Abstract
How cell numbers are determined is not understood. Hedgehog-Gli activity is involved in precursor cell proliferation and stem cell self-renewal, and its deregulation sustains the growth of many human tumours. However, it is not known whether GLI1, the final mediator of Hh signals, controls stem cell numbers, and how its activity is restricted to curtail tumourigenesis. Here we have altered the levels of GLI1 and p53, the major tumour suppressor, in multiple systems. We show that GLI1 expression in Nestin+ neural progenitors increases precursor and clonogenic stem cell numbers in vivo and in vitro. In contrast, p53 inhibits GLI1-driven neural stem cell self-renewal, tumour growth and proliferation. Mechanistically, p53 inhibits the activity, nuclear localisation and levels of GLI1 and in turn, GLI1 represses p53, establishing an inhibitory loop. We also find that p53 regulates the phosphorylation of a novel N' truncated putative activator isoform of GLI1 in human cells. The balance of GLI1 and p53 functions, thus, determines cell numbers, and prevalence of p53 restricts GLI1-driven stem cell expansion and tumourigenesis.
Keywords: brain, cancer, GLI1, p53, stem cells
Introduction
A key unresolved issue in biology is how cell numbers are determined during development, largely maintained in adulthood and deregulated in cancer. Hedgehog-Gli signalling is a key intercellular communication pathway involved in many aspects of development and cancer. During early brain development, it modulates precursor proliferation in different brain regions such as the neocortex, tectum and cerebellum (e.g., Dahmane and Ruiz i Altaba, 1999; Dahmane et al, 2001; Lien et al, 2006), while later on it regulates brain stem cell lineages (e.g., Lai et al, 2003; Palma and Ruiz i Altaba, 2004; Palma et al, 2005). It also drives rodent (Goodrich et al, 1997) and human (Dahmane et al, 2001) brain tumourigenesis and the self-renewal and survival of brain cancer stem cells (Clement et al, 2007). This dual involvement of Hh-Gli signalling in development and disease underlines its central role in controlling precursor cell numbers in the brain and other organs. It is not known, however, whether the level of GLI1 is critical to determine stem cell numbers and how GLI1 activity is restrained to prevent tumour formation.
Here we have tested two hypotheses: (1) GLI1 is a central determinant of stem cell numbers; (2) p53, the major human tumour suppressor, which regulates stem cell and precursor numbers (Gil-Perotin et al, 2006; Meletis et al, 2006), does so by negatively modulating the activity of GLI1. Testing these two ideas is important, as GLI proteins can integrate patterning and proliferative inputs in addition to HH signals (Brewster et al, 2000; Palma and Ruiz i Altaba, 2004; Kasper et al, 2006; Riobó et al, 2006; Dennler et al, 2007), as well as those from oncogenic RAS-MEK/AKT (Stecca et al, 2007); and p53 suppresses whereas GLI1 promotes tumourigenesis. We report that (1) the level of GLI1 determines the number of neural stem cells in vivo, (2) GLI1 and p53 act in an inhibitory loop, (3) endogenous GLI1 isoforms exist, and (4) a novel N'Δ GLI1 isoform is subject to regulation by phosphorylation in a p53-dependent manner. Our present and previous data suggest a critical regulation of GLI1 function in stem cell lineages by oncogenes and tumour suppressors, which is off-balance in disease.
Results
Elevation of GLI1 levels in neural progenitors results in a larger brain with expanded progenitor populations
To enhance Gli1 activity in stem cell lineages in vivo, we developed doxycycline (dox) inducible bigenic mice using the Nestin intron II driver (Nestin->rtTA-IRES-LacZ driver; Supplementary Figure 1), which is strongly expressed in embryonic neural progenitors but only very weakly postnatally (Figure 1; Supplementary Figures 1, 2, 3, 4 and not shown). As a responder, we made a bidirectional GFP<-tetO->myc-GLI1 transgene by random insertion (one line) or by targeting into the hprt locus (three lines). All bigenic lines showed the same phenotypes, and we focused on perinatal phenotypes of the first line resulting from sustained embryonic dox administration (Supplementary Table 1; Supplementary Figure 1).
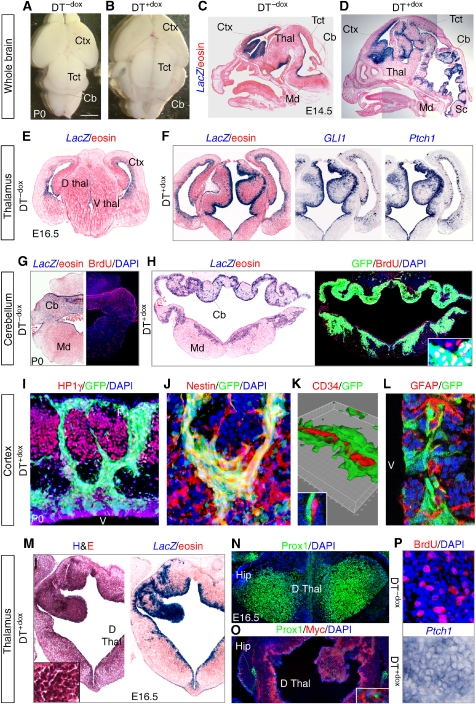
Figure 1.
Enhanced GLI1 activity induces a larger brain with hyperplasias. (A–D) DT+dox bigenic mice with strong phenotypes are perinatal lethal exhibiting a larger brain than DT−dox siblings, seen dorsally (A, B) or in sagittal sections after Xgal staining highlighting the expression of the Nestin->rtTA-LacZ driver (C, D) (see Supplementary Figure 1). (E–H) Phenotypes detected in cross section in the thalamus (Thal; E, F), cerebellum (Cb) and medulla (Md; G, H) showing LacZ (Xgal) labelling (E–H), BrdU incorporation (G, H), GLI1 and Ptch1 (F) or GFP (H) expression in DT−dox and DT+dox siblings as indicated. (I–K) HP1γ−/Nestin+/GFP+ precursors from the ventricle (v) invade the parenchyma through the cortical plate and expand near the pial (p) surface in the cingulate cortex (I, J). Invading GFP+ cells associate with CD34+ endothelial cells in vessels as seen in a Z-stack confocal reconstruction (K) and a confocal image detail (K inset). (L–P) Phenotypic analyses in cross sections of subventricular zone of the lateral ventricle (SVZ) (L) and thalamus (M–P) for the markers indicated. Myc-GLI1 was mostly cytoplasmic and Prox1 nuclear (O inset). L shows a confocal image. Tissue is counterstained with eosin (e.g., C, D, M) and nuclei with DAPI (e.g., I, L). Hip, hippocampus; Sc, spinal cord; Tct, tectum; vz, ventricular zone; Scale bar=1.5 mm (A, B), 1 mm (C, D), 0.5 mm (E–H), 100 μm (O inset, J, P), 200 μm (I), 70 μm (L; H inset), 14 μm (K), 10 μm (K inset), 0.6 mm (M, N, O), 150 μm (M inset).
Double-transgenic (DT)-treated mice (DT+dox), but not untreated (DT−dox) siblings or treated single transgenics, expressed GLI1 and GFP in precursors and displayed larger brains with hyperplasias of progenitor zones throughout from forebrain to spinal cord (Figure 1A–D; Supplementary Figure 1 and not shown). The strength of the phenotype varied within a litter. DT+dox mice with a strong phenotype (Figure 1) were embryonic lethal or died shortly after birth. Surviving animals had milder alterations (Supplementary Figure 3A). Three brain regions with strong phenotypes (cortex, thalamus and cerebellum) and the spinal cord had high levels of LacZ, GFP and Nestin and of GLI1 and endogenous Ptch1 mRNAs in progenitor cells (Figure 1E–H, M and P; Supplementary Figures 2, 3, 4). The cerebellar external germinal layer, which has the highest levels of endogenous Gli1 (Dahmane and Ruiz i Altaba, 1999), was largely unaffected, whereas the ventricular zone (VZ) was hyperplastic (Figure 1G and H; Supplementary Figure 3A, B). The tectum (colliculi) was also larger but did not show convoluted tissue as the cerebellum, thalamus or medulla (Figure 1A–D; Supplementary Figure 1). Massive and highly invasive precursor hyperplasias were also detected in the spinal cord (Supplementary Figure 4).
Transgenic GLI1 induced ∼4–13-fold the activity of the endogenous Hh-Gli pathway as assessed by the increased expression of Gli1 and Ptch1 (Figure 2A; Supplementary Figure 5). It also boosted 2–8-fold BrdU incorporation in a region-specific manner (Figure 1G, H and P; Supplementary Figure 2C, D, G, H). In the cortex and subventricular zone of the lateral ventricle (SVZ), we found high GFP+, LacZ+, Ptch1+, myc-GLI1+ groups of cells streaming from the VZ to the pial surface (Figure 1I and J; Supplementary Figure 2A–E, I). These cells were HP1γ−, a heterochromatin (neuronal and senescent cell) marker (Figure 1I); they expressed high levels of Nestin (Figure 1J; Supplementary Figure 2B, E), associated with CD34+ vessels (Figure 1K) similarly to human gliomas (Farin et al, 2006), and derived from Nestin+, GFAP− VZ cells (Figure 1L).
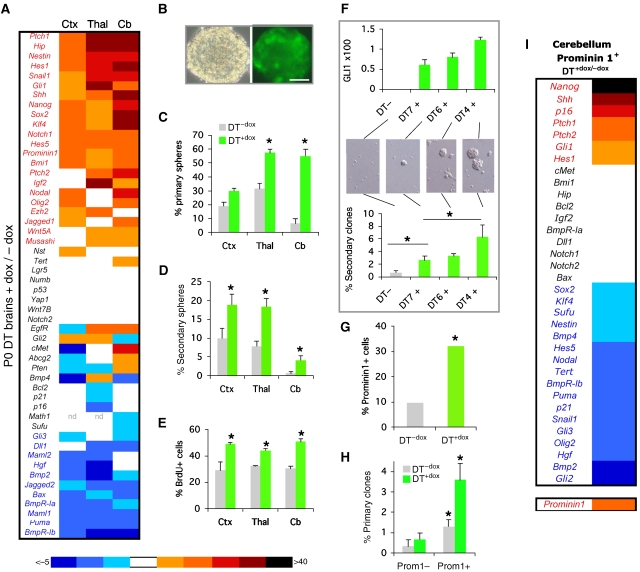
Figure 2.
Gene expression and cellular behaviour in neurospheres and stem cells. (A) RT–qPCR analyses of P0 dissected tissues from cortex (Ctx), thalamus (Thal) or cerebellum (Cb). qPCR values in all panels reflect ct values after normalisation with the geometrical mean of the values of Gapdh, βactin and EFIα and shown as the DT+dox/DT−dox value ratios. See Supplementary Figure 5. (B) Phase contrast and fluorescence images of DT+dox NS. (C–E). Histograms of the number of neurospheres (C, D) or BrdU+ cells (E) as indicated. Primary spheres were quantified per 250 000 brain cells. (F) The level of GLI1 determines NS numbers. GLI1 expression values and secondary sphere number were determined for the same samples. (G–I) Quantification of the number (G), clonogenicity after MACS (H) and gene expression analyses (I) by RT–qPCR of Prominin1+ cerebellar stem cells (see Supplementary Figure 6B). Here and in all figures asterisks denote significative changes (P<0.05) and error bars are s.e.m. Scale bar=80 μm (B).
DT+dox GFP+ regions in thalamus and cerebellum showed the highest proliferative levels (Figure 1G and H; Supplementary 2G, H) with reduced (2–3-fold) apoptosis (Supplementary Figure 3C). Massive, tumour-like hyperplasias with high nuclear density, GFP, LacZ and Ptch1 expression were prominent in these regions (Figure 1F, H and M). Hyperplasias showed little differentiation as assessed, for instance, by exclusion of neuronal Prox1 (Figure 1N and O), neuroblast NeuroD1 staining in dorsal thalamus (Supplementary Figure 3D), or astrocytic GFAP expression in cerebellum (not shown). A slight increase in the expression of the oligodendrocyte progenitor marker Pdgfrα was detected in thalamus (Supplementary Figure 3D). Unaffected regions preserved differentiation (Supplementary Figure 2J). As the strong phenotype is perinatal lethal, and we found that transgenes are silenced ∼1–2 weeks after birth, it is unclear whether such lesions could develop into frank tumours.
GLI1 upregulates an embryonic stem cell signature in the brain and modulates different signalling pathways
Gene expression analyses by qPCR of different regions of perinatal DT+dox brains showed that GLI1 induced Shh-Gli pathway components and targets including Shh, Gli1, Hip, Ptch1, Snail1 and Igf2 (Figure 2A; Supplementary Figure 5). It also upregulated a group of ‘stemness' genes, including Nanog, Sox2, Klf4, Nestin, Prominin1 (CD133) and Bmi1 (Figure 2A), which is reminiscent of that of human gliomas (Clement et al, 2007). The changes in gene expression levels were greatest in the cerebellum, consistent with phenotype strength (Figure 1; Supplementary Figures 1, 2, 3, 4), although each region displayed overlapping yet context-dependent signatures. DT+dox brains also showed an increase in Notch (Hes1, Hes5, Notch1, Jagged1) and the repression of BMP (Bmp2, Bmpr-Ia, Bmpr-Ib) activity signatures (Figure 2A), with Bmpr-Ia and -Ib being key determinants for stem cell responses to BMPs (Panchision et al, 2001). Enhanced GLI1 may thus increase the number of Nestin+ stem cells reflected in the upregulation of stemness genes involving modification of Notch, BMP and other (Igf2, Nodal, Hgf; Figure 2A) signalling pathways.
GLI1 induces an increase in the number of clonogenic neural stem cells
Given the lack of unequivocal neural stem cell markers in vivo, we tested for stem cell behaviour. Different parts of DT+dox brains showed an increase in the number of primary and secondary clonal stem cell-derived neurospheres (NS), as compared with DT−dox brains, displaying a concomitant increase in proliferation with unchanged apoptosis levels (Figure 2B–E and not shown). DT−dox and DT+dox NS remained multipotent and DT+dox produced 2–5-fold more neurons than DT−dox NS upon differentiation (Supplementary Figure 6A). Efficient NS formation from the cerebellum was only obtained from DT+dox mice (Figure 2C and D).
Importantly, we found a direct correlation between the number of NS formed from single DT+dox cerebella and the level of GLI1 (Figure 2F) so that higher levels of GLI1 produced more NS. To further address the role of GLI1 in stem cells in vivo, we analysed the number of stem cells expressing Prominin1 (Lee et al, 2005) by MACS sorting DT+dox and DT−dox cerebella. The number of cerebellar Prominin1+ stem cells in vivo increased ∼3-fold in DT+dox over DT−dox (Figure 2G), showing the in vivo enhancement of stem cell numbers by GLI1. As expected, the Prominin1+ fraction showed 4–6-fold increased clonogenicity over Prominin1− cells (Figure 2H).
Gene expression signatures of purified Prominin1+ stem cells from DT+dox and DT−dox E17.5 cerebella revealed an enrichment of Gli1, Shh, Ptch1, Ptch2, Nanog and Hes1 and low expression of BMP signalling components in DT+dox stem cells (Figure 2I; Supplementary Figure 6B), showing an action of GLI1 in stem cells. Gene expression analyses of whole E17.5 NS further supported the findings with dissected brain and purified Prominin1+ stem cells, although a number of genes showed variant expression (Supplementary Figure 5), probably reflecting different cellular compositions.
Cooperation of GLI1 and p53 knock-down on neural stem cell self-renewal and tumour growth
The downregulation of p21 and Puma, two p53 targets, in Prominin1+ stem cells of DT+dox cerebella (Figure 2I) is consistent with the finding that p53 negatively controls neural stem cell numbers (Gil-Perotin et al, 2006; Meletis et al, 2006). Because the perinatal lethality of DT+dox mice with strong phenotypes prevented us from analysing DT+dox;p53−/− mice for the possible development of frank tumours and further expansion of stem cell pools, we first tested for the role of p53 on GLI1-induced NS self-renewal.
E17.5 cortical and thalamic DT−dox and DT+dox NS, or wt NS transduced with control or GLI1-expressing lentivectors were further transduced with a control or an shRNA specific for p53 (shp53). Enhanced GLI1 or knock-down (kd) of p53 increased the number of clones as expected, but importantly, their simultaneous action was synergistic (Figure 3A). They also increased proliferation and decreased apoptosis additively (Figure 3B and C). Wt postnatal SVZ NS transduced with lentivectors expressing full-length GLI1 or shPtch1 (to endogenously enhance the Shh-Gli1 pathway) alone and in combination with shp53 yielded similar synergism in clonogenicity and additive results in proliferation and apoptosis (Figure 3D–F). p53 therefore antagonises GLI1 in the control of neural stem cell self-renewal in NS of different brain regions.
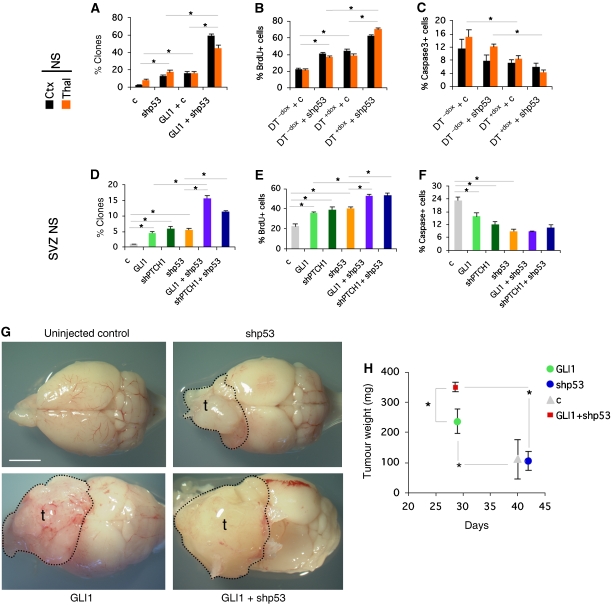
Figure 3.
Synergistic control of neural stem cell self-renewal and xenograft growth by GLI1 and kd of p53. (A–F) Quantification of the behaviour of E17.5 NS from cortex (Ctx) or thalamus (Thal) (A–C) or of first week postnatal SVZ NS (D–F) testing for clonogenicity (A, D), proliferation (B, E) and apoptosis (C, F). Cells were derived from DT mice and treated as indicated or were wt NS transduced with lentivectors as shown. (G) Dorsal views of mouse brains uninjected or injected with transduced U87 cells as indicated. Tumour contours (t) are denoted. (H) Quantification of tumour weights versus time after injection; n=5 for each condition. c=control lentivector. Scale bar=0.4 mm (G).
Following these findings, we tested whether enhanced GLI1 cooperates with p53 kd in tumour growth. As all patient-derived glioblastoma stem cell cultures that efficiently grew in our hands were p53 mutant (M Vucikevic and ARA, personal communication), and as NS from DT+dox mice extinguished transgene expression in vivo after transplantation (not shown), we used human U87 glioblastoma (p53 wt) cells. These cells show the least number of genomic changes among glioma cell lines (Li et al, 2008). Cells transduced with lentivectors expressing GLI1 or GLI1 plus shp53 were grafted intracranially and tumours removed when the mice showed signs of neurological disease. Enhanced GLI1 expression induced an acceleration of tumour development (Figure 3G and H) and an increase in tumour weight as compared with controls. p53 kd alone did not accelerate tumour development, but it synergised with enhanced GLI1 levels (Figure 3G and H). These results are consistent with the acceleration of cerebellar tumours in Ptc1+/−;p53−/− mice as compared with Ptc1+/− siblings (Wetmore et al, 2001), although the bases of such acceleration is unknown.
Balancing SHH-GLI and p53 activities in the control of tumour cell numbers
As expected, kd of HH-GLI signalling with a validated shSMOH (Clement et al, 2007; Stecca et al, 2007) or with validated 21-nt siRNAs against GLI1 (siGLI1; Sanchez et al, 2004; Stecca et al, 2007) decreased U87 cell proliferation and enhanced apoptosis (Figure 4A–C). In contrast, p53 kd increased proliferation of U87 cells (Figure 4A and B). Interestingly, epistatic analyses revealed that shp53-enhanced proliferation was blocked by both shSMOH and siGLI1 but not by control lentivectors or control siRNAs (Figure 4A and B) on the one hand, whereas on the other, cell death induced by shSMOH was blocked by shp53 (Figure 4C), suggesting mutual requirement.
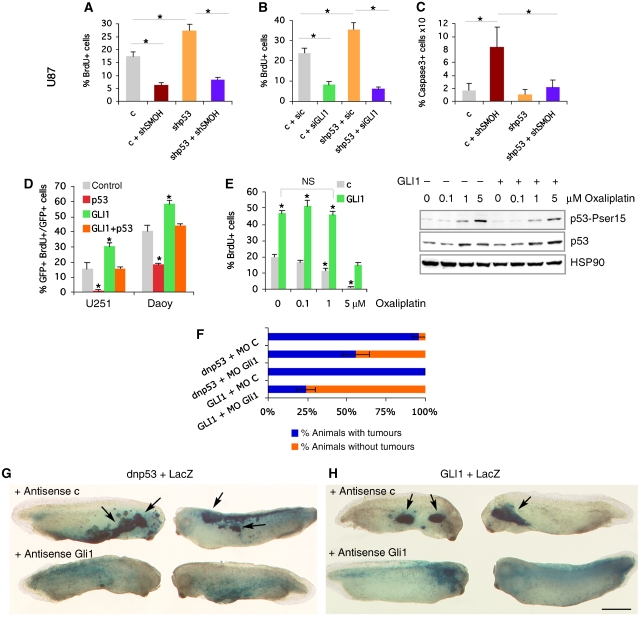
Figure 4.
Mutual requirements of p53 and GLI1 in proliferation and apoptosis. (A–C) Histograms of the changes in cell proliferation (A, B) or apoptosis (C) after altering SMOH, GLI1 or p53 levels by lentiviral transduction or siRNA lipofection in U87 cells. (D) Compensatory effects in cell proliferation in U251 and Daoy cells by GLI1 and p53. (E) Left: Rescue of the anti-proliferative effect of p53 induction by oxaliplatin (48 h) by transduced GLI1 in U87 cells. Comparisons are with untreated (0) control (c) except where noted. Right: Western blot of the levels of p53 and phospho-serine15 p53 induced by oxaliplatin with or without exogenous GLI1. Hsp90 is used as control. (F–H) Quantification (F) and whole-mount side views (G, H) of Xgal-stained stage 32 tadpoles facing the centre of each panel with epidermal hyperplasias (arrows; top rows, G, H) or the normal distribution of cells in the epidermis (bottom rows, G, H) as indicated. Scale bar=0.6 mm (G, H).
Consistent with this idea, restoration of p53 function in p53-mutant brain tumour cells (U251 glioma and Daoy medulloblastoma) reduced, and enhancement of GLI1 levels increased, their proliferation, but these effects were mutually compensated (Figure 4D). Similarly, oxaliplatin treatment of U87 cells increased endogenous p53 levels through the DNA damage response and decreased cell proliferation (Figure 4E), but this proliferative defect and the level of induced phospho-p53 were attenuated in cells with enhanced GLI1 levels (Figure 4E), further suggesting a balance of levels and activities. Enhanced GLI1 levels, however, also induced hallmarks of DNA replication stress (Gorgoulis et al, 2005) that are known to lead to enhanced p53 function in different scenarios: increasing the number of 53BP1 protein foci in 293T cells and increasing the number of phospho-Chk2+ cells in DT+dox hyperplastic thalamic and cerebellar regions (Supplementary Figure 7A, B). Together, these data suggest an endogenous antagonistic relationship between p53 and GLI1 but also the possible induction of p53 function by GLI1 through the DNA replication stress response.
Epidermal hyperplasias induced by interference with p53 function require endogenous Gli1
The blockade of shp53-mediated increased cell proliferation by interference with endogenous SMOH or GLI1 function (Figure 4A and B) suggested that interference with p53 frees endogenous GLI1 from negative regulation, leading to cell overproliferation. As an in vivo assay to analyse this idea, we tested for the role of Gli1 in tumours induced by blockade of p53. Frog embryos expressing dominant negative (Thr280) p53 (dnp53), which inhibits endogenous p53 function, develop epidermal tumours (hyperplasias) (Wallingford et al, 1997). Co-injection of a control antisense morpholino oligonucleotide (MO) along with dnp53 did not modify its tumour-inducing activity (n=18/19 tadpoles with tumours; Figure 4F–H). In contrast, coinjection with a proven MO specific for frog Gli1 (Dahmane et al, 1997, 2001; Nguyen et al, 2005) reduced the development of dnp53-induced tumours by ∼40% (n=21/37). As a control for MO specificity, injection of Gli1 MO (n=6/24), but not the control MO (n=27/27), reduced tumour development by co-injected human GLI1 by 66% (Figure 4F–H; Dahmane et al, 1997, 2001; Nguyen et al, 2005). Blockade of endogenous Gli1 changed the distribution of the descendants of the injected blastomeres from clumped in tumours to evenly in the epidermis, appropriate for the normal fate of the injected blastomeres (Figure 4G and H). Enhanced GLI1 function (through injection) therefore bypasses proliferative restrictions imposed by p53, and blockade of endogenous p53 unleashes the proliferative activity of endogenous Gli1.
p53 negatively regulates the activity, nuclear localisation and levels of GLI1
Gene expression analyses of NS expressing both GLI1 and shp53 revealed context-specific differences in the effect of p53 kd on GLI1-regulated genes (Supplementary Figure 8), for instance, increasing the expression of the stemness genes Prominin1, Bmi1 and Nanog, as well as p16, Gli1, Hes1, Hes5 and other Notch components in different regions, whereas decreasing p21 in cortex and cerebellum, all as compared with NS expressing GLI1.
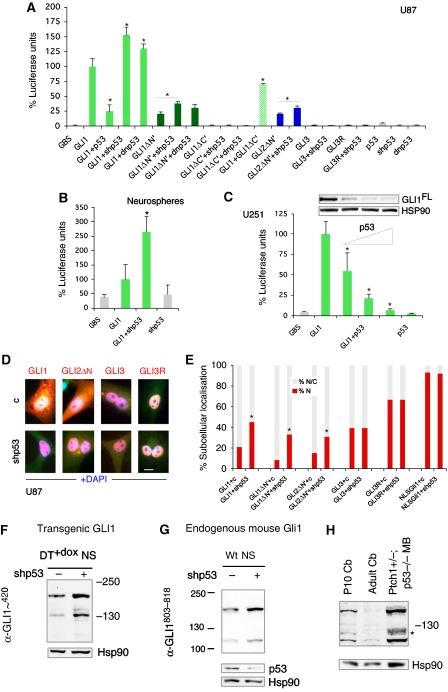
To elucidate how p53 alters the genetic program induced by GLI1, we first asked whether p53 affects GLI1-dependent transcription in GLI-dependent luciferase reporter assays. shp53 increased 2–3-fold the activity of GLI1 in wt postnatal mouse SVZ NS, and this was reproduced in human U87 and in 293T cells (Figure 5A and B; Supplementary Figure 9A). dnp53 behaved similarly and p53 kd also augmented the diminished activity of N'Δ forms of GLI1 and GLI2 (Figure 5A). shp53 did not endow GLI3 or its C'Δ repressor form (GLI3R) with activity (Figure 5A; Supplementary Figure 9A), suggesting that it does not act by inhibiting repressor function. Conversely, restoring p53 activity in U251 and Daoy cells (both p53 mutant) decreased GLI1 reporter activity and the levels of exogenous GLI1 in a concentration-dependent manner (Figure 5C; Supplementary Figure 9B). p53 did not affect the levels of GLI1 mRNA in co-transfected cells (not shown). Enhanced transcriptional activity by shp53 was correlated with two-fold enhanced nuclear localisation of GLI1, of N'Δ forms of GLI1 and GLI2 but not of GLI3, GLI3R or of a nuclear targeted NLS-Gli1 (Ruiz i Altaba, 1999; Figure 5D and E). p53 thus appears to promote the localisation of GLI1, and possibly of GLI2, in the cytoplasm, where it can be degraded by the proteasome (Huntzicker et al, 2006).
Figure 5.
Regulation of GLI1 by p53. (A–C) Quantification of GLI-dependent luciferase reporter assays (A–C) and inhibition of GLI1FL by p53 (C). Luciferase units are GLI reporter firefly/renilla control ratios with the level induced by GLI1 equated to 100%. p53 was used at 1/0.25, 1/0.5, 1/1 maintaining equal amounts of GLI1 (triangle in C). (D, E) Representative images (D) and quantification of GLI nuclear localisation in U87 cells (E). (F–H) Western blots showing the expression of transgenic GLI1 in thalamic DT+dox NS (F), endogenous Gli1 in postnatal SVZ NS (G) and endogenous Gli1 in P10 and adult cerebella as well as in a Ptch1+/−;p53−/− medulloblastoma (MB; H). Transduction with shp53 lentivectors leads to an increase in the abundance of transgenic GLI1 (F) and endogenous Gli1 (G). p53 and Hsp90 are used as controls. Scale bar=15 μm (D).
To test whether p53 generally inhibits GLI1 levels, we analysed GLI1 protein in different contexts. E17.5 thalamic NS of DT+dox brains were transduced with control or shp53 lentivectors and kept in the presence of dox. Western blots with a commercial anti-GLI1 antibody (α-GLI1∼420; Cell Signalling) showed that p53 kd induced a 30% increase in full-length GLI1 (>160 KD) and a three-fold increase of a shorter form running ±130 KD (Figure 5F), quantified by densitometry. Similarly, analysis of postnatal wt SVZ NS for endogenous mouse Gli1 with an anti-GLI1 antibody to the C-terminus (α-GLI1803−818; GeneTex) showed a 40% increase in full-length mouse Gli1 (>160 KD) and a 2.3-fold increase in a shorter form running at ∼115 KD (Figure 5G). As control, and as α-GLI1∼420 does not recognise mouse Gli1 (Figure 6C), we used α-GLI1803−818 to probe extracts of P10 wt cerebellum, adult (∼4 mo) Cb and a Ptch1+/−;p53−/− medulloblastoma (MB) (Figure 5H). As expected from the transient perinatal expansion of cerebellar GLI1+ granule neuron progenitors and the strong expression of GLI1 in MB (Dahmane and Ruiz i Altaba, 1999; Dahmane et al, 2001), the full-length and the shorted 115 KD form were clearly detected at P10 and were nearly undetectable in adult cerebellum, whereas the full-length form was 14-fold and the 115 KD form 6.5-fold more abundant in MB than at P10 (Figure 5H).
Figure 6.
GLI1 protein isoforms and endogenous repression of p53 by GLI1. (A) Transfected myc-GLI1 isoforms detected as indicated in samples treated with cycloheximide (CHX) as shown. GLI1FL migrates >160 KD. (B) Deduced relationship of GLI1130 and GLI1100 to GLI1FL, and location of Ab epitopes and of the transactivating (TA) domain. (C) Analyses of human GLI1 protein in transgenic P0 mice as indicated. All three antibodies used recognise GLI1FL and GLI1130, but only those to the domain immediately C-terminal to the zinc fingers (ZF in B) recognise GLI1100. (D) Analysis of GLI1 isoforms in human cell lines and in a GBM stem cell culture (gliomasphere). (E) GLI1 kd with a specific siRNA for 8 h, but not a control siRNA, depletes all three isoforms in U87 cells, while enhancing total and phospho-serine15 p53 (p53-P) levels. Non-specific bands are denoted by asterisks. (F) Levels of Mdm2 in the blot shown in panel C middle for GLI1 and Hsp90. (G) Comparison of the levels of GLI1130, p53, phospho-serine15 p53 and HSP90 in two gliomaspheres that proliferate at different rates.
GLI1 represses p53
The mutual compensatory effect of GLI1 and p53 (Figure 4D) and the dominance of GLI1 kd over p53 kd (Figure 4B) raised the possibility that in addition to the negative regulation of GLI1 by p53, p53 itself could be under GLI1 regulation. Analyses of p53 in GLI1 kd cells (see below) revealed that endogenous p53 is under the control of endogenous GLI1 as assessed by the levels of total p53 and of active phospho-serine15 p53 (p53-P) in cells lipofected with siGLI1 versus siC (Figure 6E). The kd of endogenous GLI1 function increased p53 ∼5-fold and phospho-serine15 p53 ∼2-fold. This indicates that in addition to the inhibition of GLI1 by p53 described earlier, endogenous GLI1 regulates the levels of endogenous p53, suggesting a negative regulatory loop.
Enhanced GLI1 levels can inhibit p53 regulation in vitro through the activation of the p53 inhibitor Hdm2 (Abe et al, 2008). We find that Mdm2, the mouse homologue of Hdm2, is greatly enhanced in DT+dox versus DT−dox brains (Figure 6F), extending this possible mechanism of action to an in vivo context.
The inhibitory loop between GLI1 and p53 is consistent with the inversely reciprocal levels of GLI1 and p53 observed in two primary glioblastoma stem cell cultures (Figure 6G; Clement et al, 2007). The level of GLI1 correlated with their proliferation rate (Figure 6G). GBM12 harbours a p53 gene with a frameshift mutation corresponding to aa 209 (M Vukicevic and ARA, unpublished) and predicted to encode an unstable protein as no p53 or phospho-serine15 p53 proteins were detected (Figure 6G). GBM13 expresses full-length p53 with an R->H substitution at aa 158 within the DNA-binding domain (M Vukicevic and ARA, unpublished) predicted to yield a protein with compromised transcriptional activity (Kato et al, 2003). Whereas full-length GLI1 protein was barely detectable (not shown), GMB12, which has no p53, showed higher levels of GLI1130 (see below) than GBM13, which expresses p53 with reduced activity.
Identification of human GLI1 isoforms
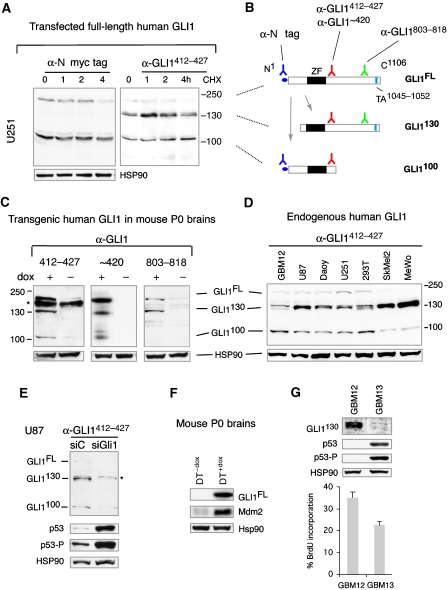
Given the existence of isoforms of GLI1 (Figure 5F–H), reminiscent of those of other Gli proteins, we investigated the possibility that p53 affects their formation or function in human cells. Transfection of U251 glioma cells with full-length myc-tagged GLI1 (Figure 6A, left) revealed the full-length form running at >160 KD (GLI1FL) plus one predominant smaller N-terminal tagged form running at ∼110 KD (GLI1110) after probing the western with α-N' myc tag antibodies. This form has been proposed to act as a C'Δ repressor (Ruiz i Altaba, 1999). Treatment with cycloheximide to block de novo protein synthesis showed increased stability of GLI1100 over GLI1FL (Figure 6A, left). The production of distinct GLI1 isoforms could be limiting in different cells or context dependent as GLI1 transfected in U87 cells yielded the full-length protein but only traces of smaller products (Supplementary Figure 9C). This variance also suggests that the stable products seen in U251 are not non-specific, ubiquitous degradation products.
To directly monitor GLI1 isoforms, we developed an affinity-purified, specific antibody (α-GLI1412−427) against a domain immediately C-terminal to the zinc fingers of human GLI1 that we had previously targeted (Dahmane et al, 1997). Reprobing the anti-myc tag westerns in human U251 cells with α-GLI1 revealed the same full-length and 110 KD isoforms plus a novel ∼130 KD form at similar exogenous levels (Figure 6A, right). This form lacks the myc tags and thus the N-terminus. Endogenous GLI1 was expressed at much lower levels (see below). Treatment with cycloheximide showed differential isoform stability: GLI1130 peaked at 1 h, GLI1100 was largely stable with a 1.6-fold increase at 4 h, and GLI1FL decayed 2.4-fold at 4 h (Figure 6A, right). As the 130 KD N'Δ species cannot give rise to the ∼100 KD C'Δ form, this suggests a precursor-product relationship between the GLI1FL and the GLI1130 form and between GLI1FL and GLI1100 (Figure 6B), with GLI1100>GLI1130≫GLI1FL stabilities, arguing against the first two being non-specific degradation products.
To further characterise the structure of the isoforms, we used two commercial polyclonal antibodies against GLI1: α-GLI1∼420 made to the same region as our α-GLI1412−427 and the C-terminal antibody α-GLI1803−818. Western blots of P0 DT+dox and DT−dox brain extracts revealed the presence of GLI1FL, GLI1130 and GLI1100 only after induction with dox with α-GLI1412−427 and α-GLI1∼420, confirming specificity (Figure 6C). α-GLI1412−427 also picked up a non-specific band in mouse brain extracts (asterisk, Figure 6C). In contrast, α-GLI1803−818 picked up GLI1FL and GLI1130 but not GLI1100 (Figure 6C) (and minor background bands), consistent with the C' truncation of GLI1100 and the N' truncation of GLI1130 (Figure 6B). Probing with a different polyclonal C-terminal antibody (α-GLI1805−820; Chemicon) made to the same region gave the same results (not shown).
To analyse whether endogenous human GLI1 isoforms exist, we probed western blots of U87 cells that are known to harbour an active HH-GLI pathway (Clement et al, 2007) with our own and seven commercial α-GLI1 antibodies (see Materials and methods). Only α-GLI1412−427 yielded the expected pattern of endogenous isoforms (Figure 6D; Supplementary Figure 9D and not shown) seen previously (Figure 6A and C). The other antibodies revealed various non-consistent patterns (not shown). Thus, whereas many α-GLI1 antibodies recognise enhanced levels of GLI1, only α-GLI1412−427 recognises endogenous human GLI1.
Analysis of a patient-derived glioblastoma stem cell culture (GMB12; Clement et al, 2007) and a panel of human tumour cell lines with α-GLI1412−427 revealed all isoforms with GLI1130>GLI1100>GLI1FL abundances (Figure 6D). GLI1130 was most prevalent in metastatic melanomas (which harbour active GLI1; Stecca et al, 2007), and it was consistently detected as a doublet (Figure 6D). Endogenous human GLI1FL and GLI1100 do not have N' myc tags and are therefore slightly smaller than their tagged exogenous counterparts (Figure 6A and C). Commercial westerns further confirmed the presence of a GLI1 ∼130 KD form in various human tissues and tumours with a possible tumour-specific alteration in GLI1 (Supplementary Figure 9E, F).
To verify the identity of the isoforms recognised by α-GLI1412−427, we used RNAi to kd endogenous GLI1. Lipofection of a specific siRNA previously proven to target GLI1 mRNA (siGLI1; Sanchez et al, 2004; Clement et al, 2007; Stecca et al, 2007) but not of a control siRNA (siC), knocked-down GLI1FL two-fold, GLI1130 13-fold and GLI1100 4.4-fold. The levels of HSP90 were unaltered (Figure 6E).
Constructs that approximate the GLI1 N' and C' isoforms have divergent functions
To address the possible functional significance of GLI1 isoforms, we analysed the activity of GLI1 constructs that approximate N'Δ GLI1130 and C'Δ GLI1100 (GLI1N'Δ and GLI1C'Δ; Ruiz i Altaba, 1999). Consistent with the presence of the transcriptional activation domain close to the C-terminus (Figure 6B; Yoon et al, 1998), these constructs behaved as weak activators or repressors, respectively, in GLI1-dependent reporter assays (Ruiz i Altaba, 1999) (Figure 5A). The strength of GLI1N'Δ activator was context dependent, as its activity was 1.2-fold weaker than GLI1FL in melanoma SkMel2 cells but 3.4-fold stronger in prostate cancer LNCaP cells (not shown), and similarly or more active in other cell types (Yoon et al, 1998; Sasaki et al, 1999).
p53 and HH signalling regulate the status of GLI1130
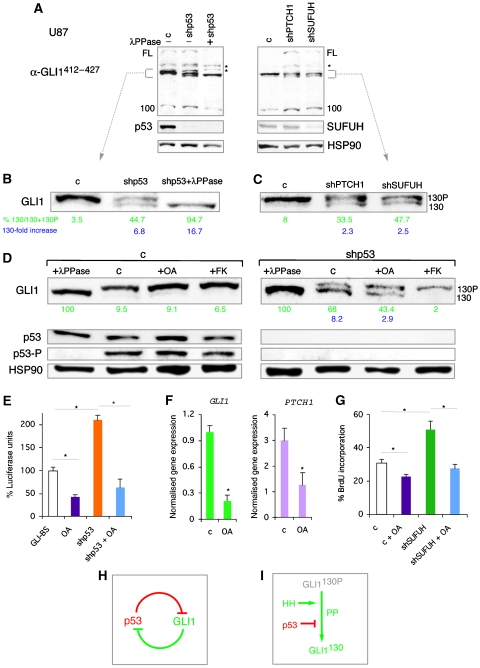
The existence of GLI1 isoforms raised the possibility that p53 could alter their genesis or levels. Treatment of U87 cell extracts in vitro with λ phosphatase followed by western blotting with α-GLI1412−427 revealed the conversion of the upper band of the GLI1130 doublet to the lower one (Figure 7A and D), indicating that the upper band is a phosphoform of GLI1130 (GLI1130P). GLI1100 decreased slightly but did not change size (Figure 7A), and the GLI1FL band became fainter (Figure 7A and not shown), suggesting that it might also be affected although its rarity made further assessment not possible. Consistently, however, the level of exogenous GLI1FL decreased with lambda phosphatase treatment, suggesting that it is phosphorylated and highly unstable in its dephosphorylated form (Supplementary Figure 9G). GLI1FL and GLI1130P, but not of GLI1130, were detected in a phosphoprotein selected pool, confirming their phosphorylated status (Supplementary Figure 9H).
Figure 7.
Regulation of GLI1 isoforms and phosphorylation status of GLI1130 by p53 and HH signalling. (A) Effects of λ phosphatase treatment (+λPPase) or of shp53, shPTCH1 or shSUFUH. Endogenous p53 and SUFUH levels are shown. (B, C) Enlargements of the ±130 KD area in (A). The %GLI1130 calculated by densitometry in relation to total GLI1130+GLI1130P values is green. The fold increase in GLI1130/control is blue. (D) Effects of shp53, and of okadaic acid (OA) and forskolin (FK) for 16 h, on GLI1130 isoforms in U87 cells. λPPase treatments and p53 and p53-P levels are shown as controls. (E) Quantification of endogenous GLI—luciferase assays in control U87 cells and after 16 h OA treatment in wt or shp53-expressing cells. (F) RT–qPCR of control and OA-treated U87 cells after 12 h showing decreased expression of GLI1 ( × 10−4) and PTCH1 ( × 10−3). (G) Quantification of BrdU incorporation in control and shSUFUH-transduced U87 cells treated or untreated with OA. (H) GLI1-p53 inhibitory loop model. (I) Proposed regulation of GLI1130 by PP2A or a similar enzyme, HH and p53 activities.
The kd of p53 in U87 cells increased the levels of GLI1130 ∼6–8-fold at the expense of GLI1130P. Treatment with λ phosphatase confirmed that GLI1130P is the phosphoform of GLI1130 (Figure 7B and D). Endogenous GLI1FL and GLI1100 were modestly increased by p53 kd, consistent with the increase in exogenous GLI1FL levels by p53 kd (Figure 5F; Supplementary Figure 9C). Similar results were obtained in glioma D54 (p53 wt) cells (not shown). The data raise the possibility that GLI1130P is a latent and stable form, the conversion of which to its unphosphorylated, labile and active form (GLI1130) is antagonised by p53.
To begin to investigate the mechanisms regulating the balance of GLI1130 and GLI1130P, we focused on phosphatase (PP) activity given the suggested involvement of PP2A in Hh signalling (Krishnan et al, 1997). U87 and U251 cells were treated with low doses of okadaic acid (OA, 10 nM, 16 h) and extracts subjected to western blotting. OA is a serine/threonine protein phosphatase inhibitor with high affinity for PP2A and much lower affinities for PP1 and PP2B. OA increased (50–80%) GLI1130P in control U87 cells (Figure 7D). In p53 kd cells, which have increased levels of GLI1130, OA treatment reversed the 130P/130 ratio (Figure 7D), decreasing GLI1130 by ∼40–50% and augmenting GLI1130P ∼2-fold. Similar results were obtained in U251 cells (not shown). The very low levels of endogenous GLI1FL prevented us from assessing changes after OA treatment (not shown).
To correlate the levels of GLI1130P with transcriptional activity, endogenous GLI-dependent reporter function was measured in OA-treated U87 cells. OA reduced reporter levels by 60–70% and abolished the increase induced by shp53 (Figure 7E), paralleling the western blot results. Moreover, OA treatment repressed the endogenous levels of both GLI1 and PTCH1 mRNAs (Figure 7F), confirming the downregulation of the endogenous HH-GLI pathway.
We also analysed GLI1 isoforms after increasing the strength of HH signalling through kd of the major negative regulators PTCH1 or SUFUH, by RNAi (with shPTCH1 or shSUFUH). Such interference resulted in a ∼2.5-fold upregulation of GLI1130 at the expense of GLI130P mimicking the effect of p53 kd (Figure 7B and C). Enhanced HH signalling also increased GLI1FL and GLI1100 levels by 3–5-fold and 2.5–3-fold, respectively (Figure 7B and C).
These results suggested that OA should inhibit the proliferative effects of enhanced HH pathway activation. The increase in BrdU incorporation in U87 cells driven by shSUFUH was reverted by OA treatment (Figure 7G). OA also inhibited untransduced U87 cell proliferation (Figure 7G), which is known to depend on an active HH-GLI pathway (Clement et al, 2007). GLI1130 thus appears to depend on the action of PP2A or a similar enzyme, being positively regulated by HH signalling and inhibited by p53 (Figure 7H).
The kinases and mechanisms that yield GLI1130P are not known. cAMP-dependent Protein Kinase A (PKA) blocks HH signalling (e.g., Epstein et al, 1996), and its enhancement with forskolin (FK), a selective activator of adenyl cyclase, inhibits GLI1 activity (Ruiz i Altaba, 1999; Kaesler et al, 2000; not shown). PKA promotes GLI1 phosphorylation and its localisation in the cytoplasm, where it can be degraded (Sheng et al, 2006). We therefore tested whether enhanced PKA activity by FK treatment could have the same effect on GLI1130 as inhibiting endogenous PP2A and/or similar phosphatase activity by OA. FK treatment of wt U87 cells yielded similar levels of GLI1130P as control or OA-treated cells (Figure 7D). However, FK treatment of cells expressing shp53 did not reverse the GLI1130P/GLI1130 ratio to the extent that OA treatment did. Rather, GLI1130 was largely eliminated by FK treatment (Figure 7D). The data raise the possibility that the mechanisms by which PKA antagonises GLI1 activity include the degradation of the GLI1130 activator form.
Discussion
Here we show that GLI1 is under negative control by p53 in multiple systems, and that GLI1 and p53 form a novel homeostatic inhibitory loop that normally controls precursor and stem cell numbers. We propose that its modification alters the behavior of stem cell lineages leading to increases or decreases in cell numbers, such as those in cancer or aging, respectively.
The level of GLI1 activity determines brain size and neural stem cell self-renewal
GLI1 is the last mediator of HH signals, acts as a nexus to integrate non-HH signals such as RAS, and is an emerging central regulator of stemness and cancer in multiple contexts (reviewed in Ruiz i Altaba et al, 2007). We find that the level of GLI1 in transgenic mice controls precursor and stem cell numbers in multiple brain regions. Consistent with this, GLI1 induces a stemness signature that includes Nanog, Klf4 and Sox2, which parallels that of glioma cancer stem cells (Clement et al, 2007) and includes genes involved in reprogramming (Takahashi and Yamanaka, 2006). As the majority of GFP+ cells in the DT+dox SVZ are not GFAP+, a marker of postnatal semiquiescent stem cells (Doetsch et al, 1999), enhanced GLI1 levels could thus increase the number of actively dividing stem cells. Such increases in brain stem cells and precursors are concomitant with the region-specific modulation of several signalling pathways. Notably, GLI1 upregulates Notch and downregulates critical BMP signalling components, in line with the requirement of Notch–Jagged signalling in neural stem cell self-renewal (Nyfeler et al, 2005), and the pro-differentiative action of BMP signalling on stem cells (Panchision et al, 2001). Moreover, the strong upregulation of Hes1 that we detect in vivo and in cerebellar stem cells is consistent with it being a Notch and Gli target (Ingram et al, 2008; V Wallace, personal communication) and is interesting in view of its recent implication in the prevention of senescence (Sang et al, 2008).
We suggest a role for endogenous Gli1 in neural stem cells as enhanced GLI1 levels expand the number of clonogenic stem cells, and Prominin1+ stem cells from DT+dox cerebella upregulate endogenous Gli1 while downregulating Gli2 and Gli3 (this work). Moreover, even though it is dispensable for mouse development (Park et al, 2000), acute interference with endogenous Gli1 in mouse hippocampal NS decreases clone numbers (Galvin et al, 2008), and SVZ Gli1−/− mutant mouse NS show a 40% reduction in secondary clonogenicity (M Vukicevic and ARA, personal communication). Gli1 may, therefore, normally contribute to the control of stem cell lineage longevity, acting to balance antagonising p53 function, consistent with the finding that loss of p21, a p53 target, transiently enhances the self-renewal of neural stem cells but then leads to their premature exhaustion (Kippin et al, 2005).
The balance of antagonistic GLI1 and p53 functions regulates cell numbers
GLI1 is tightly controlled by multiple mechanisms, and here we show that a novel critical modulator of GLI1 in precursors, stem and tumour cells is p53, a multifunctional tumour suppressor (Harris and Levine, 2005). Our data suggest a model for a novel homeostatic mechanism in which p53 and GLI1 form a regulatory loop (Figure 7H). Here, any inappropriate elevation of GLI1 would induce the DNA damage (replication stress) response, which leads to activation of p53, and elevated p53 would then decrease GLI1 activity. At the same time, however, endogenous GLI1 antagonises endogenous p53, possibly through its regulation of BMI1 or its activation of the p53 inhibitor Hdm2/Mdm2 (Abe et al, 2008; this work).
Normal organ-specific regulation of this loop may act to maintain precursors and stem cells in their appropriate numbers to insure life-long persistence and constant organ size, while preventing tumourigenesis. Interestingly, we found that synergistic enhancement of stem cell numbers by GLI1 and p53 kd increased expression of p16INK4a. As increasing p16INK4a levels mark aging neural stem cells (Molofsky et al, 2006), such synergistic action may induce neural stem cell aging, possibly leading to their eventual exhaustion, consistent with the self-renewal deficit in NS lacking Gli1 (see above). The p53-GLI1 regulatory loop might thus not only normally act to maintain size and shape, preventing cancer, but might also act to time aging.
Control of GLI1 by p53
Detailed analyses of exogenous and endogenous human GLI1 reveal that GLI1 exists in different forms: GLI1FL, GLI1130 and GLI1100 (Figure 6A), with the first two likely acting as activators and the last one as a weak repressor. GLI1130 lacks the N-terminus of GLI1FL, and as it is produced from the full-length cDNA, it is distinct from a differential splice form of GLI1 lacking 128aa from the N-terminus (Shimokawa et al, 2008). Indeed, this spliced form is exceedingly rare (∼19-fold less abundant than GLI1FL on average in cell lines (Shimokawa et al, 2008), whereas GLI1130 is >10-fold more abundant than GLI1FL. The greater abundance of GLI1130 over GLI1FL is consistent with the deduced absence in GLI1130 of two inhibitory sites in the N-terminus of GLI1FL: an N' degron (Huntzicker et al, 2006) and a key binding site for the GLI1 antagonist SUFUH (Dunaeva et al, 2003).
Our data suggest that p53 acts to inhibit GLI1 function in two ways:
(1) p53 represses the transcriptional activity of GLI1FL by reducing its nuclear localisation and protein levels. This type of regulation is seen with exogenous transfected GLI1 and may be operative with the very rare endogenous GLI1FL protein and possibly with GLI1130.
(2) p53 promotes the phosphorylation of the endogenous novel N'Δ GLI1130 isoform. We find that GLI1130 is predominantly found in a phosphorylated state (GLI1130P) and suggest that non-phosphorylated GLI1130 is a labile activator, the abundance of which correlates with activity (Figure 7I). Exogenous GLI1FL also appears to be phosphorylated and endogenous GLI1FL might behave similarly, being subjected also to this second mode of regulation.
Concerning the regulation of GLI1130, p53 could act, directly or indirectly, to antagonise a protein phosphatase, favouring the inactive and more stable GLI1130P. Treatment with low doses of OA increases GLI1130P and decreases GLI1130. As OA has a higher affinity for PP2A than to PP1 or PP2B, and the B56ɛ subunit of PP2A is required for Gli1 function in frog embryos (Rorick et al, 2007), the phosphatase responsible for the dephosphorylation of GLI1130P may be PP2A.
The mechanism of N'Δ truncation, phosphorylation site(s) and kinases involved in the genesis of GLI1130P remain to be determined. PKA is an important antagonist of HH signalling (Epstein et al, 1996), it inhibits GLI1 activity (Ruiz i Altaba, 1999; Kaesler et al, 2000), and we show that activation of endogenous PKA through FK treatment results in the degradation of GLI1130. We suggest that although the formation of GLI1130 depends on phosphatase activity, its degradation may involve PKA. We also suggest that p53, unlike PKA, acts to antagonise GLI1130 by promoting GLI1130P.
As activation of HH signalling (by inhibition of PTCH1 or of SUFUH) also modulates and appears to depend on the level and phosphorylation status of GLI1130, GLI1 is regulated transcriptionally (Lee et al, 1997) and posttranscriptionally by HH signals.
In contrast to the positive role of GLI1130, GLI1100 might act as an immediate and tonic negative feedback, as it may have repressor activity and resembles the C'Δ repressor form of GLI3, but unlike this (Pan and Wang, 2007), it is not inhibited by Hh signalling.
It is possible that GLI2 and GLI3 share similarities with the novel regulation of GLI1 we describe here, as N'Δ forms are strong transcriptional activators and Gli2 is an early mediator of Hh signals (Sasaki et al, 1999), OA treatment can inhibit Shh signalling (Krishnan et al, 1997) and FK treatment phosphorylates Gli2 (Riobó et al, 2006).
Imbalance of GLI1 and p53 activities in disease
The homeostatic balance of the GLI1-p53 regulatory loop that we have uncovered appears to be critical. Whereas its imbalance in favour of p53 could affect stem cell lineages and aging (Serrano and Blasco, 2007), imbalance in favour of GLI1 appears to be predominant in cancer and cancer stem cells. For example, glioma cancer stem cells require sustained HH-GLI function (Clement et al, 2007) and gliomas frequently lack p53 (e.g., Ohgaki and Kleihues, 2007). We suggest that loss of p53, which is a common event in human cancers, unleashes the normally restricted proliferative and self-renewing activities of GLI1, leading to an expansion of cancer stem cells and derived progenitors. Tumor suppressors may thus normally act to regulate stem cell lineages. In addition to p53 control, the positive regulation of GLI1 activity by the major human oncogenes (RAS, AKT; Stecca et al, 2007), and its negative regulation by the tumour suppressor PTEN (Stecca et al, 2007), highlights the exquisite regulation of GLI1 activity and provides a rationale for its widespread involvement in human cancer.
Materials and methods
A full account of all methods is given in Supplementary data.
Transgenic mice, neurospheres and cell lines
Doxycycline-regulated GLI1 transgenic (GFP<-tetO->GLI1) mice were crossed with Nestin->rtTA-IRES-LacZ mice to obtain bigenic mice (Supplementary Figure 1). NS were prepared from brains as in Palma and Ruiz i Altaba (2004), transduced with lentivectors and analysed 4–5 days afterwards.
Plasmids, lentiviral vectors and RNA interference
Transfections were at equimolar amounts unless otherwise specified with pCS2-Myc-tagged human GLI1, GLI1ΔN', GLI2ΔN', GLI3, GLI3R (GLI3C'ΔClaI), frog Gli1, frog NLSGli1 (Ruiz i Altaba, 1999), wt human p53, human dnp53 (p53Thr280), pCMV-EGFP or pCMV-LacZ.
Lentiviral vectors were pLV-CTH-shSMOH (Clement et al, 2007); pLV-CTH-shPTCH1 with targeting sequence 5′-GCACTATGCTCCTTTCCTC-3′; pLV-WPXL-shp53 (targeting human p53); pLV-Sico-shp53 (targeting mouse p53 Addgene); pLV-KO.1-puro-shSHFUH (Sigma); and pLV-GLI1 was in pLV-Lox-CW or pLV-TWEEN. siRNA sequences for human GLI1 and control siRNA were as described in Sanchez et al (2004).
Luciferase reporter assays and isolation of Prominin1+ cells
GLI-binding site luciferase reporter assays were as described in Sasaki et al (1999). Cortical and SVZ NS were transfected by nucleofection (Amaxa). Cells of DT+dox and DT−dox brains were dissociated, labelled with Prominin1-microbeads and purified in a magnetic-activated cell-sorting column (Miltenyi Biotec).
Intracranial xenografts and RNA microinjection
Lentivector-transduced U87 human glioblastoma cells (105) were grafted using a stereotaxic apparatus (L2,P1,D2 relative to Bregma). Synthetic RNA and MO (Genetools) microinjection into four-cell frog embryos was performed as described in Dahmane et al (1997, 2001).
Immunohistochemistry, in situ hybridisation and PCR
Cryostat sections were Xgal stained or immunostained with antibodies as noted. In situ hybridisation with digoxygenin-labelled RNA probes was as described in Dahmane et al (1997, 2001). Quantitative rtPCR with cDNA was carried out at 60°C using iQTm SYBR green mix (Bio-Rad).
Western blotting
GLI1 protein was detected in westerns with anti-myc tag or anti-GLI1 antibodies and ECL chemiluminescence (Amersham), or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) for endogenous GLI1.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure 9
Supplementary data
Acknowledgments
We are grateful to N Dahmane, T Halazonetis and all Ruiz i Altaba laboratory members for discussion; M Vukicevic for p53 sequencing data; C Mas for GLI1 lentivectors; G Zacchetti for technical help; T Halazonetis for antibodies; J Massagué for p53; M Serrano for protocols; D Trono and P Salmon for lentivectors; P Vize for dnp53; N Dahmane for U251 and Daoy cells, and DD Bigner for D54 cells. Grant support was from the NIH-NINDS, SNF, Oncosuisse, and the Louis-Jeantet Foundation to ARA.
References
- Abe Y, Oda-Sato E, Tobiume K, Kawauchi K, Taya Y, Okamoto K, Oren M, Tanaka N (2008) Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc Natl Acad Sci USA 105: 4838–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster R, Mullor JL, Ruiz i Altaba A (2000) Gli2 functions in FGF signaling during antero-posterior patterning. Development 127: 4395–4405 [DOI] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A (2007) HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 17: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A (1997) Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature 389: 876–881 [DOI] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A (1999) Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126: 3089–3100 [DOI] [PubMed] [Google Scholar]
- Dahmane N, Sánchez P, Gitton Y, Palma V, Sun T, Beyna M, Weiner H, Ruiz i Altaba A (2001) The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 128: 5201–5212 [DOI] [PubMed] [Google Scholar]
- Dennler S, André J, Alexaki I, Li A, Magnaldo T, ten Dijke P, Wang XJ, Verrecchia F, Mauviel A (2007) Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res 67: 6981–6986 [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A (1999) Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97: 703–716 [DOI] [PubMed] [Google Scholar]
- Dunaeva M, Michelson P, Kogerman P, Toftgard R (2003) Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem 278: 5116–5122 [DOI] [PubMed] [Google Scholar]
- Epstein DJ, Marti E, Scott MP, McMahon AP (1996) Antagonizing cAMP-dependent protein kinase A in the dorsal CNS activates a conserved Sonic hedgehog signaling pathway. Development 122: 2885–2894 [DOI] [PubMed] [Google Scholar]
- Farin A, Suzuki SO, Weiker M, Goldman JE, Bruce JN, Canoll P (2006) Transplanted glioma cells migrate and proliferate on host brain vasculature: a dynamic analysis. Glia 53: 799–808 [DOI] [PubMed] [Google Scholar]
- Galvin KE, Ye H, Erstad DJ, Feddersen R, Wetmore C (2008) Gli1 induces G2/M arrest and apoptosis in hippocampal but not tumor-derived neural stem cells. Stem Cells 26: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Gil-Perotin S, Marin-Husstege M, Li J, Soriano-Navarro M, Zindy F, Roussel MF, Garcia-Verdugo JM, Casaccia-Bonnefil P (2006) Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J Neurosci 26: 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Mile L, Higgins KM, Scott MP (1997) Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277: 1109–1113 [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913 [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899–2908 [DOI] [PubMed] [Google Scholar]
- Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, Oro AE (2006) Dual degradation signals control Gli protein stability and tumor formation. Genes Dev 20: 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram WJ, McCue KI, Tran TH, Hallahan AR, Wainwright BJ (2008) Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signalling. Oncogene 27: 1489–1500 [DOI] [PubMed] [Google Scholar]
- Kaesler S, Lüscher B, Rüther U (2000) Transcriptional activity of GLI1 is negatively regulated by protein kinase A. Biol Chem 381: 545–551 [DOI] [PubMed] [Google Scholar]
- Kasper M, Schnidar H, Neill GW, Hanneder M, Klingler S, Blaas L, Schmid C, Hauser-Kronberger C, Regl G, Philpott MP, Aberger F (2006) Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol 26: 6283–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C (2003) Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA 100: 8424–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippin TE, Martens DJ, van der Kooy D (2005) p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev 19: 756–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Pereira FA, Qiu Y, Chen CH, Beachy PA, Tsai SY, Tsai MJ (1997) Mediation of Sonic hedgehog-induced expression of COUP-TFII by a protein phosphatase. Science 278: 1947–1950 [DOI] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH, Schaffer DV (2003) Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci 6: 21–27 [DOI] [PubMed] [Google Scholar]
- Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, Johnson JE, Wechsler-Reya RJ (2005) Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci 8: 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Platt KA, Censullo P, Ruiz i Altaba A (1997) Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development 124: 2537–2552 [DOI] [PubMed] [Google Scholar]
- Li A, Walling J, Kotliarov Y, Center A, Steed ME, Ahn SJ, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA (2008) Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res 6: 21–30 [DOI] [PubMed] [Google Scholar]
- Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V (2006) alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science 311: 1609–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede SM, Nistér M, Lundeberg J, Frisén J (2006) p53 suppresses the self-renewal of adult neural stem cells. Development 133: 363–369 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ (2006) Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V, Chokas AL, Stecca B, Ruiz i Altaba A (2005) Cooperative requirement of the Gli proteins in neurogenesis. Development 132: 3267–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler Y, Kirch RD, Mantei N, Leone DP, Radtke F, Suter U, Taylor V (2005) Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. EMBO J 24: 3504–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P (2007) Genetic pathways to primary and secondary glioblastoma. Am J Pathol 170: 1445–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma V, Lim DA, Dahmane N, Sánchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Alvarez-Buylla A, Ruiz i Altaba A (2005) Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development 132: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma V, Ruiz i Altaba A (2004) Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 131: 337–345 [DOI] [PubMed] [Google Scholar]
- Pan Y, Wang B (2007) A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J Biol Chem 282: 10846–10852 [DOI] [PubMed] [Google Scholar]
- Panchision DM, Pickel JM, Studer L, Lee SH, Turner PA, Hazel TG, McKay RD (2001) Sequential actions of BMP receptors control neural precursor cell production and fate. Genes Dev 15: 2094–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127: 1593–1605 [DOI] [PubMed] [Google Scholar]
- Riobó NA, Lu K, Ai X, Haines GM, Emerson CP Jr (2006) Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci USA 103: 4505–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorick AM, Mei W, Liette NL, Phiel C, El-Hodiri HM, Yang J (2007) PP2A: B56epsilon is required for eye induction and eye field separation. Dev Biol 302: 477–493 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A (1999) Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development 126: 3205–3216 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Mas C, Stecca B (2007) The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol 17: 438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, Hernández AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, Beyna M, Datta MW, Datta S, Ruiz i Altaba A (2004) Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci USA 101: 12561–12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Coller HA, Roberts JM (2008) Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321: 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H (1999) Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126: 3915–3924 [DOI] [PubMed] [Google Scholar]
- Serrano M, Blasco MA (2007) Cancer and ageing: convergent and divergent mechanisms. Nat Rev Mol Cell Biol 8: 715–722 [DOI] [PubMed] [Google Scholar]
- Sheng T, Chi S, Zhang X, Xie J (2006) Regulation of Gli1 localization by the cAMP/protein kinase A signaling axis through a site near the nuclear localization signal. J Biol Chem 281: 9–12 [DOI] [PubMed] [Google Scholar]
- Shimokawa T, Tostar U, Lauth M, Palaniswamy R, Kasper M, Toftgård R, Zaphiropoulos PG (2008) Novel human glioma-associated oncogene 1 (Gli1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J Biol Chem 283: 14345–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet R, Beermann F, Ruiz i Altaba A (2007) Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA 104: 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Seufert DW, Virta VC, Vize PD (1997) p53 activity is essential for normal development in Xenopus. Curr Biol 7: 747–757 [DOI] [PubMed] [Google Scholar]
- Wetmore C, Eberhart DE, Curran T (2001) Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res 61: 513–516 [PubMed] [Google Scholar]
- Yoon JW, Liu CZ, Yang JT, Swart R, Iannaccone P, Walterhouse D (1998) GLI activates transcription through a herpes simplex viral protein 16-like activation domain. J Biol Chem 273: 3496–3501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure 9
Supplementary data