEV1) among Puerto Ricans and Mexicans (11). Age, sex, asthma duration, regular use of asthma medication, socioeconomic status (SES), and body mass index (BMI) were entered in the model as covariates. To adjust for potential environmental interactions, secondhand exposure to environmental tobacco smoke (ETS) and birthplace were also incorporated into the models as covariates. We used a forward stepwise procedure to select covariates for each model.
We found significant relationships between ancestry and asthma severity in our Mexican American population. European ancestry was associated with more severe asthma (defined by lower baseline FEV1 [Pearson r = –0.211, p = 0.0051; Spearman r = −0.228, p = 0.0024]). European ancestry remained a significant predictor of baseline FEV1 in a multivariate regression model after adjustment for age, sex, asthma duration, regular use of asthma medication, ETS exposure, birthplace, SES, recruitment site, and BMI. The forward stepwise regression model identified age and ETS exposure as the only other significant effectors of baseline FEV1. A decrease of 1.7% (95% CI, 0.6–2.8%) in baseline FEV1 was observed per 10% increase in European ancestry. Because our Mexican subjects with asthma, on average, had 45% European, 52% Native American, and only 3% African ancestry, there was a strong negative correlation between Native American and European ancestry. Therefore, any negative association with European ancestry would be expected to have a positive association with Native American ancestry. As expected, in models in which Native American ancestry was the main predictor, Native American ancestry was associated with milder asthma (defined as higher measures of baseline FEV1 [Pearson r = 0.176, p = 0.0197; Spearman r = 0.186, p = 0.0139]). After correction for the aforementioned potential confounders, Native American ancestry remained significantly associated with higher baseline FEV1 values.
We also tested the association between ancestry and asthma severity as a qualitative trait, comparing the proportion of ancestry among subjects with mild versus severe asthma, as defined by asthma medication use and clinical symptoms. We found an association between higher European ancestry and asthma severity among Mexicans with asthma. For each 10% increase in European ancestry, there was an approximate 37% risk increase for severe asthma. Adjustment for age, sex, asthma duration, ETS exposure, birthplace, and BMI did not affect the association. Conversely, Native American ancestry was associated with decreased risk of asthma severity, with an OR of 0.66 (95% CI, 0.54–0.81, p = 0.00006) for having severe asthma versus mild asthma per 10% increase in Native American admixture. Both of the associations between European ancestry and lower baseline FEV1 and increased clinical severity were significant after adjustment for multiple hypothesis testing. These associations between ancestry and asthma-related phenotypes provide supporting evidence for the use of admixture mapping method (described below) to identify genes associated with asthma-related traits among Latino populations.
INTERACTION BETWEEN ANCESTRY AND SES
Asthma prevalence varies with SES within and across racial/ethnic groups, with low-SES populations exhibiting higher rates (29, 30). In addition to individual SES status, neighborhood SES also influences the risk of asthma (31–33). Claudio and colleagues demonstrated that children who live in predominantly low SES communities had a 70% increased risk of current asthma, independent of ethnicity and individual income level, except for Puerto Rican children, who had high asthma prevalence, regardless of income (32). The higher asthma prevalence and morbidity rates experienced by Puerto Rican children cannot be explained by traditional measures of sociodemographic and other risk factors assessed in traditional epidemiologic surveys (1).
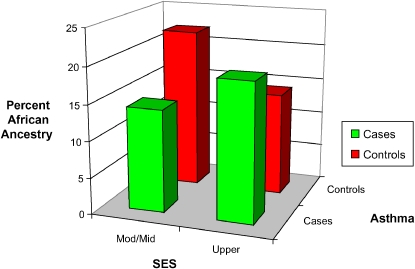
We hypothesized that, in this admixed population, the association between SES and asthma may interact with genetic ancestry. Using genetic AIMs, we found that ancestry interacts with SES to modify risk for asthma among Puerto Ricans (Figure 2) (6). Specifically, we demonstrated that, among Puerto Ricans of lower SES, European ancestry was associated with increased risk of asthma whereas African ancestry was associated with decreased risk. The opposite was true for their higher SES counterparts. These results reveal a complex interaction between SES and ancestry with respect to asthma within the Puerto Rican population. Although we have previously demonstrated gene–environment interactions for asthma in Latino populations in which the environmental exposure is secondhand tobacco smoke (7), the results reported here reflect a higher order interaction among ancestry (possibly reflecting genetic background), SES, and asthma, suggesting the complexity of gene–environment interaction for asthma. The observed interaction may help to explain the unique pattern of risk for asthma in Puerto Ricans and the lack of association with SES observed in previous studies when not accounting for varying proportions of ancestry. To our knowledge, this is the first demonstration of an interaction between ancestral proportions and environmental factors that modifies the risk of a complex disease.
Figure 2.
Percentages of African ancestry in Puerto Rican asthma cases and control subjects stratified by socioeconomic status (SES). SES was obtained using clinic recruitment site address.
ADMIXTURE MAPPING
Admixture mapping is an alternative approach to perform genomewide association analysis to identify regions harboring race/ethnicity-specific risk alleles for a disease in admixed populations. Admixture mapping capitalizes on the fact that recently admixed populations are known to have large regions of LD (or genetic blocks) across genetic markers that are informative for ancestry (34–36). Admixture mapping uses this increased LD to identify loci associated with complex disease phenotypes. The underlying premise behind admixture mapping is that if a marker increases the risk of disease and is found at a much higher frequency in one population (the high-risk population), then that marker will also be found more frequently among cases. Furthermore, that marker will be in LD with other AIMs that are specific to the high-risk population, and this LD will be spread across large regions (LD blocks) of the genome. By genotyping AIMs across the genome, one may be able to identify genomic regions in which the cases share ancestry with the high-risk population more commonly than expected. Such loci presumably harbor disease-causing variants.
Admixture mapping is especially relevant in Latino populations because their admixture is relatively recent and this results in long-range LD (37). The ideal period of admixture for admixture mapping is approximately 5 to 20 generations (38). More remote admixture would mean that LD would have decayed and therefore would require more markers. Conversely, more recent admixture (1–3 generations ago) would mean that LD would extend too far to accurately localize a genomic region (39). Historical and genetic evidence suggests that the admixture among Latinos and African Americans has occurred over the past 20 generations and thus provides an ideal situation for admixture mapping (37).
In comparison to family-based linkage studies, admixture mapping has higher statistical power to detect genes of modest effect if risk alleles in these genes are distributed differentially between subpopulations. For example, in an extreme case, less than 200 affected individuals were required for detection of a disease locus by admixture mapping, but at least 4,000 affected sibling pairs were required for detection of the disease locus at the same statistical power (40, 41). Admixture mapping is also less susceptible to allelic heterogeneity (42). The ability to detect a disease locus using an admixture mapping approach depends only on whether the pool of high-risk alleles is distributed differentially between subpopulations; it does not matter whether there are a few common risk alleles or many rare risk alleles at the locus under study. In contrast, single-nucleotide polymorphism association studies of common diseases depend on the “common disease–common variant” hypothesis.
The phenotypes that are of greatest interest for admixture mapping are those which demonstrate differences in racial/ethnic populations and which may not be explained by known environmental differences among populations (43). Thus far, there are no published results from admixture mapping studies performed for asthma or asthma-related traits. However, Zhu and colleagues recently used a panel of 269 microsatellite markers and an admixture mapping approach to estimate ancestry across the entire genome among African-American subjects with hypertension and healthy control subjects (44). They identified two regions on chromosomes 6q and 21q that had excess locus specific African ancestry in hypertensive cases versus normotensive controls, suggesting that these regions may contain genes influencing risk of hypertension in African Americans. Patterson and colleagues demonstrated that this approach can be used to map other known loci, such as the HLA locus (39). Another recent high-powered admixture scan, using 605 African-American cases and 1,043 control subjects, revealed a locus on chromosome 1 that is significantly associated with multiple sclerosis (45). These promising results indicate a strong possibility for success in well-designed admixture mapping studies for asthma and asthma-related traits in admixed populations.
MIGRANT STUDIES AND ACCULTURATION
Large migrations of various Latino ethnic groups to the United States over the last several generations facilitate migrant studies, which may provide important clues about disease etiology. For example, if a group migrating into a new country with a different rate of disease takes on the same rate of disease as the resident population with successive generations, this strongly suggests that environmental factors are responsible for the initial difference. Conversely, if the migrant group retains the same rate of disease as the country from which they migrated over several generations, a genetic difference may be responsible. Contrasting the migration effects of different Latino ethnic groups on asthma may also provide additional clues, especially regarding issues of genetic, socioeconomic, and cultural factors, and their interactions. However, migrant studies need to be interpreted with caution, particularly among Latino populations, in which the populations in the countries of origin are diverse and may have unique genetic and socioeconomic characteristics. Migrant groups may not reflect the general population of the country of origin, resulting in a selection bias. For example, Holguin and colleagues analyzed two independent national databases and confirmed that asthma prevalence is higher among U.S.-born Mexicans than Mexican-born Mexicans (46). In this case, it might be concluded that such differences in disease prevalence arise purely from environmental factors. However, the ancestry of Mexican subjects participating in these studies was based on self-report and not their genetic ancestry. Using AIMs in a population of self-identified Mexicans with asthma recruited from California and Mexico City, we demonstrated that admixture proportions differ among U.S.-born Mexicans, Mexicans who were born in Mexico and who are currently living in California, and Mexicans who were born in and are currently living in Mexico City (10). Specifically, among our study participants, the proportion of Native American ancestry is higher among Mexicans with asthma living in Mexico City than among those living in California. Furthermore, Mexicans with asthma born in Mexico who then immigrated to the United States also have lower Native American ancestry than Mexicans in Mexico City. Finally, the migrating populations may change over time due to socioeconomic and political forces.
Acculturation refers to the cultural modification of an individual, group, or population by adapting to or borrowing traits from another culture: that is, in the United States, acculturation is usually defined and measured by indicators of cultural belonging, such as nativity status (U.S.- vs. foreign-born), language preference, and length of stay in the United States. Acculturation has been shown to affect the health status of Latinos and asthma is no exception. For example, Klinnert and coworkers showed that Latino children with low-acculturated parents (Spanish speaking and foreign-born) exhibited lower prevalence of asthma than their high-acculturated counterparts (English speaking and U.S.-born) (47). Moreover, the high-acculturated Latinos had a higher prevalence of the risk variables associated with low SES. The transition and flows of the Latino population between their country of origin and the United States provide a natural experiment to study the role of cultural changes due to acculturation and the effect of a migration on asthma and related phenotypes.
The study of Latino ethnic groups affords a unique opportunity to study the effect of a migration and acculturation on the asthma phenotype and related phenotypes. Furthermore, this type of study affords the opportunity to examine the “Hispanic paradox” or health advantage of Latinos over non-Latinos on a group other than Mexican Americans and on outcomes other than mortality (48–54).
CONCLUSIONS: NEW APPROACHES TO OLD CHALLENGES
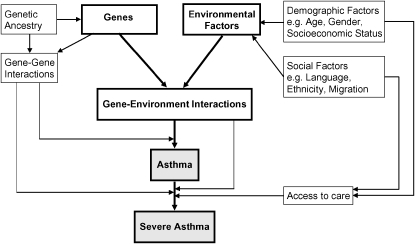
There is evidence to suggest that there are genetic, environmental, social, and economic risk factors independently and dependently influencing the development and severity of asthma (see Figure 3).
Figure 3.
Causal model for asthma.
Novel multidisciplinary approaches will be required to elucidate the complex gene–environment interactions that result in asthma and other complex diseases. Our success will require a paradigm shift with respect to traditional research. Although not new, the idea of translational and cross-disciplinary research has yet to be fully implemented for asthma. The results from the GALA study have broadened our understanding of the interactions of race, genetics, culture, and environment, and their impact on asthma. Most important, the GALA study has provided new directions for our future work.
An integrative and comprehensive approach will require “team science”; large and well-phenotyped cohorts; thorough measures of environmental, demographic, and social factors; large-scale genotyping; and complex analyses in which we test for gene–gene and gene–environment interactions. In addition, team science will require novel and innovative ways to acknowledge individual contributions and shared credit. By taking a team approach, we are likely to gain a much more thorough understanding of disease, its causes, and its distribution, which will benefit all.
Acknowledgments
The authors thank the families and the patients for their participation. The authors also thank the numerous health care providers and community clinics for their support and participation in the GALA study. The authors would like to especially thank Jeffrey M. Drazen, M.D., Scott Weiss, M.D., Ed Silverman, M.D., Ph.D., Homer A. Boushey, M.D., and Jean G. Ford, M.D., for all of their effort toward the creation of the GALA study.
Supported by the National Institutes of Health (K23 HL04464, HL078885, American Lung Association of California, RWJ Amos Medical Faculty Development Award, NCMHD Health Disparities Scholar, Extramural Clinical Research Loan Repayment Program for Individuals from Disadvantaged Backgrounds, 2001–2003, to E.G.B.), an American Thoracic Society “Breakthrough Opportunities in Lung Disease” (BOLD) Award and a Tobacco-Related Disease Research Program New Investigator Award (15KT-0008) to S.C.; the Ernest S. Bazley Grant to P.C.A., and the Sandler Center for Basic Research in Asthma and the Sandler Family Supporting Foundation.
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Lara M, Akinbami L, Flores G, Morgenstern H. Heterogeneity of childhood asthma among Hispanic children: Puerto Rican children bear a disproportionate burden. Pediatrics 2006;117:43–53. [DOI] [PubMed] [Google Scholar]
- 2.Census, race and science [Editorial]. Nat Genet 2000;24:97–98. [DOI] [PubMed] [Google Scholar]
- 3.U.S. Census Bureau. 2003.
- 4.U.S. Census Bureau. U.S. Department of Commerce; 2000.
- 5.Burchard, EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, Toscano M, Sylvia JS, Alioto M, Salazar M, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican asthmatic subjects. Am J Respir Crit Care Med 2003;169:386–392. [DOI] [PubMed] [Google Scholar]
- 6.Choudhry S, Burchard EG, Borrell LN, Tang H, Gomez I, Naqvi M, Nazario S, Torres A, Casal J, Martinez-Cruzado JC, et al. J. Ancestry–environment interactions and asthma risk among Puerto Ricans. Am J Respir Crit Care Med 2006;174:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhry S, Avila PC, Nazario S, Ung N, Kho J, Rodriguez-Santana JR, Casal J, Tsai HJ, Torres A, Ziv E, et al. CD14 tobacco gene–environment interaction modifies asthma severity and immunoglobulin E levels in Latinos with asthma. Am J Respir Crit Care Med 2005;172:173–182. [DOI] [PubMed] [Google Scholar]
- 8.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet 2006;118:652–664. [DOI] [PubMed] [Google Scholar]
- 9.Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, Torres A, Gorman JD, Salari K, Rodriguez-Santana JR, et al. Pharmacogenetic differences in response to albuterol between Puerto Rican and Mexican asthmatics. Am J Respir Crit Care Med 2005;171:563–570. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez Burchard E, Borrell LN, Choudhry S, Naqvi M, Tsai HJ, Rodriguez-Santana JR, Chapela R, Rogers SD, Mei R, Rodriguez-Cintron W, et al. Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am J Public Health 2005;95:2161–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salari K, Choudhry S, Tang H, Naqvi M, Lind D, Avila PC, Coyle NE, Ung N, Nazario S, Casal J, et al. Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet Epidemiol 2005;29:76–86. [DOI] [PubMed] [Google Scholar]
- 12.Tsai HJ, Choudhry S, Naqvi M, Rodriguez-Cintron W, Burchard EG, Ziv E. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum Genet 2005;118:424–433. [DOI] [PubMed] [Google Scholar]
- 13.Tsai HJ, Kho JY, Shaikh N, Choudhry S, Naqvi M, Navarro D, Matallana H, Castro R, Lilly CM, Watson HG, et al. Admixture-matched case-control study: a practical approach for genetic association studies in admixed populations. Hum Genet 2006;118:626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med 2003;348:1170–1175. [DOI] [PubMed] [Google Scholar]
- 15.Schmidley D. The foreign-born population in the United States: March 2002. Current Population Reports: U.S. Census Bureau; 2003. p. 20–539.
- 16.Carter-Pokras OD, Gergen PJ. Reported asthma among Puerto Rican, Mexican-American, and Cuban children, 1982 through 1984. Am J Public Health 1993;83:580–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homa DM, Mannino DM, Lara M. Asthma mortality in U.S. Hispanics of Mexican, Puerto Rican, and Cuban heritage, 1990–1995. Am J Respir Crit Care Med 2000;161:504–509. [DOI] [PubMed] [Google Scholar]
- 18.Flores G, Fuentes-Afflick E, Barbot O, Carter-Pokras O, Claudio L, Lara M, McLaurin JA, Pachter L, Gomez FR, Mendoza F, et al. The health of Latino children: urgent priorities, unanswered questions, and a research agenda. JAMA 2002;288:82–90. [DOI] [PubMed] [Google Scholar]
- 19.Freeman NC, Schneider D, McGarvey P. Household exposure factors, asthma, and school absenteeism in a predominantly Hispanic community. J Expo Anal Environ Epidemiol 2003;13:169–176. [DOI] [PubMed] [Google Scholar]
- 20.Nelson HS. Beta-adrenergic bronchodilators. N Engl J Med 1995;333:499–506. [DOI] [PubMed] [Google Scholar]
- 21.Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, Matallana H, Lilly CM, Eng CS, Kumar G, et al. Beta 2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet 2006;119:547–557. [DOI] [PubMed] [Google Scholar]
- 22.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol 1986;70:433–441. [DOI] [PubMed] [Google Scholar]
- 23.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet 2003;72:1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L, et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol 2001;114:18–29. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000;155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol 2002;3(7): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziv E, Burchard EG. Human population structure and genetic association studies. Pharmacogenomics 2003;4:431–441. [DOI] [PubMed] [Google Scholar]
- 28.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet 2003;361:598–604. [DOI] [PubMed] [Google Scholar]
- 29.Litonjua AA, Carey VJ, Weiss ST, Gold DR. Race, socioeconomic factors, and area of residence are associated with asthma prevalence. Pediatr Pulmonol 1999;28:394–401. [DOI] [PubMed] [Google Scholar]
- 30.Almqvist C, Pershagen G, Wickman M. Low socioeconomic status as a risk factor for asthma, rhinitis and sensitization at 4 years in a birth cohort. Clin Exp Allergy 2005;35:612–618. [DOI] [PubMed] [Google Scholar]
- 31.Braback L, Hjern A, Rasmussen F. Social class in asthma and allergic rhinitis: a national cohort study over three decades. Eur Respir J 2005;26:1064–1068. [DOI] [PubMed] [Google Scholar]
- 32.Claudio L, Stingone JA, Godbold J. Prevalence of childhood asthma in urban communities: the impact of ethnicity and income. Ann Epidemiol 2006;16:332–340. [DOI] [PubMed] [Google Scholar]
- 33.Beckett WS, Belanger K, Gent JF, Holford TR, Leaderer BP. Asthma among Puerto Rican Hispanics: a multi-ethnic comparison study of risk factors. Am J Respir Crit Care Med 1996;154:894–899. [DOI] [PubMed] [Google Scholar]
- 34.Chakraborty R, Kamboh MI, Ferrell RE. ‘Unique’ alleles in admixed populations: a strategy for determining ‘hereditary’ population differences of disease frequencies. Ethn Dis 1991;1:245–256. [PubMed] [Google Scholar]
- 35.Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, O'Brien SJ. Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am J Hum Genet 2001;69:1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet 2004;74:1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet 2004;68:139–153. [DOI] [PubMed] [Google Scholar]
- 38.Stephens JC, Briscoe D, O'Brien SJ. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am J Hum Genet 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O'Brien SJ, Altshuler D, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet 2004;74:979–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montana G, Pritchard JK. Statistical tests for admixture mapping with case-control and cases-only data. Am J Hum Genet 2004;75:771–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am J Hum Genet 1998;63:241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol 1998;9:578–594. [DOI] [PubMed] [Google Scholar]
- 43.Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA 1988;85:9119–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet 2005;37:177–181. [DOI] [PubMed] [Google Scholar]
- 45.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet 2005;37:1113–1118. [DOI] [PubMed] [Google Scholar]
- 46.Holguin F, et al. Country of birth as a risk factor for asthma among Mexican Americans. Am J Respir Crit Care Med 2005;171:103–108. [DOI] [PubMed] [Google Scholar]
- 47.Klinnert MD, Price MR, Liu AH, Robinson JL. Unraveling the ecology of risks for early childhood asthma among ethnically diverse families in the southwest. Am J Public Health 2002;92:792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgos AE, Schetzina KE, Dixon LB, Mendoza FS. Importance of generational status in examining access to and utilization of health care services by Mexican American children. Pediatrics 2005;115:e322–e330. [DOI] [PubMed] [Google Scholar]
- 49.Franzini L, Ribble JC, Keddie AM. Understanding the Hispanic paradox. Ethn Dis 2001;11:496–518. [PubMed] [Google Scholar]
- 50.Hayes-Bautista DE, Schink W, Chapa J. The burden of support: young Latinos in an aging society. Stanford, CA: Stanford University Press; 1988.
- 51.Hunt KJ, Williams K, Resendez RG, Hazuda HP, Haffner SM, Stern MP. All-cause and cardiovascular mortality among diabetic participants in the San Antonio Heart Study: evidence against the “Hispanic Paradox.” Diabetes Care 2002;25:1557–1563. [DOI] [PubMed] [Google Scholar]
- 52.Markides KS, Coreil J. The health of Hispanics in the southwestern United States: an epidemiologic paradox. Public Health Rep 1986;101:253–265. [PMC free article] [PubMed] [Google Scholar]
- 53.Palloni A, Arias E. Paradox lost: explaining the Hispanic adult mortality advantage. Demography 2004;41:385–415. [DOI] [PubMed] [Google Scholar]
- 54.Patel KV, Eschbach K, Ray LA, Markides KS. Evaluation of mortality data for older Mexican Americans: implications for the Hispanic paradox. Am J Epidemiol 2004;159:707–715. [DOI] [PubMed] [Google Scholar]