Abstract
The plasmin system is involved in the degradation of Aβ peptides, the accumulation of which in brain is a hallmark of Alzheimer’s disease (AD). In a North European case-control AD dataset we studied 14 common variations in the PLG, PAI-1, PLAT and PLI genes encoding components of the plasmin system. Among the four polymorphisms in the PLAT, PAI-1 and PLI genes showing nominally significant evidence for an association with AD (allele p-value = 0.01–0.00003) the strongest association was detected for the deletion allele in the Alu-repeat region of the PLAT gene. However, none of these positive results were confirmed in follow-up studies using an independent Canadian case-control cohort and two familial AD datasets of North European and Caribbean Hispanic origin. Thus, the current survey does not support the notion that common polymorphisms in the plasmin genes influence the development of AD.
Keywords: Alzheimer’s disease, plasmin, polymorphism, risk factor
1. Introduction
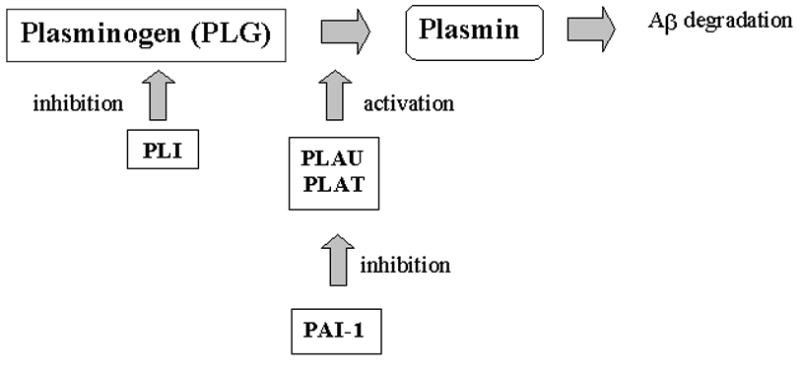
The earliest pathology seen in Alzheimer’s disease (AD) brain is the deposition of amyloid plaques, consisting mainly of Aβ40/42 peptides generated from the β-amyloid precursor protein (APP). Genes involved in the production/degradation of Aβ are strong functional candidates for AD susceptibility genes. Plasmin has been found to cleave Aβ peptides in vitro and to prevent Aβ aggregation into β-pleated sheet structures [14–17] (supplemental Figure 1). Here we conducted genetic studies to investigate the association between late-onset AD and common polymorphisms in four genes involved in the activation of plasmin: alpha-2-plasmin inhibitor (PLI), plasminogen activator inhibitor type I (PAI-1), plasminogen activator tissue (PLAT) and plasminogen (PLG).
2. Materials and methods
Fourteen single nucleotide polymorphisms (SNPs) in the PLG, PAI-1, PLAT and PLI genes were initially screened in a case-control dataset of North European origin consisting of 192 late-onset sporadic AD subjects and 195 normal controls (supplemental Table 2). A χ2 test was used to examine differences in genotype/allele frequencies. Nominally significant results (p<0.05) were followed-up in three independent late-onset AD datasets: 124 North European pedigrees, 96 Caribbean Hispanic pedigrees and a Canadian case-control dataset consisting of 189 sporadic AD subjects and 245 normal controls. The statistical analyses were computed using the software SAS release 8.02. The genotype data obtained for the familial AD datasets was analyzed using a software package for computing Family Based Association Tests (FBAT version 1.5) [10] (see supplemental material for details).
3. Results
The genotypes for the 14 variations in the PLG, PAI-1, PLAT, and PLI genes were in Hardy–Weinberg equilibrium. The calculation of pair-wise linkage disequilibrium (LD) within each gene in the North European case-control dataset revealed a range of disequilibria (D′=0.56–1.0) with the strongest LD amongst the four PLAT variations; however r2 detected only a moderate degree of LD (supplemental Table 4).
The results of the initial screening using a North European case-control dataset detected nominally significant association between AD and four SNPs (after Bonferroni correction): the rs1799889 in the PAI-1 gene; rs2287322 in the PLI gene; rs1058720 and a 289 bp insertion/deletion variation in the Alu-repeat (Alu I/D) in the PLAT gene (Table 1, supplemental Table 3). The strongest evidence of association with AD was observed for the PLAT gene, with the “C” allele of the rs1058720 and the deletion allele (“D”) of the Alu I/D over-represented in AD patients (allelic p-value is 0.01 and 0.00003, respectively). The distribution of haplotypes derived from the rs1058720 and Alu I/D polymorphisms was significantly different between cases and controls (global p=0.001), however the association was driven by the Alu I/D variation, and no specific haplotype was more significantly associated with AD than the “D” allele (data not shown).
Table 1.
Single SNP association results generated in four independent Alzheimer Disease datasets (supplemental Table 3 shows allele and genotype frequencies). Nominally significant p-values are in bold; the alleles putatively associated with AD are depicted only for SNPs generating nominal p-values< 0.05. N/D = not done.
| Case-Control datasets | Familial datasets | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| North European | Canadian | North European | Caribbean Hispanic | |||||||
| Gene (Locus) | Distance (kb) | SNP | SNP Function | p-values | Putative risk allele | P-values | FBAT p-values | |||
| genotype | Allele | genotype | Allele | |||||||
| PLG (6q26) | rs3757019 | intron 5 | 0.62 | 0.34 | N/D | N/D | N/D | N/D | ||
| 4.4 | rs1130656 | exon 8 F314F | 0.21 | 0.18 | N/D | N/D | N/D | N/D | ||
| 34.5 | rs11060 | exon 19 G762G | 0.76 | 0.5 | N/D | N/D | N/D | N/D | ||
| PAI-1 (7q22) | rs1799889 | 5′ UTR -675 4G/5G | 0.002 0.008* | 0.0003 0.001* | 4G | 0.24 | 0.54 | 0.49 | 0.46 | |
| 4.8 | rs2227662 | intron 2 | 0.36 | 0.92 | N/D | N/D | N/D | N/D | ||
| 4.7 | rs2227692 | intron 6 | 0.86 | 0.63 | N/D | N/D | N/D | N/D | ||
| 1.8 | rs11178 | 3′ UTR | 0.46 | 0.27 | N/D | N/D | N/D | N/D | ||
| PLAT (8p12) | rs2020923 | intron 10 | 0.26 | 0.16 | N/D | N/D | N/D | N/D | ||
| 2.3 | Alu I/D | intron 7 | 0.0001 0.0004* | 0.00003 0.0001* | D | 0.83 | 0.74 | 0.28 | 0.15 | |
| 5.5 | rs1058720 | exon 5 D167D | 0.006 0.02* | 0.01 0.05* | C | 0.09 | 0.17 | 0.48 | 0.93 | |
| 1 | rs2070711 | intron 3 | 0.54 | 0.35 | N/D | N/D | N/D | N/D | ||
| PLI (17p13) | rs2287322 | 5′ UTR | 0.002 0.009* | 0.0003 0.001* | G | 0.87 | 0.79 | 0.42 | 0.86 | |
| 15.2 | rs736060 | intron 8 | 0.86 | 0.81 | N/D | N/D | N/D | N/D | ||
| 1.4 | rs1057335 | exon 9 K434R | 0.06 | 0.02 0.06 | N/D | N/D | N/D | N/D | ||
Association of SNP with AD is significant after adjustment for multiple testing with Bonferroni correction.
To validate the nominally positive results, all four SNPs giving statistically significant scores were re-genotyped in three independent replication AD cohorts consisting of a case-control and two familial datasets (Table 1). These follow-up studies did not generate any significant results. Adjustment for APOE genotype and haplotype analyses had little impact on inferences about association with the SNPs in any of the datasets (data not shown).
4. Discussion
The current association study investigated 14 common variations in four genes encoding the components of the plasmin system. Four SNPs in PLAT, PAI-1 and PLI genes were found to be associated with AD in the North European case-control dataset (Table 1). The strongest evidence of association was detected in the PLAT gene, where rs1058720 and the Alu I/D are likely tagging the same LD block (supplemental Table 4). However, several lines of evidence suggest that the observed associations represent false positive findings. Our follow-up studies in three independent AD datasets did not confirm the initial association results. In addition, a recent case-control study in a Spanish dataset did not reveal association between AD and the Alu I/D polymorphism [3]. The allele frequencies in the Spanish dataset were identical to our control group, however the frequency of the deletion allele was considerably higher in the North European AD subjects (60% vs 44%). Finally, the PLAT locus, mapped to 8p12, has not been previously implicated in published genome-wide AD linkage surveys.
Supplementary Material
Acknowledgments
This work was supported by the Alzheimer Society of Canada, Japan-Canada and Canadian Institutes of Health Research Joint Health Research Program (ER), the Canadian Institutes of Health Research, Alzheimer Society of Ontario, Howard Hughes Medical Institute, Canada Foundation for Innovation (PH), Fonds de la Recherche en Santé (YM), the National Institutes of Health: R01-AG09029 and P30-AG13846 (LAF) and Mitsubishi Pharma Research Foundation (NS). National Institute of Health and the National Institute on Aging: R01-AG15473, P50-AG08702, and P01-AG07232, the Alzheimer Association, the Charles S. Robertson Memorial Gift for Alzheimer’s Disease Research from the Banbury Fund and the Blanchette Hooker Rockefeller Foundation (RM).
Footnotes
Disclosure statement. None of the authors have financial interest or conflicts of interest in the described results. Informed consent for research purposes was obtained from all individuals involved in the study approved by a research ethics board.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


