Abstract
Osteosarcoma (OS) is an aggressive bone tumor with complex abnormal karyotypes and a highly unstable genome, exhibiting both numerical- and structural-chromosomal instability (N- and S-CIN). Chromosomal rearrangements and genomic imbalances affecting 8q24 are frequent in OS. RECQL4 gene maps to this cytoband and encodes a putative helicase involved in the fidelity of DNA replication and repair. This protective genomic function of the protein is relevant because often patients with Rothmund-Thomson syndrome have constitutional mutations of RECQL4 and carry a very high risk of developing OS. To determine the relative level of expression of RECQL4 in OS, 18 sporadic tumors were studied by reverse transcription-polymerase chain reaction. All tumors overexpressed RECQL4 in comparison to control osteoblasts, and fluorescence in situ hybridization analysis of tumor DNA showed that expression levels were strongly copy number-dependent. Relative N- and S-CIN levels were determined by classifying copy number transitions within array comparative genomic hybridization profiles and by enumerating the frequency of break-apart fluorescence in situ hybridization within 8q24 using region-specific and control probes. Although there was no evidence that disruption of 8q24 in OS led to an elevated expression of RECQL4, there was a marked association between increased overall levels of S-CIN, determined by copy number transition frequency and higher levels of RECQL4.
Introduction
Osteosarcoma (OS) is the most common primary bone malignancy and is characterized by complex chromosomal abnormalities that vary widely from cell to cell. These tumors exhibit a high degree of aneuploidy, gene amplification, and multiple unbalanced chromosomal rearrangements. A combined approach of molecular cytogenetics techniques [comparative genomic hybridization (CGH), spectral karyotyping, or multicolor banding] together with classic G-banded cytogenetics analysis of OS tumors describe complex karyotypes with multiple numerical and structural chromosomal aberrations. Collectively, these studies [1–12] have highlighted the unique and highly unstable karyotype of OS.
Two distinct processes governing genome stability may be disrupted in cancer cells: those that affect numerical segregation and ploidy of chromosomes and those that affect the fidelity of DNA replication/repair and lead to structural chromosome aberrations (reviewed in [13]). The complexity of the OS genome likely arises as a consequence of chromosomal instability (CIN), generated by both numerical and structural chromosome abnormalities. Numerical patterns of chromosomal aberration have been referred to as N-CIN, whereas CIN that leads to elevated levels of structural change has been termed S-CIN. The development of high-resolution array comparative genomic hybridization (aCGH) methods provides an opportunity to analyze genomic complexity at both the N-CIN and S-CIN level using whole-genome imbalance plots. In this approach, the respective distributions of copy number alteration present within the entire aCGH profile can be used to determine the relative contributions of S-CIN [as defined by copy number transitions (CNTs) [14], within each chromosome] or N-CIN (as defined by chromosomal imbalance affecting an entire chromosome) to the overall complexity of the OS genome in a given tumor. Previously [15], we have used aCGH analysis of OS to map chromosomal regions recurrently subject to genomic changes such as gene amplification. In the current article, we further interrogate aCGH profiles to measure levels of S-CIN and N-CIN in OS in the context of genomic and molecular changes of cytoband 8q24.
Some chromosomal regions are more frequently involved in genomic aberrations in OS, namely, 1p35-p36, 6p12-p21, 8q23-q24, 17p11-p12, and 19p13 [4] (reviewed in [16]). The 8q24 region is of particular interest to OS because a number of genes both directly and indirectly implicated in OS oncogenesis map to this region. The MYC oncogene at 8q24 is highly amplified in a subset of OS [17,18], is overexpressed more frequently in relapsed and metastatic OS [19], and is often amplified in a wide variety of carcinomas [20]. RECQL4 maps to this same cytoband and the gene encodes a putative helicase involved in the fidelity of DNA replication, and this protective genomic function of the protein is provocative because patients with constitutional mutations of RECQL4 have Rothmund-Thomson syndrome (RTS) and carry a very high risk of developing an OS [21]. Approximately two-thirds of patients with a clinical diagnosis of RTS will have RECQL4 mutations. The other one third likely represents genetic heterogeneity and have mutations in another gene(s). The RTS patients with RECQL4 mutations have a much higher risk of developing OS compared with the RTS patients without RECQL4 mutations [22]. Moreover, the 8q24 cytoband is now of particular interest to cancer biologists because recent genome-wide association studies have identified multiple neighboring regions within a 600-kb segment of chromosome 8q24 that harbors variants associated with predisposition to prostate, colon, and bladder cancers [23–25]. The most likely candidate gene within the 8q24 region that could contribute directly to the CIN phenotype of OS is the RECQL4 gene, which encodes a helicase member of the RecQ family [26].
In this study, we address the hypothesis that deregulation of RECQL4 expression, caused by the 8q24 rearrangements, could underlie the high rate of CIN observed in OS. Complex genomic alterations and amplifications at 8q24 were of particular interest because the affected regions are relatively small; they have been found to be aberrant in multiple OS samples and they are located in a region of the human genome strongly implicated in tumorigenesis and DNA repair. If genomic alterations occur near the RECQL4 region, it is conceivable that such changes lead to deregulation of the locus, and this may compromise its repair and DNA maintenance functions, with consequences for the entire genome's integrity [26–30]. In the present study, we used 18 OS tumor samples to investigate whether RECQL4 gene expression levels were linked to the extent and type of CIN (N-CIN and S-CIN) throughout the OS genome and specifically at 8q24.
Materials and Methods
Patient Tumors
The collection of frozen tissue specimens (n = 18), archival formalinfixed, paraffin-embedded OS sections (n = 12), and clinicopathologic data was obtained and handled in accordance with the Hospital for Sick Children Research Ethics guideline (Toronto, Canada). The OS specimens and corresponding paraffin-embedded specimens consisted of resected or biopsy tumor tissue obtained at diagnosis. Hematoxylin and eosin-stained sample sections were subjected to standard histopathologic evaluation to determine the tumor content and the pathologic grade according to the World Health Organization [31]. All samples presented a tumor content higher than 90%. The clinical and histologic features of presented OS cohort are detailed in Table 1.
Table 1.
Clinical Data of the 18 OSs.
| No. | Sex | Age (years) | Localization | Sample | Subtype | Grade |
| OS1 | F | 9 | Tibia | Biopsy | Osteoblastic | High |
| OS2 | M | 14 | Femur | Lesion | Osteoblastic | High |
| OS3 | M | 13 | Femur | Biopsy | Osteoblastic | Intermediate |
| OS4 | M | 7 | Humerus | Biopsy | Poorly differentiated | High |
| OS5 | F | 12 | Femur | Biopsy | Poorly differentiated | High |
| OS6 | M | 14 | Femur | Biopsy | Osteoblastic | High |
| OS7 | M | 7 | Femur | Biopsy | Osteoblastic | High |
| OS8 | M | 13 | Femur | Resection | Osteoblastic | High |
| OS9 | F | 13 | Humerus | Biopsy | Osteoblastic | High |
| OS10 | M | 8 | Tibia | Resection | Osteoblastic | High |
| OS11 | M | 13 | Femur | Biopsy | Osteoblastic | High |
| OS12 | M | 17 | Lung | Resection | Osteoblastic | High |
| OS13 | M | 9 | Leg | Biopsy | Poorly differentiated | High |
| OS14 | F | 12 | Femur | Biopsy | Osteoblastic | Intermediate |
| OS15 | F | 14 | Femur | Biopsy | Osteoblastic | High |
| OS16 | F | 17 | Tibia | Resection | Osteoblastic | High |
| OS17 | M | 15 | Tibia | Resection | Osteoblastic | High |
| OS18 | M | 15 | Femur | Biopsy | Poorly differentiated | High |
F indicates female; M, male.
Cell Culture
The RTS primary fibroblasts were obtained from the Corriell Institute for Medical Research (AG18371; Camden, NJ) [32]. These fibroblasts do not express RECQL4 because of an 11-bp intronic deletion, which disrupts the splicing and compromises its expression [32,33]. They were cultured in alpha-minimum essential medium (Invitrogen, Burlington, Ontario, Canada) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen). Osteoblasts were obtained from PromoCell (Heidelberg, Germany) and cultured in osteoblast culture medium (PromoCell).
RNA Extraction and Semiquantitative Reverse Transcription-Polymerase Chain Reaction
The mRNA level of RECQL4 was examined by semiquantitative reverse transcription-polymerase chain reaction (RT-PCR), using the housekeeping gene PBGD as a calibration control. Total RNA from snap-frozen OS tumors (18 samples) and from cultured cells (RTS fibroblasts and osteoblasts) were isolated using the TRIzol Reagent (Invitrogen). The RNA quality was assessed by BioAnalyzer RNA 600 Nano Kit (Agilent Technologies, Palo Alto, CA). Total RNA from kidney and testis (Ambion, Foster City, CA) were used as positive control detection for low and high RECQL4 expressions, respectively [34]. Total RNA from RTS fibroblasts were used as negative control [33,35]. Coamplification of RECQL4 and PBGD genes was performed on 150 ng of total RNA by applying the one-step RT-PCR method (Superscript One-Step RT-PCR III; Invitrogen) according to the manufacturer's instructions. RECQL4 forward (5′-CTCATCTAAGGCATCCACCC-3′) and reverse (5′-CTGTGACATCGCTGTAACCA-3′) primers were designed to amplify a 188-bp fragment (Accession Number NM_004260). PBGD primers were designed to amplify a 127-bp fragment [36]. Primers for RECQL4 and PBGD were combined as follows: 0.2 µM of each forward and reverse RECQL4 primers and 0.15 µM of each forward and reverse PBGD primers. The reverse transcription and amplification conditions were performed using the MJ Research PTC200 thermocycler and consisted of an initial reverse transcription reaction at 57°C for 30 minutes, followed by denaturation (2 minutes at 94°C) and 30 cycles of amplification (94°C for 30 seconds; 57°C for 30 seconds; 68°C for 30 seconds). The final elongation reaction was performed at 68°C for 10 minutes. Genomic DNA contamination in every sample was excluded by omitting the reverse transcriptase in the RT-PCR.
The measurement and quantification of the 188-bp (RECQL4) and 127-bp (PBGD) coamplified fragments were performed using the DNA 1000 LabChip Kit (Agilent 2100 Bioanalyzer; Agilent Technologies) and the 2100 Expert Software (Agilent Technologies), respectively. PBGD was used for calibration, and the mRNA fold change of RECQL4 in OS cohort was compared with the normal osteoblasts. The SD from triplicate RT-PCR experiments was calculated for each sample.
Genome-wide Analysis of Chromosomal Instability
Ten OS samples for which sufficient total DNA could be extracted were hybridized against Human Genome CGH 44k microarrays (Agilent Technologies), spanning the entire human genome at a median resolution of 75 kb as described previously [15]. These aCGH data files have also been used to map the distribution of recurrently deleted and amplified regions in OS [15] and they are deposited in National Center for Biotechnology Information's Gene Expression Omnibus (GEO) Web site (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through the GEO Series accession number GSE9654. For each tumor, the normalized data were run through the Circular Binary Segmentation (CBS) algorithm [37], with a threshold of 0.5 (CGH Analytics, 3.5.14; Agilent Technologies). The CBS algorithm provides a list of aberrations (imbalanced genomic region with a unique abnormal CGH ratio), each determined by two CNTs, as defined by Ferreira et al. [14]. The distribution along the chromosome of these CNTs for each aberration was used to determine the respective contribution of N-CIN and S-CIN (Table W1). Briefly, an aberration with the starting and ending CNTs positioned within an arm of a chromosome was considered to be caused by copy number change arising from an unbalanced structural alteration and was scored as S-CIN. An aberration, which CNTs were positioned in telomeric or centromeric regions, was considered to result from copy number change affecting an entire chromosome or chromosome arm and was scored as N-CIN. All the aberrations were classified as N- or S-CIN-related and scored for each of the 10 OS tumors studied by aCGH. The aberrations on X and Y chromosomes were excluded from the analysis to eliminate the sex mismatching bias.
Fluorescence In Situ Hybridization
Dual-color fluorescence in situ hybridization (FISH) method was applied to the 5-µm thick archival formalin-fixed, paraffin-embedded tissue sections. Bacterial artificial chromosome (BAC) genomic clone RP11-349C2 was identified in the Resources for Molecular Cytogenetics database (www.biologia.uniba.it) by its location at the RECQL4 locus (chr8:145,707,623-145,713,976, UCSC genome browser, www.genome.ucsc.edu, version March 2006). The BAC clone was obtained from the Centre for Applied Genomics (Toronto, Ontario, Canada). The presence of the RECQL4 sequence and the correct chromosome location of the BAC clone were verified by PCR and by hybridization to metaphase spreads from normal peripheral lymphocytes, respectively. The BAC probe was labeled either with ULYSIS-Alexa-594 or with ULYSIS-Oregon-green-488 (Molecular Probes, Invitrogen) and was combined with the alpha satellite centromeric probe of chromosome 8, cen(8) (CEP 8 SpectrumGreen; Abbott Molecular, Des Plaines, IL) or with a commercial MYC-containing probe (LSI C-MYC-SpectrumOrange; Abbott Molecular), respectively. Dual-color FISH was performed according to standard procedures [38]. Either RECQL4 and cen(8) or RECQL4 and MYC were evaluated by spot visualization and enumeration for each probe in a range from 50 to 100 nonoverlapped, intact interphase nuclei per tumor tissue using a Zeiss Imager.Z1 microscope equipped with a digital camera AxioCam MRm and AxioVision 4.3 capturing software (Carl Zeiss Canada, Ltd., Toronto, Canada). If fluorescent signals could not be seen in at least 80% of cells, the result was considered to be noninterpretable (6/12). The most represented pattern of signal was recorded for the probe combinations mentioned above. The establishment of a cutoff value of >10% of tumor nuclei for the different probes used and for all signal patterns was defined considering the truncation artifacts, aneusomy, nuclear size, and chromatin condensation [39]. The relative gene copy number of RECQL4 was calculated by adding the number of RECQL4 signals to the RECQL4/cen(8) ratio. This calculation allowed to enhance the distinction between a ratio of 1 with two copies of each probe (2:2), and a ratio of 1 with three to five copies of each probe (3:3, 4:4, and 5:5). The pattern of signals (contiguous vs scattered) for the dual-color FISH experiment using RECQL4 and MYC probes was evaluated nucleus by nucleus to document the S-CIN of the 8q24 region. The observation of scattered signals was interpreted as the manifestation of S-CIN.
Chromosome copy number analysis was performed using centromeric enumeration probes for cen(3) (CEP 3 SpectrumRed), cen(7) (CEP 7 SpectrumGreen), and cen(17) (CEP 17 SpectrumAqua; Abbott Molecular). Sequential three-color FISH method was applied to the paraffin-embedded tissue sections according to the manufacturer's instructions. Chromosome enumeration was determined by scoring the number of signals for cen(3), cen(7), and cen(17) in 200 nonoverlapped intact interphase nuclei (10 of the 12 samples reached these criteria). The estimation of the overall ploidy of each sample is described case by case in the supplementary material (Table W2). Briefly, adapted from Rossi et al. [40], the OS cohort was classified by ploidy FISH as follows: 1) diploid, >50% of cells showing two signals for all probes; 2) near-triploid, >20% of cells showing three signals for at least two probes; 3) near-tetraploid, >20% of cells showing four signal for at least two probes; 4) polyploidy, >50% of cells showing more than five signals for at least one probe [40].
Results
RECQL4 Expression Levels in OS Determined by RT-PCR Analysis
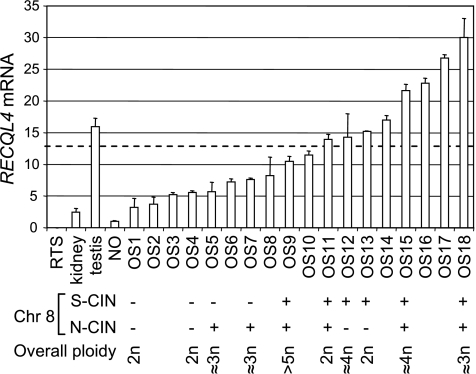
To establish the expression level of RECQL4 in OS samples, we performed semiquantitative RT-PCR using total RNA extracted from patient tumors. We demonstrated that RECQL4 was overexpressed in all the tested tumors compared with normal osteoblasts. The mean expression levels observed for all 18 OS tumors was ∼13-fold higher than osteoblasts (range, x3 to x30; Figure 1). In keeping with the published literature, RECQL4 could not be detected in RNA-derived RTS fibroblasts [33,35]. OS1 exhibited the lowest level of RECQL4 expression, which was comparable to the low expression level observed in normal kidney tissue. In contrast, OS18 was characterized by an expression twice higher than that of the normal testis sample, which has been shown to be one of the most RECQL4-rich tissues [34]. We conclude that the expression level of RECQL4 is deregulated in OS.
Figure 1.
RECQL4 is overexpressed in OS and is associated with S-CIN. The histogram is showing the fold change of RECQL4 mRNA compared with normal osteoblasts (NO) measured by semiquantitative RT-PCR. Rothmund-Thomson fibroblasts (RTS) and normal tissues (kidney and testis) were used as negative and positive controls, respectively. Each value is the result of three independent experiments. The SD is shown above each bar. The dashed line represents the mean value calculated from OS sample data. The lower panel part describes the chromosomal 8 instability (CIN) in the context of overall ploidy for 10 samples for which material was available for FISH studies. The absence of N-CIN or S-CIN is shown as (-). Identification of S-CIN or N-CIN is shown as (+). The overall ploidy status was established according to the FISH data using cen(3), cen(7), and cen(17). OS tumors that are showing an RECQL4 expression value close or more than the mean are showing S-CIN, whereas the ones less than the mean do not. The N-CIN and S-CIN are independent, and a high level of RECQL4 expression is associated with the appearance of S-CIN on the 8q24 region.
Association of RECQL4 Expression and Genome-wide Chromosomal Instability Levels
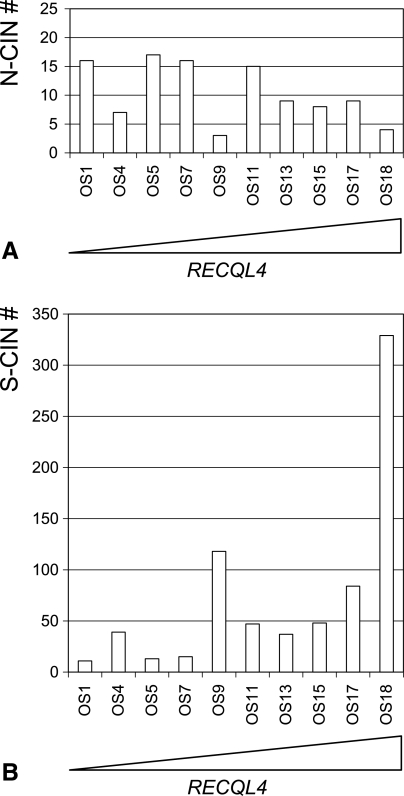
To determine whether there was a relationship between expression levels of RECQL4 and overall levels of CIN, we analyzed aCGH profiles to distinguish between N-CIN and S-CIN using 10 of the 18 tumors. Analysis of CNT distributions within each chromosome and the pattern of overall chromosomal imbalance present in each aCGH profile provided an objective overview of the variation in N-CIN and S-CIN levels that characterized each of the 10 OS analyzed (Table W1 and Figure 2). The mean number of N-CIN changes was 10.4 (SD, 5) for the study group, with no apparent relationship between the levels of RECQL4 expression and N-CIN (Figure 2A). In contrast, there was a clear trend showing an increase in S-CIN with higher expression levels of RECQL4. For example, OS18 had the highest expression level and exhibited >300 S-CIN aberrations and OS1 showed the lowest-expressing tumor for RECQL4 and had only 11 S-CIN aberrations. The mean number of S-CIN for the study group was 74.1 (SD, 95; Figure 2B). Collectively, these data indicate that elevated RECQL4 expression is associated with a greater incidence of CNTs and concomitant elevation in S-CIN frequency. We were not able to demonstrate any relationship between N-CIN levels and varying levels of gene expression of RECQL4.
Figure 2.
Association between RECQL4 expression levels and varying N-CIN and S-CIN frequencies. (A) Differing frequencies of N-CIN (y-axis) as determined by analyzing imbalance profiles of whole chromosomes or entire arms for the 10 OS tumors (x-axis) after aCGH analysis. (B) Differing frequencies of S-CIN (y-axis) as determined by analyzing CNT distributions across each genomic profile derived from the ten OS tumors (x-axis) after aCGH analysis. The OS samples are ordered from left to right according to their RECQL4 expression level (lowest to highest) and are represented as a triangle in which the adjacent side represents the highest RECQL4 expression level.
Influence of RECQL4 Gene Copy Number on Its Expression in OS
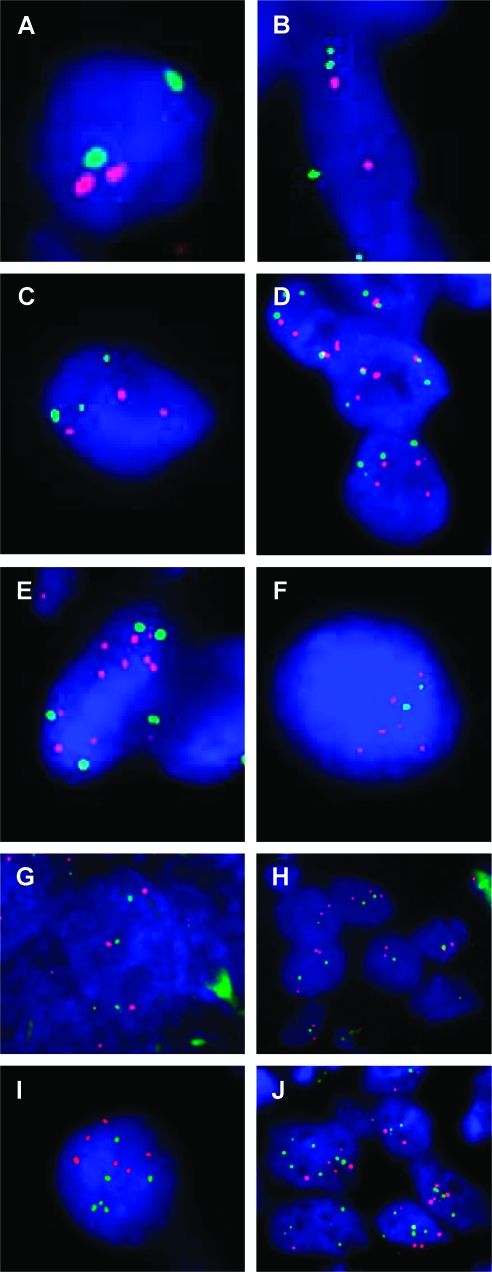
The variation of the RECQL4 expression in OS could be the result of genomic copy number changes of the locus at 8q24. Dual-color interphase FISH analysis, using the RECQL4 and the cen(8) probes, was therefore performed on formalin-fixed, paraffin-embedded tissue derived from 12 OS tumors. As illustrated in Figure 3, A–D, and summarized in Table 2, we show that the increase of RECQL4 expression followed the genomic status of the RECQL4 gene. To determine copy number changes, cohybridization of RECQL4 and cen(8) probes and systematic analysis of the dual-color patterns allowed us to quantify numerical chromosome 8 abnormalities. OS samples with the lowest expression of RECQL4 (such as OS1 and OS4) have two normal chromosome 8s each bearing two copies of RECQL4 and cen(8) (Figure 3A). In OS2, four cen(8)s were present with only two copies of the RECQL4 gene, suggesting that a tetrasomy of chromosome 8 also involved RECQL4 loss (Figure 3B). OS5, OS7, and OS9 had a higher expression of RECQL4 (below the mean value) and three to five copies for each RECQL4 and cen(8) probes (Figure 3, C and D). This pattern could be interpreted as an acquisition of extra copies of an intact chromosome 8. Lastly, OS samples with the highest RECQL4 expression, (above the mean) all exhibited a copy number gain of RECQL4, which was observed as nine RECQL4 signals with four cen(8) for OS15 (Figure 3E) or six RECQL4 signals for two cen(8) as in OS13 (Figure 3F). Collectively, these findings indicate that of RECQL4 gene expression is largely copy number-dependent (Table 2).
Figure 3.
(A–F) RECQL4/cen(8) dual-color interphase FISH experiment on OS tumors samples' fixed sections. RECQL4 probe is shown in red, whereas the cen(8) is in green. OS samples exhibited a normal pattern [OS1 (A)], a loss [OS2 (B)], an abnormal pattern associated with a 1:1 ratio [OS7 (C) and OS9 (D)], or a gain of RECQL4 [OS15 (E) and OS13 (F)]. (G–J) Structural CIN of the 8q24 cytoband. Dual-color interphase FISH was done on fixed tissue sections using the MYC probe (red) located on 8q24.21 and the RP11-349C2 probe (green) located on 8q24.3. The signal pattern of contiguous red and green signals on OS7 (G) and OS1 (H) identifies these samples as no S-CIN for the 8q24 cytoband. In contrast, for OS13 (I) and OS15 (J), the red and green signals are scattered all around the nuclei, characterizing these two samples as S-CIN for the 8q24 region.
Table 2.
RECQL4 Relative Gene Copy Number and Expression Analysis.
| No. | RECQL4 Copy Number (Genomic Status) | cen(8) Copy Number | RECQL4 Relative Copy Number* | RECQL4 Expression (Fold Change)† |
| OS1 | 2 (N) | 2 | 3 | 3 |
| OS2 | 2 (L) | 4 | 2.5 | 4 |
| OS4 | 2 (N) | 2 | 3 | 6 |
| OS5 | 4 (AN) | 4 | 5 | 6 |
| OS7 | 3 (AN) | 3 | 4 | 8 |
| OS9 | 5 (AN) | 5 | 6 | 10 |
| OS11 | 6 (G) | 4 | 7.5 | 14 |
| OS12 | 3 (G) | 2 | 4.5 | 14 |
| OS13 | 6 (G) | 2 | 9 | 15 |
| OS15 | 8 (G) | 4 | 10 | 22 |
| OS16 | 9 (G) | 2 | 13.5 | 23 |
| OS18 | 5 (G) | 3 | 6.6 | 30 |
AN indicates aneusomy; G, gain; L, loss; N, normal.
RECQL4 relative number is calculated as described in the Materials and Methods section.
Expression of RECQL4 is normalized against the PBGD housekeeping gene expression, and shown as a fold change compared with the expression level measured in normal osteoblasts.
Influence of Structural Alterations of Cytoband 8q24 on RECQL4 Expression in OS
We used dual-color interphase FISH to study S-CIN levels within 8q24.21 (MYC probe) and 8q24.4 (RECQL4 probe) cytobands. Because both probes are closely linked within 8q24, paired two-color signals within nuclei can be used to determine whether disruption of the cytoband has taken place. In four OS tumors (OS1, OS4, OS5, and OS7), there was no evidence of disruption between MYC and the RECQL4 probes within nuclei (Table 3 and Figure 3, G and H). For six OS tumors, disruption of 8q24 was apparent. In OS13 (Figure 3I), six copies of 8q24 were apparent, but the red and green signals were no longer paired, indicative of structural rearrangement between the MYC and RECQL4 loci. In OS15 (Figure 3J), we observed the most complex signal pattern in the series; no pairing of green and red signals was apparent, and there was also evidence of numerical change. Thus, a range of structural aberrations affecting 8q24, varying from simple to complex, was apparent within this series of OS tumors. The varying levels of both N-CIN and S-CIN in the 10 OS as determined by FISH analyses of 8q24 with probes are detailed in Table 3. OS1 and OS4 did not exhibit numerical or 8q24 structural aberrations. OS5 and OS7 were both characterized by N-CIN only, whereas OS12 and OS13 had S-CIN only. The largest group of tumors, OS9, OS11, OS15, and OS18 had more complex FISH patterns, with both N- and S-CIN occurring together at 8q24. We then compared RECQL4 expression findings in tumors with N- or S-CIN at 8q24 to determine whether expression levels were associated with numerical or structural alterations of this region of chromosome 8. OS tumors with RECQL4 expression values that were close or higher than the mean value (13.5) across the 18 OS samples had S-CIN for the 8q24 region. This relationship held regardless of the N-CIN level or ploidy for chromosome 8 (Figure 1). Moreover, OS with the highest S-CIN levels determined by aCGH (shown above) also had the highest S-CIN level with the 8q24 cytoband region. These findings do not indicate that disruption of 8q24 leads to elevated expression of RECQL4 per se; rather, elevated RECQL4 is strongly associated with a greater overall frequency of S-CIN.
Table 3.
Relative Levels of Numerical and Structural Chromosomal Instability in 10 OSs.
| No. | Numerical Aberration | Structural Aberration of Chromosome 8 | Type of CIN for Chromosome 8 | |||
| Overall Ploidy* | cen(8) | RECQL4 Copy Number | MYC Copy Number | Pattern of RECQL4 and MYC FISH Signals | ||
| OS1 | Diploid | 2 | 2 | 3 | Not rearranged | — |
| OS4 | Diploid | 2 | 2 | 2 | Not rearranged | — |
| OS5 | Near-triploid | 4 | 4 | 4 | Not rearranged | N-CIN |
| OS7 | Near-triploid | 3 | 3 | 3 | Not rearranged | N-CIN |
| OS9 | Polyploid (>5n) | 5 | 4 | 4 | Scattered | S- and N-CIN |
| OS11 | Diploid | 4 | 6 | 4 | Scattered | S- and N-CIN |
| OS12 | Near-tetraploid | 2 | 3 | 2 | Scattered | S-CIN |
| OS13 | Diploid | 2 | 6 | 6 | Scattered | S-CIN |
| OS15 | Near-tetraploid | 4 | 8 | 5 | Scattered | S- and N-CIN |
| OS18 | Near-triploid | 3 | 5 | 3 | Scattered | S- and N-CIN |
CIN indicates chromosomal instability; N-CIN, numerical chromosomal instability; S-CIN, structural chromosomal instability.
The overall ploidy has been estimated by the enumeration of cen(3), cen(7), and cen(17) (Table W2) as described in the Materials and Methods section.
Discussion
Previous studies of the RECQL4 gene have shown a strong association between constitutional mutations of the locus and predisposition to OS with 32% of RTS patient developing OS [22]. Moreover, fibroblasts and lymphocytes from RTS exhibit both N- and S-CIN [41–45]. Sequence analyses of RECQL4 in a large series of sporadic OS tumors failed to detect mutations in a significant proportion (only 3 samples of 71 had mutation/deletion) [46], but no study to date has determined the RECQL4 genomic status or analyzed expression levels in the context of CIN in OS.
The biologic function of the RECQL4 protein is poorly understood. The extensive studies of RTS, BLM and WRN, and other members of the RecQ helicase family, have not provided conclusive information concerning the specific and diverse functions of the RECQL4 protein. It is clear that for all three syndromes, germ line mutation of a specific RecQ helicase gene is associated with premature aging, CIN, and predisposition to diverse types of cancer [47]. However, there are important clinical and biologic differences between these genes. For example, conserved areas of the RECQL4 and BLM helicase motif do not have functional equivalence in vitro [27]. Each syndrome has distinct clinical features, and the associated cancer risk involves a different spectrum of tumors [48–50]. Moreover, the prominent class of CIN observed in the lymphocytes or fibroblasts of each syndrome varies, with RTS being characterized by chromatid breaks and isochromosomes, BLM by triradial and quadriradial figures, and WRN by multiple clones with distinctive balance translocations [50–52]. Interestingly, recent characterization of the overexpression of the RecQ helicases has shown it to be linked with the deregulation of the Rb pathway and the RAS activation. These data provide additional support to our supposition that there is an association between oncogenesis and RecQ helicases' overexpression [53]. In the proposed model, RecQ helicases' overexpression would promote and facilitate the DNA synthesis and telomere maintenance, processes that are mandatory for any transformed cells. However, similar to the different effects of the WRN, BLM, or RECQL4 mutations, the regulation of the RecQ helicase seems to be specific for each of the member [53]. In the last few years, several groups identified a more specific function for the RECQL4 helicase, specially its involvement in the early steps of replication fork machinery [27–30]. In an animal model, Sangrithi et al. [28] showed that the Xenopus laevi RECQL4 homolog is associated with chromatin during replication initiation and makes the origin of replication accessible to the replication factors. This function acts on the initiation and the unraveling of single-stranded DNA at the origin of replication. An altered expression of RECQL4 could disturb the processes governing the duration and extent of single-strand DNA exposure. Therefore, these single-stranded DNA regions would be the targets of DNA fragility and breaks. This abnormal single configuration of DNA is likely to promote interchromosome exchanges in a pseudo homolog recombination way. Because RECQL4 is activated by single-strand DNA and it promotes annealing of single-strand DNA to its complementary sequence, an excess of RECQL4 could force unmatched DNA annealing sequence, whereas a lack of RECQL4 could impair proper reannealing of separated strands of DNA [27]. Thus, the deregulation of RECQL4 could have profound consequences on overall genomic integrity and CIN in terms of structural complexity and heterogeneity.
By mapping the distribution of regions of imbalance and, in particular, determining the location of CNTs (Table W1) within each chromosome, it is possible to evaluate the relative contributions of N-CIN and S-CIN to destabilizing the OS genome. In this study, we have found that elevated RECQL4 expression is associated with a greater overall frequency of structural chromosomal change, but there was no obvious relationship between expression and N-CIN levels (Figure 2). Moreover, S-CIN changes were also apparent when FISH was used to determine the frequency of structural alteration at cytoband 8q24. It is possible that the elevated levels of RECQL4 would keep the DNA in a prolonged single stranded vulnerable stage, and promote the S-CIN. A result of this deregulation would be the perpetual initiation of S-CIN, leading to a polyclonal population of cells within one tumor. Indeed,OS tumor can exhibit highly polyclonal population where the most represented clone could account for only 30% of the cell [2–4].
Overexpression of RECQL4 has also been reported in other sarcomas (leiomyosarcoma, liposarcoma, and synovial sarcoma) and in some carcinomas (breast, colon, cervix, and laryngeal squamous cells), and its elevated expression has been correlated with metastasis or a later stage of disease [54–58]. Of these tumors, none have been studied systematically to determine whether S-CIN levels are elevated. However, in liposarcoma, the presence of a supernumerary giant chromosomes that varies structurally from cell to cell suggests ongoing instability [59]. These observations would give RECQL4 a strong impact in oncogenesis in general. In the present study, the MYC amplicon (8q24.21) seems to be independent of from the RECQL4 locus (8q24.3). The study by Mai and Mushinski [60] suggests that amplification and deregulation of MYC lead to instability characterized by gene amplification rather than by elevated frequencies of structural chromosomal rearrangement. This would be consistent with our finding: we observed a disruption of the 8q24 cytoband rather than an over-replication/amplification pattern (Figure 3, I and J), as one would expect as a result of an MYC-driven S-CIN in a given tumor. Furthermore, MYC is a multifunctional protein that acts on cell cycle, apoptosis, and cellular transformation through its transcription factor activity [20]. Conversely, RECQL4 seems to be more involved in specific DNA metabolism. We therefore suggest that the imbalances of the MYC oncogene and RECQL4 are two independent processes of oncogenesis.
In this study, we have investigated the relationship between S-CIN/N-CIN in OS and the RNA expression levels of RECQL4. We find that whereas ploidy changes and elevated N-CIN is common in OS, the more structurally abnormal tumors have higher levels of RECQL4. OS with the highest S-CIN levels determined by aCGH also had the highest S-CIN level with the 8q24 cytoband region. We found no evidence that disruption of 8q24 led to an elevated expression of RECQL4; rather, elevated RECQL4 is strongly associated with a greater overall frequency of S-CIN that characterizes the OS genome.
Supplementary Material
Acknowledgments
The authors thank Olga Ludkovski (Ontario Cancer Institute, Toronto, Canada) for her technical assistance for the enumeration FISH experiments.
Footnotes
This research is supported by the Canadian Cancer Society 016215. G.M. is the recipient of the Helena Lam award.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Bridge JA, Nelson M, McComb E, McGuire MH, Rosenthal H, Vergara G, Maale GE, Spanier S, Neff JR. Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer Genet Cytogenet. 1997;95:74–87. doi: 10.1016/s0165-4608(96)00306-8. [DOI] [PubMed] [Google Scholar]
- 2.Zielenska M, Bayani J, Pandita A, Toledo S, Marrano P, Andrade J, Petrilli A, Thorner P, Sorensen P, Squire JA. Comparative genomic hybridization analysis identifies gains of 1p35 approximately p36 and chromosome 19 in osteosarcoma. Cancer Genet Cytogenet. 2001;130:14–21. doi: 10.1016/s0165-4608(01)00461-7. [DOI] [PubMed] [Google Scholar]
- 3.Squire JA, Pei J, Marrano P, Beheshti B, Bayani J, Lim G, Moldovan L, Zielenska M. High-resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes Chromosomes Cancer. 2003;38:215–225. doi: 10.1002/gcc.10273. [DOI] [PubMed] [Google Scholar]
- 4.Bayani J, Zielenska M, Pandita A, Al-Romaih K, Karaskova J, Harrison K, Bridge JA, Sorensen P, Thorner P, Squire JA. Spectral karyotyping identifies recurrent complex rearrangements of chromosomes 8, 17, and 20 in osteosarcomas. Genes Chromosomes Cancer. 2003;36:7–16. doi: 10.1002/gcc.10132. [DOI] [PubMed] [Google Scholar]
- 5.Ozaki T, Neumann T, Wai D, Schafer KL, van Valen F, Lindner N, Scheel C, Bocker W, Winkelmann W, Dockhorn-Dworniczak B, et al. Chromosomal alterations in osteosarcoma cell lines revealed by comparative genomic hybridization and multicolor karyotyping. Cancer Genet Cytogenet. 2003;140:145–152. doi: 10.1016/s0165-4608(02)00685-4. [DOI] [PubMed] [Google Scholar]
- 6.Lim G, Karaskova J, Vukovic B, Bayani J, Beheshti B, Bernardini M, Squire JA, Zielenska M. Combined spectral karyotyping, multicolor banding, and microarray comparative genomic hybridization analysis provides a detailed characterization of complex structural chromosomal rearrangements associated with gene amplification in the osteosarcoma cell line MG-63. Cancer Genet Cytogenet. 2004;153:158–164. doi: 10.1016/j.cancergencyto.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Lim G, Karaskova J, Beheshti B, Vukovic B, Bayani J, Selvarajah S, Watson SK, Lam WL, Zielenska M, Squire JA. An integrated mBAND and submegabase resolution tiling set (SMRT) CGH array analysis of focal amplification, microdeletions, and ladder structures consistent with breakage-fusionbridge cycle events in osteosarcoma. Genes Chromosomes Cancer. 2005;42:392–403. doi: 10.1002/gcc.20157. [DOI] [PubMed] [Google Scholar]
- 8.Atiye J, Wolf M, Kaur S, Monni O, Bohling T, Kivioja A, Tas E, Serra M, Tarkkanen M, Knuutila S. Gene amplifications in osteosarcoma-CGH microarray analysis. Genes Chromosomes Cancer. 2005;42:158–163. doi: 10.1002/gcc.20120. [DOI] [PubMed] [Google Scholar]
- 9.Man TK, Lu XY, Jaeweon K, Perlaky L, Harris CP, Shah S, Ladanyi M, Gorlick R, Lau CC, Rao PH. Genome-wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer. 2004;4:45. doi: 10.1186/1471-2407-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Dartel M, Cornelissen PW, Redeker S, Tarkkanen M, Knuutila S, Hogendoorn PC, Westerveld A, Gomes I, Bras J, Hulsebos TJ. Amplification of 17p11.2 approximately p12, including PMP22, TOP3A, and MAPK7, in high-grade osteosarcoma. Cancer Genet Cytogenet. 2002;139:91–96. doi: 10.1016/s0165-4608(02)00627-1. [DOI] [PubMed] [Google Scholar]
- 11.Selvarajah S, Yoshimoto M, Maire G, Paderova J, Bayani J, Squire JA, Zielenska M. Identification of cryptic microaberrations in osteosarcoma by high-definition oligonucleotide array comparative genomic hybridization. Cancer Genet Cytogenet. 2007;179:52–61. doi: 10.1016/j.cancergencyto.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet. 2003;145:1–30. [PubMed] [Google Scholar]
- 13.Bayani J, Selvarajah S, Maire G, Vukovic B, Al-Romaih K, Zielenska M, Squire JA. Genomic mechanisms and measurement of structural and numerical instability in cancer cells. Semin Cancer Biol. 2007;17:5–18. doi: 10.1016/j.semcancer.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira BI, Alonso J, Carrillo J, Acquadro F, Largo C, Suela J, Teixeira MR, Cerveira N, Molares A, Gomez-Lopez G, et al. Array CGH and geneexpression profiling reveals distinct genomic instability patterns associated with DNA repair and cell-cycle checkpoint pathways in Ewing's sarcoma. Oncogene. 2008;27:2084–2090. doi: 10.1038/sj.onc.1210845. [DOI] [PubMed] [Google Scholar]
- 15.Selvarajah S, Yoshimoto M, Ludkovski O, Park PC, Bayani J, Thorner P, Maire G, Squire JA, Zielenska M. Genomic signatures of chromosomal instability and osteosarcoma progression detected by high resolution array CGH and interphase FISH. Cytogenet Genome Res. 2008;122:5–15. doi: 10.1159/000151310. [DOI] [PubMed] [Google Scholar]
- 16.Baudis M. Online database and bioinformatics toolbox to support data mining in cancer cytogenetics. Biotechniques. 2006;40:269–270. doi: 10.2144/000112102. 272. [DOI] [PubMed] [Google Scholar]
- 17.Ladanyi M, Park CK, Lewis R, Jhanwar SC, Healey JH, Huvos AG. Sporadic amplification of the MYC gene in human osteosarcomas. Diagn Mol Pathol. 1993;2:163–167. [PubMed] [Google Scholar]
- 18.Stock C, Kager L, Fink FM, Gadner H, Ambros PF. Chromosomal regions involved in the pathogenesis of osteosarcomas. Genes Chromosomes Cancer. 2000;28:329–336. doi: 10.1002/1098-2264(200007)28:3<329::aid-gcc11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 19.Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Pompetti F, Merli M, Magagnoli G, Balladelli A, et al. C-myc and c-fos in human osteosarcoma: prognostic value of mRNA and protein expression. Oncology. 1998;55:556–563. doi: 10.1159/000011912. [DOI] [PubMed] [Google Scholar]
- 20.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 21.Wang LL, Levy ML, Lewis RA, Chintagumpala MM, Lev D, Rogers M, Plon SE. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am J Med Genet. 2001;102:11–17. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 22.Wang LL, Gannavarapu A, Kozinetz CA, Levy ML, Lewis RA, Chintagumpala MM, Ruiz-Maldanado R, Contreras-Ruiz J, Cunniff C, Erickson RP, et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 23.Poynter JN, Figueiredo JC, Conti DV, Kennedy K, Gallinger S, Siegmund KD, Casey G, Thibodeau SN, Jenkins MA, Hopper JL, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 24.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, Gudmundsson J, Jakobsdottir M, Bergthorsson JT, Sigurdsson A, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Kumata Y, Tada S, Yamanada Y, Tsuyama T, Kobayashi T, Dong YP, Ikegami K, Murofushi H, Seki M, Enomoto T. Possible involvement of RecQL4 in the repair of double-strand DNA breaks in Xenopus egg extracts. Biochim Biophys Acta. 2007;1773:556–564. doi: 10.1016/j.bbamcr.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Capp C, Feng L, Hsieh TS. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev Biol. 2008;323:130–142. doi: 10.1016/j.ydbio.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schajowicz F, Sissons HA, Sobin LH. The World Health Organization's histologic classification of bone tumors. A commentary on the second edition. Cancer. 1995;75:1208–1214. doi: 10.1002/1097-0142(19950301)75:5<1208::aid-cncr2820750522>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 32.Wang LL, Worley K, Gannavarapu A, Chintagumpala MM, Levy ML, Plon SE. Intron-size constraint as a mutational mechanism in Rothmund-Thomson syndrome. Am J Hum Genet. 2002;71:165–167. doi: 10.1086/341234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 34.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 35.Yin J, Kwon YT, Varshavsky A, Wang W. RECQL4, mutated in the Rothmund-Thomson and RAPADILINO syndromes, interacts with ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Hum Mol Genet. 2004;13:2421–2430. doi: 10.1093/hmg/ddh269. [DOI] [PubMed] [Google Scholar]
- 36.Finke J, Fritzen R, Ternes P, Lange W, Dolken G. An improved strategy and a useful housekeeping gene for RNA analysis from formalin-fixed, paraffin-embedded tissues by PCR. Biotechniques. 1993;14:448–453. [PubMed] [Google Scholar]
- 37.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 38.Sirvent N, Coindre JM, Maire G, Hostein I, Keslair F, Guillou L, Ranchere-Vince D, Terrier P, Pedeutour F. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am J Surg Pathol. 2007;31:1476–1489. doi: 10.1097/PAS.0b013e3180581fff. [DOI] [PubMed] [Google Scholar]
- 39.Ventura RA, Martin-Subero JI, Jones M, McParland J, Gesk S, Mason DY, Siebert R. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn. 2006;8:141–151. doi: 10.2353/jmoldx.2006.050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi E, Ubiali A, Balzarini P, Cadei M, Alpi F, Grigolatoi PG. High-level detection of gene amplification and chromosome aneuploidy in extracted nuclei from paraffin-embedded tissue of human cancer using FISH: a new approach for retrospective studies. Eur J Histochem. 2005;49:53–58. doi: 10.4081/927. [DOI] [PubMed] [Google Scholar]
- 41.Ying KL, Oizumi J, Curry CJ. Rothmund-Thomson syndrome associated with trisomy 8 mosaicism. J Med Genet. 1990;27:258–260. doi: 10.1136/jmg.27.4.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Der Kaloustian VM, McGill JJ, Vekemans M, Kopelman HR. Clonal lines of aneuploid cells in Rothmund-Thomson syndrome. Am J Med Genet. 1990;37:336–339. doi: 10.1002/ajmg.1320370308. [DOI] [PubMed] [Google Scholar]
- 43.Orstavik KH, McFadden N, Hagelsteen J, Ormerod E, van der Hagen CB. Instability of lymphocyte chromosomes in a girl with Rothmund-Thomson syndrome. J Med Genet. 1994;31:570–572. doi: 10.1136/jmg.31.7.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miozzo M, Castorina P, Riva P, Dalpra L, Fuhrman Conti AM, Volpi L, Hoe TS, Khoo A, Wiegant J, Rosenberg C, et al. Chromosomal instability in fibroblasts and mesenchymal tumors from 2 sibs with Rothmund-Thomson syndrome. Int J Cancer. 1998;77:504–510. doi: 10.1002/(sici)1097-0215(19980812)77:4<504::aid-ijc5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 45.Lindor NM, Devries EM, Michels VV, Schad CR, Jalal SM, Donovan KM, Smithson WA, Kvols LK, Thibodeau SN, Dewald GW. Rothmund-Thomson syndrome in siblings: evidence for acquired in vivomosaicism. Clin Genet. 1996;49:124–129. doi: 10.1111/j.1399-0004.1996.tb03270.x. [DOI] [PubMed] [Google Scholar]
- 46.Nishijo K, Nakayama T, Aoyama T, Okamoto T, Ishibe T, Yasura K, Shima Y, Shibata KR, Tsuboyama T, Nakamura T, et al. Mutation analysis of the RECQL4 gene in sporadic osteosarcomas. Int J Cancer. 2004;111:367–372. doi: 10.1002/ijc.20269. [DOI] [PubMed] [Google Scholar]
- 47.Risinger MA, Groden J. Crosslinks and crosstalk: human cancer syndromes and DNA repair defects. Cancer Cell. 2004;6:539–545. doi: 10.1016/j.ccr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 48.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- 49.Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 50.Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 51.Kuhn EM, Therman E. Cytogenetics of Bloom's syndrome. Cancer Genet Cytogenet. 1986;22:1–18. doi: 10.1016/0165-4608(86)90132-9. [DOI] [PubMed] [Google Scholar]
- 52.Hoehn H, Bryant EM, Au K, Norwood TH, Boman H, Martin GM. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15:282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, El-Naggar S, Clem B, Chesney J, Dean DC. The Rb/E2F pathway and Ras activation regulate RecQ helicase gene expression. Biochem J. 2008;412:299–306. doi: 10.1042/BJ20070975. [DOI] [PubMed] [Google Scholar]
- 54.Linton KM, Hey Y, Saunders E, Jeziorska M, Denton J, Wilson CL, Swindell R, Dibben S, Miller CJ, Pepper SD, et al. Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br J Cancer. 2008;98:1403–1414. doi: 10.1038/sj.bjc.6604316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-9927-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 56.Saglam O, Shah V, Worsham MJ. Molecular differentiation of early and late stage laryngeal squamous cell carcinoma: an exploratory analysis. Diagn Mol Pathol. 2007;16:218–221. doi: 10.1097/PDM.0b013e3180d0aab5. [DOI] [PubMed] [Google Scholar]
- 57.Buffart TE, Coffa J, Hermsen MA, Carvalho B, van der Sijp JR, Ylstra B, Pals G, Schouten JP, Meijer GA. DNA copy number changes at 8q11-24 in metastasized colorectal cancer. Cell Oncol. 2005;27:57–65. doi: 10.1155/2005/401607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Durst M, Schneider A, Pothuri B, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer. 2007;46:373–384. doi: 10.1002/gcc.20418. [DOI] [PubMed] [Google Scholar]
- 59.Coindre JM, Hostein I, Maire G, Derre J, Guillou L, Leroux A, Ghnassia JP, Collin F, Pedeutour F, Aurias A. Inflammatory malignant fibrous histiocytomas and dedifferentiated liposarcomas: histological review, genomic profile, and MDM2 and CDK4 status favour a single entity. J Pathol. 2004;203:822–830. doi: 10.1002/path.1579. [DOI] [PubMed] [Google Scholar]
- 60.Mai S, Mushinski JF. c-Myc-induced genomic instability. J Environ Pathol Toxicol Oncol. 2003;22:179–199. doi: 10.1615/jenvpathtoxoncol.v22.i3.30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.