Abstract
Neuronal activity orchestrates the proper development of the neuronal circuitry by regulating both transcriptional and post-transcriptional gene expression programmes. How these programmes are coordinated, however, is largely unknown. We found that the transcription of miR379–410, a large cluster of brain-specific microRNAs (miRNAs), is induced by increasing neuronal activity in primary rat neurons. Results from chromatin immunoprecipitation and luciferase reporter assays suggest that binding of the transcription factor myocyte enhancing factor 2 (Mef2) upstream of miR379–410 is necessary and sufficient for activity-dependent transcription of the cluster. Mef2-induced expression of at least three individual miRNAs of the miR379–410 cluster is required for activity-dependent dendritic outgrowth of hippocampal neurons. One of these miRNAs, the dendritic miR-134, promotes outgrowth by inhibiting translation of the mRNA encoding for the translational repressor Pumilio2. In summary, we have described a novel regulatory pathway that couples activity-dependent transcription to miRNA-dependent translational control of gene expression during neuronal development.
Keywords: dendritogenesis, Mef2, microRNA, neuronal activity, Pumilio
Introduction
The development and refinement of neuronal circuitry are regulated by both intrinsic and activity-dependent programmes of gene expression (Flavell and Greenberg, 2008). The latter is particularly important at later stages of neuronal development, such as dendritogenesis (Chen and Ghosh, 2005) and synapse formation (Waites et al, 2005). An important layer of regulation is new transcription induced by activity-regulated transcription factors (Hong et al, 2005). For example, CREB has a key function in activity-dependent dendritic outgrowth of central neurons (Lonze and Ginty, 2002; Redmond et al, 2002). On the other hand, the activity-dependent transcription factor, myocyte enhancing factor 2 (Mef2), functions as a negative regulator of excitatory synapse number (Flavell et al, 2006; Shalizi et al, 2006). Morphological abnormalities in dendritic arbourization and postsynaptic structure are common hallmarks of a number of cognitive diseases, for example, mental retardation (Bagni and Greenough, 2005). Activity-dependent regulation of gene expression involves, in addition to the activation of new transcriptional programmes within the nucleus, also post-transcriptional control of pre-existing mRNAs (Steward, 2002). The expression of pre-existing mRNAs is often regulated locally in dendrites close to synaptic contacts (Sutton and Schuman, 2006). Important post-transcriptional regulatory molecules are RNA-binding proteins (i.e. CPEB, Pumilio (Pum), etc.) that regulate transport, stability or translation of the mRNAs in response to activity (Kiebler and Bassell, 2006; Richter, 2007). MicroRNAs (miRNAs) are another class of key post-transcriptional regulators that can bind to the 3′UTR of target mRNAs to downregulate their expression by inducing either mRNA degradation or translational suppression (Kosik, 2006; Fiore et al, 2008). Little is known, however, on how these two mechanisms of activity-dependent gene expression, global transcriptional control and local post-transcriptional control, are coordinated within a neuron.
We have previously shown that miR-134 controls spine morphogenesis in rat hippocampal neuron by repressing local translation of the LimK1 mRNA within dendrites (Schratt et al, 2006). BDNF, which is secreted in response to neuronal activity, relieves the translational block of the LimK1 mRNA thereby allowing spine growth. It is still an open question whether miR-134 function is also controlled globally within a neuron at the transcriptional level. Intriguingly, the miR-134 gene is clustered together with more than 50 other miRNAs within the Gtl2/Dlk1 locus (referred hereafter as the miR379–410 cluster) (Seitz et al, 2004). Clustered miRNA genes are often co-expressed, a prerequisite for the coordinated control of related biological processes (He et al, 2005). In this study, we provide evidence that the entire miR379–410 cluster is co-regulated at the transcriptional level by neuronal activity in a Mef2-dependent manner. Importantly, activity-dependent regulation of multiple miRNAs from the cluster, including miR-134, is necessary for the correct elaboration of the dendritic tree. Furthermore, we show that the RNA-binding protein Pum2 is a direct miR-134 target and a key mediator of the miR-134 growth-promoting effect on dendritogenesis. Our results point to the miR379–410 cluster, in particular miR-134, as a key component of a mechanism that couples transcriptional and local control of gene expression in response to neuronal activity during the development of neural circuitry.
Results
The miR379–410 cluster is co-regulated by neuronal activity
Recently, we provided evidence that miR-134 regulates activity-dependent control of dendritic mRNA translation in response to BDNF. We decided to investigate whether neuronal activity also regulates miR-134 and possibly the entire miR379–410 cluster globally within the cell at the transcriptional level.
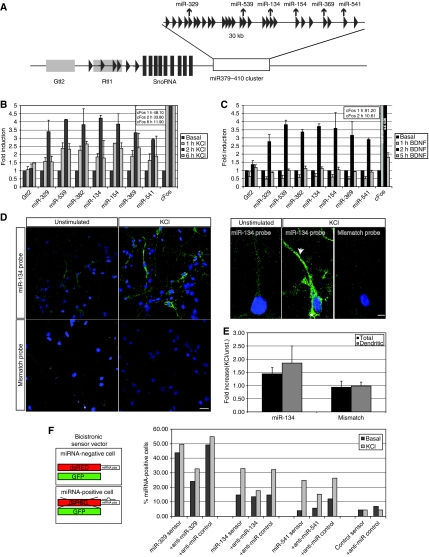
We first tested whether expression of candidate miRNAs of the miR379–410 cluster was coordinately induced by neuronal activity. We used either membrane-depolarizing concentrations of KCl, which leads to Ca2+ influx, or the application of BDNF, a growth factor released by synaptic stimulation, as paradigms to mimic neuronal activity in a culture of dissociated neurons (Redmond et al, 2002; Wayman et al, 2006). Primary cortical neurons cultured for 5 days in vitro (5DIV) were treated with either BDNF or KCl for up to 6 h. After isolation of total RNA, the expression of pre-miRNAs that are encoded at different positions within the GTl2/DLK1 locus was analysed by quantitative RT–PCR (Figure 1A). Consistent with our earlier findings, miRNAs from the miR379–410 cluster (including miR-134) are expressed at very low levels in unstimulated neurons at this early developmental stage. Strikingly, all of the tested pre-miRNAs located within miR379–410 were robustly induced by both BDNF and KCl stimulation. Similar to the known activity-regulated cFos gene, miR379–410 pre-miRNA induction was both rapid and transient, peaking at 2 h and lasting for at least 6 h. The level of the neighbouring Gtl2 transcript was not affected by KCl and BDNF (Figure 1A–C), demonstrating that our treatment led to a specific induction of the miR379–410 domain.
Figure 1.
The miR379–410 cluster is co-regulated by neuronal activity. (A) Schematic representation of the mouse GTL2/RTL1 locus on distal chromosome 12. miRNA genes are indicated by triangles, small nucleolar RNAs (SnoRNA) by filled bars, the non-coding RTL1 and GTL2 genes by grey rectangles and the miR379–410 cluster by an open rectangle. Arrows point to miRNAs analysed by RT–PCR and sensor assays. Diagram is not drawn to scale. (B) Membrane depolarization increases miR379–410 precursor expression. Quantitative RT–PCR analysis of total RNA extracted from KCl-stimulated primary cortical neurons. DIV5 cortical neurons were treated for 6 h with 16 mM KCl, and total RNA was isolated at different time points during the stimulation period and analysed by real-time PCR with primers for different miRNA precursors located within the GTL2/RTL1 locus, cFos (positive control) and GTL2 (negative control). The data are normalized to β3-tubulin and presented as relative to the basal. Data represent the average of three independent experiments+s.d. cFos induction values are out of scale and indicated in the insert. (C) BDNF treatment increases miR379–410 precursor expression. Real-time PCR analysis of total RNA extracted from BDNF-stimulated primary cortical neurons. DIV5 cortical neurons were treated for 6 h with 40 ng/ml BDNF; total RNA was isolated at different time points during the stimulation and analysed as in (B). Data represent the average of three independent experiments+s.d. cFos induction values are out of scale and indicated in the insert. (D) Effect of membrane depolarization on the subcellular localization of miR-134 in hippocampal neurons. DIV7 rat hippocampal neurons were stimulated for 6 h with 16 mM KCl, fixed and analysed by fluorescent in situ hybridization. A DIG-labelled LNA probe directed against miR-134 (miR-134 probe) and a DIG-labelled control probe of unrelated sequence (mismatch probe) were used (5 pmol each). Representative images for unstimulated cells (left panels) and KCl-treated neurons (right panels) are shown. Higher panels show the robust increase in miR-134 signal in both the neuronal soma (asterisks) and dendrites (arrowheads) upon KCl stimulation. Scale bars: 20 and 5 μm. (E) Quantification of miR-134 levels obtained by ISH analysis. Ten pictures for each experimental condition were measured to calculate the average intensity of the fluorescent signal obtained with the indicated probes. Data are presented as the fold increase in average intensity in KCl-treated versus unstimulated whole cells (total) and dendrite only (dendritic). Error bars represent the average of two independent experiment+s.d. (F) Membrane depolarization increases functional miR379–410 miRNAs. An miRNA sensor assay was performed in KCl-stimulated hippocampal neurons. The principle of an miRNA sensor assay is described on the left. Right: hippocampal neurons (DIV4) were transfected with the indicated sensors (50 ng) alone or in combination with the indicated anti-miRs (50 nM). After KCl incubation (DIV7), cells were fixed and miRNA-positive cells were scored based on the fluorescent sensor signal. Results from one representative out of three independent experiments are shown.
We next investigated the effect of membrane depolarization on the levels and subcellular localization of one of the miR379–410 miRNAs, miR-134, by in situ hybridization (ISH) of primary hippocampal neurons (DIV7). We used a probe that was able to recognize both mature and pre-miR-134. (Figure 1D). Low levels of miR-134 were detectable in unstimulated neurons and KCl led to a robust and specific increase in miR-134 ISH signal, confirming our results obtained with quantitative RT–PCR (Figure 1B). The KCl-mediated increase was completely abolished by pretreatment of neurons with actinomycin D, demonstrating that the increase was due to de novo miR-134 transcription (data not shown). The increase of miR-134 upon depolarization was not restricted to the soma but the miR-134 signal was also evident in dendrites (Figure 1D, higher magnification panels). Quantification of the in situ signal confirmed the depolarization-induced increase of miR-134 in both the somatic and dendritic compartments (Figure 1E). The robust increase of miR-134 in dendrites suggests a local function in this compartment. So far, our expression analysis did not directly address whether our activity paradigm induces functional miR379–410 miRNAs. To verify that neuronal activation induces the expression of mature and functional miRNAs from the cluster, we used a previously described single cell sensor assay (Mansfield et al, 2004). We used bicistronic GFP/dsRED expression vectors (‘sensor'), the expression of which is controlled by miR379–410 miRNAs due to two perfectly complementary miRNA-binding sites in the 3′UTR of the dsRED gene. Neurons that express functional miRNA (miRNA positive) are identified by the lack of dsRED due to RISC-mediated cleavage of the dsRED mRNA (Figure 1F). The number of miRNA-positive neurons increased upon depolarization for all tested miR379–410 miRNAs (Figure 1F), validating that neuronal activity induces functional miRNAs. Co-transfection of the sensors with specific 2'O-methyl antisense oligonucleotides (anti-miRs) confirmed that the increase in miRNA-positive cells was due to a specific elevation of miRNA activity. These results also demonstrate that anti-miRs can be used to specifically interfere with the function of the KCl-induced miRNAs miR-329, -134 and -541.
Mef2 is necessary for activity-dependent regulation of the miRNA-379–410 cluster
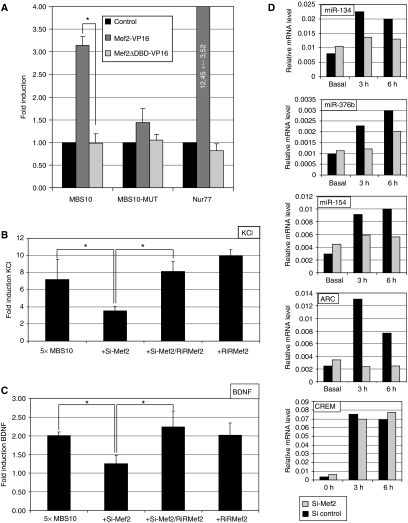
We devised a comparative genomic approach based on sequence conservation to identify regulatory elements that could mediate activity-dependent transcription of the miR379–410 cluster. Highly conserved regions within 20 kb upstream of the cluster were screened for potential binding sites for activity-regulated transcription factors (Figure 2A). Thereby, we identified 10 potential binding sites for Mef2, a known activity-regulated transcription factor that negatively regulates synapse number in mature hippocampal neurons (Shalizi and Bonni, 2005). The occupancy of the putative Mef2-binding sites (MBSs) in vivo was assessed by chromatin immunoprecipitation (ChIP) using DIV5 primary cortical neurons (Figure 2B and data not shown). A PCR product encompassing one of the potential MBS, MBS10, could be specifically amplified from immunoprecipitates of formaldehyde-fixed chromatin using a Mef2-specific antibody (Figure 2B, upper panel), in a similar manner as the known Mef2 target gene Nur77 (Figure 2B, upper and lower panels). In contrast, Mef2 antibodies were unable to enrich chromatin from the β-globin locus that lacks a MBS, confirming the specificity of our ChIP protocol. Therefore, Mef2 is bound to MBS10 in neurons in vivo. To begin to assess the relevance of MBS10 in activity-dependent transcription of miR379–410, we first monitored the presence of a transcript between the miR-379 gene and MBS10. Using RT–PCR with a set of overlapping primers located within the region from MBS10 to miR-379, we were able to detect a continuous transcript except for a small gap (Figure 2A and C, primer pairs 6 and 7, and data not shown). This gap consists of highly repetitive sequence that is likely resistant to PCR amplification. In further support of the existence of a long, continuous transcript spanning the region from MBS10 all the way to the end of the miR379–410 cluster, we found that the detected PCR fragments, similar to the adjacent miRNA genes, were robustly induced by depolarization (Figure 2C). Taken together, our transcript analysis supports the idea that the miR379–410 cluster is transcribed as a single polycistronic unit starting in the proximity of MBS10.
Figure 2.
A Mef2-binding site (MBS10) is located upstream of the miR379–410 cluster. (A) Alignment of the 20 kb directly upstream of the miR379–410 cluster. Peaks represent conserved regions, the position of MBS10 and the sequence conservation of the consensus are shown. The different PCR fragments that were amplified are indicated by black numbers, fragments that failed to be amplified due to the repetitive nature of the sequence are indicated by blue numbers. (B) MBS10 is bound by Mef2 in native chromatin in vivo. ChIP was performed in primary cortical neurons (5DIV) using an anti-Mef2 antibody or IgG as a control. Primers that specifically amplify the genomic region of MBS10, β-globin or Nur77 promoter were used for PCR amplification. Input corresponds to genomic DNA isolated before immunoprecipitation. One representative out of three independent experiments is shown. (C) A continuous transcript spans the region between MBS10 and miR-379. Semiquantitative RT–PCR analysis was performed on cortical neurons (5DIV) that were either left untreated or membrane depolarized (16 mM KCl, 6 h). Overlapping primer pairs specific for the genomic location indicated in (A) were used. Only the amplification products of a few selected PCR reactions are shown.
To test the functionality of MBS10, we cloned either the wild-type or a mutated MBS10 upstream of a minimal promoter driving the firefly luciferase reporter gene (pGL3-MBS10). The expression of a constitutively active mutant of Mef2 (Mef2-VP16) in cortical neurons induced luciferase activity from MBS10 reporter construct (Figure 3A) to a similar extent as a promoter construct derived from a known Mef2 target gene (pGL3-Nur77). The DNA-binding deficient mutant Mef2ΔDBD-VP16 failed to increase the activity of both the Nur77 and MBS10 luciferase reporters, suggesting that induction is dependent upon Mef2 binding. This is further supported by the lack of induction of the pGL3-MBS10mut upon co-transfection with Mef2-VP16 (Figure 3A). We next investigated the role of endogenous Mef2 in activity-dependent transcriptional control of the cluster. KCl or BDNF stimulation induced expression of pGL3-MBS10, with a more prominent effect observed in membrane-depolarized neurons (Figure 3B and C). Importantly, both KCl and BDNF (Figure 3B and C)-mediated induction were strongly attenuated upon Mef2 knockdown using a previously published Mef2 shRNA construct (Flavell et al, 2006). Simultaneous introduction of an RNAi-resistant Mef2 expression vector (RiRMef2) completely abolished the inhibitory effect of Mef2 knockdown, demonstrating the specificity of the siRNAs. Taken together, results from reporter assays suggest that endogenous Mef2 is required for the depolarization-induced regulation of the miRNA cluster. To confirm that activity-regulated transcription of the endogenous miRNAs is Mef2 dependent, we infected primary neurons using a lentivirus expressing the described Mef2 siRNA. Infected cultures were stimulated with KCl and the levels of selected endogenous miR379–410 pre-miRNAs were assessed by quantitative RT–PCR. For all the miRNAs from the cluster analysed (miR-134 -154, and -376b), Mef2 knockdown significantly reduced the magnitude of KCl-mediated induction (Figure 3D). This effect was specific, as Mef2 knockdown did not affect activity-dependent induction of CREM, a gene that is not regulated by Mef2. Taken together, our data indicate that Mef2 activates miRNA expression in response to neuronal activity by binding to a site located 20 kb upstream of the miRNA cluster.
Figure 3.
Mef2 is necessary for activity-dependent regulation of the miR379–410 cluster. (A) Binding of Mef2 to MBS10 activates transcription. Reporter genes containing either a wt or mutant MBS10 (MBS10-luc and MBS10-mut-luc, 50 ng) upstream of the luciferase coding region were transfected into cortical neurons (5DIV) along with expression plasmids for constitutively active Mef2 (Mef2-VP16) or a DNA-binding deficient mutant (Mef2-ΔDBD-VP16, 200 ng each). The Mef2 responsive Nur77 reporter was used as a positive control. Luciferase activity was determined and normalized to the internal Renilla control. Fold inductions were derived by dividing the normalized luciferase activity of Mef2-expressing neurons to that of control-transfected neurons. Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test). (B) Mef2 is required for activity-dependent transcription of a MBS10-driven reporter gene. MBS10-luc (100 ng) was transfected into cortical neurons (5DIV) along with a Mef2 shRNA construct (Si-Mef2, 5 ng) and/or an RNAi-resistant Mef2 expression construct (RiRMef2, 100 ng). Neurons were either left untreated or membrane depolarized (57 mM KCl, 6 h). Luciferase activity was determined and normalized to the internal Renilla control. Fold inductions were derived by dividing the normalized activity of KCl-treated neurons to that of untreated neurons. Data represents the mean of three independent experiments+s.d. *P<0.05 (t-test). (C) Mef2 is required for BDNF-dependent transcription of a MBS10-driven reporter gene. Neurons were transfected and treated as in B). Data represents the mean of at least three independent experiments+s.d. *P<0.05 (t-test). (D) Regulation of activity-dependent transcription of the endogenous miRNAs of the miR379–410 cluster by Mef2. Hippocampal neurons were infected with lentiviruses expressing either an shRNA directed against Mef2 (Si-Mef2) or against an unrelated sequence (Si control). Neurons were stimulated with KCl for up to 6 h, total RNA was isolated at the indicated time points and analysed by quantitative RT–PCR using primers specific for miR379–410 precursors, Arc (positive control) and CREM (negative control). For miR-134, one representative out of three independent experiments is shown.
Activity-dependent expression of miR379–410 cluster miRNAs is necessary for dendritic development
A large number of studies have provided evidence that activity-dependent transcription during early stages of synaptic development has a critical function in dendritic outgrowth (Whitford et al, 2002). To address the physiological relevance of activity-dependent transcription of the miR379–410 cluster, we therefore tested whether perturbation of miR379–410 members affected the ability of neurons to elaborate the dendritic tree in response to activity. To mimic activity-dependent dendritogenesis in vitro, cultured hippocampal neurons (DIV7) were treated for 6 h with KCl or BDNF and analysed by Sholl analysis (see Materials and methods) at DIV10 (Wayman et al, 2006). Both membrane depolarization and BDNF treatment increased the complexity of the dendritic arbour (Figure 4A, left panel) as indicated by a higher number of branches and an increase in the total dendritic length. To obtain a quantitative estimate of activity-dependent changes in complexity, we calculated an induction index by dividing the total number of intersections derived from the Sholl analysis of stimulated neurons to that of unstimulated neurons. Treating neurons with a specific miR-134 anti-miR completely abolished the KCl- and BDNF-mediated increase in dendritic complexity, but had no significant effect on dendrites under basal conditions (Figure 4A–C; Supplementary Figure S1). An antisense oligonucleotide of unrelated sequence (anti-miR control) had no effect on dendritic complexity under both basal and stimulated conditions, demonstrating the specificity of the anti-miR-134. Furthermore, an miR-134 antisense inhibitor of a different chemistry (LNA-modified nucleotides) had a similar effect on the induction index after both KCl and BDNF stimulation (Supplementary Figure S2). We next extended our analysis to other miRNAs from the cluster. Of the four miRNAs considered, we found that anti-miR inhibition of two of them (miR-381 and -329), also blocked activity-dependent dendritogenesis (Figure 4D). Interestingly, inhibition of two other cluster members (miR-495 and -541) had no effect on dendritic complexity under our experimental conditions (Figure 4D). The lack of a dendritic phenotype is unlikely due to inefficient inhibition of these miRNAs, as anti-miRs effectively interfered with three different miRNAs in our sensors assays including miR-541 (Figure 1F). Thus, activity-dependent expression of multiple, but not all miRNAs from the miR379–410 cluster is required for activity-dependent dendritic growth. The requirement of activity-induced expression of the miR379–410 cluster is further supported by our observation that knock down of its upstream activator Mef2 phenocopies miRNA loss of function (Figure 4E). The negative effect of Mef2 siRNA on KCl-dependent dendritogenesis can be rescued by co-transfection of RiRMef2. These data show a previously unknown function of Mef2 in activity-dependent dendritogenesis and identify miRNAs from the miR379–410 cluster as one of the key mediators of this new role of Mef2.
Figure 4.
Several miRNAs of the miR371–410 cluster are necessary for activity-dependent dendritogenesis. (A) Hippocampal neurons (4DIV) were transfected with GFP together with the indicated anti-miRs (50 nM). At DIV7, neurons were incubated with 16 mM KCl or 40 ng/ml BDNF for 6 h and dendritic complexity was analysed at DIV10 using Sholl analysis (see Materials and methods for details). Scale bar: 20 μm. (B) Quantification of the dendritic complexity of KCl-treated neurons after miR-134 inhibition. Dendritic complexity was calculated by Sholl analysis and the effect of the KCl stimulation on the dendritic tree is expressed as induction index (total number of intersection in stimulated neurons divided by the total number of intersection under basal condition). Here, 10–16 cells for condition were analysed in each experiment. Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test). (C) Quantification of the dendritic complexity of BDNF-treated neurons after miR-134 inhibition. Conditions and data analysis are the same as in (B). Data represent the mean of at least three independent experiments+s.d. *P<0.05 (t-test). (D) Multiple miR379–410 members are necessary for activity-dependent dendritogenesis. Quantification of dendritic complexity of KCl-treated neurons transfected with the indicated anti-miRs (50 nM) was basically as described in (B). Here, 10–16 cells for condition were analysed in each experiment. Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test). (E) Mef2 knockdown phenocopies miR-134 loss of function in dendritogenesis. Hippocampal neurons were transfected with GFP together with a Mef2 shRNA (2 ng) and/or a construct expressing an RNAi-resistant form of Mef2 (RiRMef2, 100 ng). Treatment of neurons and data analysis were performed as described in (B). Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test).
Pum2 is an miR-134 target
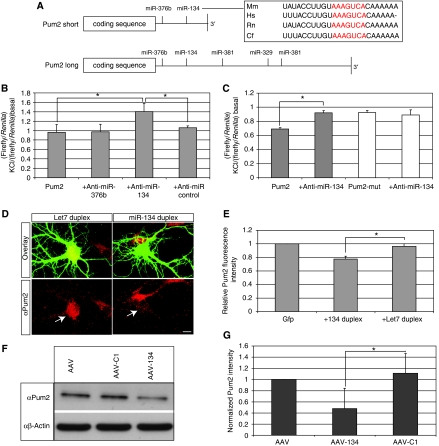
To gain insight into the mechanism by which members of the miR379–410 cluster regulate dendritogenesis, we searched for target mRNAs using the Target Scan prediction algorithm. Initially, we focused on predictions that contained multiple sites for individual members of the miR379–410 cluster. Intriguingly, we found that the RNA-binding protein Pum2, a protein that has been recently implicated in the control of dendrite morphogenesis in Drosophila melanogaster (Ye et al, 2004), contains in its 3′UTR multiple potential binding sites for functionally important miR379–410 miRNAs (Figures 4D and 5A). Two Pum2 3′UTR of different lengths have been predicted. The shorter one has been experimentally isolated and includes putative binding site for miR-134 and miR-376b. The longer 3′UTR includes, in addition, sites for miR-381 and miR-329. To test whether the predicted miRNA-binding sites are functional, we cloned both Pum2 3′UTRs downstream of the coding region of a firefly luciferase reporter gene, and monitored luciferase activity in the presence of specific anti-miRs in rat cortical neurons. Under conditions of high neuronal activity, inhibition of miR-134 led to a robust increase in relative luciferase activity of the constructs containing the wild-type Pum2 3′UTR (Figure 5B; Supplementary Figure S3). Importantly anti-miR-134 did not affect luciferase activity of the reporter under basal condition (Supplementary Figure S3A), suggesting that after depolarization, newly transcribed miR-134 binds to and downregulates the expression of the Pum2 mRNA. A number of anti-miRs of different sequence, including three directed against members of the miR379–410 cluster (anti-miR-376b, -381 and -329) had no effect demonstrating the specificity of anti-miR-134 (Figure 5B and data not shown). Importantly, miR-134 is the most conserved of the five putative binding sites (Figure 5A) we tested, and is present in both predicted isoforms of the Pum2 3′UTR. Both observations suggest a functionally significant role of the miR-134 site in the regulation of Pum2 translation. To verify that inhibition of reporter gene activity was dependent on a functional miR-134-binding site in the 3′UTR, we cloned a luciferase reporter where the critical seed region in the miR-134 responsive element was mutated. Comparison of the mutated and wild-type Pum2 luciferase reporter activity in cortical neurons after KCl stimulation showed that, first, the mutant reporter is not downregulated after depolarization and, second, co-transfection of pGL3-Pum2mut with anti-miR-134 does not result in an increase in the activity of the reporter (Figure 5C). These results indicate that binding of miR-134 to the 3′UTR of Pum2 is critical for the repression of the reporter after KCl stimulation. Induction of other miRNAs from the cluster appears to be less relevant for the regulation of Pum2 mRNA translation, at least under our experimental conditions.
Figure 5.
Pum2 is a new miR-134 target gene. (A) Schematic representation of the two predicted 3′UTRs of the Pum2 mRNA. The putative miR379–410 miRNA-binding sites in the two predicted 3′UTRs are indicated. The conservation of the miR-134 site is shown in the box, the seed sequence is in red. (B) Validation of Pum2 as miR-134 target mRNA by dual-luciferase reporter assay in membrane-depolarized cortical neurons. Cortical neurons were transfected with a luciferase reporter gene containing the shorter form of the Pum2 3′UTR downstream of the luciferase coding region (Pum2-luc, 100 ng) together with the indicated anti-miRs (60 nM). At DIV7, neurons were either left untreated or membrane depolarized (16 mM KCl, 6 h). Luciferase activity was determined and normalized to the internal Renilla control. Fold inductions were derived by dividing the normalized activity of KCl-treated neurons to that of untreated neurons. Data represent the mean of at least three independent experiments+s.d. *P<0.05 (t-test). (C) Downregulation of the Pum2 luciferase reporter by miR-134 requires a functional miRNA-binding site. Cortical neurons were transfected with Pum2-luc or a reporter gene containing mutations in the seed region of the Pum2 miR-134-binding site (Pum2-mut-luc, 100 ng each), together with anti-miR-134 (60 nM) when indicated. Data analysis and presentation were performed as in (B). Data represent the mean of at least three independent experiments+s.d. *P<0.05 (t-test). (D) Overexpression of miR-134 in hippocampal neurons downregulates the endogenous Pum2 protein. Hippocampal neurons (4DIV) were transfected with GFP together with either miR-134 or Let7 duplex RNAs (10 nM each). Neurons were fixed at DIV7 and analysed by immunocytochemistry using a rabbit polyclonal anti-Pum2 antibody. Arrows depict cell bodies of representative cells transfected either with Let-7 (left) or miR-134 (right). Note reduced Pum2 staining in miR-134-transfected neurons. Scale bar: 10 μm. (E) Quantification of the Pum2 fluorescence intensities derived from (D). Signal intensities of Pum2 staining in transfected, GFP-positive neurons were measured for a total of 8–10 cells per experiment. Average intensities for each condition were normalized to control cells transfected with GFP only. Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test). (F) AAV-mediated miR-134 overexpression in cortical neurons downregulates Pum2 protein levels. Protein extracts from cortical neurons (DIV10–18) transduced with either AAV, AAV expressing a control shRNA (AAV-C1) or AAV expressing miR-134 (AAV-134) were analysed by western blotting using an anti-Pum2 antibody (upper panel). Probing the same blot with an anti-β-actin antibody served as a loading control (lower panel). (G) Quantification of western blot results shown in (F). The intensity of the Pum2 band was normalized to the respective signal from the β-actin blot, and the normalized Pum2 intensity of the AAV only condition was set to one. Data represent the mean of at least three independent experiments+s.d. *P<0.05 (t-test).
We next investigated whether miR-134 is able to downregulate the endogenous Pum2 protein. First, overexpression of miR-134 duplex RNA in hippocampal neurons resulted in a significant decrease in endogenous Pum2 protein levels as assessed by immunocytochemistry (Figure 5D and E). As a control, overexpression of an unrelated miRNA (Let-7) did not affect Pum2 levels in transfected neurons (Figure 5D and E).
Second, transduction of rat hippocampal neurons with an adeno-associated virus (AAV) overexpressing miR-134 led to a significant decrease in Pum2 protein levels compared with control conditions as assessed by western blotting. β-Actin levels remained unchanged (Figure 5F and G).
Co-transfection of the Pum2 luciferase reporter together with either Si-Mef2 or Mef2-VP16 confirmed that Mef2 contributes to KCl-mediated inhibition of the reporter (Supplementary Figure S4A) and that activated Mef2 on its own is sufficient to induce a significant decrease in Pum2 reporter gene activity (Supplementary Figure S4B). In summary, our results indicate that miR-134 is a negative regulator of Pum2 protein expression following membrane depolarization. They further suggest that Mef2 is an important component of this pathway, presumably through induction of miR-134.
Downregulation of Pum2 by miR-134 is necessary for activity-dependent dendritogenesis
The Drosophila Pum homologue is essential for dendrite morphogenesis in peripheral neurons, suggesting that the Pum2–miR-134 interaction might also be involved in this process in mammals. We used RNA interference to probe the function of Pum2 in miR-134-regulated activity-dependent dendritogenesis. Two siRNAs (Si-Pum2-1 and Si-Pum2-2) directed against the rat Pum2 mRNA were able to efficiently downregulate the expression of recombinant Pum2 in HEK293T cells (Figure 6A). Immunocytochemistry revealed a significant and specific decrease in the endogenous protein in hippocampal neurons upon transfection of one of the active siRNAs, Si-Pum2-2 (Figure 6B). To determine whether Pum2 is a physiologically relevant miR-134 target for activity-dependent dendritogenesis, we asked whether siRNA-mediated reduction of Pum2 levels was able to rescue the reduced dendrite complexity observed upon miR-134 inhibition in membrane-depolarized neurons. To this end, hippocampal neurons were transfected with anti-miR-134 together with either Si-Pum2-2 or a control siRNA (Si control). We found that knock down of Pum2 using two independent siRNAs (Si-Pum2-1 and Si-Pum2-2) specifically rescues the miR-134 loss of function phenotype whereas the Si control had no effect (Figure 6C and D; Supplementary Figure S5). Thus, downregulation of Pum2 protein expression by miR-134 is necessary for activity-dependent dendritogenesis.
Figure 6.
Pum2 is a physiological target of miR-134 during activity-dependent dendritogenesis. (A) Two independent Pum2 shRNAs efficiently downregulate the expression of recombinant Pum2. Western blot analysis of HEK293T cell lysates transfected with the indicated amounts of GFP–Pum2 expression vectors, two Pum2 shRNAs as well as a control shRNA (4 ng each) were analysed by western blot using an anti-Pum2 antibody and an anti-eIF4E antibody as a loading control. (B) Si-Pum2-2 efficiently downregulates the expression of the endogenous Pum2 protein. Hippocampal neurons were transfected at DIV4 with GFP together with either Si-Pum2-2 (left) or a control siRNA (right, 2 ng each). The cells were fixed at DIV7 and analysed by immunocytochemistry using an anti-Pum2 antibody. Arrows point to cell bodies of representative transfected neurons. Note the reduced Pum2 signal in the cell transfected with Si-Pum2-2 (left) compared with the control neuron (right). Scale bar: 10 μm. (C) Pum2 knockdown by siRNA rescues the miR-134 loss-of-function phenotype in membrane-depolarized hippocampal neurons. Hippocampal neurons were transfected with GFP in conjunction with the indicated anti-miRs (50 nM) and shRNA constructs (2 ng). At 7DIV, neurons were incubated with 16 mM KCl for 6 h and dendritic complexity was evaluated 3 days later. Scale bar: 20 μm. (D) Quantitative analysis of the dendritogenesis assay shown in (C). The calculation of dendritic complexity and the data presentation are as in Figure 4B. Here, 10–16 cells for condition were analysed in each experiment. Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test). (E) Overexpression of miR-134 duplex RNA perturbs membrane depolarization-induced dendritic outgrowth. Hippocampal neurons (4DIV) were transfected with GFP together with the indicated duplex RNAs (10 nM), and depolarization was performed at 7DIV followed by the assessment of dendritic complexity at 10DIV, as described in (C). Here, 10–16 cells for condition were analysed in each experiment. Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test). (F) Both knockdown and overexpression of Pum2 perturb membrane depolarization-induced dendritic outgrowth. Hippocampal neurons (4DIV) were transfected with GFP together with the indicated shRNA (2 ng) or Pum2 expression (100 ng) constructs, and depolarization was performed at 7DIV followed by the assessment of dendritic complexity, as described in (C). Here, 10–16 cells for condition were analysed in each experiment. Data represent the mean of three independent experiments+s.d. *P<0.05 (t-test). (G) Schematic diagram of the fine-tuning of Pum2 levels by miR-134 in membrane-depolarized neurons, see main text for details. (H) Model for the miR379–410 activity-dependent dendritogenesis pathway. Neuronal activity activates Mef2 which in turn induces the expression of the miR379–410 cluster. Several members of the cluster are necessary for activity-dependent dendritogenesis. Among them is miR-134, which binds to the Pum2 mRNA and downregulates its translation. Lower levels of the translational repressor Pum2 allow for the translation of a set of so far unknown mRNAs that are likely positive regulators of dendrite outgrowth.
We next investigated whether expression of miR-134 or downregulation of Pum2 is sufficient to trigger dendritic outgrowth. Transfection of hippocampal neurons with either miR-134 duplex or Si-Pum2 did not increase basal dendritic complexity (data not shown). Surprisingly, membrane depolarization also failed to induce dendritic outgrowth in cells overexpressing miR-134 or the Si-Pum2-2 (Figure 6E and F). Thus, both decreasing and increasing Pum2 expression by means of miR-134 perturbation are detrimental to activity-dependent dendritogenesis. Our results are consistent with a role for miR-134 in fine-tuning gene expression during dendritogenesis. Consistent with such a fine-tuning relationship between miR-134 and Pum2, overexpression of Pum2 similarly precludes an activity-dependent increase in dendritic complexity (Figure 6F). Taken together, our results suggest that miR-134 buffers Pum2 levels within a narrow range critical for activity-dependent dendritogenesis (Figure 6G).
Discussion
Neurons respond to changes in their activity status by remodelling the dendritic tree and the number and strength of synapses. This morphological and functional plasticity is necessary for correct neuronal development and modulates numerous behavioural adaptations such as memory formation and addiction (Kandel, 2001; Calabresi et al, 2007). We have previously shown that several miRNAs enriched in the dendritic compartment control local protein synthesis and are necessary for spine morphogenesis (Schratt et al, 2006; Siegel et al, 2009). Importantly, the function of one of them, miR-134, is modulated locally by neuronal activity. We now show that activity-dependent regulation of miR-134 is not limited to the dendritic compartment but is controlled globally within the neuron by an activity-dependent transcriptional programme. The miR-134 gene is located within the Gtl2/Dlk1 locus clustered together with more than 50 other miRNA genes. Membrane depolarization or BDNF treatment enhances the expression of all the miRNAs within the cluster that we tested. We validated these miRNAs as new targets for the activity-regulated transcription factor Mef2, a regulator of synapse number and differentiation, and show that activity-dependent, Mef2-mediated expression of the miRNA cluster is necessary for the development of the dendritic tree in primary neurons. MiR-134 specifically regulates activity-dependent dendritogenesis by a novel pathway, namely the downregulation of the RNA-binding protein and translational repressor Pum2 (Figure 6H). Our results, together with existing data, show that miRNA function is regulated at multiple levels by neuronal activity, and suggest that miRNAs are critical components of the relay that coordinates nuclear programmes of gene expression with local changes in dendritic and synaptic morphology in response to neuronal activity.
Activity-dependent regulation of the miR379-410 cluster
Several lines of evidence suggest that the miR379–410 cluster is co-regulated by neuronal activity and might be transcribed as a single polycistronic unit. First, all the miRNAs located in the distal part of the GTL2/DLK1 locus show a very similar brain-specific expression pattern (Seitz et al, 2004). Second, BDNF and KCl treatment lead to a very similar induction profile of pre-miRNAs from the miR379–410 cluster (Figure 1B and C). Third, knock down of Mef2 greatly reduces the stimulation-induced expression of all the miR379–410 miRNAs we tested (Figure 3D). Finally, we could detect an overlapping transcript by RT–PCR spanning about 20 kb upstream of the first miRNA within the cluster. Northern blot analysis will be necessary to confirm the existence of a large continuous transcript spanning the entire miR379–410 cluster. We identified a Mef2-binding element (MBS10) about 20 kb upstream of the miR379–410 cluster. Our results show that MBS10 is occupied by Mef2 in vivo and is sufficient to induce upregulation of the expression of a luciferase gene reporter upon Mef2 binding following KCl and BDNF stimulation. Despite its functional importance, it is unclear whether MBS10 is located within a promoter or a long-range enhancer sequence. Fine mapping of the transcriptional start site should allow discrimination between these two possibilities.
miRNAs of the miR379–410 cluster are necessary for activity-dependent neuronal development
Anti-miR-mediated inhibition of several members of the miR379–410 showed that at least three of them (miR-134, -329 and -381) are necessary for dendritic outgrowth triggered by KCl in hippocampal neurons. The functional consequences of activity-dependent expression of the other members of miR379–410 remain to be determined. A variety of programmes are triggered by neuronal activity, such as cell survival or synapse remodelling. Thus, it is conceivable that multiple aspects of activity-dependent development are controlled by the miR379–410 cluster. Consistently, the sequence of the mature miRNAs is significantly divergent and a recent bioinformatic study revealed a limited degree of overlap in the GO annotations of the different miR379–410 miRNAs (Glazov et al, 2008). Recently, altered biogenesis of miR-134 and other members of the miR379–410 cluster has been described in a mouse model of the human microdeletion of the 22q11.2 chromosomal locus (Stark et al, 2008). Mice hemizygous for the deletion showed several behavioural and cognitive deficits accompanied by abnormal dendritic arbourization in the hippocampus. This animal model further indicates that the miR379–410 cluster has a crucial function in dendritic morphogenesis.
Mef2 negatively regulates synapse number in mature hippocampal neurons and miR-134, the best characterized member of the miR379–410 cluster, negatively regulates dendritic spine size. This suggests that the miR379–410 cluster could also be involved in the Mef2-dependent control of synapse number at later stages of neuronal development. The role of Mef2 in dendritogenesis was previously unknown, presumably as Mef2 is required only for dendrite outgrowth upon neuronal activation. The dual function of Mef2 as a positive regulator of dendritogenesis and negative regulator of synapse number appears at first glance paradoxical. One possible explanation is that at different stages of neuronal development Mef2 might regulate different sets of target genes. Indeed, known Mef2 targets include both dendrite growth-promoting factors, such as BDNF, and negative regulators of synapse formation and strength, such as Arc and SynGAP (Flavell and Greenberg, 2008). A recent in vivo study suggests that Mef2 is a key component of a compensatory mechanism that couples structural plasticity with sensitized response to cocaine in striatal neurons (Pulipparacharuvil et al, 2008). It is thus tempting to speculate that the apparently opposing effects of Mef2 on hippocampal neurons are in fact components of the same homoeostatic circuit that controls the overall neuronal excitability, by both decreasing synapse number and promoting dendritic outgrowth (Turrigiano, 2007). This is consistent with the observation that Mef2 targets such as miR-134 display a similar dichotomy. MiR-134 is necessary for activity-dependent dendritogenesis and negatively regulates spine growth by regulating Pum2 and LimK1, respectively. Strikingly, BDNF upregulates miR-134 transcription, whereas, within the dendrite, BDNF relieves the inhibition of the known miR-134 target LimK1 to promote spine development. Therefore, synaptic stimulation might elevate miR-134 expression throughout the cell while inactivating miR-134 locally within stimulated spines. This could be a mechanism to permit spine growth specifically at active synapses without changing the overall excitability of the neuron.
In recent studies, the CREB-regulated miR-132 was shown to control activity-dependent dendritic growth by regulating the Rho family GTPase-activating protein p250GAP (Vo et al, 2005; Cheng et al, 2007; Wayman et al, 2008). Thus, at least two independent miRNA pathways coordinately regulate dendritic plasticity. Further studies are necessary to analyse the extent of cross-talk between CREB- and Mef2-regulated miRNA pathways in dendrite development.
Pum2 is a new miR-134 target necessary for activity-dependent dendritogenesis
We have shown that the RNA-binding protein Pum2 is a new bona fide miR-134 target. The presence of predicted binding sites for other members of the miR379–410 cluster in the Pum2 3′UTR suggested that multiple miRNAs are necessary to robustly downregulate Pum2 after membrane depolarization. However, miR-134 alone appears to be sufficient to fine-tune Pum2 levels within a functionally critical window. Therefore, we favour the idea that other miR379–410 miRNAs might be involved in the regulation of different aspects of activity-dependent dendritogenesis (e.g. membrane trafficking, cytoskeletal dynamics, etc.) by fine-tuning critical regulators of these pathways. Interestingly, a number of predicted miR379–410 target mRNAs have been associated with dendrite morphogenesis. For example, members of the SMAD and kinesin families (Guo et al, 2001; Satoh et al, 2008) are predicted targets of miR-329 and -381, respectively (Targetscan 3.0). Future studies will reveal whether fine-tuning of these genes is a common regulatory function of the miR379–410 cluster.
The epistasis experiments in hippocampal neurons validate the functional relevance of the Pum2–miR-134 interaction. Both inhibition and overexpression of miR-134 and Pum2 compromise activity-dependent dendritogenesis. Similarly, in Drosophila peripheral neurons, both depletion and ectopic expression of Pum have a detrimental effect on dendrite morphogenesis. Taken together, these findings support the idea that although Pum2 expression is necessary for activity-dependent dendritogenesis, a careful control of its levels by miR-134 is equally critical for the correct elaboration of the dendritic tree (Ye et al, 2004). Thus, the miR-134–Pum2 relationship in membrane-depolarized neurons might be best described by the recently proposed tuning model (Hobert, 2007; Karres et al, 2007). In this model, an miRNA buffers the target protein levels within a critical window that is optimal for the cell (Figure 6G). An important implication of the tuning model is that it takes into account the spatial regulation of miRNA targets, for example, during local protein synthesis in dendrites (Tsang et al, 2007). As both the Pum2 mRNA and protein are localized in dendrites (Vessey et al, 2006; Zhong et al, 2006), it is possible that Pum2 and miR-134 functionally interact within this compartment. Pum2 is a translational repressor and a component of somatic and dendritic RNA granules, in particular stress granules (Vessey et al, 2006). Therefore, miR-134, by downregulating the Pum2 protein, might induce the redistribution of Pum2 target mRNAs to the local protein synthesis machinery. Interesting candidates are known Drosophila Pum targets, such as eukaryotic initiation factor 4E (eIF4E), PSD-95 homologues and a voltage-gated sodium channel (Mee et al, 2004; Menon et al, 2004; Chen et al, 2008).
In summary, we have identified the miR379–410 cluster as a new activity-regulated Mef2 target gene, the expression of which is necessary for dendritic plasticity in hippocampal neurons.
These results represent a road map to investigate the function of these miRNAs in activity-dependent processes in vivo and to understand how a neuron coordinates local regulation of protein synthesis with global control of gene expression to properly adapt to its environment.
Materials and methods
DNA constructs
Single cell fluorescent sensor constructs were generated by subcloning of an EcoRI–NotI flanked dsRED cassette (Clontech) together with a NotI–XbaI flanked oligonucleotide containing two perfectly complementary sites to the miRNA of interest, into EcoRI–XbaI linearized pTracer-CMV (Invitrogen).
The Pum2 3′UTR was amplified from a rat brain cDNA library and cloned into the XbaI site of pGL3promoter vector (Promega). To generate pGL3-Pum2mut, the miR-134 seed sequence was replaced with an XhoI site by overlapping PCR, resulting in three point mutations in the miR-134-binding site.
Luciferase sensor constructs, pGL3basic-MBS10, MBS10-mut and NUR77, were generated by ligation of annealed oligonucleotides to XhoI–NheI linearized pGL3basic (Promega). poFluc-3 × MRE, pcDNA3-Mef2-VP16 (constitutively active Mef2), pcDNA3-Mef2ΔDBD-VP16 (mutant Mef2), pcDNA3-Mef2D-RiR (RNAi-resistant Mef2D) and pSuper-Mef2D were kindly provided by ME Greenberg (Harvard Medical School, Boston, USA; Flavell et al, 2006). poFluc-5 × MBS10 was generated by substituting HindIII–BglII flanked 3 × MRE from poFluc-3 × MRE with 5 × MBS10, whereas poFluc control was obtained from self-blunt-end ligation after 3′ end filling with Klenow enzyme (Roche Diagnostics). GFP–Pum2 expression vector is a kind gift of Michael Kiebler (Medical University of Vienna, Austria; Vessey et al, 2006).
Cell culture, transfection and virus infection of primary neurons
The culture and transfection of dissociated primary cortical and hippocampal neurons from embryonic day 18 (E18) Sprague–Dawley rats (Charles River Laboratories, Sulzfeld, Germany) was as described (Schratt et al, 2004). For AAV-mediated overexpression of miR-134, the miR-134 or a control sequence embedded in the miR-30 hairpin was inserted into the 3′UTR of GFP within AAV-6P-SEWB (kind gift of Martin Schwarz, MPIMF Heidelberg; Shevtsova et al, 2005). A detailed description and characterization of AAV-134 will be published elsewhere (MC and GS, manuscript in preparation). Lentiviruses expressing Mef2A/D shRNA or a control shRNA have been described previously (Flavell et al, 2006).
For stimulation experiments, neurons were either treated with depolarization solution (NaCl 140 mM, KC l55–165 mM, CaCl2 2.5 mM, MgCl2 1.6 mM, Hepes 10 mM, glucose 24 mM, pH=7.4) at a final concentration of 16–55 mM KCl or with hBDNF (Peprotech) at 40–60 ng/ml as indicated for up to 6 h.
Single cell fluorescent sensor assay
Hippocampal neurons (DIV4) were transfected with bicistronic reporter constructs (pTracer-CMV-dsRED, 50 ng) and 2′O-Me-oligonucleotides (50 nM) as indicated. pTracer-CMV-dsRED contains a perfect binding site for miR-134, -329 or -541 at the 3′UTR of dsRED, which leads to degradation of dsRED RNA in the presence of the respective miRNAs. Cells in which the dsRED signal did not exceed background levels were scored as ‘miRNA positive'.
ISH
ISH of dissociated hippocampal neurons using DIG-labelled LNA probes (5 pmol; Exiqon) was basically as described with slight modifications (Schratt et al, 2006). Signal detection was performed with FITC-conjugated anti-digoxigenin FAB fragments (Roche) and an Alexa Fluor 488 signal amplification kit (Molecular Probes).
ChIP assay
A detailed ChIP protocol can be found in Supplementary data.
Quantitative real-time PCR
Total RNA was isolated by using Qiazol (Qiagen) and genomic DNA contamination was eliminated with TURBO DNase (Ambion). Reverse transcription of RNA was carried out with iScript™ cDNA synthesis kit (Bio-Rad), according to the manufacturer's protocol. Relative quantification of gene expression was conducted with the Applied Biosystems 7300 Real Time PCR System (Applied Biosystems) using the iTaq SybrGreen Supermix with ROX (Bio-Rad). PCR results were normalized to the expression of β-3-tubulin.
Semiquantitative RT–PCR (‘transcript walking')
Reverse transcription of 1 μg RNA with 50 ng random hexamers per reaction was carried out with SuperScript III™ first strand synthesis system (Invitrogen), according to the manufacturer's protocol. Advantage®2 PCR Enzyme System (Clontech) was used for RT–PCR (PCR parameters 95 °C for 2 min and 30 cycles of 95 °C for 30 s, 59 °C for 30 s, 68°C for 1 min 20 s and final extension at 68°C for 5 min). Oligonucleotide sequences are listed in Supplementary data.
Luciferase assay
Cortical or hippocampal neurons were transfected with pGL3 reporter constructs (100 ng per 48 well) together with anti-miRs (60 nM) at 4–5 DIV and luciferase assays were performed 1 or 2 days later with the Dual-Luciferase Reporter Assay System (Promega).
Dendritogenesis assay, immunocytochemistry and image analysis
Hippocampal neurons (4DIV) were transfected with eGFP (100 ng) alone or in combination with anti-miRs (50 nM), pSuper shRNA constructs (4 ng), pcDNA3 expression constructs (100 ng) or miRNA duplexes (10 nM) as indicated. After stimulation (7DIV), neurons were fixed at 10DIV in 4% paraformaldehyde/sucrose and subjected to fluorescence microscopy analysis.
For immunostaining of the endogenous Pum2 protein, hippocampal neurons were fixed in paraformaldehyde/sucrose, rinsed in PBS and incubated with rabbit polyclonal anti-Pum2 antibody (dilution 1:2000; Acris, Hiddenhausen, Germany, gift of Michael Kiebler) and cyanine-3-conjugated anti-rabbit IgG secondary antibodies (dilution 1:1000), both diluted in 0.02% gelatine–0.5% Triton X-100–PBS.
For image analysis, XY scans ( × 20, 1024 × 1024 pixels) taken with a confocal laser-scanning microscope (LSM 5 Pascal; Zeiss, Germany) were used. A grid of concentric circles spaced by 20 μm was placed on the cell bodies of GFP-positive neurons, and the numbers of dendrites crossing each circle was counted. A total of 12–16 random cells chosen in a blinded manner were analysed per experimental condition.
To quantify Pum2 protein levels by immunofluorescence, a total of 8–10 random GFP-positive cells per condition were imaged ( × 40 objective, 1024 × 1024 pixels) and the signal intensity in the red channel (anti-Pum2) was measured using ImageJ software.
Western blotting
Western blotting was performed as described previously (Schratt et al, 2004). The following primary antibodies were used: rabbit anti-Pum2-antibody (dilution 1:5000; Acris), mouse anti-β-actin (1:15000; MMS-435P; Covance), rabbit anti-eIF4E (1:10000; no. 9742; Cell Signaling). Primary antibodies were recognized either by an HRP-conjugated goat anti-rabbit antibody (1:20 000; 401315; Calbiochem) or an HRP-conjugated rabbit anti-mouse antibody (1:20 000; 402335; Calbiochem).
Bioinformatics
The prediction of MBSs was carried out with MAPPER (Marinescu et al, 2005), JASPAR (Sandelin et al, 2004) and rVISTA 2.0 (Loots and Ovcharenko, 2004) web servers. The sequence conservation between mouse and human was determined using BlastN.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure Legends
Supplementary Material
Acknowledgments
We thank M Kiebler for generously providing reagents and M Schwarz for initial help with AAV production. The excellent technical assistance of T Wüst is greatly acknowledged. This study was supported by grants from the DFG (SFB488) and HFSP (CDA) to GS and from the NIH (NS028829) to MEG.
References
- Bagni C, Greenough WT (2005) From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci 6: 376–387 [DOI] [PubMed] [Google Scholar]
- Calabresi P, Picconi B, Tozzi A, Di Filippo M (2007) Dopamine-mediated regulation of corticostriatal synaptic plasticity. Trends Neurosci 30: 211–219 [DOI] [PubMed] [Google Scholar]
- Chen G, Li W, Zhang QS, Regulski M, Sinha N, Barditch J, Tully T, Krainer AR, Zhang MQ, Dubnau J (2008) Identification of synaptic targets of Drosophila pumilio. PLoS Comput Biol 4: e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ghosh A (2005) Regulation of dendritic development by neuronal activity. J Neurobiol 64: 4–10 [DOI] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K (2007) MicroRNA modulation of circadian-clock period and entrainment. Neuron 54: 813–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G (2008) MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta 1779: 471–478 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME (2008) Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci 31: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, McWilliam S, Barris WC, Dalrymple BP (2008) Origin, evolution, and biological role of miRNA cluster in DLK-DIO3 genomic region in placental mammals. Mol Biol Evol 25: 939–948 [DOI] [PubMed] [Google Scholar]
- Guo X, Lin Y, Horbinski C, Drahushuk KM, Kim IJ, Kaplan PL, Lein P, Wang T, Higgins D (2001) Dendritic growth induced by BMP-7 requires Smad1 and proteasome activity. J Neurobiol 48: 120–130 [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O (2007) miRNAs play a tune. Cell 131: 22–24 [DOI] [PubMed] [Google Scholar]
- Hong EJ, West AE, Greenberg ME (2005) Transcriptional control of cognitive development. Curr Opin Neurobiol 15: 21–28 [DOI] [PubMed] [Google Scholar]
- Kandel ER (2001) The molecular biology of memory storage: a dialogue between genes and synapses. Science 294: 1030–1038 [DOI] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM (2007) The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell 131: 136–145 [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ (2006) Neuronal RNA granules: movers and makers. Neuron 51: 685–690 [DOI] [PubMed] [Google Scholar]
- Kosik KS (2006) The neuronal microRNA system. Nat Rev Neurosci 7: 911–920 [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD (2002) Function and regulation of CREB family transcription factors in the nervous system. Neuron 35: 605–623 [DOI] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I (2004) rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res 32: W217–W221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT (2004) MicroRNA-responsive ‘sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 36: 1079–1083 [DOI] [PubMed] [Google Scholar]
- Marinescu VD, Kohane IS, Riva A (2005) The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res 33: D91–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee CJ, Pym EC, Moffat KG, Baines RA (2004) Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J Neurosci 24: 8695–8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K (2004) The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron 44: 663–676 [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, Greengard P, Nairn AC, Nestler EJ, Cowan CW (2008) Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron 59: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A (2002) Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron 34: 999–1010 [DOI] [PubMed] [Google Scholar]
- Richter JD (2007) CPEB: a life in translation. Trends Biochem Sci 32: 279–285 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh D, Sato D, Tsuyama T, Saito M, Ohkura H, Rolls MM, Ishikawa F, Uemura T (2008) Spatial control of branching within dendritic arbors by dynein-dependent transport of Rab5-endosomes. Nat Cell Biol 10: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME (2004) BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci 24: 9366–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J (2004) A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 14: 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A (2006) A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311: 1012–1017 [DOI] [PubMed] [Google Scholar]
- Shalizi AK, Bonni A (2005) Brawn for brains: the role of MEF2 proteins in the developing nervous system. Curr Top Dev Biol 69: 239–266 [DOI] [PubMed] [Google Scholar]
- Shevtsova Z, Malik JM, Michel U, Bahr M, Kugler S (2005) Promoters and serotypes: targeting of adeno-associated virus vectors for gene transfer in the rat central nervous system in vitro and in vivo. Exp Physiol 90: 53–59 [DOI] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Khudayberdiev S, Leuschner PF, Busch CJL, Kane C, Hübel K, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt G (2009) A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA (2008) Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet 40: 751–760 [DOI] [PubMed] [Google Scholar]
- Steward O (2002) mRNA at synapses, synaptic plasticity, and memory consolidation. Neuron 36: 338–340 [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127: 49–58 [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G (2007) Homeostatic signaling: the positive side of negative feedback. Curr Opin Neurobiol 17: 318–324 [DOI] [PubMed] [Google Scholar]
- Vessey JP, Vaccani A, Xie Y, Dahm R, Karra D, Kiebler MA, Macchi P (2006) Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci 26: 6496–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S (2005) A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A 102: 16426–16431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites CL, Craig AM, Garner CC (2005) Mechanisms of vertebrate synaptogenesis. Annu Rev Neurosci 28: 251–274 [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S (2008) An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA 105: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR (2006) Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50: 897–909 [DOI] [PubMed] [Google Scholar]
- Whitford KL, Dijkhuizen P, Polleux F, Ghosh A (2002) Molecular control of cortical dendrite development. Annu Rev Neurosci 25: 127–149 [DOI] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN (2004) Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol 14: 314–321 [DOI] [PubMed] [Google Scholar]
- Zhong J, Zhang T, Bloch LM (2006) Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure Legends
Supplementary Material