Abstract
African American (AA) men have a higher incidence and significantly higher mortality rates from prostate cancer than white men, but the biological basis for these differences are poorly understood. Few studies have been carried out to determine whether there are areas of allelic loss or gain in prostate cancers from AA men that are overrepresented in or specific to this group. To better understand the molecular mechanisms of prostate cancer in AA men, we have analyzed 20 prostate cancers from AA men with high-density single-nucleotide polymorphism arrays to detect genomic copy number alterations. We identified 17 regions showing significant loss and 4 regions with significant gains. Most of these regions had been linked to prostate cancer by previous studies of copy number alterations of predominantly white patients.We identified a novel region of loss at 4p16.3, which has been shown to be lost in breast, colon, and bladder cancers. Comparison of our primary tumors with tumors from white patients from a previously published cohort with similar pathological characteristics showed higher frequency of loss of at numerous loci including 6q13-22, 8p21, 13q13-14, and 16q11-24 and gains of 7p21 and 8q24, all of which had higher frequencies in metastatic lesions in this previously published cohort. Thus, the clinically localized cancers from AA men more closely resembled metastatic cancers from white men. This difference may in part explain the more aggressive clinical behavior of prostate cancer in AA men.
Introduction
Prostate cancer is the most common visceral malignancy in American men and the third leading cause of cancer deaths. It is well established that African American (AA) men have both a higher incidence and significantly higher mortality rates from prostate cancer than white men. Indeed, AA men have the highest incidence of prostate cancer in the world and are twice as likely to die of prostate cancer as white Americans. The reason for this difference in incidence and mortality is unclear (for a recent review, see Freedland and Isaacs [1]). Whereas some of the difference in mortality can be attributed to socioeconomic factors, a number of studies have shown that there is a still a higher mortality rate from prostate cancer in AA men even after adjustment for socioeconomic factors [2,3]. Thus, as concluded by Freedland and Isaacs [1], it is likely that in addition to socioeconomic and cultural factors, biological differences account for some of the disparity in incidence and mortality for prostate cancer in AA men in comparison to white men.
High-density mapping of genetic losses and gains can reveal potential tumor suppressor or oncogene loci and might be useful for clinical classification of individual tumors. Therefore, high-throughput methods such as comparative genomic hybridization (CGH) arrays and single-nucleotide polymorphism (SNP) arrays have been introduced for genome-wide screening for copy number alterations (CNAs) in prostate cancer. A large number of prostate cancers have been analyzed using CGH, and these studies have identified a number of consistent areas of chromosomal loss and gain [4–7]. A combined analysis of multiple CGH studies encompassing a total of 872 cancers by Sun et al. [4] have highlighted loss of 2q21-22, 5q13-21, 6q14-21, 8p21-23, 10q23-25, 13q14-22, 16q13-24, and 18q12-23 as well as gain of 3q23-33, 7q21-33, 8q12-23, 17q24-25, and Xq11-23 in 10% or more of prostate cancers analyzed. SNPs may occur at more than 2 million sites in the genome, making it possible to place SNPs at high density along the genome for high-resolution whole genome allelotyping with accurate copy number measurements. Several studies of allelic gain and loss in prostate cancer using SNP arrays have been reported using SNP arrays with 50 to 500K SNPs per array, which have shown multiple areas of gain and loss, that are broadly similar to many of the common areas detected with array CGH, although unique areas were also identified in these studies as well, perhaps reflecting the higher resolution of later generation SNParrays [8–11]. To better understand the molecular mechanisms of prostate cancer in AA men, it will be necessary to better define patterns of allelic gain and loss and ultimately identify the genes underlying these chromosomal abnormalities.
Very few studies have been carried out to determine whether there are areas of allelic loss or gain in prostate cancers from AA men that are overrepresented in or specific to this group. Cher et al. [12] compared primary prostate cancers from 16 AA and 16 white Americans using cytogenetic CGH. They did not observe any significant differences in overall loss or gain of chromosomal regions between these two groups, although they did observe increased rates of loss at 12q21, 15q21, and 17p12-13 in AA men. However, there are limitations to this study. Cytogenetic CGH, as opposed to current techniques such as array CGH or SNP arrays, has a relatively low resolution for detecting losses. Cytogenetic CGHhas a resolution of approximately 10Mbp unless high-level amplification is present [13]. In contrast, the current SNPand array CGHtechniques have a resolution of less than 0.1 Mbp. We therefore carried out a study of allelic loss and gain in 20 prostate cancers from AA men using Affymetrix 500K SNP arrays to define regions of recurrent copy number gain and loss in clinically localized prostate cancers.
Materials and Methods
Tissue Specimens and DNA Extraction
DNA were extracted from freshly frozen prostate cancer tissues and matched benign tissue from radical prostatectomy specimens as described previously [14]. Cancer tissues contained at least 70% cancer and benign tissues were free of cancer or high-grade prostatic intraepithelial neoplasia as confirmed by frozen section before DNA extraction. All tissues were from self-identified AA men. The pathological characteristics of these cases are summarized in Table 1. This study was performed with institutional review board approval.
Table 1.
Pathological Characteristics of Prostate Cancers.
| Baylor | Stanford | |
| Gleason score | ||
| 5 and 6 | 9 (45) | 17 (38) |
| 7 | 8 (40) | 17 (38) |
| 8 and 9 | 3 (15) | 11 (24) |
| Pathological stage | ||
| T2 | 9 (45) | 21 (47) |
| T3a | 8 (40) | 19 (42) |
| T3b | 3 (15) | 5 (11) |
Number and percentage of all cases (in parentheses) are shown. Baylor cases are the 20 cases from AA men analyzed in this study. Stanford cases are primary cancers from white men analyzed by Lapointe et al. [7].
SNP Array Profiling
Affymetrix Human Mapping 500K Array Sets (Affymetrix, Santa Clara, CA), which consisted of two chips (Nsp and Sty) with ∼250K SNPs each, were used for genotyping each patient, according to Affymetrix protocols. Approximately 250 ng of genomic DNA was digested with restriction enzyme NspI or StyI and then ligated to adaptors and polymerase chain reaction-amplified for each enzymedigested sample. Fragment polymerase chain reaction products were then labeled, denatured, and hybridized to the arrays. Arrays were washed, stained, and scanned with the upgraded Affymetrix GeneChip Scanner 3000. Cell intensity (CEL) and data (DAT) files were generated using the Gene Chip Operation System (GCOS) version 1.4 software.
Data Analysis
Profiles were generated on two Affymetrix array chips: Nsp and Sty, each with ∼250K SNPs represented. In all, 20 tumors and 20 paired normal samples were profiled, 40 profiles in all. Affymetrix CEL and TXT files were processed using dChip [15] (with “Invariant Set” normalization and “Average” modeling). In addition, total intensity scale normalization was carried out for each patient, dividing the sum of intensities for the normal sample profile by the sum of intensities for the tumor sample profile, and multiplying each of the tumor values by the resulting scaling factor. To define top genomic regions of gain or loss by average copy number change, the average log ratio of each SNP was computed across the tumors; cytobands with an overrepresentation of SNPs with high averages (>0.2) were called as gain, and cytobands with overrepresentation of SNPs with low average (<-0.2) were called as loss (enrichment assessed by χ2). Copy number alterations were visualized as color maps using the Cluster [16] and Java TreeView [17] software.
For the genome-wide heat map and frequency plot analyses, the 500K SNP probe sets were first collapsed into the 818 cytoband loci represented by those genes. By integrating information across neighboring genes, binning provides a useful balance between minimizing noise and maximizing mapping resolution [7,18]. For each profile, the tumor: normal log ratios were averaged by cytoband, and each profile was then centered cytoband-wise by the average of the cytoband averages. For defining gain or loss events within each cytoband for the purposes of the frequency plot analysis, the SD of the tumor profile with the smallest SD across cytobands (patient 58, which showed little widespread gain or loss; Figure 1) was used as the reference. Cytobands with average values greater than +3SD were called as gain, and cytoband values less than -3 SD were called as loss. For the frequency plot analysis, the Lapointe CGH data set was treated in the same way as the SNP array data set (using PT187 as the reference profile, cytoband boundaries being defined by Ensembl v50).
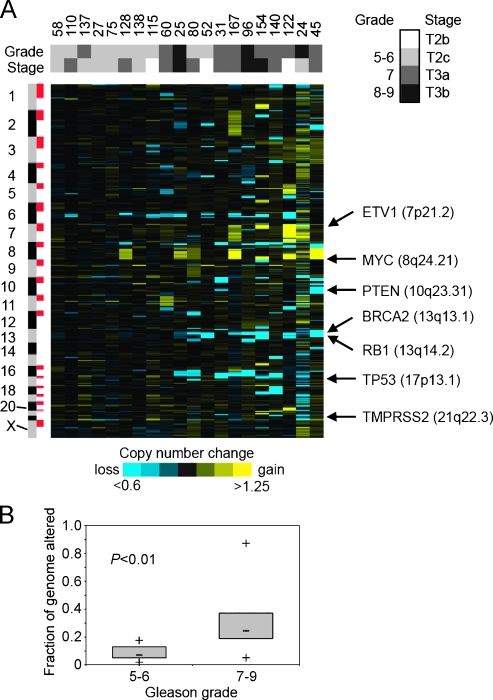
Figure 1.
Genome-wide CNAs in prostate cancer specimens from AA patients. (A) Heat map representation of the 20 cancer specimens (columns) across the 818 cytobands represented in the profiling, ordered by genome position (rows). Mean log2 ratios for cytobands comparing cancer to paired normal sample are depicted by color gram (yellow indicates gain; blue, loss). Shaded boxes above the heat map indicate grade and stage of tumor samples. Gleason grade: light gray indicates 5–6; dark gray, 7; black, 8–9. Pathological stage: white indicates T2b; light gray, T2c; dark gray, T3a; black, T3b. Selected genes in recurrent regions of gain or loss are indicated. (B) Fraction of genome (cytobands) harboring CNA for tumor with low Gleason grade (5–6) versus high Gleason grade (7–9). Box-and-whisker plots indicate median, lower and upper quartiles, and the smallest and largest values. P value by two-sided t-test.
Results
To investigate DNA CNAs in prostate cancer from a cohort of AAs, we profiled 20 tumors (each with paired normal) for 500K SNPs. A heat map representation of the entire data set (SNPs averaged by cytoband) revealed widespread CNA in most tumor specimens (Figure 1A) as well as extensive heterogeneity between tumors. Numerous genes relevant to prostate cancer biology were located in recurrent regions of copy number gain (including ETV1 at 7p21.2, and MYC at 8q24.21) and copy number loss (including PTEN at 10q23.31, RB1 at 13q14.2, TP53 at 17p13.1, and TMPRSS2 at 21q22.3). The regions of loss and gain are broadly similar to those reported previously for CGH studies of clinically localized prostate cancer [4–12]. Tumor profiles in the heat map were ordered (left to right) by their SD across cytobands, which was one measure of the amount of CNA in each tumor. When the Gleason grade and pathological stage of each patient were viewed alongside the corresponding genome copy profiles, a trend was evident where tumors having more genomic alterations tended to have higher grade and stage. On average, higher-grade tumors (Gleason 7–9) had more CNA events compared with lower-grade tumors (Gleason 5–6) with statistical significance (P < .01; Figure 1B). A similar association of increased CNAs inmore poorly differentiated cancers has been noted by others [10,11]. It is of interest to note that approximately 20% of the cancers had only small numbers of CNAs. This was not associated with a lower tumor percentage in these samples. Examination of the array CGH data of LaPointe et al. [7] reveals a similar phenomenon. Thus, there seems to be a subset of prostate cancers with low levels of genomic instability.
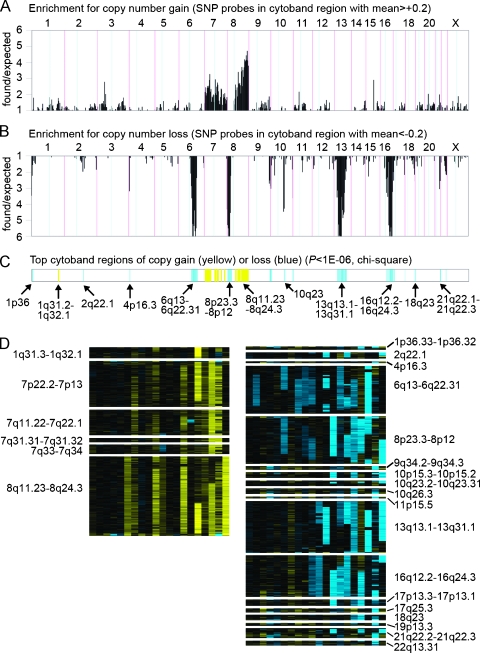
We went on to define the top CNA regions by average copy number change in our tumor cohort (Figure 2). For each SNP, the average fold difference between tumor and normal samples was computed across the 20 tumors; to assess regions of high or recurrent gain, the overrepresentation of SNPs having high averages (>0.2) was determined for each cytoband region (A); similarly, the overrepresentation of SNPs having low averages in each cytoband (<-0.2) was used to assess recurrent loss (Figure 2B). Overrepresentation was statistically assessed for each cytoband to determine the top CNA regions (Figure 2C), and these cytoband regions were viewed as heat maps (Figure 2D). By this approach, regions with recurrent but moderate gain or loss in multiple tumors and regions with gain in only a single tumor but with very high copy number (most notably 1q31.3-1q32.1; Figure 2D, left panel) could be identified, both patterns of which are relevant in cancer [19]. The regions of loss and gain identified are broadly similar to those reported previously for CGH studies of clinically localized prostate cancer [4]. Comparison of our results to the consensus CGH data reported by Sun et al. [4] is shown in Table 2, in which we compare our regions with significant loss and gain to the consensus regions of loss or gain seen in at least 10% prostate cancers identified in the meta-analysis by this group. In agreement with the consensus CGH data, we observed loss of 2q21, 6q13-22, 8p12-23, 10q23, 13q13-31, 16q12-24, and 18q23 as well as gain of 7q31-34 and 8q11-24. Overall, we found loss or gain at 9 of the 13 consensus regions identified by Sun et al. [4]. These findings argue strongly for the technical validity of our analysis.
Figure 2.
Top genomic regions of gain or loss by average copy number change in prostate tumors from the AA cohort. (A) For each of the 818 cytobands represented, the number of SNPs with high averages (>0.2) across the 20 tumors, as a ratio of the number of chance expected. High found/expected ratios may indicate significant regions of copy gain. (B) Number of SNPs with low averages (< -0.2), as a ratio of the number of chance expected. High found/expected ratios here may indicate regions of loss. (C) Top cytoband regions of CNA (yellow indicates gain; blue, loss), as assessed by χ2 statistic (which was a function of both the found/expected ratios of parts A and B, as well as the absolute number of SNPs represented in each cytoband). (D) Heat map representation of regions of gain (left panel) or loss (right panel) from part C, across the 20 tumors (same patient ordering as for Figure 1A).
Table 2.
Comparison of Copy Number Alterations in AA Men with Previously Reported Genomic Loci Implicated in Prostate Cancer.
| Altered Region | Consensus CGH* | Altered Advanced PCa† | Familial PCa Locus | Comments | |
| Loss | 1p36 | [20] | |||
| 2q21 | 2q21-22 | [4] | |||
| 4p16.3 | |||||
| 5q13-15 | [4] | ||||
| 6q13-22 | 6q14-21 | [4,5] | |||
| 8p12-23 | 8p21-23 | [4,5] | NKX3.1 locus | ||
| 9q34 | [21] | ||||
| 10p15 | [25] | Implicated by functional studies [27] | |||
| 10q23 | 10q23-25 | [4,5,7,25] | PTEN locus | ||
| 10q26 | [4,25] | [22] | |||
| 13q13-31 | 13q14-22 | [4,5,26] | RB1 and BRCA2 loci | ||
| 16q12-24 | 16q13-24 | [4] | [22] | ||
| 17p13 | [7,8] | p53 locus | |||
| 17q21 | [8] | [23] | |||
| 18q23 | 18q12-23 | [4] | |||
| 19p13 | [6] | [24] | Implicated by functional studies [28] | ||
| 21q22 | [11] | TMPRSS2/ERG fusion | |||
| 22q13 | [7] | ||||
| Gain | 1q31 | [7] | |||
| 3q23-26 | |||||
| 7p13-22 | [4,5] | ||||
| 7q31-34 | 7q21-33 | [4,5,7] | |||
| 8q11-24 | 8q21-24 | [4,5,7] | [29] | ||
| 17q24-25 | |||||
| Xq11-21 | [4] |
Consensus CGH is based on the meta-analysis of Sun et al. [4] of CGH studies summarizing regions with changes in at least 10% of all tumors analyzed.
Loci were considered to be altered in advanced prostate cancer if alterations were identified in at least 20% of advanced cancers in the individual study referenced. The 20% threshold was chosen owing to the higher background rate of genomic alterations in such cancers.
In addition, we identified loss at an additional 10 loci not among the consensus sites identified by Sun et al. [4]. Interestingly, five of these loci (1p36, 9q34, 10q26, 17q21, and 19p13) have been identified previously as sites of potential familial prostate cancer susceptibility and/or aggressiveness genes [20–24]. Three of these have also been identified as areas of loss in at least 20% advanced prostate cancer tissues in individual studies [4,6,8,25]. Other loci that we have identified as lost (10p15, 17p13, 21q22, and 22q13) have also been identified as lost in at least 20% of advanced prostate cancer in individual studies [4,7,8,11,25] but were not among the consensus regions identified by Sun et al. [4]. In particular, we noted loss at 17p13, the site of the p53 locus and 21q22, the site of the TMPRSS2/ERG fusion gene that has recently been shown to occur in approximately 50% of human prostate cancers. Finally, we noted gain of 1q31 and 7p13-22, two regions that have been shown to be gained in advanced prostate cancer [4,5,7]. The only unique CNA identified in our analysis that has not been previously linked to prostate cancer is loss at 4p16.3.
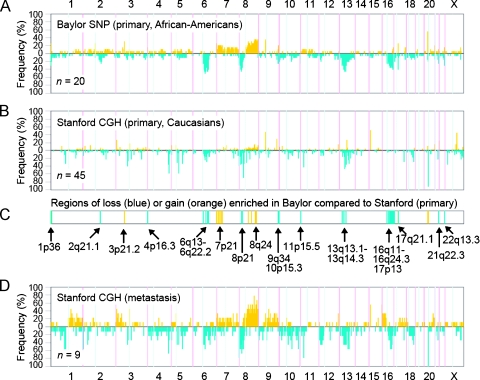
We wished to compare the CNA patterns as observed in our data set with those of previously published genomic profiling studies. Recently, Lapointe et al. [7] profiled 55 primary tumors and 9 unmatched therapy-naive pelvic lymph node metastases, using CGH on cDNA microarrays representing ∼40K cDNA. Of the primary tumors in this series, 45 were fromwhites and 2 were fromAA; for the remaining 8, the race was not known (Jonathan Pollack and James Brooks, personal communication). We compared the pathological data of the primary tumors from white patients in this study with our tumors, and there was very close concordance in Gleason scores and pathological stage (Table 1). For each of the 767 cytoband regions represented in both our data set and the Lapointe data set (hereafter referred to as Stanford), we represented the number of tumors in our cohort with copy number gain or loss, using a frequency plot (Figure 3A). Similarly, we generated a frequency plot for the primary tumors from white patients in the Stanford data set for comparison (Figure 3B).
Figure 3.
Tumors from AA patients in the Baylor cohort exhibit both similar and distinct patterns of CNA, compared with tumors from an independent cohort of predominantly white patients. (A) Frequency plot summarizing the distribution of CNAs in the Baylor cohort. Yellow indicates gain; blue, loss. Number (n) of specimens represented is indicated. (B) Frequency plot summarizing the distribution of CNAs in white patients from an independent cohort of patients (primaries only), from a previously published CGH array data set by Lapointe et al. [7] (“Stanford” cohort). (C) Regions of copy gain (yellow) and loss (blue) nominally overrepresented in the Baylor cohort compared with the white patients from the Stanford cohort (P < .01, one-sided Fisher exact test). (D) Frequency plot summarizing the distribution of CNAs in lymph node metastases from the Lapointe study.
Many of the frequent aberrations previously reported in the study of Lapointe et al. [7] were also frequent in ours, including gains at 8q (35% of our cases at 8q21) and losses at 13q (40% at 13q14), 8p (40%), and 6q (45% at 6q21). Of note, 4p16.3 loss was not seen in the Stanford cohort. We went on to compare the relative frequencies of gain or loss between our primary tumors cohort and the white patients in Stanford cohort (Figure 3C). We found 17 cytobands with a statistically higher frequency of gain (P < .01, one-sided Fisher exact test) in our cohort over the Stanford cohort; although some of these cytobands may have appeared nominally significant due to multiple testing of 767 cytobands (chance expected = 8), we did see a number of these enriched cytobands in regions of potential interest, including 8q24 and 7p21. Conversely, we found 33 cytobands of loss that were enriched in our cohort (P < .01), which well-exceeded chance expected (∼8) and which included regions spanning 6q13-6q22.3, 8p23-8p12, 13q13-13q31, and 16q12-16q24. We also analyzed gains or losses that were more frequent in the Stanford cohort compared with our cohort, but we did not find many more than expected by chance (gains, 3; losses, 9; and chance expected, 8).
We also determined the frequency of CNAs for the nine prostate cancer metastases profiled in the Lapointe study (Figure 3D). Eight of the nine metastatic tumors were from white patients, and in one case, the race was unknown. Comparison of this plot with the plots from our primary tumors and the primary tumors from white patients analyzed by the Stanford group revealed a number of regions where our primary tumors more closely resembled the metastatic lesions from the Stanford study. These included a higher frequency of loss of 6q13-22, 8p21, 10p15, 13q13-14, and 16q11-24 and gains of 7p21 and 8q24.
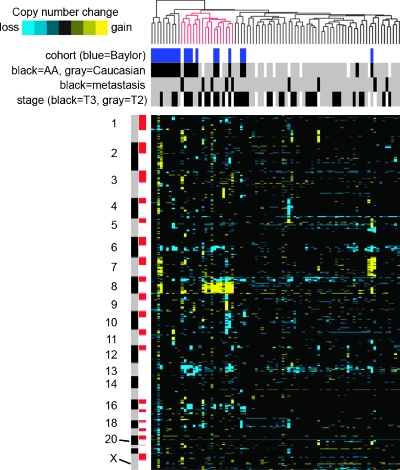
To further explore the patterns of gains and losses in the two cohorts we carried out hierarchical clustering (complete linkage method) of copy gain/loss profiles of prostate tumors from Baylor cohort (N = 20) and the entire Stanford cohort including metastatic lesions (N = 64). Interestingly, 17 of the 20 Baylor cases (and 1 of 2 Stanford AA cases) were in two major clusters (Figure 4, left side). A subtree of the dendrogram (highlighted in red) contained 7 of the 10 Baylor cases, 6 of which were T3 cancers, and 5 of 9 metastatic tumors. Both the Stanford metastasis samples and the Baylor T3 samples were overrepresented (P = .02 and P = .01, respectively, one-sided Fisher exact test). Thus, this subtree consisted of aggressive cancers and is characterized by loss of 8p, 13q, and 16q and gain of 8q. Of note, the white T3 cancers were not enriched in this subtree. A second major subtree contained 10 of the Baylor cases, including 7 of 9 T2 cancers. Thus, this subtree was enriched with the less aggressive AA cancers. Notable features of cancers in this subtree were loss of 6q and infrequent loss 13q. Therefore, it seems that prostate cancers in AA men can be divided into more aggressive and less aggressive subgroups based on the pattern on CNAs.
Figure 4.
Hierarchical clustering (complete linkage method) of copy gain/loss profiles of prostate tumors from Baylor (N = 20) and Stanford cohorts (N = 64). Gain or loss events (from Figure 3) are depicted by heat map color gram (yellow indicates gain). Shaded boxes above the heat map indicate cohort (Baylor or Stanford), race, primary versus metastasis, and stage. A subtree of the dendrogram is highlighted in red, for which both the Stanford metastasis samples and the Baylor T3 samples are overrepresented (P = .02 and P = .01, respectively, one-sided Fisher exact test).
Discussion
To date, most studies of genomic CNAs in prostate cancer have analyzed prostate cancer tissues from white patients. Thus, there is only limited data regarding CNAs in prostate cancer in AA men. Our studies indicate that the regions of loss and gain are similar in AA men and white men. Most of the loci that we have identified as being lost or gained in prostate cancers from AA men have previously been identified as being altered in prostate cancer in studies consisting predominantly or exclusively of white patients. Several of the loci have also been linked to familial prostate cancer susceptibility. Thus, the regions of loss and gain in prostate cancer in AA and white men appear to be broadly similar. The only unique CNA identified in our analysis that has not been previously linked to prostate cancer is loss of 4p16.3. Loss of this region has been previously noted in breast, bladder, and colon cancers [30–32]. The region of loss contains 46 identified genes, several of which are candidate tumor suppressor genes. MXD4 (also known as hMAD4) is a MAX-binding protein and a transcriptional repressor that can block myc-dependent cell transformation and may play a role in cellular senescence [33]. CRIPak [34] encodes a protein that is a negative regulator Pak1 (p21-activated kinase). Pak1 promotes cellular motility and has been shown to have antiapoptotic activities in prostate cancer cells [35], so loss of its inhibitor would potentially be tumor-promoting. Larger studies will be needed to validate whether this loss occurs in other prostate cancers from AA and/or white men.
Although our studies indicate that the loci of loss and gain are broadly similar between AA and white patients, there are indications of significant quantitative differences. When we compared our clinically localized cancers from our AA cohort to the white patients from the cohort examined by Lapointe et al. [7], we found multiple regions that had significantly higher rates of loss (1p36, 2q21, 4p16, 6q13-22, 8p21, 9q34, 10p15, 11p15, 13q13-14, 16q11-24, 17p13, 17q21, 21q22, and 22q13) and gain (3p21, 7p21, 8q24, 20p11, and 20q11). Examination of the losses and gains detected by LaPointe et al. [7] indicates that there are multiple regions where the CNAs in AA prostate cancer are more similar to metastatic disease than the clinically localized disease cohort. Particularly striking are gains of chromosome 7 and 8q and loss of 16q. The gain of 8q, particularly the 8q24 region, has been repeatedly linked to aggressive disease in prostate cancer [5,7], and there is a risk allele for prostate cancer in AA men in this region [29]. However, other regions that are altered in the metastatic tumors analyzed by LaPointe et al. [7] are not altered in the clinically localized prostate cancers in AA men, such as losses on chromosome 4 and 12p and gains on broad regions of 9q. Thus, the increased CNAs in prostate cancers from AA men are specific and do not reflect nonspecific chromosomal loss or gain. Cluster analysis of the two cohorts confirms this analysis and reveals that T3 lesions in AA men seems more closely related to the Stanford metastases than to the Stanford T3 samples.
In addition to the more frequent gains and losses we noted in primary cancers from AA men when compared with the primary cancers in the Stanford cohort, we also noted that we did not see significant loss on 5q. LaPointe et al. [7] have correlated CNA and expression profiles in their cohort. This group had previously identified three subtypes of prostate cancer based on expression profiling [36]. Subtype 1 was associated with better prognosis, and one of the defining CNAs associated with this subtype is loss of 5q [7]. Our finding suggests that subtype 1 prostate cancers may be less common in AA men. Given that the grade and stage of our cohort and that of Lapointe et al. [7] were very similar, this finding cannot be explained by differences in standard pathological markers of disease aggressiveness between the two groups. All of our findings suggest that at a similar stage, prostate cancers in AA men may harbor patterns of CNAs that are associated with more aggressive disease.
Finally, it is of interest to note that theAApatients havemore frequent loss at 21q22.3 than the white patients. This locus corresponds to the recently described TMPRSS2/ERG fusion gene that is found in the 40% to 80% of prostate cancers analyzed to date [37–39]. This fusion results in juxtaposition of the androgen-regulated TMPRSS2 promoter and the ERG oncogene. The ERG oncogene is expressed at variable levels in prostate cancers with the gene fusion [39] and has pleiotropic tumorpromoting activities [40]. As summarized by Freedland and Isaacs [1], AA men have been shown to have variations in various components of the androgen signaling pathway including higher serum androgen levels, 5-alpha reductase isoenzyme activity, testosterone biosynthesis or degradation pathways, androgen receptor levels, and trinucleotide repeats affecting androgen receptor signaling. The net effect of all these alterations would be higher androgen receptor activity in AA men [1]. Thus, it is possible that AA men express higher levels of the TMPRSS2/ERG fusion gene transcript when the fusion gene is present and thus are more likely to develop clinically significant cancers bearing this gene fusion. Further work is needed to explore this hypothesis.
Our results are consistent with the results of Cher et al. [12], in that almost all the regions of CNA in prostate cancers from AA men were also lost in white men. We also observed increased loss at the p53 locus at 17p13 in our AA cohort compared with the white patients from the Stanford cohort, consistent with the prior observation of Cher et al. [12]. However, we did find an increased frequency of CNAs in multiple regions in prostate cancer tissues from AA men, in contrast to the results of Cher et al. [12]. One potential confounding variable in the studies of Cher et al. [12] was that 4 of their 16 cases from whites were poorly differentiated (Gleason 8–9) versus only 1 of 16 cases from AA men. Given that poorly differentiated cancers tend to have more CNAs than better differentiated lesions (Figure 1 and references [10] and [11]), this could have masked differences in CNA frequency between the two groups. Clearly, more extensive studies are needed to resolve this question.
In summary, our study has found that although CNAs in prostate cancers from AA men are broadly similar to those found white men, there are significant differences in the frequency of particular CNAs. Prostate cancers from AA men tended to have lower rates of loss of 5q, loss of which is associated with less aggressive disease, and more frequent losses and gains of several regions associated with metastatic disease such as gain of 8q24. It should be noted that although these observations are important, there are limitations to our study including the relatively small size of our AA cohort and lack of exact clinical and pathological matching to the Stanford cohort. Another variable is that the analytical platforms differed in the two groups. However, given the extremely high concordance of our CNA results with previously reported studies in white men, it seems unlikely that this impacts our overall result. It is possible that our more dense arrays may detect losses in very small regions not represented in less dense arrays; at the same time, however, it seems unlikely that the differences noted in broad regions such as chromosome 6q13-22, 8p21, 8q24, 13q13.1-14.3, and 16q11-24.3 would be impacted by the analytical platform. Furthermore, the differences between our cohort and the Stanford cohort were confined to specific regions, not all regions of CNA, again arguing for the analytical platform not representing a major determinant in the differences we observed. In the future, it will be critical to replicate these studies in larger groups of AA and white men who are carefully matched using existing clinical and pathological variables and profiled by the same analytical platform. Such studies may give critical new insights into the biological basis of aggressive prostate cancer in AA men.
Acknowledgments
The authors thank the assistance of Jonathan Pollack and James Brooks (Stanford University) who provided us with unpublished data regarding the race of patients analyzed in their previous studies cited in this paper.
Abbreviations
- AA
African American
- CGH
comparative genomic hybridization
- SNP
single-nucleotide polymorphism
Footnotes
This work was supported by grants from the P30 Cancer Center support grant (P30 CA125123) and the Baylor Prostate Cancer SPORE (P50CA058204; M.I.) the Dept of Veterans Affairs Merit Review program (M.I.), an ASCO Career Development Award (M.M.), and by the use of the facilities of the Michael E. DeBakey Veterans Affairs Medical Center.
References
- 1.Freedland SJ, Isaacs WB. Explaining racial differences in prostate cancer in the United States: sociology or biology? Prostate. 2005;62:243–252. doi: 10.1002/pros.20052. [DOI] [PubMed] [Google Scholar]
- 2.Robbins AS, Whittemore AS, Thom DH. Differences in socioeconomic status and survival among white and black men with prostate cancer. Am J Epidemiol. 2000;151:409–416. doi: 10.1093/oxfordjournals.aje.a010221. [DOI] [PubMed] [Google Scholar]
- 3.Evans S, Metcalfe C, Ibrahim F, Persad R, Ben-Shlomo Y. Investigating black-white differences in prostate cancer prognosis: a systematic review and meta-analysis. Int J Cancer. 2008;123:430–435. doi: 10.1002/ijc.23500. [DOI] [PubMed] [Google Scholar]
- 4.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB, et al. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 5.Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, Kumar-Sinha C, Cao X, Dash A, Wang L, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M, Li M, Fu X, Huang Y, Qian H, Sun R, Mao Y, Xie Y, Li Y. Simultaneously detection of genomic and expression alterations in prostate cancer using cDNA microarray. Prostate. 2008;68:1496–1509. doi: 10.1002/pros.20756. [DOI] [PubMed] [Google Scholar]
- 7.Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 8.Dumur CI, Dechsukhum C, Ware JL, Cofield SS, Best AM, Wilkinson DS, Garrett CT, Ferreira-Gonzalez A. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics. 2003;81:260–269. doi: 10.1016/s0888-7543(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 9.Lieberfarb ME, Lin M, Lechpammer M, Li C, Tanenbaum DM, Febbo PG, Wright RL, Shim J, Kantoff PW, Loda M, et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res. 2003;63:4781–4785. [PubMed] [Google Scholar]
- 10.Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, Li T, Yan G, Sun J, Sun J, Adams TS, et al. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer. 2006;45:1018–1032. doi: 10.1002/gcc.20369. [DOI] [PubMed] [Google Scholar]
- 11.Tørring N, Borre M, Sørensen KD, Andersen CL, Wiuf C, Ørntoft TF. Genome-wide analysis of allelic imbalance in prostate cancer using the Affymetrix 50K SNP mapping array. Br J Cancer. 2007;96:499–506. doi: 10.1038/sj.bjc.6603476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cher ML, Lewis PE, Banerjee M, Hurley PM, Sakr W, Grignon DJ, Powell IJ. A similar pattern of chromosomal alterations in prostate cancers from African-Americans and Caucasian Americans. Clin Cancer Res. 1998;4:1273–1278. [PubMed] [Google Scholar]
- 13.Bentz M, Plesch A, Stilgenbauer S, Dohner H, Lichter P. Minimal sizes of deletions detected by comparative genomic hybridization. Genes Chromosomes Cancer. 1998;21:172–175. [PubMed] [Google Scholar]
- 14.Ittmann M, Wieczorek R, Heller P, Dave A, Provet J, Krolewski J. Alterations in the p53 and MDM-2 genes are infrequent in clinically localized, stage B prostate adenocarcinomas. Am J Pathol. 1994;145:287–293. [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 18.Bergamaschi A, Kim Y, Wang P, Sorlie T, Hernadez-Boussard T, Lonning P, Tibshirani R, Borresen A, Pollack J. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 19.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbs M, Stanford JL, McIndoe RA, Jarvik GP, Kolb S, Goode EL, Chakrabarti L, Schuster EF, Buckley VA, Miller EL, et al. Evidence for a rare prostate cancer-susceptibility locus at chromosome 1p36. Am J Hum Genet. 1999;64:776–787. doi: 10.1086/302287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Gillanders EM, Isaacs SD, Chang BL, Wiley KE, Zheng SL, Jones M, Gildea D, Riedesel E, Albertus J, et al. Genome-wide scan for prostate cancer susceptibility genes in the Johns Hopkins hereditary prostate cancer families. Prostate. 2003;57:320–325. doi: 10.1002/pros.10306. [DOI] [PubMed] [Google Scholar]
- 22.Witte JS, Suarez BK, Thiel B, Lin J, Yu A, Banerjee TK, Burmester JK, Casey G, Catalona WJ. Genome-wide scan of brothers: replication and fine mapping of prostate cancer susceptibility and aggressiveness loci. Prostate. 2003;57:298–308. doi: 10.1002/pros.10304. [DOI] [PubMed] [Google Scholar]
- 23.Lange EM, Robbins CM, Gillanders EM, Zheng SL, Xu J, Wang Y, White KA, Chang BL, Ho LA, Trent JM, et al. Fine-mapping the putative chromosome 17q21-22 prostate cancer susceptibility gene to a 10 cM region based on linkage analysis. Hum Genet. 2007;121:49–55. doi: 10.1007/s00439-006-0274-2. [DOI] [PubMed] [Google Scholar]
- 24.Kammerer S, Roth RB, Reneland R, Marnellos G, Hoyal CR, Markward NJ, Ebner F, Kiechle M, Schwarz-Boeger U, Griffiths LR, et al. Large-scale association study identifies ICAM gene region as breast and prostate cancer susceptibility locus. Cancer Res. 2004;64:8906–8910. doi: 10.1158/0008-5472.CAN-04-1788. [DOI] [PubMed] [Google Scholar]
- 25.Ittmann M. Allelic loss on chromosome 10 in prostate adenocarcinoma. Cancer Res. 1996;56:2143–2147. [PubMed] [Google Scholar]
- 26.Melamed J, Einhorn JM, Ittmann M. Allelic loss on chromosome 13q in human prostate carcinoma. Clin Cancer Res. 1997;3:1867–1872. [PubMed] [Google Scholar]
- 27.Fukuhara H, Maruyama T, Nomura S, Oshimura M, Kitamura T, Sekiya T, Murakami Y. Functional evidence for the presence of tumor suppressor gene on chromosome 10p15 in human prostate cancers. Oncogene. 2001;20:314–319. doi: 10.1038/sj.onc.1204079. [DOI] [PubMed] [Google Scholar]
- 28.Gao AC, Lou W, Ichikawa T, Denmeade SR, Barrett JC, Isaacs JT. Suppression of the tumorigenicity of prostatic cancer cells by gene(s) located on human chromosome 19p13.1-13.2. Prostate. 1999;38:46–54. doi: 10.1002/(sici)1097-0045(19990101)38:1<46::aid-pros6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shivapurkar N, Maitra A, Milchgrub S, Gazdar AF. Deletions of chromosome 4 occur early during the pathogenesis of colorectal carcinoma. Hum Pathol. 2001;32:169–177. doi: 10.1053/hupa.2001.21560. [DOI] [PubMed] [Google Scholar]
- 31.Sibley K, Bell S, Knowles MA. Redefining a critical region of LOH on 4p16.3 in bladder cancer. Genes Chromosomes Cancer. 2000;29:378–379. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1045>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 32.Shivapurkar N, Sood S, Wistuba II, Virmani AK, Maitra A, Milchgrub S, Minna JD, Gazdar AF. Multiple regions of chromosome 4 demonstrating allelic losses in breast carcinomas. Cancer Res. 1999;59:3576–3580. [PubMed] [Google Scholar]
- 33.Marcotte R, Chen JM, Huard S, Wang E. c-Myc creates an activation loop by transcriptionally repressing its own functional inhibitor, hMad4, in young fibroblasts, a loop lost in replicatively senescent fibroblasts. J Cell Biochem. 2005;96:1071–1085. doi: 10.1002/jcb.20503. [DOI] [PubMed] [Google Scholar]
- 34.Talukder AH, Meng Q, Kumar R. CRIPak, a novel endogenous Pak1 inhibitor. Oncogene. 2006;25:1311–1319. doi: 10.1038/sj.onc.1209172. [DOI] [PubMed] [Google Scholar]
- 35.Sastry KS, Karpova Y, Kulik G. Epidermal growth factor protects prostate cancer cells from apoptosis by inducing BAD phosphorylation via redundant signaling pathways. J Biol Chem. 2006;281:27367–27377. doi: 10.1074/jbc.M511485200. [DOI] [PubMed] [Google Scholar]
- 36.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 39.Narod SA, Seth A, Nam R. Fusion in the ETS gene family and prostate cancer. Br J Cancer. 2008;99:847–851. doi: 10.1038/sj.bjc.6604558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]