Abstract
Bone undergoes continuous remodelling throughout adult life, and the equilibrium between bone formation by osteoblasts and bone resorption by osteoclasts defines the final bone mass. Here we show that Snail1 regulates this balance by controlling osteoblast differentiation. Snail1 is necessary for the early steps of osteoblast development, and it must be downregulated for their final differentiation. At the molecular level, Snail1 controls bone mass by repressing the transcription of both the osteoblast differentiation factor Runx2 and the vitamin D receptor (VDR) genes in osteoblasts. Sustained activation of Snail1 in transgenic mice provokes deficient osteoblast differentiation, which, together with the loss of vitamin D signalling in the bone, also impairs osteoclastogenesis. Indeed, the mineralisation of the bone matrix is severely affected, leading to hypocalcemia-independent osteomalacia. Our data show that the impact of Snail1 activity on the osteoblast population regulates the course of bone cells differentiation and ensures normal bone remodelling.
Keywords: bone remodelling, osteoblasts, osteoclasts, Runx2, snail, VDR
Introduction
The integrity of the bone depends on the balance between bone resorption by osteoclasts and bone formation by osteoblasts (Karsenty and Wagner, 2002). Indeed, imbalances between these two processes lead to a number of diseases (Rodan and Martin, 2000). Although increased bone mass may be due to osteoclast dysfunction (osteopetrosis) or to high osteoblast activity (osteosclerosis), a reduction in bone mass is usually caused by increased osteoclast number or activity, sometimes accompanied by suppression of osteoblast activity (osteoporosis). Defective mineralisation of the bone matrix associated with defects in the vitamin D signaling leads to osteomalacia. In addition, osteoclast differentiation directly depends on osteoblast activity (Boyle et al, 2003), as they secrete osteoclastogenic factors, such as the receptor for activation of nuclear factor kappa B ligand (RANKL), macrophage colony stimulating factor (M-CSF) and osteoprotegerin (OPG) (Lagasse and Weissman, 1997; Lacey et al, 1998). Thus, a tight control of osteoblast differentiation is necessary to ensure the appropriate balance between bone formation and bone resorption in the adult.
Runx2 is the main transcriptional regulator of osteoblast differentiation. It is required for both the commitment of mesenchymal cells to the osteoblast lineage and the late stages of osteoblast differentiation and bone mineralisation (Ducy et al, 1997; Komori et al, 1997; Otto et al, 1997; Nakashima and de Crombrugghe, 2003; Xiao et al, 2005). Although the mechanisms that control Runx2 activity at the post-translational level are already known (Bialek et al, 2004; Jones et al, 2006), regulators of its transcription in osteoblasts remain to be identified.
Snail genes are transcriptional repressors best known by their ability to trigger the epithelial to mesenchymal transition (EMT), endowing epithelial cells with motility and invasive properties (Barrallo-Gimeno and Nieto, 2005; Peinado et al, 2007). In addition to the regulation of cell movements and adhesion, Snail factors have a broad spectrum of biological functions, including the regulation of cell proliferation and survival (Vega et al, 2004). Interestingly, Snail also functions in non-epithelial cells, such as chondrocytes, where it is unable to induce EMT but still controls proliferation. Indeed, its deregulated expression in the developing bone leads to achondroplasia in transgenic mice, the most common form of dwarfism in humans (de Frutos et al, 2007). Achondroplasias are associated with activating mutations in FGFR3, which signal in a ligand-independent manner to impair chondrocyte proliferation and differentiation (Ornitz and Marie, 2002). Snail1 is the transcriptional effector of FGFR3 signaling during bone development and disease, and its activity can be inversely correlated with the length of the long bones (de Frutos et al, 2007). As in other tissues, Snail1 expression is very tightly regulated in the bone, and thus, we wondered whether its aberrant activation in the adult had any impact on bone homeostasis. Indeed, our data highlight the fundamental role of Snail1 in controlling bone mass by acting as a repressor of both Runx2 and VDR transcription during osteoblast differentiation.
Results
Sustained Snail1 activation in osteoblasts leads to osteomalacia in transgenic mice
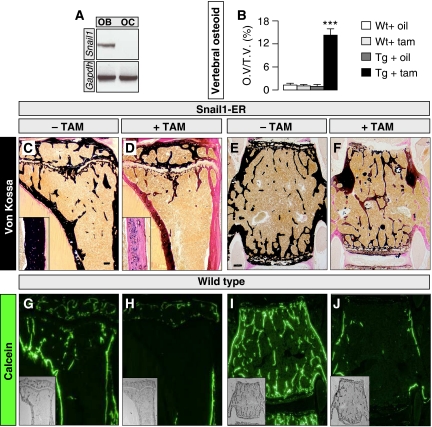
We showed recently that Snail1 activity regulates longitudinal bone growth by controlling chondrocyte proliferation and differentiation (de Frutos et al, 2007). However, Snail1 transcripts are also found in the perichondral area (Supplementary Figure 1A), suggesting that it might also be expressed by osteoblasts. Indeed, we have found intense Snail1 expression in osteoblasts from mouse calvaria (Figure 1A). As Snail transcription factors are pleiotropic proteins involved in different cellular processes depending on the cell context (Barrallo-Gimeno and Nieto, 2005), we examined whether Snail1 might also fulfil additional roles in bone formation. To address this issue, we took advantage of our conditional transgenic mouse model in which Snail1 could be activated in the developing bones (Snail1-ER; de Frutos et al, 2007; see also Materials and methods and Supplementary Figure 1). To avoid interference with the effects of Snail1 on longitudinal bone growth (de Frutos et al, 2007), we have only analysed adult animals here. In this model, constitutively expressed exogenous Snail1 protein only becomes active on nuclear translocation induced by tamoxifen administration. Accordingly, the transgenic Snail1-ER protein was expressed in the osteoblasts of both the trabecular and the cortical bones, and it was efficiently translocated to the nucleus on tamoxifen administration (Supplementary Figure 1B). On Snail1 activation, cartilage-bone staining highlighted deficiencies in the ossification of the long bones in these mice as assessed by Von Kossa staining (Figure 1C and D and Supplementary Figure 2). The images shown in Figure 1C and D are representative of the phenotype observed in around two thirds of the Snail1-ER tamoxifen-treated mice (20 mice analysed per condition in four independent experiments). Cortical thickness was not affected as assessed in Van Gieson stained transverse sections of the tibias (not shown). In the vertebrae, around 15% of the matrix was not calcified (Figure 1B, E and F). The histomorphometric analysis also revealed that when total bone volume was considered as both mineralised bone and osteoid, as established previously (Parfitt et al, 1987), the total bone volume did not significantly change (Supplementary Figure 3). Similarly, the trabeculae were similar in number and thickness (Supplementary Figure 3). The deficient mineralisation was more severe in the tibia than in the vertebrae and indeed, in around 20% of the treated animals the majority of the cortex and some trabeculae were composed of only osteoid tissue 2 months after Snail1 activation commenced. As the mice had well mineralised bones before tamoxifen administration, it appears that bone remodelling is very rapid. To confirm this, we analysed the rate of remodelling in wild-type mice by injecting calcein for 8 consecutive days into wild-type mice, and assessing the labelling that remained at different time points. This labelling had decreased 1 month after the injection (not shown), and it was greatly diminished after 2 months, particularly in the tibia and the vertebrae (Figure 1G–J). Hence, the strong phenotype that we observed in our transgenic mice was compatible with Snail1 inducing a defect in bone formation, and on the other hand, it is very reminiscent of osteomalacia or defective bone mineralisation (Karsenty and Wagner, 2002).
Figure 1.
Snail1 activation induces a bone mineralisation defect in adult transgenic mice. (A) RT–PCR showing Snail expression in osteoblasts (OB) but not in osteoclasts (OC). (B) Percentage of osteoid volume versus total bone volume (OV/TV) determined histomorphometrically in vertebrae (n=5). (C, D) Von Kossa staining of sections of long bones from 16-week-old Snail1-ER transgenic mice shows defective mineralisation as assessed by the deficit in calcium salt deposits (black staining) after 8 weeks of tamoxifen administration. Insets show higher magnification of the cortical bone regions. (E, F) Von Kossa staining of vertebral sections. Scale bars, 1 mm, ANOVA analysis (***) P<0.001. (G–J) (G, H) Calcein labelling of WT tibiae showing that virtually all trabecular and cortical bone was labelled after 8 days of injections, whereas 2 months after the end of the treatment, labelling was not detectable in the trabeculae and in cortical area located below the growth plate. (I, J) Similar results were obtained in vertebrae in which only scarce calcein labelling was detectable in cortical bone and in the trabecular area 2 months after the end of the treatment.
Snail1 represses VDR transcription in osteoblasts and impairs osteoclastogenesis
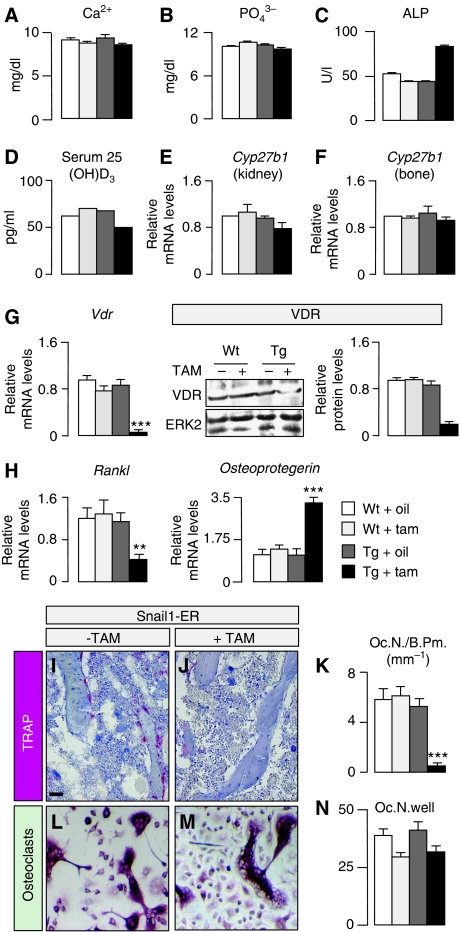
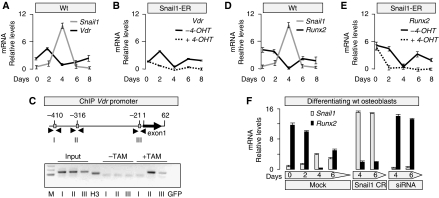
Osteomalacia has been associated with a deficit in vitamin D or its signalling through the VDR receptor (Li et al, 1997; Yoshizawa et al, 1997). However, the primary role of vitamin D signalling in bone mineralisation is to stimulate intestinal absorption of calcium. Indeed, dietary restoration of mineral ion homeostasis prevents osteomalacia in VDR mutant mice (Li et al, 1998; Amling et al, 1999). As Snail1 directly represses the transcription of the Vdr gene in human colon cancer cells (Palmer et al, 2004; Pena et al, 2005), the osteomalacia phenotype of Snail1-ER mice could be due to a deficient VDR expression in the intestine and low calcium absorption. However, as the Snail1-ER transgenic protein was not expressed in the intestine, this does not appear to be the case (Supplementary Figure 4A–E). Consequently, VDR protein expression and distribution as well as the morphology of the intestine were unaffected by tamoxifen administration (Supplementary Figure 4F–M). Consistent with this finding, calcium and phosphate serum levels were normal in the Snail1-ER tamoxifen-treated mice (Figure 2A and B). Moreover, serum levels of creatinine and urea (not shown) were also unaffected, indicating that tamoxifen administration to Snail1-ER mice did not affect the kidneys, as might be expected from the very low levels of transgenic protein in the kidney (not shown). Although the serum levels of alkaline phosphatase were elevated in the tamoxifen-treated mice (Figure 2C), this increase was very modest compared with that observed in VDR mutants (Yoshizawa et al, 1997; Panda et al, 2004). Interestingly, we also found normal serum levels of the vitamin D3 precursor 25(OH)D3 (Figure 2D) and normal expression levels of its hydroxylase Cyp27b1 in both kidney and bone (Figure 2E and F).
Figure 2.
Aberrant Snail1 activation impairs osteoclastogenesis. Calcium (A), phosphate (B), alkaline phosphatase (ALP, C) and 25(OH)D3 (D) concentrations in serum samples from 16-week-old mice after 8 weeks of corn oil or tamoxifen administration. The results are expressed as the means±s.d. from five samples. (E, F) The 25(OH)D3 hydroxylase Cyp271b is unaltered in both the kidney and the bone of Snail1-ER transgenic mice regardless of whether tamoxifen is administered. (G) Snail1 activation inhibits vitamin D receptor mRNA expression and protein accumulation. (H) Snail1 activation is also accompanied by a decrease in the expression of the Rankl osteoclastogenic factor and an increase in that of the osteoclastogenesis inhibitor Osteoprotegerin. The results are expressed as the mean±s.d. from three independent experiments carried out on limbs obtained from 16-week-old Snail1-ER mice after 8 weeks of treatment with tamoxifen. (I, J) Osteoclasts are revealed by tartrate-resistant acid phosphatase (TRAP) staining. Sections are counterstained with alcian blue. (K) Quantification of the osteoclast population by histomorphometric analysis of bones from 16-week-old wild-type and Snail1-ER mice (n=10). (L–N) Osteoclast differentiation in culture is not affected by Snail1 activation. Oc.N., osteoclast number; B.Pm., bone perimeter. Scale bar, 100 μm. ANOVA analysis, **P<0.01 and ***P<0.001.
Although an ion-rich diet can rescue osteomalacia in VDR mutant mice, the number of osteoclasts remains low because VDR signalling is also required in osteoblasts for osteoclast differentiation (Panda et al, 2004). As such, VDR is also expressed in osteoblasts, where it directly activates the promoter of the osteoclast differentiation factor RANKL and downregulates the expression of OPG, a decoy RANKL receptor that inhibits osteoclastogenesis (Simonet et al, 1997; Kitazawa et al, 2003; Kondo et al, 2004). We examined whether Snail1 could repress Vdr transcription in the bone as it does in colon cancer cells, and we found that Snail1 activation on tamoxifen administration did indeed repress of both Vdr transcription and protein accumulation (Figure 2G). This inhibition of Vdr expression impairs the activation of Rankl expression and increases Osteoprotegerin levels (Figure 2H), which could explain the reduction observed in the osteoclast population (Figure 2I–K). We also examined the chondrocytes, as the deletion of VDR in these cells also provokes a reduction in osteoclastogenesis due to decreased RANKL expression (Masuyama et al, 2006). Tamoxifen did not affect Rankl expression (not shown) in chondrocytes differentiated from dissociated 14.5 dpc hindlimb cells obtained from transgenic embryos as described previously (de Frutos et al, 2007). Finally, although endogenous Snail1 is not expressed in the osteoclast population (Figure 1A), we examined whether the transgenic protein might be expressed in these cells, possibly contributing to the impaired osteoclast differentiation observed in the Snail1-ER mice treated with tamoxifen. Transgenic osteoclasts do not express Snail1-ER (not shown) and their differentiation in cultures was similar in wild-type and transgenic animals, although tamoxifen seemed to slightly decrease the efficiency of the process in both cases (Figure 2L–N). Together, these data indicate that on the one hand, the impaired osteoclastogenesis was due to the defects in VDR signalling in osteoblasts that diminished the availability of RANKL, and on the other hand, that there is no cell-autonomous defect in the osteoclast population or defects in VDR signalling in chondrocytes. Thus, with respect to VDR signalling, the Snail1-ER mice represent an osteoblast-specific model for VDR deficiency.
Transient Snail1 expression is required for osteoblast differentiation
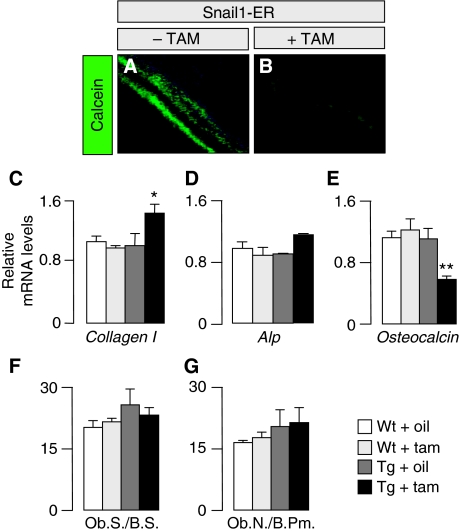
Although bone-specific repression of VDR transcription by Snail1 can explain the defects in osteoclastogenesis in the transgenic mice, it cannot account for the defective mineralisation given that ion homeostasis is maintained. Thus, we decided to assess the rate of bone formation by examining calcein incorporation after its injection in vivo. The strong impairment in calcein apposition observed was compatible with the defective mineralisation observed in these bones (Figure 3A and B), and suggesting that the osteoblasts failed to fully differentiate. Indeed, the bones from Snail1-ER tamoxifen-treated mice expressed higher levels of Collagen I transcripts and lower levels of Osteocalcin expression in Snail1-ER bones (Figure 3C and E), markers of early and late osteoblast differentiation, respectively (Nakashima and de Crombrugghe, 2003), while the number of osteoblasts remained similar (Figure 3F and G).
Figure 3.
Aberrant Snail1 activation impairs calcein apposition. (A, B) Mineral apposition was visualised after calcein injection (n=5 per group). (C–E) Snail1 activation increases the transcription of the early differentiation osteoblast markers Type I Collagen and alkaline phosphatase (Alp; only a slight increase) and decreases that of the differentiated population marker Osteocalcin. Real-time PCR was carried out on limbs obtained from 16-week-old Snail1-ER mice after 8 weeks of treatment with tamoxifen. The results are expressed as mean±s.d. from three independent experiments. (F, G) The number of osteoblasts per bone perimeter is unaltered on Snail1 activation. Histomorphometric analysis of bones from 16-week-old wild-type and Snail1-ER mice (n=10). Ob.S., osteoblast surface; Ob.N., osteoblast number; B.S., bone surface; B.Pm., bone perimeter. ANOVA analysis, *P<0.1 and **P<0.01.
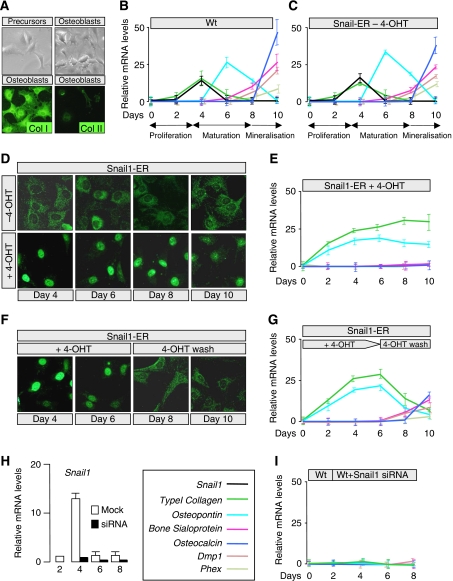
To examine the role of Snail1 in osteoblast differentiation, we developed an in vitro system from embryonic bones in which the stages of proliferation, differentiation and mineralisation could be followed. This culture system accurately reproduced the differentiation pattern described previously (Maes et al, 2007), in terms of osteoblasts differentiated from wild-type embryos or from transgenic Snail1-ER embryos in the absence of tamoxifen (Figure 4A–C). We found that Snail1 was transiently expressed in a profile that closely resembled that of Collagen I, probably reflecting the indirect induction of Collagen I expression by Snail1 as occurs in kidney epithelial cells (Boutet et al, 2006). The expression of phospho-histone 3, p21 or p27 was not altered when cells from transgenic embryonic bones differentiated to osteoblasts in the presence of tamoxifen, indicating that unlike in chondrocytes (de Frutos et al, 2007), Snail1 does not affect osteoblast proliferation (Supplementary Figure 5).
Figure 4.
Snail1 is necessary for the early steps of osteoblast differentiation, and its downregulation is required for terminal differentiation. (A) Primary cultures of wild-type mesenchymal limb cells differentiate into osteoblasts, as confirmed by the expression of the Type I Collagen osteoblast marker and the absence of the Type II Collagen chondrocyte marker. (B, C) Relative mRNA expression of Snail1 and the Type I Collagen, Osteopontin, Bone Sialoprotein, Osteocalcin, Dentin matrix protein 1 (Dmp1) and the transmembrane endopeptidase Phex during osteoblast differentiation in culture of wild-type and Snail1-ER transgenic mesenchymal cells, respectively. (D, E) Snail1 activation prevents osteoblast differentiation in culture. Note the nuclear translocation and ensuing Snail1 activation on tamoxifen administration. (F, G) Osteoblast differentiation occurs in vitro in cultures in which tamoxifen was washed out after 6 days of administration, showing the reversibility of the treatment. Note that Snail1 nuclear translocation is only observed in the presence of 4-OH-Tamoxifen. (H) Snail1 siRNA transfection almost completely blocks Snail1 expression as assessed by real-time RT–PCR. (I) Snail1 silencing prevents osteoblast differentiation, as assessed by the absence of early differentiation markers. Snail1 (black), Type I Collagen (green), Osteopontin (sky blue), Bone Sialoprotein (fuchsia), Osteocalcin (navy blue), Dmp1 (light brown) and Phex (light green).
Transgenic Snail1 activation by nuclear translocation on tamoxifen administration (Figure 4D) was correlated with cell commitment to the osteoblast lineage. Mesenchymal cells were converted into immature osteoblasts as they expressed Osteopontin and upregulated Collagen I expression (Komori, 2008). However, sustained Snail1 activation prevented them from continuing their differentiation (Figure 4E). Significantly, the normal maturation process was recovered on Snail1 inactivation due to its cytoplasmic relocalisation after tamoxifen removal. Collagen I and Osteopontin expression were downregulated, and the transcription of the Bone Sialoprotein, Osteocalcin, Dentin matrix protein 1 (Dmp1) and the transmembrane endopeptidase Phex began to increase (Figure 4F and G; see also Supplementary Figure 6). The expression of ATF4, an osteoblast marker not regulated at the transcriptional level, was unaltered regardless of the state of Snail1 activity (Supplementary Figure 6). In summary, our data indicate that although Snail1 downregulation was necessary for osteoblast differentiation, it might be required for the initial steps in the differentiation process. To assess whether this was indeed the case, Snail1 siRNA was transfected into wild-type primary cultures derived from 14.5 dpc embryonic hindlimbs at the time of endogenous Snail1 induction. Snail1 was almost totally silenced (Figure 4H) and differentiation was completely abolished (Figure 4I). Thus, Snail1 is necessary at early stages of osteoblast differentiation and must be downregulated for differentiation and mineralisation to proceed. Given the role of Snail1 in osteoblast differentiation, it is likely that it also contributes to intramembranous ossification.
Snail1 directly controls Vdr and Runx2 transcription during osteoblast differentiation
Having determined that Snail1 regulates osteoblast differentiation in culture, and given that it is a transcription factor, we set out to define its targets. Having shown that VDR expression was inhibited in bones when Snail1 was activated, we analysed Vdr expression in our osteoblast differentiation system. As expected, Vdr expression inversely correlated with that of Snail1 (Figure 5A) and, furthermore, its transcription remained repressed in the presence of activated Snail1 (Figure 5B). As Snail1 directly inhibits Vdr transcription in colon cancer cells as assessed in promoter activity assays (Palmer et al, 2004), we examined whether it could also bind to its promoter in osteoblasts. Three consensus E-boxes for Snail binding exist in a 0.5-kb fragment upstream of the Vdr coding region (Cano et al, 2000; Figure 5C). Chromatin immunoprecipitation (ChIp) analyses of osteoblasts from Snail1-ER transgenic mice confirmed that Snail1-ER can bind to boxes II and III and as expected, only in the presence of tamoxifen (Figure 5C). In these ChIp experiments, the ER-antibody was used to detect only the exogenous Snail1-ER fusion protein and not the endogenous Snail1 that would bind to the promoter in the presence or absence of tamoxifen administration. These data confirm that Snail1 also binds directly to the Vdr promoter in osteoblasts.
Figure 5.
Snail1 directly regulates Vdr and Runx2 expression during osteoblast differentiation in culture (A) The levels of Snail1 and Vdr expression are inversely correlated during osteoblast differentiation. (B) Sustained activation of Snail1 after 4-OHT administration maintains low Vdr expression. (C) Chromatin immunoprecipitation (ChIP) assays carried out on osteoblasts differentiated from Snail1-ER transgenic mice confirm that Snail1 only binds to boxes II and III in the Vdr promoter on tamoxifen administration. M, fragment length markers; input material was tested for each primer set (boxes I, II and III); H3, positive control of the immunoprecipitate, the sample was immunoprecipitated with anti-H3 antibody and amplified with Gapdh primers; GFP, negative control of the immunoprecipitate, the sample was immunoprecipitated with anti-GFP antibody and amplified with Gapdh primers. (D, E) The levels of Snail1 and Runx2 expression are also inversely correlated during osteoblast differentiation, and similarly, sustained Snail1 activation maintains Runx2 expression inhibited. (F) When osteoblast primary cultures were transfected with a plasmid containing the coding region of Snail1 (Snail1 CR), Runx2 expression was very weak, whereas transfection with a Snail1 siRNA led to stable and strong Runx2 expression.
Besides VDR, Runx2 was a clear candidate target to examine, as it is considered the main regulator of osteoblast differentiation, and it is first required for the commitment of mesenchymal cells to the osteogenic and osteoblast lineage (Ducy et al, 1997; Komori et al, 1997; Otto et al, 1997; Nakashima and de Crombrugghe, 2003). Later on, Runx2 is also required for the conversion of these cells to mature osteocytes and for the expression of mineralisation proteins (e.g., osteocalcin; Xiao et al, 2005). We analysed Runx2 expression in our osteoblast differentiation system and as for Vdr, Runx2 transcription was inversely correlated with that of Snail1 (Figure 5D). This relationship suggested that Snail1 might repress Runx2, compatible with the repression of Runx2 observed in transgenic osteoblasts cultured in the presence of tamoxifen (Figure 5E). Furthermore, Snail1 transfection in differentiating osteoblasts induced sustained repression of Runx2 transcription (Figure 5F), although Runx2 was expressed strongly after transfection of a Snail1 siRNA (Figure 5F). These data indicate that Snail1 may act as a Runx2 repressor, and this led us to assess whether Snail1 could directly repress Runx2 promoter activity.
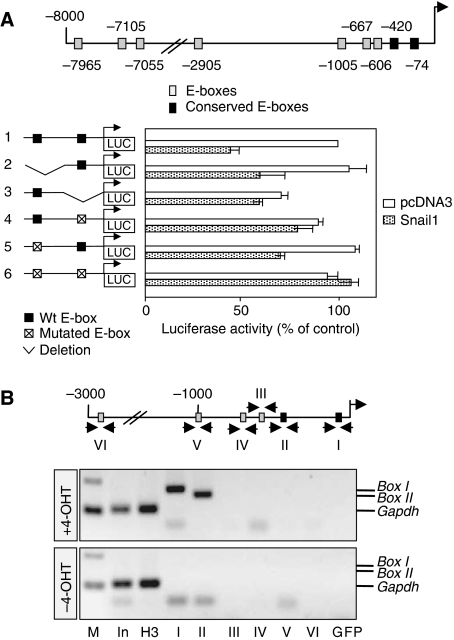
Nine consensus E-boxes for Snail binding exist in an 8-kb fragment upstream of the Runx2 coding region (Figure 6A). However, only the two most proximal boxes were conserved between human and mouse. Using a reporter construct that included these two boxes, we found that Snail1 decreased the activity of the Runx2 promoter in osteoblasts (Figure 6A; construct 1). Although deletion or mutations of the distal E-box of this pair (constructs 2 and 5) did not affect repressor activity, deletion or mutations in the most proximal box significantly relieved the repression of the Runx2 promoter (Figure 6A; constructs 3 and 4). Moreover, simultaneous mutations in the two boxes completely abrogated inhibition. The sequences deleted in construct number 3 (Figure 6A) are likely to contain binding sites for Runx2 activators, as its activity in the absence of Snail1 is much lower than that of the control fragment (construct 1). Together, these data indicate that the binding of Snail1 to the most proximal box is sufficient to inhibit Runx2 promoter activity and that the second box cooperates in this repression.
Figure 6.
Snail1 is a direct repressor of Runx2 transcription. (A) Diagram of the 8-kb region upstream of the translational initiation site in the mouse Runx2 gene showing the putative sites for Snail1 binding (E-boxes). Two of these boxes are conserved in the human (black squares). Luciferase reporter constructs carrying the wild-type Runx2 promoter or deletions/mutations in the two conserved E-boxes were assayed in osteoblasts together with either the mouse Snail1 expression vector or an empty vector as a control (pcDNA3). Luciferase activity was measured 24 h after transfection, and the activity was expressed relative to that of the wild-type construct. The results are expressed as the mean values±s.e. of duplicates from four independent experiments. Snail1 repressed the activity of the wild-type Runx2 promoter, but it did not affect the promoter constructs in which the most proximal conserved E-box was deleted or mutated. (B) Chromatin immunoprecipitation (ChIP) assays carried out in osteoblasts differentiated from Snail1-ER transgenic mice confirm that Snail1 only binds to these two conserved proximal E-boxes and only on tamoxifen administration. M, fragment length markers; In (the input material), immunoprecipitation positive control H3 (sample immunoprecipitated with anti-H3 antibody) and immunoprecipitation negative control GFP (sample immunoprecipitated with anti-GFP antibody) were tested with GAPDH primers. The input material was also tested for each primer set corresponding to the fragments containing all the putative binding sites (see Supplementary Figure 7).
A similar ChIp analysis of osteoblasts from Snail1-ER transgenic mice confirmed that like the Vdr promoter, Snail1-ER can also only bind to these two conserved boxes in the Runx2 promoter in the presence of tamoxifen (Figure 6B). Interestingly, Snail1 is unable to bind to four additional consensus boxes or to the three perfect-match E-boxes contained in 3 kb or between 7 and 8 Kb fragments upstream of the Runx2 coding region, respectively (Figure 6B and Supplementary Figure 7).
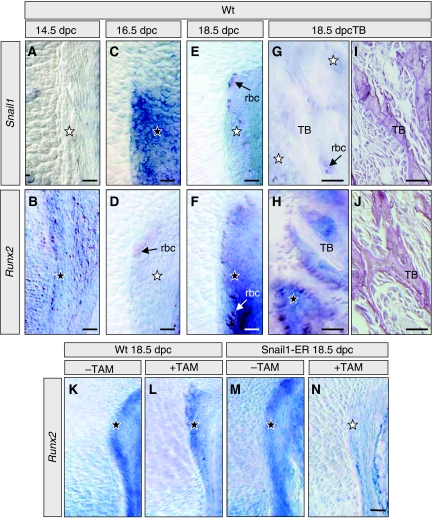
Although our culture system reproduced the described pattern of osteoblast differentiation, we decided to analyse whether Snail1 also behaved as a Runx2 repressor in vivo. We first examined the endogenous pattern of Snail1 expression in the perichondral area of mouse embryos throughout fetal development. We found that Snail1 expression was very dynamic, and that it followed a similar profile to that found in our cultures, with a peak of high expression at 16.5 dpc (Figure 7C). Interestingly, Snail1 expression was inversely correlated with that of Runx2 both in the perichondral area and in the trabecular bone (Figure 7A–J). These data are compatible with the repression of Runx2 by Snail1. As such, we also found that Runx2 is repressed in Snail1-ER transgenic bones on Snail1 activation in vivo (Figure 7K–N). In addition, the direct downstream target of Runx2, Osterix (Nakashima et al, 2002; Nishio et al, 2006), was also inhibited on Snail1 activation in these bones (not shown). These data validate the specificity of the transfection and ChIp assays and confirm that Snail1 directly binds to the Runx2 promoter and represses its transcription during osteoblast differentiation.
Figure 7.
Snail1 activation represses Runx2 expression in vivo. (A–J) Snail1 and Runx2 show excluding expression patterns during osteoblast differentiation in vivo in wild-type mice. (A, B) At 14.5 dpc, only Runx2 is expressed in the perichondrium and in the periosteum (not shown). (C, D) Snail1 is expressed in the perichondrium at 16.5 dpc while Runx2 transcripts are absent. (E, F) At 18.5 dpc, Snail1 expression has decreased in the perichondrium, whereas that of Runx2 has significantly increased. (G, J) At the same stage, Runx2 is expressed in the osteoblasts surrounding the trabecular bone (TB) in the absence of Snail1 transcripts. (K–N) Snail1 activation by tamoxifen administration inhibits Runx2 expression in the perichondrium of Snail1-ER transgenic mice. Black and white stars indicate high and low expression levels, respectively. Arrows point to unspecific signal observed in red blood cells (rbc). Black stars indicate endogenous Runx2 expression, and white star indicate repression of endogenous Runx2 expression by Snail1 activation.
Discussion
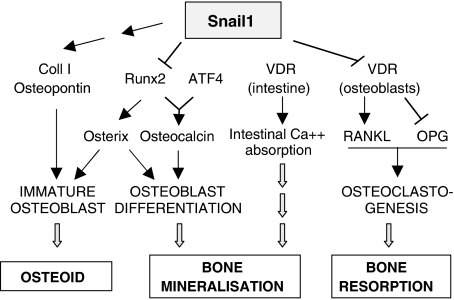
We have shown that Snail1 directly represses both VDR and Runx2 transcription during osteoblast differentiation. The transitory expression of Snail1 in osteoblasts can control the course of differentiation by facilitating an early and a late phase of Runx2 activity. Sustained activation of Snail1 in mice stabilises the normally transient repression of transcriptional cascades downstream of Runx2 and Vdr in osteoblasts, inducing defective mineralisation and impairing osteoclastogenesis, respectively. When coupled with the Snail1-mediated induction of Collagen I, these alterations generate uncalcified matrix or osteoid and thus, osteomalacia independent of hypocalcemia and hypophosphatemia.
Despite the in vivo data obtained in this study come from a transgenic mouse model overexpressing Snail1, we believe that it significantly helps understand the mechanisms that control bone formation and resorption. It reveals an unexpected and important role of Snail1 in osteoblast differentiation in culture, which is compatible with the data obtained in transgenic mice (Figure 8). Although our model is not osteoblast specific, we have ruled out the cell-autonomous contribution of chondrocytes or osteoclasts in these processes in our mice. On the one hand, Snail1-ER protein is not expressed in osteoclasts, and on the other hand, we have not seen any defect in chondrocytes that could explain the observed phenotype. We show that Snail1 controls Vdr expression in osteoblasts. It is known that impaired Vdr signalling in the intestine leads to osteomalacia due to defective calcium absorption in the intestine. However, Snail1 cannot be associated with VDR signalling or calcium uptake in the intestine in healthy conditions, as Snail1 is not endogenously expressed in normal intestinal tissue. Similarly, this transgenic line of Snail1-ER mice does not express Snail1 in the intestine either and maintained normal VDR expression and serum ion levels. Thus, with respect to VDR signalling, Snail1-ER mice provide an osteoblast-specific defective model, where Snail1-mediated repression of Vdr transcription in osteoblasts leads to defects in osteoclastogenesis due to the consequent deregulation of Rankl and Osteoprotegerin. Interestingly, Snail1 is aberrantly activated in colon cancer cells, where it represses Vdr expression (Palmer et al, 2004), as it normally occurs in osteoblasts (this work). Perhaps the benefits of a higher calcium intake associated with a reduced risk of colon cancer detection (Wu et al, 2002) can be related to a compensation of deficient calcium uptake in patients with impaired VDR signalling after aberrant activation of Snail1 as part of the transformation process.
Figure 8.
Diagram showing the central position of Snail1 in the bone remodelling pathways. In wild-type mice, Snail1 is necessary for the first steps of osteoblast differentiation in a dual manner. It activates the expression of the early differentiation markers Collagen I and Osteopontin while it inhibits Runx2 expression, necessary for differentiation to proceed. The transient nature of Snail1 expression allows a second wave of Runx2 transcription needed for bone mineralisation. On the other hand, Snail also directly represses Vdr transcription in osteoblasts providing another link between osteoblastogenesis and osteoclastogenesis. Consequently, sustained Snail1 activation in adult Snail1-ER mice supports the formation of the osteoid but inhibits osteoblast and osteoclast terminal differentiation, leading to a defective mineral deposition.
Importantly, our studies also help to clarify the temporal regulation of Runx2 transcription during normal bone development and homeostasis. Undifferentiated mesenchymal cells have low levels of Snail1, allowing the expression of Runx2 transcripts. However, Runx2 activity is low at these early stages due to its interaction with Twist proteins, which prevent the Runx2 protein from binding to its target promoters (Bialek et al, 2004). When Twist is downregulated, Runx2 becomes functional and by blocking adipocyte and chondrocyte differentiation, it activates osteoblast formation. It is known that Runx2 must be later suppressed for immature osteoblasts to continue differentiating (Komori, 2008), and later on, it needs to be re-expressed as it is essential for terminal differentiation into osteocytes and complete mineralisation (Stein et al, 2004; Xiao et al, 2005). Our data showing the transient nature of Snail1 expression during osteoblast differentiation and its ability to act as a direct transcriptional repressor of Runx2, provide an explanation for this complex dynamics of Runx2 expression throughout the differentiation process. As such, at the time when Runx2 has specified the osteoblast phenotype, Snail1 is activated and transiently represses Runx2 so that differentiation to the immature osteoblast can proceed. In addition to inhibiting Runx2 expression, Snail1 actively favours the formation of immature osteoblasts, as it is necessary for Collagen I and Osteopontin expression (Figure 4H) and represses the expression of the mineralisation genes Phex and Dmp1. Subsequent Snail1 downregulation releases Runx2 transcriptional repression and allows mineralisation to proceed, as this second wave of Runx2 expression is necessary to activate Bone Sialoprotein and Osteocalcin (Stein et al, 2004). On mineralisation, the levels of Runx2 protein are controlled by Schnurri-3, which promotes its degradation in the proteasome (Jones et al, 2006).
In conclusion, we have revealed Snail1 to be the first transcriptional repressor of Runx2, which together with the post-translational regulation co-ordinated by Twist and Schnurri-3 (Bialek et al, 2004; Jones et al, 2006), controls the complex dynamics of Runx2 protein activity during osteoblast differentiation. Snail1 also favours the synthesis of unmineralised matrix by indirectly activating Collagen I expression and it controls osteoclastogenesis by directly repressing Vdr transcription. Thus, Snail1 lies upstream of three fundamental pathways in bone remodelling and the control of adult bone mass (Figure 8). Our data indicate that aberrant expression of Snail1 would lead to osteomalacia even under normal ion homeostasis conditions as it occurs in the presence of vitamin D and a wild-type Vdr gene. In addition, this study provides further insight into the complex regulation of bone formation and resorption.
Materials and methods
Mice
The Snail-ERT2 construct (de Frutos et al, 2007) was microinjected into fertilised C57 × CBA hybrid eggs to generate transgenic mice according to Hogan et al (1994). A line was selected that expressed significant levels of the transgenic protein in the adult bone, immunohistochemically detecting the transgenic protein with a human estrogen-receptor antibody (Santa Cruz). All mice were killed by cervical dislocation.
Induction with tamoxifen or 4-OH-Tamoxifen
Tamoxifen (Sigma) was dissolved in corn oil at a concentration of 30 mg/ml. Corn oil, alone or containing tamoxifen, was injected intraperitoneally every 2 days. Tamoxifen was injected at a concentration of 200 μg or 75 μg/g of body weight to 8-week-old animals for 8 weeks or to pregnant females at 12.5 and 14.5 dpc, respectively. A 1-mM stock solution of 4-OH-Tamoxifen (Sigma) was prepared in ethanol and further diluted to the appropriate concentrations before its use in cultures.
Embryo dissection and in situ hybridisation
Embryos were dissected at 12.5–18.5 dpc and fixed overnight in 4% paraformaldehyde. Hindlimbs were decalcified in 22.5% formic acid and 10% sodium citrate at 4°C for 24 h and subsequently, they were gelatin-embedded before obtaining 30–50 μm vibratome sections. In situ hybridisation was performed as previously described in Cano et al (2000), and the plasmids used to obtain digoxigenin riboprobes corresponded to the following cDNA sequences: Snail1 (1–1600 bp) and Runx2 (484–650 bp).
Skeleton staining
The hindlimbs were dissected and the skin was removed. The limbs were fixed in 10% formalin, and the cartilage was stained with Alcian Blue. After washing, the limbs were trypsinised and stained with Alizarin Red S (for calcium salts), cleared by KOH treatment and stored in glycerol.
Histology and histomorphometric analyses
Non-decalcified bones were embedded in methylmethacrylate according to standard protocols (Parfitt et al, 1987) before processing them for Von Kossa, tartrate resistant acid phosphatase (TRAP) staining or histological analysis. Mice were injected with 100 μl of a calcein solution (5 mg/ml), both 6 and 2 days before they were killed, and they were then processed for plastic embedding as described above. In the bone-remodelling assay, injections were given on 8 consecutive days, and mice were killed 1 week (to confirm labelling efficiency), 1 or 2 months after the last injection. Measurements were performed with the Osteo Measure Analysis System (Osteometrics) using a 3CCD color video DXC-390 camera (Sony) coupled to a microscope (DMLB; Leica). Statistical differences between groups were assessed by the t-test.
Immunohistochemistry
Histological sections were prepared from hindlimbs fixed in 10% formalin that were decalcified with 22.5% formic acid in 10% sodium citrate at room temperature for 16 h, and embedded in gelatin. Gelatin sections (100 μm) were stained using an anti-hER antibody (Santa Cruz) diluted 1:200, and they were developed with the ABC reagent (Pierce) according to the manufacturer's instructions. Other histological sections were prepared from tissues fixed in 10% formalin and embedded in paraffin. Paraffin sections (5 μm) were stained with haematoxylin and eosin or subjected to immunohistochemistry with antibodies against either the anti-human estrogen receptor (α-hER, 1:200; Santa Cruz) or anti-VDR (α-VDR, 1:200; Chemicon) antibodies. The appropriate Alexa 488 secondary antibodies were used diluted 1:5000 in 1% BSA in PBS.
Calvaria primary cultures
Osteoblasts were isolated from enzymatically dissociated calvaria from 3-day-old mice, and they were plated in differentiation medium: α-MEM (Invitrogen) containing 10% heat-inactivated serum (Sigma–Aldrich), 50 μg/ml ascorbic acid and 10 mM sodium β-glycerophosphate. After 24 h, the cells were harvested with 0.01% trypsin in PBS, and they were then plated in 35 mm culture dishes at 2 × 104 cells/dish and grown for 22 days in the differentiation medium. Total cellular mRNA was extracted from primary osteoblasts with RNA-Now (Eurobio, Les Ulis, France).
Primary osteoclast cultures
Osteoclast cell cultures were established with spleen cells from 6- to 8-week-old OF1 male mice or Snail1-ER mice, whereby 2500 or 4000 cells/mm2 were seeded and cultured for 8 days. Bone marrow mononuclear cells from 8-week-old WT or Snail1-ER mice were plated on 96-wells plate at the density ranging from 104 cells to 5 × 104 per well, and the following day (D1) treated with 200 nM tamoxifen (Sigma) or vehicle for 6 days. All cultures were carried out in the presence of α-MEM (Invitrogen) containing 10% (v/v) foetal calf serum (FCS, Biowest, Nuaille, France), M-CSF (20 ng/ml) and RANKL (50 ng/ml) from R&D system. Osteoclast differentiation was evaluated by TRAP staining using the leukocyte acid phosphatase kit from Sigma.
Primary osteoblast cultures
Hindlimbs from 14.5 dpc transgenic or wild-type mouse embryos were dissected in medium (α-MEM, 1% BSA, 0.1% L-Glutamine, 0.1% penicillin/streptomycin) and left overnight at 37°C and 5% CO2. The following day, the bones were trypsinised for 10–15 min at 37°C and digested for 2 h in 3 mg/ml Collagenase P in DMEM with 10% FCS at 37°C. The reaction was inactivated by adding primary culture media (50% F-12, 50% DMEM, 10% FCS, 0.1% L-Glutamine, 0.1% penicillin/streptomycin), and the cells were plated at a density of 1.5 × 106 cells/P100 in primary culture media (Woods and Beier, 2006). After 5 days in culture, the cells were seeded at a density of 1 × 105 per well and differentiated to osteoblasts in differentiation media (50% F-12, 50% DMEM, 0.1% L-Glutamine, 0.1% penicillin/streptomycin, 10 mM β-glycerophosphate, 100 nM dexamethasone, 0.2 mM ascorbic acid (Reyes et al, 2001) in the presence or absence of 200 nM 4-OH-Tamoxifen, and the medium was changed every 2 days. Cells were transfected after 2 days in differentiation medium using Lipofectamine (Invitrogen) with Snail1 siRNA or the random negative control, and/or an expression plasmid containing the Snail1-coding region (CR), RNA was isolated every 2 days using the SIGMA Genelute Mammalian Total RNA Kit according to the manufacturer's instructions. The siRNA duplex oligonucleotide used was described previously (de Frutos et al, 2007). Immunofluorescence was carried out using antibodies against either anti-collagen I or II (1:50, Calbiochem), anti-hER (1:200, Santa Cruz) or anti-PH3 (1:100, Upstate) antibodies and the appropriate Alexa 488 secondary antibodies.
Western blotting
For immunoblotting, cells were lysed in 50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% SDS, 0.5% Deoxycholate and 1% Triton X-100 supplemented with a standard protease inhibitor standard mix (1 mM NaF, 1 mM β-glycerophosphate, 5 mM NaPPi, 5 μg/ml Leupeptine, 1 mM Vanadate and 100 μg/ml PMSF). Total protein (50 μg per lane) was separated on a denaturing 12% SDS–PAGE gel and then transferred to PVDF membranes, which were then blocked with 0.1% Tween-20 and 3% low-fat milk in PBS. The membranes were then incubated in the same solution with anti-hER (1:200; Santa Cruz) or anti-total ERK2 (1:500; Santa Cruz) antibodies, and finally with the appropriate horseradish peroxidase-conjugated secondary antibody. Quantification was performed with the L Process v2.0 software from Bioimager Fujifilm.
Quantitative PCR
Quantitative RT–PCR was carried on an ABI PRISM® 7000 sequence detection system using the Syber Green® method. RNA expression was calculated using the comparative Ct method normalised to GAPDH. In experiments on complete bones, data were expressed relative to a calibrator (wild-type mice without tamoxifen) using the 2−(ΔΔCt)±s.d. formula. For cell culture experiments, data were normalised using the C0±s.d. formula to compare the relative expression between the different genes. Data are represented as the mean±s.d. of triplicates from a representative experiment (n=3). ANOVA analysis, *P<0.1, **P<0.01, ***P<0.001.
RT–PCR
Total mRNA (1 μg) was reverse-transcribed using a random primer and Super Script III RT (Promega). PCR was performed with specific primers for mouse Snail1, Runx2 and Gapdh. The products amplified were separated in agarose gels and quantified with the L Process v2.0 software from Bioimager Fujifilm.
Promoter analyses
Runx2 promoter activity was measured by cotransfecting primary osteoblast cultures with 50 ng of pcDNA3-Snail1 or empty pcDNA3 vectors and 400 mg of pcL2-Runx2 promoter fragments-Luc. A Renilla reniformis luciferase plasmid (phRL-CMV-Luc from Promega) was also cotransfected as a control for efficiency. Firely and renilla luciferase activity were measured 48 h after transfection using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The results are presented as the percentage of Luciferase activity relative to control (Luciferase values in cells cotransfected with an empty vector).
ChIP assays
Cells were crosslinked with formaldehyde before DNA sonication. Chromatin was immunoprecipitated with antibodies against rabbit anti-hER (Santa Cruz), anti-H3 (Abcam), as a positive control, or anti-GFP (Invitrogen), as a negative control. In all cases, chromatin was sheared to an average length of 0.5–1 kb. Primers were designed to amplify fragments of 100–200-bp from the mouse Runx2 promoter containing one of the nine E-boxes or from the mouse Vdr promoter containing one of the three E-boxes identified by sequence analysis. The control samples (immunoprecipitated with H3 or GFP antibodies) were amplified with Gapdh primers.
Supplementary Material
Supplementary Figures 1–7
Acknowledgments
We thank C Lopez for excellent technical assistance, A Muñoz for kindly providing Vdr promoter constructs, MJ Mayol for her help with the serum analyses, all members from M.A. Nieto's lab for helpful discussions and comments and M Sefton for editorial assistance. This work has been supported by the Spanish Ministry of Education and Science (Grants BFU2005-05772, BFU2008-01042, NAN2004-09230-C04-04 and CONSOLIDER-INGENIO 2010 CSD2007-00017) and from the Generalitat Valenciana (Prometeo 2008/049) to MAN. RD is recipient of an INSERM grant.
References
- Amling M, Priemel M, Holzmann T, Chapin K, Rueger JM, Baron R, Demay MB (1999) Rescue of the skeletal phenotype of vitamin D receptor-ablated mice in the setting of normal mineral ion homeostasis: formal histomorphometric and biomechanical analyses. Endocrinology 140: 4982–4987 [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA (2005) The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132: 3151–3161 [DOI] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G (2004) A twist code determines the onset of osteoblast differentiation. Dev Cell 6: 423–435 [DOI] [PubMed] [Google Scholar]
- Boutet A, de Frutos CA, Maxwell PH, Mayol MJ, Romero J, Nieto MA (2006) Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J 25: 5603–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL (2003) Osteoclast differentiation and activation. Nature 423: 337–342 [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol 2: 76–83 [DOI] [PubMed] [Google Scholar]
- de Frutos CA, Vega S, Manzanares M, Flores JM, Huertas H, Martinez-Frias ML, Nieto MA (2007) Snail1 is a transcriptional effector of FGFR3 signaling during chondrogenesis and achondroplasias. Dev Cell 13: 872–883 [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89: 747–754 [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Constantini F, Lacy E (1994) Manipulating the mouse embryo A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH (2006) Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science 312: 1223–1227 [DOI] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF (2002) Reaching a genetic and molecular understanding of skeletal development. Dev Cell 2: 389–406 [DOI] [PubMed] [Google Scholar]
- Kitazawa S, Kajimoto K, Kondo T, Kitazawa R (2003) Vitamin D3 supports osteoclastogenesis via functional vitamin D response element of human RANKL gene promoter. J Cell Biochem 89: 771–777 [DOI] [PubMed] [Google Scholar]
- Komori T (2008) Regulation of bone development and maintenance by Runx2. Front Biosci 13: 898–903 [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764 [DOI] [PubMed] [Google Scholar]
- Kondo T, Kitazawa R, Maeda S, Kitazawa S (2004) 1 alpha,25 dihydroxyvitamin D3 rapidly regulates the mouse osteoprotegerin gene through dual pathways. J Bone Miner Res 19: 1411–1419 [DOI] [PubMed] [Google Scholar]
- Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V et al. (1998) Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93: 165–176 [DOI] [PubMed] [Google Scholar]
- Lagasse E, Weissman IL (1997) Enforced expression of Bcl-2 in monocytes rescues macrophages and partially reverses osteopetrosis in op/op mice. Cell 89: 1021–1031 [DOI] [PubMed] [Google Scholar]
- Li YC, Pirro AE, Amling M, Delling G, Baron R, Bronson R, Demay MB (1997) Targeted ablation of the vitamin D receptor: an animal model of vitamin D-dependent rickets type II with alopecia. Proc Natl Acad Sci USA 94: 9831–9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Pirro AE, Demay MB (1998) Analysis of vitamin D-dependent calcium-binding protein messenger ribonucleic acid expression in mice lacking the vitamin D receptor. Endocrinology 139: 847–851 [DOI] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Kronenberg HM (2007) A novel transgenic mouse model to study the osteoblast lineage in vivo. Ann N Y Acad Sci 1116: 149–164 [DOI] [PubMed] [Google Scholar]
- Masuyama R, Stockmans I, Torrekens S, Van Looveren R, Maes C, Carmeliet P, Bouillon R, Carmeliet G (2006) Vitamin D receptor in chondrocytes promotes osteoclastogenesis and regulates FGF23 production in osteoblasts. J Clinical Inves 116: 3150–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, de Crombrugghe B (2003) Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet 19: 458–466 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29 [DOI] [PubMed] [Google Scholar]
- Nishio Y, Dong Y, Paris M, O'Keefe RJ, Schwarz EM, Drissi H (2006) Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene 372: 62–70 [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Marie JP (2002) FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev 16: 1446–1465 [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771 [DOI] [PubMed] [Google Scholar]
- Palmer HG, Larriba MJ, Garcia JM, Ordonez-Moran P, Pena C, Peiro S, Puig I, Rodriguez R, de la Fuente R, Bernad A, Pollan M, Bonilla F, Gamallo C, de Herreros AG, Munoz A (2004) The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nat Med 10: 917–919 [DOI] [PubMed] [Google Scholar]
- Panda DK, Miao D, Bolivar I, Li J, Huo R, Hendy GN, Goltzman D (2004) Inactivation of the 25-hydroxyvitamin D 1alpha-hydroxylase and vitamin D receptor demonstrates independent and interdependent effects of calcium and vitamin D on skeletal and mineral homeostasis. J Biol Chem 279: 16754–16766 [DOI] [PubMed] [Google Scholar]
- Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR (1987) Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2: 595–610 [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428 [DOI] [PubMed] [Google Scholar]
- Pena C, Garcia JM, Silva J, Garcia V, Rodriguez R, Alonso I, Millan I, Salas C, de Herreros AG, Munoz A, Bonilla F (2005) E-cadherin and vitamin D receptor regulation by SNAIL and ZEB1 in colon cancer: clinicopathological correlations. Hum Mol Genet 14: 3361–3370 [DOI] [PubMed] [Google Scholar]
- Reyes M, Lund T, Lenvik T, Aguiar D, Koodie L, Verfaillie CM (2001) Purification and ex vivo expansion of postnatal human marrow mesodermal progenitor cells. Blood 98: 2615–2625 [DOI] [PubMed] [Google Scholar]
- Rodan GA, Martin TJ (2000) Therapeutic approaches to bone diseases. Science 289: 1508–1514 [DOI] [PubMed] [Google Scholar]
- Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Lüthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, Shimamoto G, DeRose M, Elliott R, Colombero A, Tan HL, Trail G, Sullivan J, Davy E, Bucay N, Renshaw-Gegg L et al. (1997) Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89: 309–319 [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM (2004) Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23: 4315–4329 [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocaña OH, Valdes F, Fabregat I, Nieto MA (2004) Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 18: 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Beier F (2006) RhoA/ROCK signaling regulates chondrogenesis in a context-dependent manner. J Biol Chem 281: 13134–13140 [DOI] [PubMed] [Google Scholar]
- Wu K, Willett WC, Fuchs CS, Colditz GA, Giovannucci EL (2002) Calcium intake and risk of colon cancer in women and men. J Natl Cancer Inst 94: 437–446 [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT (2005) Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem 280: 30689–30696 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Handa Y, Uematsu Y, Takeda S, Sekine K, Yoshihara Y, Kawakami T, Arioka K, Sato H, Uchiyama Y, Masushige S, Fukamizu A, Matsumoto T, Kato S (1997) Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat Genet 16: 391–396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–7