Abstract
Transforming mutations in NRAS and KRAS are thought to play a causative role in the development of numerous cancers, including myeloid malignancies. Although mutations at amino acids 12, 13, or 61 account for the majority of oncogenic Ras variants, we hypothesized that less frequent mutations at alternate residues may account for disease in some patients with cancer of unexplained genetic etiology. To search for additional, novel RAS mutations, we sequenced all coding exons in NRAS, KRAS, and HRAS in 329 acute myeloid leukemia (AML) patients, 32 chronic myelomonocytic leukemia (CMML) patients, and 96 healthy individuals. We detected 4 “noncanonical” point mutations in 7 patients: N-RasG60E, K-RasV14I, K-RasT74P, and K-RasA146T. All 4 Ras mutants exhibited oncogenic properties in comparison with wild-type Ras in biochemical and functional assays. The presence of transforming RAS mutations outside of positions 12, 13, and 61 reveals that alternate mechanisms of transformation by RAS may be overlooked in screens designed to detect only the most common RAS mutations. Our results suggest that RAS mutations may play a greater role in leukemogenesis than currently believed and indicate that high-throughput screening for mutant RAS alleles in cancer should include analysis of the entire RAS coding region.
Introduction
The Ras proto-oncogene belongs to the small GTPase family and exists in 3 distinct isoforms, N-Ras, K-Ras, and H-Ras. Mutant alleles of all 3 isoforms of RAS have been implicated in numerous types of tumors, including cancer of the lung, skin, thyroid, bladder, breast, pancreas, gastrointestinal tract, and kidney as well as multiple forms of leukemia.1 Myeloid leukemia represents one malignancy with a high prevalence of RAS mutations with approximately 1 in 4 cases exhibiting mutations in NRAS or KRAS.2–4 Interestingly, mutations in HRAS are extremely rare in myeloid leukemia.5,6 Point mutation at 3 canonical residues, 12, 13, and 61, is generally thought to account for most Ras-mediated oncogenesis across this broad spectrum of malignancies.
Recent evidence has emerged that suggests additional mutations may also contribute to RAS-associated disease. Large-scale sequencing efforts of tumors from colorectal cancer patients revealed novel mutations in K-Ras, particularly at residue 146.7,8 In addition, juvenile myelomonocytic leukemia (JMML), a blood malignancy often associated with Noonan syndrome, is thought to be uniformly associated with mutations that activate the Ras pathway. Although some of these mutations occur at the canonical N-Ras and K-Ras residues, genetic aberrations have also been found in the Ras regulatory proteins, NF-1 and PTPN11.9–15 In addition, it has recently been reported that 5 noncanonical mutations in KRAS, V14I, P34R, T58I, D153V, and F156L, were found in individuals with the JMML-associated Noonan and cardio-facio-cutaneous syndromes.16–18
Thus, we hypothesized that additional RAS mutations outside of codons 12, 13, and 61 may account for oncogenesis in myeloid malignancies. To test this hypothesis, we sequenced all coding exons of NRAS, KRAS, and HRAS in 192 patients with acute myeloid leukemia (AML), 32 patients with chronic myelomonocytic leukemia (CMML), and 96 healthy individuals for comparison. We identified 7 patients with 4 noncanonical mutations, N-RasG60E, K-RasV14I, K-RasT74P, and K-RasA146T. Functional evaluation revealed that all mutations conferred transforming capacity in comparison with wild-type Ras, indicating these mutant proteins may play a causative role in leukemogenesis.
Methods
Approval was obtained from the institutional review boards of Oregon Health and Science Cancer Institute, Harvard Medical School, Inserm (Paris, France), Hopital Necker (Paris, France), and Heinrich-Heine-Universität for research use of deidentified, archived patient samples.
Patient sample collection
Informed consent was obtained in accordance with the Declaration of Helsinki from all patients. Three-hundred twenty-nine peripheral blood or marrow samples were obtained from patients with AML and 32 from patients with CMML/atypical chronic myelogenous leukemia (aCML). Peripheral blood was isolated from 96 healthy, randomly selected volunteers for comparison purposes. Genomic DNA was prepared from peripheral blood or bone marrow aspirate specimens using the QIAamp DNA Blood Maxi Kit (QIAGEN, Hilden, Germany) as previously described.
Cell culture
A31 cells (derived from embryonic fibroblasts from BALB/c mice) were obtained from ATCC (Manassas, VA). HEK 293T/17 cells (a human embryonic kidney cell line) were provided by Dr Rick Van Etten (Tufts University, Boston, MA). All cells were maintained in DMEM medium supplemented with 10% (HEK 293T/17) or 5% (A31) FBS (Atlanta Biologicals, Lawrenceville, GA), l-glutamine, and penicillin/streptomycin (Invitrogen, Carlsbad, CA). HEK 293T/17) cells were cotransfected with the EcoPack plasmid (kindly provided by Dr Rick Van Etten) using Fugene6 transfection reagent (Roche, Indianapolis, IN) according to the manufacturer's protocols. Retrovirus-containing supernatants were harvested after 48 hours. HEK 293T/17 whole-cell extracts were lysed in 1× cell lysis buffer (Cell Signaling Technology, Danvers, MA) supplemented with aprotinin, 4-(2-aminoethyl)-benzene-sulfonyl fluoride hydrochloride (AEBSF; Sigma-Aldrich, St Louis, MO), and MgCl2.
RAF pull-down assay
One milligram of HEK293T/17 whole-cell extracts was incubated with Ras Assay binding reagent (Millipore, Billerica, MA) according to the manufacturer's instructions and separated on SDS–polyacrylamide gel electrophoresis (PAGE) for analysis of Ras-GTP levels. Fifty micrograms of whole-cell extracts was directly separated on SDS-PAGE for analysis of levels of total Ras expression.
Immunoblotting
Whole-cell extracts from HEK 293T/17 cells expressing wild-type or mutant Ras were subjected to SDS-PAGE analysis and immunoblots were incubated overnight with antibodies specific for Ras (Millipore), total or phospho-MEK, total or phospho-ERK (Cell Signaling), or β-actin (EMD Biosciences, Darmstadt, Germany).
Focus formation assays
A31 cells (105) were plated in 60-mm culture dishes and allowed to adhere overnight. The following day, cells were infected with equivalent multiplicity of infection of retrovirus-expressing empty vector, N-RasG12D, N-RasWT, N-RasG60E, K-RasG12D, K-RasWT, K-RasT74P, and K-RasA146T in the presence of polybrene (10 μg/mL final). Cells were cultured for 12 days, washed with DMEM, fixed with methanol, and stained with a solution of 0.5% crystal violet in 25% methanol for visualization of foci. Foci were counted manually.
Colony-forming assays
Murine bone marrow cells were harvested from C57BL/6 mouse femurs and tibias and plated in media containing interleukin-3, interleukin-6, stem cell factor, and FBS. Cells were infected twice with retrovirus-expressing empty vector, wild-type KRAS or NRAS, as well as each RAS mutant allele. Cells were washed twice with IMDM and plated in triplicate (5 × 104 cells per plate) in methylcellulose without cytokines with the exception of various doses of GM-CSF (0-10 ng/mL). After 7 days, colonies were quantified on a bright-field microscope. The use of animals in this study was approved by OHSU and performed according to the OHSU and State of Oregon Institutional Animal Care and Use Committee Guidelines.
Flow cytometry and cell sorting
Nonneoplastic T cells and neoplastic myeloid cells were sorted from viably frozen patient samples (approximately 107 cells) using a FACSAria cell sorter (BD Biosciences, San Jose, CA). Prior to cell sorting, cells were stained according to the manufacturer's instructions with the following antibody combination: CD3-FITC, CD33-PE, CD45-Percp-Cy5.5, CD34-APC (all from BD Biosciences).
Sequencing analysis
All coding exons of NRAS, KRAS, and HRAS were sequenced with primers in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) according to previously described protocols.8,19
Results
High-throughput sequencing of NRAS, KRAS, and HRAS
In an effort to identify novel oncogenic RAS alleles in patients with myeloid malignancies, we undertook a high-throughput sequencing screen of all coding regions of NRAS, KRAS, and HRAS in genomic DNA from cohorts of 329 AML patients, 32 CMML patients, and 96 healthy individuals. In addition to identifying several patients with mutations at codons 12, 13, and 61, we also identified 7 patients with mutations at alternate residues, specifically K-RasV14I and N-RasG60E in 2 separate AML patients, K-RasT74P in a patient with atypical CML, and K-RasA146T in 2 AML and 2 CMML patients (Table 1; Figure S1). Of note, 1 AML patient with a KRASA146T allele exhibited homozygosity at this locus, whereas all other mutations detected were heterozygous. Noncanonical mutations appeared more prevalent in KRAS where 6 of 16 mutations were found outside of residues 12, 13, or 61. In addition, CMML/aCML patients appeared to carry these noncanonical mutations more frequently than AML patients, in that one-third of identified RAS mutations in CMML/aCML were noncanonical compared with approximately one-tenth in AML. Finally, the overall RAS mutations clustered in AML French-American-British (FAB) subtypes in a similar fashion as has been seen previously20 with monocytic subtypes (M4/M5) accounting for approximately half of cases for which a subtype was available. Frozen viable material was available for 3 of the 7 patients with noncanonical mutations, thus we analyzed whether these mutations were somatic or germline in these individuals by flow cytometric sorting of neoplastic cells (CD34+ and/or CD33+ myeloid cells) and nonneoplastic cells (CD3+ T cells) and sequencing of the respective RAS exons in each population. We found that only the neoplastic cell populations exhibited the mutations in each case, indicating that these mutations were acquired and not germline. Interestingly, this was the case even in the homozygous K-RasA146T specimen, suggesting that this mutation was not only acquired, but that the neoplastic clone also exhibited loss of the wild-type allele (Figure S1). Since the KRASV14I mutation has been characterized as a transforming allele elsewhere17,18 and none of the mutations were detected in 96 healthy persons, we sought to functionally evaluate these mutant alleles to understand their potential contribution to oncogenesis.
Table 1.
Incidence of RAS mutations and patient characteristics
| Age | Sex | Mutation | FAB/WHO classification | Cytogenetic studies | Clinical course |
|---|---|---|---|---|---|
| AML cohort (incidence, %) | |||||
| N-Ras G12 (4.3) | |||||
| 83 y | F | G12D | M5B | 46, XX | Initial diagnosis, deceased in induction |
| 6 y | F | G12D | M1 | 46XX | n/a |
| 20 y | M | G12D | AML, NOS | 46, XY | n/a |
| 10 mo | F | G12D | AML, NOS | failure | n/a |
| 61 y | M | G12D | M1 | n/a | Initial diagnosis, no f/u |
| 14 y | F | G12D | M2 | 46, XX, add(6)(q?21), add(16)(q?22), add(17)(q?21), add(18)(q?21)45, X,t(X;1)(p11.2; p32), del(3)(p2/2),der(5)t(5;14)(p15;11.2), add(6)(q?22), del(10)(q?23), del(11)(p?14), −15, add(16)(q10), add(17)(q?21),−18, add(20)(q?11.2), +mar46, XX | Outside specimen for flow cytometry at diagnosis, BMT, relapse at 1 y, no additional f/u |
| 35 y | F | G12D | M4 | n/a | Outside specimen for flow cytometry at diagnosis, no additional f/u |
| 48 y | M | G12D | M4 | n/a | n/a |
| 30 y | F | G12D | M5 | n/a | Outside specimen for flow cytometry at diagnosis, no additional f/u |
| 59 y | M | G12D | M1 | n/a | Relapse at 1 y after induction therapy, no additional f/u |
| 24 y | F | G12D | M5 | n/a | Outside specimen for flow cytometry at diagnosis, no additional f/u |
| n/a | M | G12S | relapse M2 | t(8;21) | n/a |
| 67 y | F | G12A | M0 | n/a | Outside specimen for flow cytometry at diagnosis, no additional f/u |
| n/a | M | G12V | AML, NOS | n/a | Outside specimen for flow cytometry at diagnosis, no additional f/u |
| N-Ras G13 (0.6) | |||||
| n/a | n/a | G13D | AML, NOS | n/a | n/a |
| 35 y | M | G13R | M1 | n/a | Outside specimen for flow cytometry at diagnosis, no additional f/u |
| N-Ras Q61 (1.8) | |||||
| 22 y | F | Q61R | M4 | 46, XX | Initial diagnosis, relapse at 6 mo, no f/u |
| n/a | n/a | Q61R | M5 | n/a | n/a |
| n/a | M | Q61R | M5 | n/a | Outside specimen for flow cytometry at diagnosis, BMT, remission at day 110, no additional f/u |
| 37 y | M | Q61K | M5 | 46, XY | n/a |
| n/a | M | Q61K | M5 | n/a | n/a |
| n/a | M | Q61H | M5 | n/a | Relapse sample, no f/u |
| N-Ras G60 (0.3) | |||||
| n/a | M | G60E | AMLL | n/a | n/a |
| K-Ras G12 (1.5) | |||||
| 51 y | F | G12D | M4Eo | 46, XX, inv(16) | Initial diagnosis, remission, no f/u |
| n/a | n/a | G12D | M4 | n/a | n/a |
| n/a | F | G12D | M2 | n/a | n/a |
| 9 y | F | G12D | RAEB-T | n/a | BMT, relapse at 1 y after induction therapy, no additional f/u |
| 79 y | F | G12A | M2 | Normal | n/a |
| K-Ras G13 (0.6) | |||||
| n/a | n/a | G13D | AML, NOS | n/a | n/a |
| 67 y | F | G13D | M0 | n/a | Outside specimen for flow cytometry at diagnosis, no additional f/u |
| K-Ras V14 (0.3) | |||||
| 70 y | F | V14I | M1 | very complex 44, −5, add13p, add14q, −17, +22 | n/a |
| K-Ras A146 (0.6) | |||||
| 63 y | F | A146T | AML, NOS | 46XX del 5q11q32 | Initial diagnosis, complete remission at 14 mo, no f/u |
| 23 y | F | A146T | M1 | 46, XX, −1, inv(3)(q?21q?27), del(5)(q?13q31), add(5)(p?12), add(7)(p11), −10, add(10)(p?13), add(11)(q13), del(12)(p11), der(15)t(/1;15)(p?22; q2?4), add(16)(q?22), −18, +marAll 22 | Outside specimen for flow cytometry at diagnosis, relapse at 1 y after induction therapy, BMT w/ immediate relapse, no additional f/u |
| Other canonical (2.7)* | |||||
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| n/a | n/a | G12, G13, Q61 | AML, NOS | n/a | n/a |
| CMML cohort (incidence, %) | |||||
| N-Ras G12 (9.4) | |||||
| 72 | M | G12D | CMML | 46, XY[20] | n/a |
| 65 | M | G12D | CMML | 46, XY[20] | n/a |
| 80 | M | G12D | CMML | BCR-ABL–negative (FISH) | n/a |
| K-Ras G12 (6.3) | |||||
| n/a | F | G12D | aCML | BCR-ABL–negative (FISH) | n/a |
| 53 | M | G12A | CMML | 46, XY[20] | n/a |
| K-Ras G13 (3.1) | |||||
| 65 | M | G13D | CMML | NA | n/a |
| K-Ras T74 (3.1) | |||||
| n/a | F | T74P | aCML | BCR-ABL–negative (FISH) | n/a |
| K-Ras A146 (6.3) | |||||
| 71 | M | A146T | CMML | n/a | n/a |
| 48 | M | A146T | CMML | n/a | n/a |
The estimated proportion of mutant allele is given in parentheses after the mutation type.
aCML indicates atypical chronic myelogenous leukemia; AML, acute myeloid leukemia; F, female; M, male; n/a, not available; BMT, bone marrow transplantation; and RAEB, refractory anemia with excess blasts.
Known canonical mutation, but patient data unavailable.
Biochemical evaluation of RAS mutant alleles
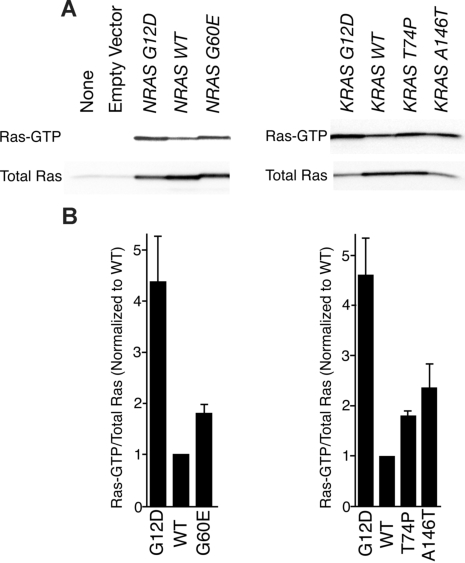
To assess the biochemical activity of each mutant RAS allele in a cellular context, we transfected HEK 293T/17 cells with MSCV-IRES-GFP (MIG)–NRAS or MIG-KRAS constructs expressing either wild-type or mutant alleles. We precipitated Ras-GTP from whole-cell extracts with a peptide corresponding to the Ras binding domain of RAF-1 and compared levels of Ras-GTP with total Ras by immunoblot analysis. As has been previously shown, the G12D substitution in both N-Ras and K-Ras induces higher levels of the GTP-bound state of the enzyme. Similarly, we observed increased levels of Ras-GTP in all 3 novel mutants compared with wild type (Figure 1A). Densitometry revealed that Ras G12D was most abundantly bound to GTP in each case with the hierarchy of G12D>A146T>G60E∼T74P for the mutant proteins (Figure 1B). Hence, each mutant appeared to exist in a GTP-bound state in greater abundance than wild type based on greater affinity to the RAF-1 peptide; however, we next wanted to correlate this with relevant increases in downstream signaling cascades.
Figure 1.
RAF-1 pull-down assay reveals that novel mutations confer increased Ras-GTP. (A) Wild-type NRAS and KRAS as well as each mutant were expressed in HEK 293T/17 cells for 48 hours. Whole-cell extracts were incubated with RAF-1 peptides conjugated with agarose beads for precipitation of Ras-GTP. Precipitates were subjected to SDS-PAGE analysis and immunoblotted with an antibody specific for total Ras. (B) Densitometry was performed on blots in panel A. Densitometric units for Ras-GTP were normalized to their respective total Ras counterparts, and all Ras-GTP/total Ras ratios were divided by the ratio for WT N-Ras or K-Ras. Values represent mean plus or minus SEM (n = 3).
After Ras binds RAF, this complex induces phosphorylation of MEK, which subsequently phosphorylates ERK.21–25 To assess the capacity of mutant Ras proteins to activate these downstream signaling events in cells, we immunoblotted for phospho-MEK as well as phospho-ERK in whole-cell extracts of HEK 293T/17 cells transfected with each wild-type and mutant RAS allele. Results mirrored the RAF-1 pull-down experiment with G12D exhibiting strongest phosphorylation of these downstream proteins in the setting of both N-Ras and K-Ras resulting in a similar hierarchy of G12D>A146T>G60E∼T74P for the mutant proteins (Figure 2A). Again, densitometry corroborated the evident immunoblot results (Figure 2B).
Figure 2.
Immunoblotting reveals that novel mutations confer increased signaling of downstream Ras pathways. (A) Wild-type NRAS and KRAS as well as each mutant were expressed in HEK 293T/17 cells for 48 hours. Whole-cell extracts were subjected to SDS-PAGE analysis and immunoblotted with antibodies specific for total and phospho-MEK and ERK as well as β-actin. (B) Densitometry was performed on blots in panel A. Densitometric units for phosphorylated proteins were normalized to their respective total protein counterparts, and all phospho/total ratios were divided by the ratio for WT N-Ras or K-Ras. Values represent mean plus or minus SEM (n = 3).
Functional characterization of RAS mutant alleles
Since all RAS mutations identified in this screen conferred an evident increase in transformative capacity by biochemical and signaling readouts, we next sought to characterize the functional consequences of this dysregulation. To address this question, we performed focus formation experiments using the murine fibroblast cell line, A31 (BALB-3T3). Cells were infected with equivalent multiplicity of infection of retrovirus expressing each wild-type or mutant allele of RAS and allowed to culture for 10 days. Parental A31 cells or those transfected with MIG empty vector exhibited contact inhibition and, thus, ceased proliferating upon reaching a monolayer of cells (Table 2; Figure S2). Similarly, A31 cells expressing NRAS or KRAS wild-type exhibited very few foci, indicating a contact inhibited growth. However, all mutant alleles of RAS efficiently transformed A31 cells to induce the cells to grow past contact inhibition thus forming numerous foci (Table 2; Figure S2). This indicates that the transformative capacity observed by biochemical and signaling assays correlates with an increase in functional transformation by the mutant RAS alleles. Quantification of foci indicated that the mutant alleles followed a similar hierarchy of G12D∼A146T>G60E∼T74P, where KRASA146T appeared equivalent to the G12D mutants and NRASG60E appeared equivalent to KRAST74P in transformative capacity.
Table 2.
Results of focus formation assays comparing wild-type RAS and various RAS mutations
| Gene/allele | Average foci per plate | Standard error | Paired t test with WT, P |
|---|---|---|---|
| None | 0 | 0 | — |
| Empty vector | 0 | 0 | — |
| NRAS | |||
| WT | 25 | 2 | — |
| G12D | 1250 | 101 | .007 |
| G60E | 698 | 20 | .001 |
| KRAS | |||
| WT | 26 | 2 | — |
| G12D | 957 | 53 | .003 |
| T74P | 659 | 54 | .007 |
| A146T | 1012 | 48 | .002 |
Finally, to assess the transformative capacity of each mutant RAS allele when expressed in a cellular setting relevant to leukemia, we introduced each wild-type or mutant RAS allele into murine bone marrow cells. A colony-forming assay in methylcellulose in the presence of a dose gradient of GM-CSF (0-10 ng/mL) revealed that each mutant allele exhibited increased ability to form colonies compared with wild type at several concentrations of GM-CSF (Figure 3). Interestingly, in this setting, NRASG60E appeared to have equivalent transformative capacity as NRASG12D. As seen with the focus formation assay, KRASA146T exhibited similar colony formation as KRASG12D, whereas KRASV14I and KRAST74P showed slightly less transformative capacity, but still far greater than wild type. These results yield the similar hierarchy of G12D∼A146T∼G60E>T74P∼V14I (Table S2).
Figure 3.
RAS mutant alleles confer growth factor hypersensitivity to murine bone marrow cells. (A) Murine bone marrow cells were infected with retrovirus-expressing empty vector, wild-type KRAS, as well as each KRAS mutant allele. Cells were plated in triplicate in methylcellulose containing various doses of GM-CSF (0-10 ng/mL). After 7 days, colonies were quantified on a bright-field microscope. Values represent mean plus or minus SEM. (B) Murine bone marrow cells were infected with retrovirus-expressing empty vector, wild-type NRAS, and each NRAS mutant allele. Cells were plated in triplicate in methylcellulose containing various doses of GM-CSF (0-10 ng/mL). After 7 days, colonies were quantified on a bright-field microscope. Values represent mean plus or minus SEM.
Discussion
Our high-throughput sequencing screen identified 4 noncanonical RAS mutations in 7 patients with myeloid malignancies. Three mutant alleles were detected in single patients, whereas one was found in 4 patients. All of these mutations exhibited properties similar to canonical oncogenic RAS mutations in several different functional assays including RAF-1 pull-down, phosphorylation of downstream signaling events, and focus and colony formation assays. One of the mutated residues, G60, lies very near the previously characterized mutated residues of T58 and Q61, which have been shown to exhibit impaired GTP hydrolysis.18 The other novel mutant, K-RasT74P, occurs in the normally flexible chain of residues (71-77) found at the C-terminus of helix a2. The substitution of proline at this residue may disrupt the terminal unraveling of this helix and subsequent extension of loop L5 observed upon GTP hydrolysis in the switch II region, perhaps impairing conversion from GTP-bound to GDP-bound Ras.26
Two of these mutant alleles were previously identified. The KRASV14I mutant was identified in 3 patients with Noonan syndrome and characterized as a transforming allele.17,18 Mutations involving the K-RasA146 residue have been previously detected in 12 tumors from patients with colorectal cancers, as well as several cell lines including one derived from AML.7,8,27 In addition, the KRASA146T and HRASA146V mutants were identified as oncogenic alleles in random mutagenesis screens in mice and bacteria.28,29 The presence of this mutation in 4 patients with myeloid malignancies as well as at least 12 patients with colorectal cancer, coupled with our characterization of this mutation as conferring strongly increased RAS transformative capacity, indicate that this allele may occur at high frequency in cancer. Comparison of the frequency of A146 mutations with that of mutations at codons 13 and 61 in our study as well as those in colorectal cancer reveals a similar incidence of mutations at codon 146 as seen at codons 13 and 61.
In summary, we have demonstrated that RAS mutations outside those observed at codons 12, 13, and 61 occur in patients with myeloid leukemia and have the capacity to contribute to leukemogenesis. Although the NRASG60E and KRAST74P mutations seem to occur at very low frequency, the KRASV14I and KRASA146T alleles have now been detected in 4 and 16 patients, respectively, suggesting that mutations in NRAS and KRAS are contributing to leukemogenesis at a higher incidence than was previously understood. Hence, tests aimed at detecting oncogenic RAS alleles in patients need to include ways of identifying these less frequent, but equally oncogenic, noncanonical mutations. As targeted therapeutics aimed at activated Ras pathways are developed and adopted into clinical use,30–33 it will be important to apply these drugs in all patients for whom they may be effective.
Supplementary Material
Acknowledgments
M.M.L. is supported by the Leukemia & Lymphoma Society (White Plains, NY) and the T. J. Martell Foundation (New York, NY). J.W.T. is supported by a cancer biology training grant from the National Institutes of Health (NIH, Bethesda, MD). M.C.H. is supported in part by a VA Merit Review Grant. D.G.G. and B.J.D. are Howard Hughes Medical Institute investigators.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.W.T., H.E., and M.M.L. designed and performed research, analyzed data, and wrote the paper; S.G.W. and C.A.E. performed research; and R.L.L., M.W.N.D., M.C.H., N.G., D.G.G., and B.J.D. contributed vital reagents, designed research, and revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marc M. Loriaux, Department of Pathology, Oregon Health & Science University, Portland, OR 97239; e-mail: loriauxm@ohsu.edu.
References
- 1.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 2.Bartram CR. Mutations in ras genes in myelocytic leukemias and myelodysplastic syndromes. Blood Cells. 1988;14:533–538. [PubMed] [Google Scholar]
- 3.Janssen JW, Lyons J, Steenvoorden AC, Seliger H, Bartram CR. Concurrent mutations in two different ras genes in acute myelocytic leukemias. Nucleic Acids Res. 1987;15:5669–5680. doi: 10.1093/nar/15.14.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen JW, Steenvoorden AC, Lyons J, et al. RAS gene mutations in acute and chronic myelocytic leukemias, chronic myeloproliferative disorders, and myelodysplastic syndromes. Proc Natl Acad Sci U S A. 1987;84:9228–9232. doi: 10.1073/pnas.84.24.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neubauer A, Dodge RK, George SL, et al. Prognostic importance of mutations in the ras proto-oncogenes in de novo acute myeloid leukemia. Blood. 1994;83:1603–1611. [PubMed] [Google Scholar]
- 6.Shen WP, Aldrich TH, Venta-Perez G, Franza BR, Jr, Furth ME. Expression of normal and mutant ras proteins in human acute leukemia. Oncogene. 1987;1:157–165. [PubMed] [Google Scholar]
- 7.Edkins S, O'Meara S, Parker A, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–932. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 9.Birnbaum RA, O'Marcaigh A, Wardak Z, et al. Nf1 and Gmcsf interact in myeloid leukemogenesis. Mol Cell. 2000;5:189–195. doi: 10.1016/s1097-2765(00)80415-3. [DOI] [PubMed] [Google Scholar]
- 10.Lauchle JO, Braun BS, Loh ML, Shannon K. Inherited predispositions and hyperactive Ras in myeloid leukemogenesis. Pediatr Blood Cancer. 2006;46:579–585. doi: 10.1002/pbc.20644. [DOI] [PubMed] [Google Scholar]
- 11.Le DT, Kong N, Zhu Y, et al. Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder. Blood. 2004;103:4243–4250. doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- 12.Loh ML, Vattikuti S, Schubbert S, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004;103:2325–2331. doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- 13.Miles DK, Freedman MH, Stephens K, et al. Patterns of hematopoietic lineage involvement in children with neurofibromatosis type 1 and malignant myeloid disorders. Blood. 1996;88:4314–4320. [PubMed] [Google Scholar]
- 14.Schubbert S, Lieuw K, Rowe SL, et al. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. 2005;106:311–317. doi: 10.1182/blood-2004-11-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Side LE, Emanuel PD, Taylor B, et al. Mutations of the NF1 gene in children with juvenile myelomonocytic leukemia without clinical evidence of neurofibromatosis, type 1. Blood. 1998;92:267–272. [PubMed] [Google Scholar]
- 16.Niihori T, Aoki Y, Narumi Y, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 17.Schubbert S, Bollag G, Lyubynska N, et al. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007;27:7765–7770. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubbert S, Zenker M, Rowe SL, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 19.Loriaux MM, Levine RL, Tyner JW, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–4796. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen DT, Frew ME, Hills R, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106:2113–2119. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 21.Gardner AM, Vaillancourt RR, Johnson GL. Activation of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase by G protein and tyrosine kinase oncoproteins. J Biol Chem. 1993;268:17896–17901. [PubMed] [Google Scholar]
- 22.Kolch W, Heidecker G, Lloyd P, Rapp UR. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- 23.Macdonald SG, Crews CM, Wu L, et al. Reconstitution of the Raf-1-MEK-ERK signal transduction pathway in vitro. Mol Cell Biol. 1993;13:6615–6620. doi: 10.1128/mcb.13.11.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas SM, DeMarco M, D'Arcangelo G, Halegoua S, Brugge JS. Ras is essential for nerve growth factor- and phorbol ester-induced tyrosine phosphorylation of MAP kinases. Cell. 1992;68:1031–1040. doi: 10.1016/0092-8674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- 25.Wood KW, Sarnecki C, Roberts TM, Blenis J. ras mediates nerve growth factor receptor modulation of three signal-transducing protein kinases: MAP kinase, Raf-1, and RSK. Cell. 1992;68:1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- 26.Stouten PF, Sander C, Wittinghofer A, Valencia A. How does the switch II region of G-domains work? FEBS Lett. 1993;320:1–6. doi: 10.1016/0014-5793(93)81644-f. [DOI] [PubMed] [Google Scholar]
- 27.Orita S, Higashi T, Kawasaki Y, et al. A novel point mutation at codon 146 of the K-ras gene in a human colorectal cancer identified by the polymerase chain reaction. Virus Genes. 1991;5:75–79. doi: 10.1007/BF00571733. [DOI] [PubMed] [Google Scholar]
- 28.Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan SR, Newcomb EW, Pellicer A. Neutron radiation can activate K-ras via a point mutation in codon 146 and induces a different spectrum of ras mutations than does gamma radiation. Mol Cell Biol. 1990;10:405–408. doi: 10.1128/mcb.10.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blum R, Cox AD, Kloog Y. Inhibitors of chronically active ras: potential for treatment of human malignancies. Recent Patents Anticancer Drug Discov. 2008;3:31–37. doi: 10.2174/157489208783478702. [DOI] [PubMed] [Google Scholar]
- 31.Blum R, Kloog Y. Tailoring Ras-pathway–inhibitor combinations for cancer therapy. Drug Resist Updat. 2005;8:369–380. doi: 10.1016/j.drup.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Viciana P, Tetsu O, Oda K, Okada J, Rauen K, McCormick F. Cancer targets in the Ras pathway. Cold Spring Harb Symp Quant Biol. 2005;70:461–467. doi: 10.1101/sqb.2005.70.044. [DOI] [PubMed] [Google Scholar]
- 33.Smith RA, Dumas J, Adnane L, Wilhelm SM. Recent advances in the research and development of RAF kinase inhibitors. Curr Top Med Chem. 2006;6:1071–1089. doi: 10.2174/156802606777812077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.