Abstract
The hypoxia-inducible factor (HIF) activates transcription via binding to the highly variable hypoxia-responsive elements (HREs). All hypoxia-inducible constructs described to date utilize multimers of naturally occurring HREs. Here, we describe the rational design of minimal hypoxia-inducible enhancers, conceptually equivalent to using an optimized HIF-binding site (HBS) as the building block. Optimizations of the HBS, spacing between HBSs, the distance from the minimal promoter, and orientation of HBSs allowed us to design constructs with high hypoxic activity. Activation of the 4xopt HBS (36 bp) construct by hypoxia or HIF-1α and HIF-2α was comparable with that of the 4xEPO HRE (208 bp) construct. The strong synergism between the properly arranged optimized HBSs was due to stimulation of high affinity HIF binding. Our data prove, for the first time, that it is possible to assemble artificial hypoxia-inducible enhancers from a single type of regulatory element-optimized HBS.
Keywords: Hypoxia, Hypoxia-inducible factor, Hypoxia responsive element, HIF-binding site
The hypoxia-responsive element (HRE) is a minimal cis-regulatory element mediating transactivation by the hypoxia-inducible factor (HIF) [1]. The data from over 70 genes suggest that endogenous HREs are composite regulatory elements comprising the conserved HIF-binding site (HBS) with an A/GCGTG core sequence and a highly variable flanking sequence. A single HBS is necessary but not sufficient for activation by hypoxia and the flanking sequence provides binding sites for transcription factors that are not necessarily hypoxia-inducible but are required to amplify the hypoxic response or make the HRE tissue-specific [1]. In contrast to our knowledge of regulation of the HIF pathway, relatively little progress has been made towards understanding the fundamental structural features of HREs. There are only a few reports relating the sequence of HRE with activity, e.g. mutational analysis of the −2 position of HBS revealed that hypoxic induction decreases in the T ≫ G > C order [2,3].
Direct consequence of our limited understanding of HRE is that all hypoxia-inducible constructs for therapeutic or monitoring purposes described to date are based on multimers of naturally occurring HREs, ranging from 24 bp for murine phosphoglycerate kinase-1 (mPGK1) [4], 35 [5] or 44 [6] bp for erythropoietin (EPO), and 37 bp for vascular endothelial growth factor (VEGF) [5] HREs. Apparently, the progress towards optimized HREs has been hampered by two major factors: the fact that HREs come in different “flavors”, significantly diverging from each other [1] and the complexity of regulation by hypoxia, highlighted in the mLDHA promoter where mutations at three separate sites abolished hypoxic induction [7]. Moreover, concatamerization of any of the three functionally critical sites did not confer high-level hypoxic induction, leading to the conclusion that each site is necessary but not sufficient on its own for hypoxic induction [7].
Our goal in this study was to rationally design minimal hypoxia-inducible enhancers, defined as the minimal number of the shortest possible building blocks. The basic structural organizations of HRE were inferred from the sequences of naturally occurring HREs with the highest hypoxic induction. This information, combined with optimizations at several levels, allowed us to assemble short, highly inducible artificial enhancers exclusively from HBSs.
Materials and methods
Plasmid constructions
Wild type and mutant HREs (Table 1) were cloned into the TATA-box containing pLuc-MCS vector (Stratagene) as double-stranded oligonucleotides and verified by sequencing. The 4xEPO HRE construct contains 4 copies of the EPO long sequence (Table 1), separated by a linker sequence, in pLuc-MCS. The CMV-luc construct has the immediate-early CMV promoter in pGL2 basic (Promega). The HIF-1α(P402A, P564A) and HIF-2α(P405A, P531A) mutants were generated by PCR and cloned into the pcDNA3 expression vector (Invitrogen).
Table 1.
Sequences used in this study, written in the 5′;–3′ direction

|
Mutations against wt sequences are indicated in underlined bold, small cap letters indicate insertions, TATA-box and other cis-acting elements are in italics.
Cell culture and transient transfection assay
Saos-2 cell line was maintained as described previously [8]. The Chinese hamster ovary (CHO) cell lines C 4.5 (control) and Kal 3.5 (HIF-α deficient), kindly provided by Dr. P. J. Ratcliffe, were grown as described in [9]. Cells, seeded in triplicates, were co-transfected with an HRE-driven firefly luciferase reporter construct and pRL-CMV (Promega), expressing Renilla luciferase (internal control for transfection efficiency), using the Effectene reagent (QIAGENE) as described [8]. Activity of each construct was expressed as the average ratio of firefly to Renilla luciferase activities (+S.D.).
Biotinylated probe pull down
For high stringency pull down [6] nuclear extract was prepared from normoxic and hypoxic (0.5% O2) Saos-2 cells with NE-PER kit (Pierce). Streptavidin–Sepharose (Stratagene) was washed three times in phosphate- buffered saline (pH 7.4)/0.1% BSA and two times with Tris–EDTA (TE, pH 8.0)/1M NaCl and used (40 μl of 50% slurry) to immobilize 40 pmol double-stranded probes (top strand biotinylated at the 5′-end Table 1) in TE/1M NaCl (500 μl). After rotating for 20 min (all incubations were at room temperature), Sepharose was washed three times with the same buffer and two times with 1× binding buffer (10 mM Tris (pH 7.6), 50 mM KCl, 1 mM MgCl2, 1 mM EDTA, 5 mM DTT, 5% glycerol, .03% Nonidet P-40). Nuclear proteins (60 μg), preincubated for 5 min with 2.5 μg of poly (dI–dC) in 100 μl 1× binding buffer/1 mM sodium orthovanadate, were rotated with 10 μl Sepharose/probe for 20 min. Sepharose was then washed three times with 1× binding buffer/0.5 μg/ml poly(dI–dC), resuspended in 1× sample buffer, boiled, and bound proteins were separated by SDS–PAGE.
Western blotting and staining of biotinylated probes
Expression of the HIF-1α(P402A, P564A) and HIF-2α(P405A, P531A) mutants in transiently transfected cells was probed by Western blotting as described previously [8], using HIF-1α monoclonal (BD Biosciences) and HIF-2α polyclonal (Novus) antibody, respectively. β-Actin (Sigma) was used as internal control. HIF-1α in pull down experiments was also detected by Western blotting. Equal amount of biotinylated double-stranded probes was confirmed by electrophoresis in 12% non-denaturing gel, blotting onto Hybond N+ membrane (Amersham Biosciences), and detection with Streptavidin-Peroxidase Polymer (Sigma) and Enhanced Chemiluminescence (ECL) kit (Pierce).
Results and discussion
Hypoxic induction of selected endogenous HREs
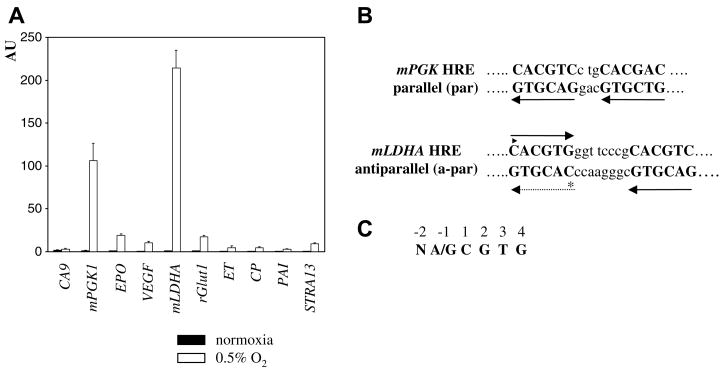
Initially, we studied hypoxic induction of a single copy of endogenous HREs upstream of a minimal TATA-box in transiently transfected Saos-2 cells. We used 29 bp-long sequences (23 bp in the case of mPGK1 HRE), excluding thus contribution of transcription factors that cooperate with HIF in the longer forms of some HREs [7,10,11]. Among the tested HREs, mLDHA and mPGK1 HREs yielded by far the strongest hypoxic induction (199 and 80×, respectively, Fig. 1A). Interestingly, the HBS containing fragment from the gene coding for carbonic anhydrase IX (CA IX), one of the intrinsic markers of cellular hypoxia [12], displayed a remarkably low induction (1.7×), one of the lowest among the HREs tested (Fig. 1A). The CCCGCAC sequence in mLDHA HRE (Table 1) closely resembles the SP1/SP3-binding GC-box [13]. Although its functionality has not been formally confirmed, the synergistic cooperation between the SP1 site and HBS in CA9 HRE [11] prompted us to test its role in mLDHA HRE function. Mutations eliminating SP1/SP3-binding (Table 1) [13] failed to affect mLDHA HRE activity (data not shown), meaning that not only the GCbox-like motif does not contribute to hypoxic induction but also that mutations between the two HBSs do not significantly affect HRE activity. Reversing the orientation did not affect activity of either mLDHA or mPGK1 HREs (data not shown). In conclusion, hypoxic induction of mLDHA and mPGK1 HREs indicated that their cis-acting element(s) are sufficient for effective recognition by the HIF system and therefore these two HREs were selected for further study.
Fig. 1.
(A) Hypoxic induction of selected wild type HREs. HREs in pLucMCS were co-transfected with pRL-CMV into Saos-2 cells for 16 h, followed by exposure to 0.5% O2 for 24 h. Promoter activities are expressed as the ratio of firefly/Renilla activity in arbitrary units (AU), each of the bars representing the mean value (X ± SD) from three experiments. CA9, carbonic anhydrase 9; mPGK1, murine phosphoglycerate kinase-1; EPO, erythropoietin; VEGF, vascular endothelial growth factor; mLDHA, murine lactate dehydrogenase-A; rGlut1, rat glucose transporter-1; ET, endothelin; CP, ceruloplasmin; PAI, plasminogen activator inhibitor-1; STRA13, stimulated with retinoic acid-13 (B) Schematic outline of HBS arrangements in mLDHA and mPGK1 HREs and their designation. Arrows indicate the orientation of HBSs. (C) Numbering of positions in HBS.
Structural organization of mLDHA and mPGK1 HREs
mLDHA and mPGK1 HREs have in common two HBSs (GTCGTG in mPGK1 HRE will be considered as an imperfect HBS). In mPGK1 HRE, the arrangement of HBSs is parallel (par) (Fig. 1B). Formally, because of the symmetry of CACGTG, HBSs in mLDHA HRE could be par, with increased spacing between HBSs compared to mPGK1 HRE (solid and dashed arrows in Fig. 1B), or antiparallel (a-par) (solid arrows in Fig. 1B). In order to find the functional arrangement, we mutagenized the −2 positions (Fig. 1C) of CACGTG. As magnitude of hypoxic induction decreases in the T ≫ G > C order [2], we reasoned that introduction of T into the weak CACGTG would enhance activity only in the −2 position of the functional arrangement. Relative to the proximal HBS, TACGTG in mut1 (asterisk in Fig. 1B) is par, but it becomes CACGTA (disrupting the core HBS sequence) in the a-par arrangement, whereas in mut2 (triangle in Fig. 1B) the situation is reversed. Considerably lower activity of mut1, compared to the wild type or mut2 (Fig. 2A), confirmed that the functional arrangement of HBSs in mLDHA HRE is a-par. We conclude that HIF can effectively transactivate two fundamentally different arrangements of HBSs: par and a-par.
Fig. 2.
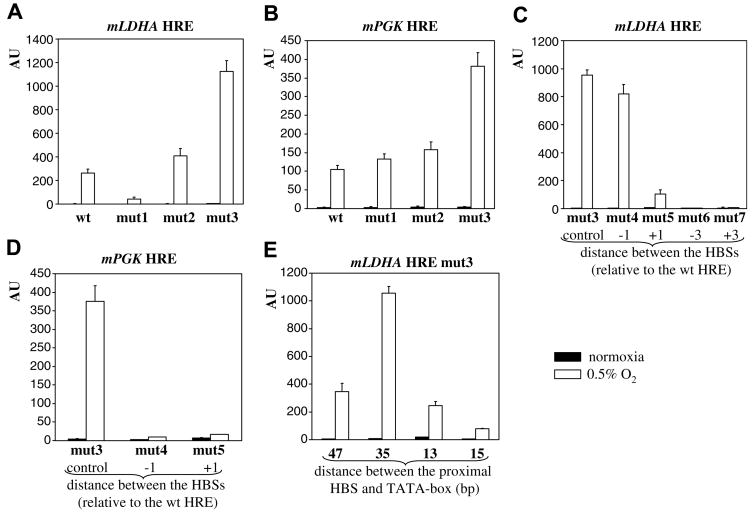
Optimization of mLDHA and mPGK1 HREs. Optimization of the −2 positions of mLDHA (A) and mPGK1 HREs (B). The effect of HBS spacing on activity of mLDHA (C) and mPGK1 (D) HRE. The effect of spacing between HRE and TATA-box on activity of mLDHA mut3 (E). Transfection and expression of promoter activities were as described in Fig. 1.
Optimization of mLDHA and mPGK1 HREs
Next, we optimized mLDHA and mPGK1 HREs. As all HBSs in these two HREs have a suboptimal G or C nucleotide at the −2 position (Fig. 1B), we introduced T into these positions plus A at the −1 position of the proximal HBS in mPGK1 HRE (Table 1). Mutations indeed stimulated hypoxic activity and HREs with the double T replacement (mut3) were the most active (Fig. 2A and B). Thus, optimization of HBSs stimulates hypoxic induction of both a-par (mLDHA HRE) and par (mPGK1 HRE) arrangements.
Deletion/insertion mutants were used to evaluate the effect of spacing between HBSs. In mLDHA HRE mut3 (a-par), deletion of a single nucleotide (mut4) was tolerated, whereas insertion of a single nucleotide (mut5) decreased activity considerably (Table 1, Fig. 2C). Deletion (mut6) and insertion (mut7) of 3 nucleotides abrogated the synergy between HBSs (Table 1, Fig. 2C). In contrast, mPGK1 HRE mut3 (par) appears to be significantly more sensitive to altered spacing between HBSs as deletion (mut4) and insertion (mut5) of a single nucleotide drastically reduced hypoxic induction (Table 1, Fig. 2D). Apparently, HBSs in mPGK1 HRE (and presumably other par arrangements) are “locked”, as the slightest alteration of spacing had a profound inhibitory effect on hypoxic activity. The a-par arrangement appears to be more flexible as it tolerated some alterations. Based on these data, the optimal spacing between HBSs is 10 bp for the par and 16 bp (from positions 1) for the a-par arrangement (Fig. 1B). Structurally, par HBSs positioned 10 bp apart are on the same side of DNA and the larger distance in the a-par arrangement is presumably required for similar favorable alignment when HBSs reside on separate strands. Previously, we noted a remarkably low hypoxic activity of a construct with three copies of the EPO HBS (par) 20 bp apart (Kaluz, unpublished). In our opinion, the low activity of this construct, mLDHA HRE mut1 (this study), and multimerized mLDHA HBSs in the earlier report [7] reflects the suboptimal (longer) spacing between the HBSs. Therefore, spacing between HBSs is a critical determinant of HRE activity and, for the purpose of constructing minimal enhancers, spacing between HBSs in mLDHA and mPGK1 HREs is optimal (providing maximal activity).
Finally, we studied the effect of spacing between the TATA-box and the HRE. Constructs described so far had 35 (mLDHA) and 34 (mPGK1) bp spacing between the proximal HBS and the TATA-box (Table 1). Interestingly, increased (47 bp) as well as decreased (33 and 15 bp) (Table 1) spacing invariably downregulated hypoxic induction of mLDHA HRE mut3 (Fig. 2E). The lowest activity of the mutant with 15 bp spacing (about two helical turns shorter than in the optimal construct) was relatively unexpected and suggested that cooperation between HREs and the TATA-box is more complex and may involve additional features, e.g., DNA bending. Increased spacing (46 bp) had a similar negative effect on mPGK1 HRE mut3 (data not shown). Therefore, the distance between the HRE and the TATA-box also affects hypoxic activation and there is an optimal spacing between the two.
Design and characterization of minimal hypoxia-inducible enhancers
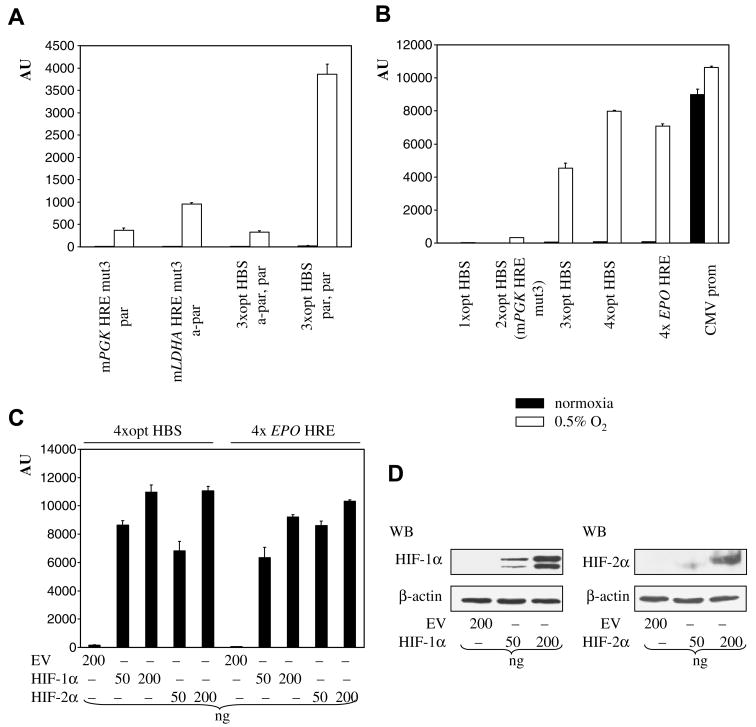
Optimizations enhanced hypoxic induction of both types of HREs, but the overall activity generated by two optimized HBSs was still rather low. Having established the optimal spacing between HBSs, we considered ways to assemble artificial minimal enhancers exclusively from optimized HBSs. Three copies can be arranged a-par, par (the a-par arrangement gave consistently higher activity) or par, par (Table 1). Surprisingly, the construct with the former arrangement, assembled from mLDHA HRE mut3 and a TACGTGcag HBS (Table 1), produced even lower activity than the parental mLDHA HRE mut3 construct (Fig. 3A). In contrast, the par, par construct, assembled from 3 copies of the TACGTGcag HBS (Table 1), provided robust hypoxic induction, considerably higher than constructs with two HBSs (Fig. 3A), confirming that even odd numbers of properly arranged HBSs can generate a strong synergistic effect. This proves that the most efficient way of arranging HBSs is par.
Fig. 3.
Design and activity of minimal hypoxia-inducible enhancers. The effect of arrangement of three HBSs (A), increasing number of HBSs in par arrangement (B), cotransfection of HIF-1α(P402A, P564A) and HIF-2α(P405A, P531A) mutants (C) on reporter activity. Transfection and expression of promoter activities were as described in Fig. 1. Production of HIF-1α(P402A, P564A) and HIF-2α(P405A, P531A) mutants was evaluated by Western blotting (D).
Next, we compared constructs with increasing numbers of par arranged optimized HBSs. Strong synergistic effects were observed with addition of each HBS, resulting in high hypoxic activity of constructs with 3 and 4 HBSs (Fig. 3B). The 4xopt HBS construct (total 36 bp) performed comparably to the 4xEPO HRE construct (total 208 bp) (Fig. 3B), confirming that HIF activates properly multimerized optimized HBSs in a way similar to multimers of the much longer endogenous EPO HRE. Activity of the 4xopt HBS construct in hypoxia was almost as high as activity of the CMV promoter (~800 bp, Fig 3B). HIF-1α and HIF-2α differ in transactivation of some hypoxia-inducible genes, e.g., expression of glycolytic enzymes (such as PGK1) is preferentially activated by HIF-1α [14]. Par arrangement of HBSs in our enhancers, originally present in mPGK1 HRE, prompted us to test the transactivation potential of HIF-1α and HIF-2α on the 4xopt HBS and 4xEPO HRE constructs. HIF-1α(P402A, P564A) and HIF- 2α(P405A, P531A) mutants comparably activated both constructs (Fig. 3C), suggesting that HIF-1α and HIF-2α are equally efficient in transactivation of multimers of optimized HBSs and endogenous EPO HREs.
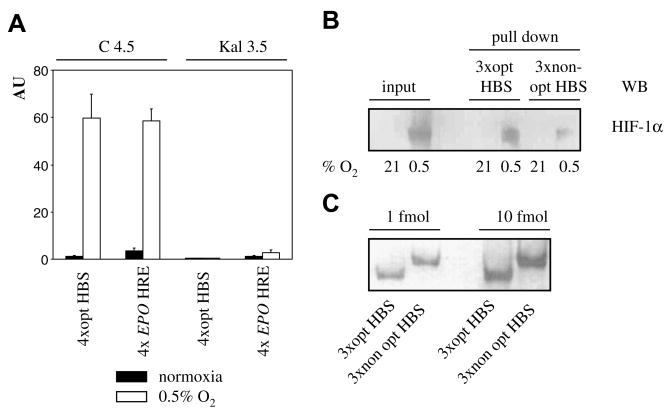
Comparison of the 4xopt HBS and 4xEPO HRE constructs in the HIF-α deficient Kal 3.5 and control C 4.5 CHO cell lines [9] confirmed activation by hypoxia only in C 4.5 cells and considerably lower normoxic activity in Kal 3.5 cells (Fig. 4A). While induction of both constructs by hypoxia was comparable in C 4.5 cells, the 4xEPO HRE construct generated higher activity in Kal 3.5 cells and higher normoxic activity in C 4.5 cells (Fig. 4A). Together, these data confirm that hypoxic induction of both constructs strictly depends on HIF and that HIF activity is responsible for increased normoxic activity.
Fig. 4.
(A) Activity of 4xopt HBS and 4xEPO HRE constructs in HIF-α deficient Kal 3.5 and control C 4.5 CHO cells. Transfection and expression of promoter activities were as described in Fig. 1. (B) Pull-down with biotinylated double-stranded probes comprising three copies of HBSs with optimal and non-optimal (+3) spacing. Bound nuclear proteins were washed under high stringency and analyzed for HIF-1α by Western blotting. (C) Indicated amounts of biotinylated double-stranded probes were subjected to electrophoresis, blotted, and detected with streptavidin-peroxidase and ECL.
To understand why spacing between the optimized HBSs has such a profound effect on hypoxic induction, we performed pull down experiments with biotinylated double-stranded probes with three HBSs, spaced either optimally (par, par opt) or non-optimally, due to insertions of three nucleotides (par, par, non-opt) (Table 1). Using high stringency washing [6], we found that optimally spaced HBSs pull down significantly more HIF-1α than the non-optimally spaced HBSs from nuclear extract prepared from hypoxic cells (Fig. 4B). Detection with streptavidin-peroxidase confirmed comparable amounts of biotinylated probes used (Fig. 4C). Thus, properly aligned HBSs facilitate cooperative interaction between HIF complexes, resulting in high affinity HIF binding and strong synergistic cooperation.
We had invested a considerable effort into developing hypoxiainducible vectors based on the exceptionally tightly regulated CA9 promoter [11]. We reasoned that the combination of the TATA-less CA9 promoter (tight control in normoxia) and minimal hypoxiainducible enhancers (high hypoxic induction) could generate a construct with superior activity. The CA9 HBS, located immediately upstream of the transcription start site [11], allows assembling a chimeric promoter that consists entirely of a single type of regulatory element—HBS. However, regardless of the arrangement of HBSs, activity of all of the CA9 chimeric promoter constructs was always considerably lower than that of the TATA-box containing mLDHA HRE mut3 construct (data not shown). This presumably reflects that HBSs on their own are deficient in activation of transcription and reiterates the importance of the synergistic cooperation between HBSs and the TATA-box.
In addition to using optimized HBSs, the fundamental difference between the minimal hypoxia-inducible enhancers described in this study and the multimers of endogenous HREs lies in the building blocks used. By definition, the building block in multimerized HRE is the whole endogenous HRE of varying length, linked together via spacer sequences [4]. Resulting increased spacing (longer than the optimal 10 bp for the par arrangement) prevents/decreases synergism between HBSs in the neighboring HREs. In minimal hypoxia-inducible enhancers, the building block is the optimized HBS that is multimerized at the same, optimal distance, ensuring maximal synergism between any two neighboring HBSs. This effectively generates a series of overlapping HREs and maximizes activity for any given number of HBSs. Although in this study, we sought maximal activity, we can also envisage fine-tuning of activity of these enhancers by mutating (“de-optimizing”) individual positions in HBSs.
In conclusion, by using the Occam’s razor, we have designed artificial minimal hypoxia-inducible constructs, consisting of a minimal TATA-box and optimally arranged optimized HBSs. These constructs are highly inducible by hypoxia, proving, contrary to the previous reports, that properly arranged HBSs are sufficient for hypoxic activation. Our work improves understanding of the basic structural determinants of HREs and pioneers the combinatorial approach that will have implications for designing new generation of hypoxiainducible vectors.
Abbreviations
- A-par
antiparallel
- CA9
carbonic anhydrase 9
- CHO
Chinese hamster ovary
- CP
ceruloplasmin
- EPO
erythropoietin
- ECL
enhanced chemiluminescence
- ET
endothelin
- HBS
HIF-binding site
- HIF
hypoxia-inducible factor
- HRE
hypoxia-responsive element
- mLDHA
murine lactate dehydrogenase-A
- mPGK1
murine phosphoglycerate kinase-1
- PAI
plasminogen activator inhibitor-1
- Par
parallel
- rGlut1
rat glucose transporter-1
- STRA13
stimulated with retinoic acid-13
- VEGF
vascular endothelial growth factor
References
- 1.Wenger RH, Stiehl DP, Camenish G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;306:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 2.Ameri K, Burke B, Lewis CE, Harris AL. Regulation of a rat VL30 element in human breast cancer cells in hypoxia and anoxia: role of HIF-1. Br J Cancer. 2002;87:1173–1181. doi: 10.1038/sj.bjc.6600576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura H, Weisz A, Ogura T, Hitomi Y, Kurashima Y, Hashimoto K, D’Acquisto F, Makuuchi M, Esumi H. Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J Biol Chem. 2001;276:2292– 2298. doi: 10.1074/jbc.M008398200. [DOI] [PubMed] [Google Scholar]
- 4.Boast K, Binley K, Iqball S, Price T, Spearman H, Kingsman S, Kingsman A, Naylor S. Characterization of physiologically regulated vectors for the treatment of ischemic disease. Hum Gene Ther. 1999;10:2197–2208. doi: 10.1089/10430349950017185. [DOI] [PubMed] [Google Scholar]
- 5.Post DE, Van Meir EG. Generation of bi-directional hypoxia/HIF-responsive expression vectors to target gene expression to hypoxic cells. Gene Ther. 2001;8:1801–1807. doi: 10.1038/sj.gt.3301605. [DOI] [PubMed] [Google Scholar]
- 6.Ebert BL, Bunn HF. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol Cell Biol. 1998;18:4089–4096. doi: 10.1128/mcb.18.7.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firth JD, Ebert BL, Ratcliffe PJ. Hypoxic regulation of lactate dehydrogenase A: interaction between hypoxia inducible factor 1 and cAMP response elements. J Biol Chem. 1995;270:21021–21027. doi: 10.1074/jbc.270.36.21021. [DOI] [PubMed] [Google Scholar]
- 8.Kaluzová M, Kaluz S, Lerman MI, Stanbridge EJ. DNA damage is a prerequisite for p53-mediated proteasomal degradation of HIF-1alpha in hypoxic cells and downregulation of the hypoxia marker carbonic anhydrase IX. Mol Cell Biol. 2004;24:5757–5766. doi: 10.1128/MCB.24.13.5757-5766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood SM, Wiesener MS, Yeates KM, Okada N, Pugh CW, Maxwell PH, Ratcliffe PJ. Selection and analysis of a mutant cell line defective in the hypoxiainducible factor-1α-subunit (HIF-1 α) J Biol Chem. 1998;273:8360–8368. doi: 10.1074/jbc.273.14.8360. [DOI] [PubMed] [Google Scholar]
- 10.Galson DL, Tsuchiya T, Tendler DS, Huang LE, Ren Y, Ogura T, Bunn HF. The orphan receptor hepatic nuclear factor 4 functions as a transcriptional activator for tissue-specific and hypoxia-specific erythropoietin gene expression and is antagonized by EAR3/COUP-TF1. Mol Cell Biol. 1995;15:2135–2144. doi: 10.1128/mcb.15.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaluz S, Kaluzová M, Stanbridge EJ. Expression of the hypoxia marker carbonic anhydrase IX is critically dependent on SP1 activity. Identification of a novel type of hypoxia-responsive enhancer. Cancer Res. 2003;63:917–922. [PubMed] [Google Scholar]
- 12.Potter C, Harris AL. Hypoxia inducible carbonic anhydrase IX, marker of tumour hypoxia, survival pathway and therapy target. Cell Cycle. 2004;3:64–167. [PubMed] [Google Scholar]
- 13.Shi Q, Le X, Abbruzzese JL, Peng Z, Qian C-N, Tang H, Xiong Q, Wang B, Li X-C, Xie K. Constitutive Sp1 activity is essential for differential constitutive expression of vascular endothelial growth factor in human pancreatic adenocarcinoma. Cancer Res. 2001;61:4143–4154. [PubMed] [Google Scholar]
- 14.Hu CJ, Wang LJ, Choodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]