Abstract
The Hsc/Hsp70 co-chaperones of the BAG (Bcl-2-associated athanogene) protein family are modulators of protein quality control. We examined the specific roles of BAG1 and BAG3 in protein degradation during the aging process. We show that BAG1 and BAG3 regulate proteasomal and macroautophagic pathways, respectively, for the degradation of polyubiquitinated proteins. Moreover, using models of cellular aging, we find that a switch from BAG1 to BAG3 determines that aged cells use more intensively the macroautophagic system for turnover of polyubiquitinated proteins. This increased macroautophagic flux is regulated by BAG3 in concert with the ubiquitin-binding protein p62/SQSTM1. The BAG3/BAG1 ratio is also elevated in neurons during aging of the rodent brain, where, consistent with a higher macroautophagy activity, we find increased levels of the autophagosomal marker LC3-II as well as a higher cathepsin activity. We conclude that the BAG3-mediated recruitment of the macroautophagy pathway is an important adaptation of the protein quality control system to maintain protein homeostasis in the presence of an enhanced pro-oxidant and aggregation-prone milieu characteristic of aging.
Keywords: BAG1, p62, proteasome, SQSTM1, ubiquitination
Introduction
Molecular chaperones play a major role in protein quality control (PQC) and the maintenance of protein homeostasis (proteostasis; Balch et al, 2008). These specialised proteins, among them members of the heat-shock protein (Hsp) family, form a complex chaperone network that supports the correct folding and re-folding of proteins (Frydman, 2001; Young et al, 2004). Molecular chaperones also sense misfolded proteins and direct them to protein degradation systems when a native folding state cannot be reached. In this case, the misfolded protein is marked for degradation, generally by polyubiquitination (Rubinsztein, 2006). The main cellular system that degrades polyubiquitinated proteins (polyUb-proteins) is the ubiquitin-proteasome system (UPS). In this pathway, polyUb-proteins are transferred to and degraded by the proteasome, a large barrel-shaped protein complex (Breusing and Grune, 2008). In recent years, it became clear that the autophagic-lysosomal pathway can also degrade polyUb-proteins, a process referred to as macroautophagy (hereafter referred to as autophagy) (Ding and Yin, 2008). During autophagy, protein substrates are first packaged together into inclusion bodies and then engulfed by a double-layered membrane. The resultant vesicle, the autophagosome, then fuses with lysosomes, whereupon the protein content is degraded by lysosomal proteases (Pankiv et al, 2007).
Recent studies implicated several members of the BAG (Bcl-2-associated athanogene) protein family in cellular PQC. The BAG family contains at least six members that can bind through their BAG domain to chaperones of the Hsc/Hsp70 family, thereby modulating chaperone function (Takayama and Reed, 2001). BAG1 has been described to catalyse nucleotide exchange on Hsc/Hsp70 (Hohfeld and Jentsch, 1997). However, it has also been shown that BAG1 can act as a negative regulator of Hsc/Hsp70 by uncoupling nucleotide hydrolysis from substrate release (Bimston et al, 1998). Furthermore, BAG1 has been shown to link Hsc/Hsp70 to the proteasome through its ubiquitin-like (UBL) domain (Luders et al, 2000). The UBL domain is also present in BAG6, suggesting UPS-related function. BAG2 was identified as a specific inhibitor of the chaperone-associated ubiquitin ligase CHIP (Arndt et al, 2005). BAG3 was reported to inhibit proteasomal degradation of Hsp90 client proteins (Doong et al, 2003) and to enhance degradation of polyQ aggregates by autophagy (Carra et al, 2008). BAG5 was identified as an inhibitor of parkin, an E3 ubiquitin ligase associated with genetic forms of Parkinson disease (Kalia et al, 2004). These findings suggest that BAG proteins are modulators of PQC systems and may have a role in degenerative diseases.
Aberrant protein aggregation is characteristic of several age-associated diseases referred to as proteinopathies. In these protein conformation disorders, polyUb-positive aggregates accumulate (Rubinsztein, 2006), indicating a change of the PQC system during aging and/or disease pathology. In this study, we investigated BAG-controlled PQC systems during the aging process. In particular, we focus on the widely expressed BAG isoforms, BAG1 and BAG3, and their specific roles in protein degradation. We show that BAG1 and BAG3 regulate proteasomal and autophagic degradation pathways, respectively, and that a switch from BAG1 to BAG3 during aging triggers degradation of insoluble, aggregated quality control substrates by autophagy. These findings might have important implications for age-related proteinopathies.
Results
BAG1 and BAG3 levels are reciprocally regulated during cellular aging
Earlier studies suggested that members of the BAG protein family are involved in PQC (Takayama and Reed, 2001). To investigate potential alterations of BAG-controlled PQC systems during aging, we used the well-established I90 cell model of aging (Nichols et al, 1977). These primary human cells exhibit the hallmarks of aging over time and become senescent after a finite number of divisions in culture, a process referred to as replicative senescence. Aged cells were identified by the typical large, flat cell morphology and the induction of the aging marker caveolin1 (Supplementary Figure S1A and B). Real-time PCR-based expression profiling of human BAG genes revealed a significant down-regulation of BAG1 and BAG2 as well as an up-regulation of BAG3 during cellular aging (Figure 1B). Transcript levels of other BAG isoforms were unchanged. Immunoblot analysis using an antibody directed against the conserved BAG domain (cBAG) revealed prominent expression of BAG1 and BAG3 in I90 cells (Figure 1A). It should be noted that BAG1 is expressed as four isoforms by alternative translation initiation from one mRNA. Weak bands were observed for BAG1M (46 kDa) and BAG1S (29 kDa), but strong signals for BAG1L (50 kDa) and BAG1 (36 kDa). Consistent with the PCR analysis, we found decreased BAG1 and increased BAG3 protein levels in aged I90 cells (Figure 1A). A similar regulation of BAG1 and BAG3 was also found in the WI38 aging model (Supplementary Figure S1C), indicating a general effect of cellular aging on BAG expression changes. Next, we analysed the expression levels of Hsc/Hsp70 and Hsp90, known to be functionally modulated by BAG proteins (Takayama and Reed, 2001; Doong et al, 2003). We observed a slight decline of both chaperone levels in aged cells (Figure 1A). Transcript levels of Hsp90 were moderately down regulated, whereas Hsc70 mRNA levels were unaltered (Figure 1B).
Figure 1.
(A) Immunoblot analysis of BAG1, BAG3, Hsc/Hsp70 and Hsp90 in young and old I90 cells. For detection of BAG proteins, an antibody directed against the conserved BAG domain was used. (B) Real-time PCR analysis of human BAG family members and Hsc70 and Hsp90 in young and old I90 cells. Depicted is the expression ratio (log2) of target genes in old cells relative to young cells. *P<0.05 and **P<0.01 versus young, n=3. (C) Co-IP analysis of Hsc/Hsp70 interaction with BAG1 and BAG3 in young (Y) and old (O) I90 cells. Upper panel shows relative amounts of proteins in cell lysates used for Co-IP (Input). Middle panel shows levels of BAG1 and BAG3 found in Hsc/Hsp70 immunoprecipitates. Lower panel shows levels of Hsc/Hsp70 co-sedimented upon immunoprecipitation of BAG3. (D, E) 293 cells were treated with the indicated agents for 8 h followed by immunoblot analysis of indicated proteins.
The altered BAG expression profile suggested an altered interaction of BAG proteins with Hsc/Hsp70 in aged cells. Indeed, co-immunoprecipitation (Co-IP) studies revealed that more BAG3 was associated with Hsc/Hsp70 in old cells compared with young cells (Figure 1C). In contrast, association of BAG1 with Hsc/Hsp70 was decreased. It should be noted that the BAG1L isoform could not be appropriately analysed because of cross-reactivity of the secondary antibody with the IgG heavy chain of the Hsc/Hsp70 antibody that co-migrated exactly with BAG1L. Moreover, when BAG3 Co-IP was performed, higher levels of Hsc/Hsp70 co-precipitated in lysates from old cells (Figure 1C). These data suggest that, due to the altered BAG expression, the interaction of Hsc/Hsp70 with BAG proteins is shifted from BAG1 to BAG3 during cellular aging.
Oxidative stress and the occurrence of oxidised, cross-linked proteins is a hallmark of aging (Brunk and Terman, 2002). We were interested whether oxidative stress induces the shift from BAG1 to BAG3. Indeed, the BAG shift was observed in 293 cells treated with 4-hydroxy-2-nonenal or H2O2 at concentrations that lead to accumulation of polyUb-proteins (Figure 1D and E). Together, these data may suggest that the shift from BAG1 to BAG3 is an adaptation of the PQC system to handle elevated levels of oxidised and cross-linked proteins. We also overexpressed polyQ-huntingtin in 293 cells, but the BAG shift was not observed despite aggregation of the mutant protein (Supplementary Figure S10 and Supplementary data).
BAG1 is essential for effective proteasomal degradation
To elucidate the specific roles of BAG1 and BAG3 in PQC, we performed knockdown experiments in 293 cells. BAG1 and BAG3 were efficiently depleted by short interfering RNAs (siRNAs) (Figure 2A). Strikingly, knockdown of BAG1 provoked an increase of BAG3 levels. Conversely, BAG1 expression, in particular BAG1L, was elevated upon BAG3 depletion (Figure 2A). This was most likely due to altered gene expression as reciprocally regulated mRNA levels were found (Figure 2B). These data suggest an adaptive response of the cell in which the expression of BAG1 is induced to compensate for the deprivation of BAG3 and vice versa. This view was supported by the finding that both knockdowns resulted in the accumulation of polyUb-proteins (Figure 2A), suggesting a functional relation of both BAG isoforms in the regulation of polyUb-protein levels. Because BAG1 has been described to link Hsc/Hsp70 to the proteasome (Luders et al, 2000), we examined whether polyUb-protein accumulation was a result of an impaired UPS. To monitor UPS activity in living cells, we used a GFP-based UPS reporter (d2GFP) containing a PEST sequence (Matsuda and Cepko, 2007). In a cycloheximide chase experiment, the degradation of d2GFP was completely blocked in the presence of the proteasome inhibitor MG132 (Supplementary Figure S2), showing the functionality of the reporter. Furthermore, d2GFP accumulated in cells treated with MG132 (Figure 2D). When d2GFP was expressed in BAG1 or BAG3 knockdown cells, the UPS reporter accumulated only in BAG1-depleted cells (Figure 2A). Knockdown of BAG3 resulted only in a negligible increase of d2GFP compared with the observed rise of polyUb-proteins. The possibility that increased BAG3 levels were responsible for UPS inhibition in BAG1-depleted cells could be ruled out on the basis of the observation that in BAG1/BAG3 double knockdown cells d2GFP accumulation even exceeded that of the single knockdown (see Supplementary data for discussion of this aspect and Supplementary Figure S3A and B). These results imply a role in the UPS exclusively for BAG1.
Figure 2.
(A) 293 cells were transfected with bag1, bag3 or nonsense (nons) siRNAs, as indicated. After 48 h, cells were transfected with d2GFP expression plasmid together with half the amounts of the indicated siRNAs. After additional 24 h, levels of indicated proteins were detected by immunoblot analysis. (B) 293 cells were transfected with indicated siRNAs for 48 h followed by real-time PCR analysis of BAG1 and BAG3 mRNA levels. Depicted is the mean relative expression ratio (log2) ±s.e.m. *P<0.05 and ***P<0.001 versus nons, n=3. (C) BAG1 knockdown was performed in 293 cells for 48 h. Thereafter, cells were transfected with GFP or GFP-based UPS reporter genes as indicated. After additional 24 h, levels of GFP-positive proteins were detected by western-blot analysis using a GFP antibody. (D, E) Immunoblot analysis of indicated proteins from d2HEK cells treated with different concentrations of MG132 for 5 h (D) or transfected with indicated amounts of BAG1S plasmid for 24 h (E).
To strengthen this conclusion, we analysed the influence of BAG1 on two UPS reporters bearing other degradation signals. Substrates of the N-end rule and ubiquitin-fusion degradation pathways, Ub-R-GFP and Ub-G76V-GFP (Dantuma et al, 2000), respectively, also accumulated upon knockdown of BAG1 (Figure 2C). In contrast, levels of GFP without a degradation signal were unaltered. Moreover, BAG1 knockdown showed effects similar to pharmacological proteasome inhibition including up-regulation of BAG3 as well as accumulation of d2GFP and polyUb-proteins (Figure 2D). Overexpression studies in a proteasome-reporter cell line (d2HEK) indicated a stimulation of the UPS by BAG1S (Figure 2E) and BAG1L (Supplementary Figure S4 and Supplementary data). Together, these results strongly indicate that BAG1 is essential for efficient degradation of polyUb-proteins by the UPS.
BAG3 promotes the autophagic degradation pathway
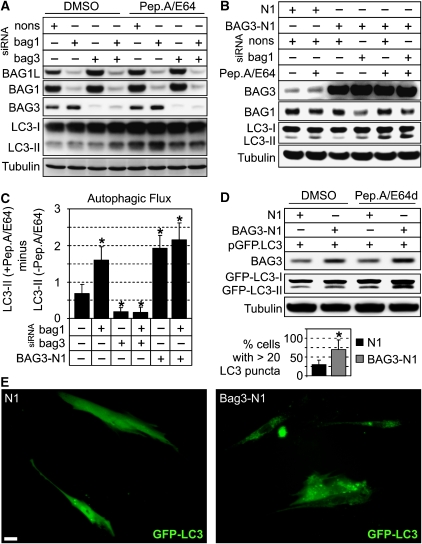
What then could be the role of BAG3 in PQC? BAG3 has been described to promote the degradation of polyQ-aggregates by autophagy (Carra et al, 2008), and autophagy has been implicated in the turnover of polyUb-proteins in addition to the UPS (Ding and Yin, 2008). We therefore analysed whether BAG3 is involved in autophagic processes. A well-established marker for autophagy is LC3-II, and the autophagic flux can be measured by monitoring LC3-II accumulation in the presence of lysosomal inhibitors (Mizushima and Yoshimori, 2007). In BAG3 knockdown cells, the accumulation of LC3-II upon lysosomal inhibition by Pepstatin A and E64 (Pep.A/E64) was significantly diminished (Figure 3A and C). This was the case in BAG3 as well as in BAG1/BAG3 double knockdown cells. Interestingly, knockdown of BAG1 alone, which resulted in the induction of BAG3, caused an increased accumulation of LC3-II (Figure 3A and C). Similar results were obtained when BAG1 and BAG3 were depleted in a LC3 overexpression background (Supplementary Figure S5). Furthermore, overexpression of BAG3 enhanced the flux of LC3-II through the lysosomal pathway (Figure 3B and C). When BAG3 was overexpressed in a BAG1 knockdown background, we measured consistently a further moderate increase of lysosomal LC3-II degradation (Figure 3B and C).
Figure 3.
(A) 293 cells were transfected with nonsense (nons), bag1 or bag3 siRNAs for 48 h and then treated for 2 h with the lysosomal inhibitors pepstatinA and E64 (both 10 μg/ml; Pep.A/E64) or DMSO as control, followed by immunoblot analysis of the indicated proteins. (B) 293 cells were transfected for 48 h with the indicated siRNAs and BAG3 expression plasmid (BAG3-N1) or vector control (N1) followed by the same analysis as in (A). (C) Diagram shows the autophagic flux of 293 cells with differently modulated BAG1 and BAG3 levels as described in (A) and (B). Autophagic flux was determined by the strength of LC3-II accumulation in a 2-h treatment period with Pep.A/E64. Therefore, normalised LC3-II levels in the absence of inhibitors were subtracted from corresponding levels obtained in the presence of Pep.A/E64. Values are expressed as mean±s.e.m. *P<0.05 versus control-transfected cells, n=3. (D) I90 cells were transfected with GFP fused to LC3 (pGFP.LC3) and co-transfected either with BAG3-N1 or N1. After transfection for 48 h, cells were treated as in (A) and levels of indicated proteins were detected by immunoblot analysis. (E) I90 cells, transfected as in (D) for 24 h, were microscopically analysed for GFP-LC3 fluorescence. Representative pictures are shown. Bar: 20 μm. Values expressed in diagram are mean±s.e.m. from three independent experiments (50 cells were counted per experiment). *P<0.05 versus N1, n=3.
Next, we expressed LC3 fused to GFP (GFP-LC3; Jackson et al, 2005) to visualise autophagosomes. However, we could not detect autophagosomes in 293 cells, which may be attributed to the high metabolic activity of 293 cells and the rapid disappearance of these transient structures. We therefore switched to the I90 cell model, in which GFP-LC3-positive autophagosomes could be clearly detected as green fluorescent punctuated structures. BAG3 overexpression led to a clear increase in the number of GFP-LC3 puncta per cell (Figure 3E). This observation could be corroborated by Western-blot analysis showing that BAG3 overexpression resulted in increased levels of GFP-LC3-II that further increased upon Pep.A/E64 treatment (Figure 3D). Together, these data indicate a BAG3-mediated forward regulation of LC3 lysosomal turnover and, in consequence, a role of BAG3 in stimulating the autophagy pathway.
BAG3 acts in concert with the polyUb-binding protein SQSTM1
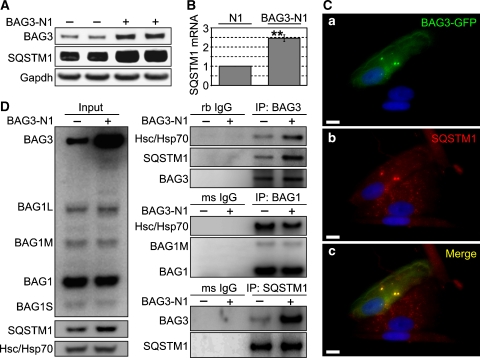
To gain insight into the role of BAG3 in autophagy, we investigated a key player of the autophagy pathway involved in degradation of polyUb-proteins, the ubiquitin-binding protein p62/SQSTM1 (hereafter referred to as SQSTM1). Owing to its ability to bind simultaneously LC3 and polyUb-proteins, SQSTM1 is suggested to control the sequestration of polyUb-proteins into inclusion bodies for autophagic degradation (Pankiv et al, 2007). Hence, we examined whether SQSTM1 is involved in the BAG3-regulated autophagy pathway. Interestingly, BAG3 overexpression induced up-regulation of SQSTM1 at the protein (Figure 4A) and transcriptional level (Figure 4B), suggesting that autophagy stimulation by BAG3 involves the recruitment of SQSTM1. It has been reported that both, BAG3 and SQSTM1, are up-regulated in response to proteasome inhibition, which is known to induce autophagy (Kuusisto et al, 2001; Ding and Yin, 2008; Wang et al, 2008). The co-regulation of these two genes implies a possible functional relationship of both proteins in autophagic processes. To test this possibility, we fused GFP to BAG3 and performed co-localisation analyses of BAG3-GFP with SQSTM1. Staining of SQSTM1 in BAG3-GFP-transfected I90 cells showed punctuated and spheric structures that were also positive for BAG3-GFP (Figure 4C). Moreover, in BAG3-GFP-positive cells, SQSTM1 appeared to form larger spheric structures compared with surrounding cells. This suggests that BAG3 might stimulate the sequestering of proteins into inclusion bodies in concert with SQSTM1. To support this view, we examined whether BAG3 physically interacts with SQSTM1. Indeed, SQSTM1 could be pulled-down when BAG3 was immunoprecipitated and vice versa, at endogenous BAG3 levels as well as when BAG3 was overexpressed (Figure 4D). We also examined whether BAG1 binds to SQSTM1. Although Hsc/Hsp70 efficiently co-precipitated, similar to the BAG3-IP, we could not detect SQSTM1 in BAG1-immunocomplexes (Figure 4D). Moreover, in SQSTM1-IPs, we could only detect BAG3 with the cBAG antibody but not BAG1. These data suggest a specific interaction of BAG3 with SQSTM1, which does not involve Hsc/Hsp70 or the BAG domain.
Figure 4.
(A) I90 cells were transfected with BAG3 (BAG3-N1) or vector control. After transfection for 48 h, levels of indicated proteins were detected by western-blot analysis. (B) Transcript levels of SQSTM1 in I90 cells transfected as in (A) were compared by real-time PCR analysis. Depicted is the fold-increase of SQSTM1 mRNA levels in BAG3-N1-transfected cells relative to control (N1)-transfected cells. Values are expressed as mean±s.e.m. **P<0.01 versus N1 cells, n=3. (C) I90 cells were transfected with a BAG3-GFP fusion plasmid for 48 h followed by indirect immunofluorescence staining of endogenous SQSTM1. (a) Direct fluorescence of BAG3-GFP (green), (b) indirect immunofluorescence of SQSTM1 (red) and (c) the stainings of (a) and (b) overlapped. DAPI (blue) was used to stain DNA. Representative pictures are shown. Bar: 10 μm. (D) Co-IP studies testing the interaction of SQSTM1 and BAG3. I90 cells were transfected as in (A). After 24 h, SQSTM1 and BAG3 were immunoprecipitated followed by analysis of co-sedimented proteins, as indicated. As negative control purified rabbit (rb) and mouse (ms) IgG was used. Left panel shows relative amounts of proteins in lysates used for Co-IP (Input).
SQSTM1 is subject to autophagic degradation (Ding and Yin, 2008). Therefore, we asked whether BAG3 is also degraded by autophagy. When autophagy was induced by amino acid starvation, the levels of both, BAG3 and SQSTM1 markedly decreased after 1 h and BAG3 levels, but not SQSTM1, were rapidly replenished (Supplementary Figure S6A). Unexpectedly and in contrast to SQSTM1, BAG3 degradation could only be blocked by proteasome inhibitors but not lysosomal inhibitors (Supplementary Figure S6B). Currently, we have no explanation for the transient proteasomal degradation of BAG3 upon amino acid starvation. Nevertheless, our results suggest a possible function of BAG3 in promoting the sequestration of proteins into inclusion bodies for subsequent autophagic degradation in association with SQSTM1. Moreover, our results show that in contrast to SQSTM1, BAG3 itself is not subject to lysosomal degradation.
Protein degradation by autophagy is enhanced during cellular aging
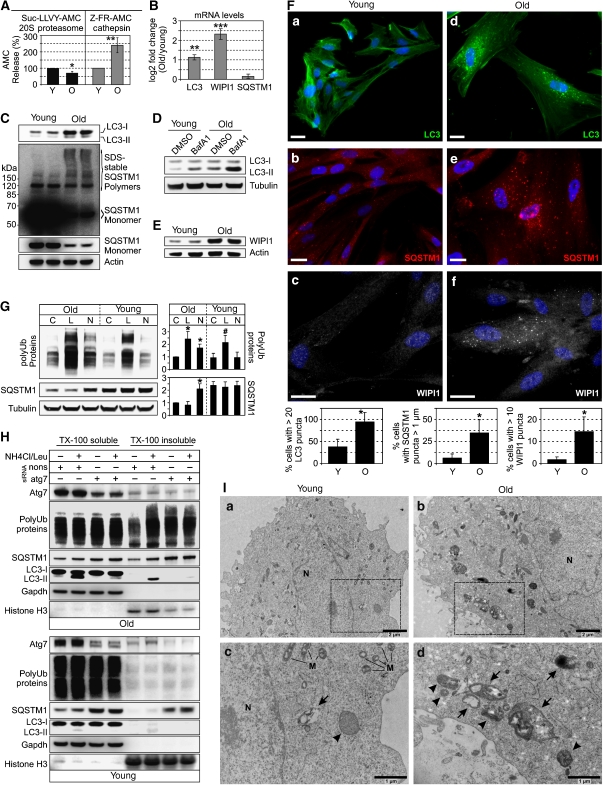
Our results so far indicate specific roles for BAG1 and BAG3 in PQC by enhancing proteasomal and autophagic pathways, respectively. The shift from BAG1 to BAG3 during cellular aging (Figure 1A) suggests possible alterations of proteasomal and autophagic activity in aged cells. Accordingly, we found reduced proteasomal chymotrypsin-like activity in lysates from old I90 cells (Figure 5A). Conversely, the activity of lysosomal proteases, the cathepsins, was enhanced (Figure 5A). Thus, we analysed whether the increased cathepsin activity in old cells was associated with an increased autophagy activity. Analysis of LC3 revealed increased protein and mRNA levels in aged cells (Figure 5B and C). Immunofluorescence analysis of endogenous LC3 revealed a higher number of LC3-positive autophagosomes in old cells (Figure 5F). Moreover, autophagic flux was elevated during aging, as LC3-II levels increased more prominently in aged cells upon lysosomal inhibition by bafilomycin A1 (Figure 5D) and Pep.A/E64 (Supplementary Figure S7A). Furthermore, we found increased mRNA and protein levels of the early autophagosome marker WIPI1 (Atg18) in old cells (Figure 5B and E) and a significantly higher proportion of aged cells showed WIPI1-positive autophagosomes (Figure 5F). In addition, ultra-structural analysis revealed numerous autophagosomes and autolysosomes containing amorphous electron-dense granular or multilamellar material in old cells, whereas in young cells, autophagic structures were only sporadically observed (Figure 5I). Together, these results strongly suggest an enhanced autophagic activity during cellular aging.
Figure 5.
(A) Proteasomal chymotrypsin and cathepsin activity in lysates from young (Y) and old (O) I90 cells was determined using specific fluorescence probes, as described in the Material and methods section. Values are expressed are mean±s.e.m. *P<0.05 and **P<0.01 versus young, n=3. (B) Real-time PCR analysis of LC3, WIPI1 and SQSTM1 mRNA levels in young and old I90 cells. Depicted is the mean expression ratio (log2) ±s.e.m. of target genes in old cells relative to young cells. **P<0.01 and ***P<0.001 versus young, n=3. (C) Immunoblot analysis of LC3 and SQSTM1 in young and old I90 cells. (D) Young and old I90 cells were treated for 2 h with bafilomycin A1 (BafA1, 2 μM) or DMSO as control followed by immunoblot analysis of indicated proteins. (E) Immunoblot analysis of WIPI1 expression in young and old I90 cells. (F) Indirect immunofluorescence staining of endogenous LC3 (green), SQSTM1 (red) and WIPI1 (white) in I90 cells of young and old age. DAPI (blue) was used to stain DNA. Representative pictures are shown. Bar: 20 μm. Diagrams show percentage of cells with indicated characteristics counted as in Figure 3E. (G) Old and young I90 cells were treated for 1 h with DMSO as control (C), lactacystin (L, 2 μM) or NH4Cl (20 mM) plus leupeptin (Leu, 5 μM) (N, NH4Cl/Leu). Western-blot analyses were performed for detection of indicated proteins. In the diagram (right panel), levels of polyUb-proteins and SQSTM1 are depicted after normalisation to corresponding Tubulin levels. Values are expressed as mean±s.e.m. *P<0.05 versus old control, #P<0.05 versus young control, n=3. (H) I90 cells of old (upper panel) and young (lower panel) age were transfected with atg7 or nonsense (nons) siRNA. After transfection for 4 days , the cells were treated with NH4Cl/Leu or DMSO for 1 h followed by fractioning of cell lysates in TritonX-100 (TX-100) soluble and insoluble material. Equal protein amounts of both fractions were directed to immunoblot analysis for analysis of indicated proteins. Gapdh and Histone H3 were used as loading controls of soluble and insoluble fractions, respectively. (I) Transmission electron microscopic analysis of young (a) and old (b) I90 cells. Magnifications of marked areas in (a) and (b) are shown in (c) and (d), respectively. An arrow indicates an autolysosome, and arrow heads indicate autophagosomes. M, mitochondrion. N, nucleus.
As a next step, we analysed the levels of SQSTM1. SQSTM1 mRNA levels were slightly increased, but this effect was not statistically significant (Figure 5B). Steady-state protein levels seemed to be down-regulated, however, a more detailed analysis revealed increased levels of SDS-resistant high molecular weight SQSTM1 polymers in aged cells (Figure 5C). These polymers likely represent inclusion bodies of aggregated proteins. Accordingly, a higher number of old cells showed large SQSTM1-positive globular structures up to 2 μm in diameter in the cytoplasm as well as in the nucleus (Figure 5F). In contrast, SQSTM1 staining in young cells was dispersed throughout the cytoplasm and concentrated in few smaller (<1 μm) punctuated structures. These data suggest an increased formation of inclusion bodies in aged cells. Accordingly, the SQSTM1 bodies in old cells co-localised with polyUb and LC3 (Supplementary Figure S7B), and numerous LC3-positive autophagosomes were positive for polyUb (Supplementary Figure S7B). Thus, during aging, the cells could have been recruited the autophagy pathway to maintain PQC. This view could be substantiated by monitoring polyUb-protein accumulation in young and old cells treated with proteasome and lysosomal inhibitors. Young cells did respond to the proteasome inhibitor lactacystin, but not to lysosomal inhibition by NH4Cl/Leu (Figure 5G), suggesting that young cells used preferentially or exclusively the UPS for polyUb-protein degradation. In contrast, old cells accumulated polyUb-proteins as well as SQSTM1 upon lysosomal inhibition (Figure 5G), suggesting that old cells might use a SQSTM1-dependent autophagy pathway to remove polyUb-proteins. Note that both aged and young cells responded strongly to lactacystin treatment, suggesting that basal proteasomal flux is unaltered, although the in vitro analysis (Figure 5A) revealed a lower overall proteasomal capacity in old cells (see Supplementary data for further discussion).
A recent study showed the existence of two distinct quality control compartments. Soluble proteins are sequestered near proteasomes, whereas insoluble, terminally aggregated proteins are sequestered in LC3-positive inclusion bodies (Kaganovich et al, 2008). Thus, we analysed whether aged cells use the autophagy pathway to dispose insoluble polyUb-proteins. Knockdown of Atg7 in old cells, which strongly suppressed autophagy (see LC3 blot in Figure 5H), as well as NH4Cl/Leu treatment led to an accumulation of polyUb-proteins and SQSTM1 predominantly in the TX-100 insoluble fraction (Figure 5H). Effects of NH4Cl/Leu were suppressed in the Atg7 knockdown background, confirming that NH4Cl/Leu blocked an autophagic degradation pathway. In contrast to old cells, Atg7 depletion in young cells did not result in accumulation of polyUb-proteins, neither in the soluble nor insoluble fraction, despite an accumulation of SQSTM1 (Figure 5H). These results may indicate that young cells degrade SQSTM1 by autophagy but, in contrast to old cells, did not form SQSTM1 inclusion bodies containing polyUb-proteins.
BAG3 determines enhanced autophagy activity in aged cells
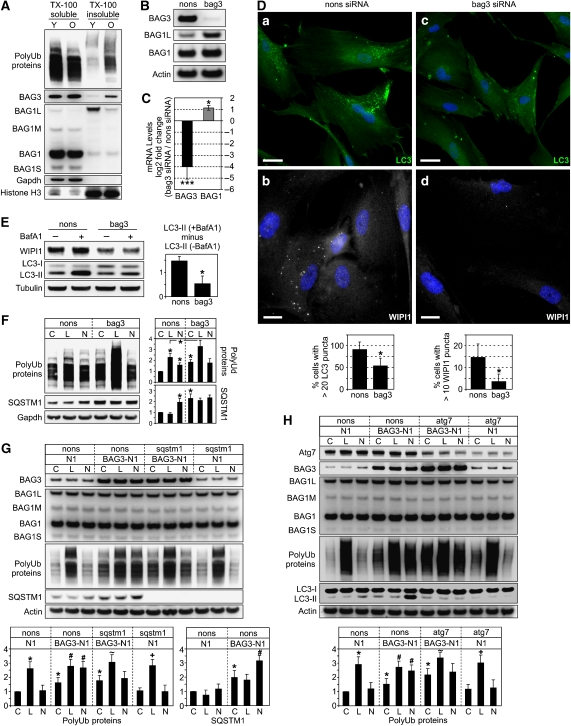
The results shown in Figure 5H suggest that during aging, the cell is increasingly stressed by insoluble quality control substrates that have to be degraded by autophagy. Accordingly, in old cells, we found a significantly higher proportion of polyUb-proteins in the TX-100 insoluble fraction (Figure 6A). Interestingly, in this fraction, we also found BAG3 (note that BAG1L as a nuclear protein was also found in this fraction). Thus, we analysed whether the autophagic turnover of polyUb-proteins in aged cells was mediated by BAG3. Therefore, BAG3 was depleted by siRNA. As seen in 293 cells, knockdown of BAG3 was accompanied by the induction of BAG1 (Figure 6B and C). BAG3-depletion also led to a decreased autophagic flux of LC3-II and decreased levels of WIPI1 (Figure 6E; Supplementary Figure S8A). Moreover, immunofluorescence analysis revealed less LC3- and WIPI1-positive autophagosomes in BAG3-depleted cells (Figure 6D). These data suggest that autophagosome formation and turnover depended on BAG3. Accordingly, the accumulation of SQSTM1 and polyUb-proteins in the presence of NH4Cl/Leu was also completely suppressed (Figure 6F). Moreover, in BAG3-depleted cells, the basal levels of polyUb-proteins were elevated, suggesting that impairment of autophagy led to the failure of PQC. Interestingly, accumulation of polyUb-proteins in the presence of the proteasome inhibitor lactacystin was enhanced (Figure 6F), suggesting that the knockdown of BAG3 forced old cells to use the UPS more extensively, possibly by the concomitant induction of BAG1.
Figure 6.
(A) Immunoblot analysis of indicated proteins of young (Y) and old (O) I90 cells upon fractionation as described in Figure 5H. (B, C) After transfection for 96 h of old I90 cells with bag3 or nonsense (nons) siRNA, BAG1 and BAG3 protein and mRNA levels were analysed by immunoblot (B) and real-time PCR (C) analysis, respectively. Transcript levels in bag3 siRNA cells are depicted as the mean log2 expression ratio ±s.e.m. relative to nons siRNA cells. *P<0.05 and ***P<0.001 versus nons, n=3. (D) Indirect immunofluorescence analysis of endogenous LC3 (green) and WIPI1 (white) in old I90 cells transfected with nons (a, b) and bag3 siRNA (c, d) for 96 h. DAPI (blue) was used as a nuclear marker. Representative pictures are shown. Bar: 20 μm. Diagrams show percentage of cells counted as in Figure 3E. (E) Same analysis as in Figure 5D but old cells transfected as in (A) were used. Values expressed in the diagram are mean±s.e.m. *P<0.05 versus nons, n=3. (F) Same analysis as in Figure 5G but old I90 cells transfected as in (A) were used. C, control; L, lactacystin, N, NH4Cl/Leu. Values are expressed as mean±s.e.m. *P<0.05 versus nons control, or as indicated, n=3. (G, H) Same analysis as in (F) but young I90 cells 48 h after transfection with a BAG3 expression plasmid (BAG3-N1) or vector control (N1) together with nons or sqstm1 siRNA (G) and nons or atg7 siRNA (H), as indicated, were used. *, #, ∼ and +, P<0.05 versus C N1, C BAG3-N1, C BAG3-N1 sqstm1 or atg7 and C N1 sqstm1 or atg7, n=3.
Expression profiles of BAG3 and SQSTM1 overlapped strictly in 293 and young I90 cells, as overexpression and knockdown of BAG3 resulted in increased and decreased levels of SQSTM1, respectively (Supplementary Figure S9A and B). In contrast, BAG3 depletion in aged I90 cells induced the expression of SQSTM1 (Figure 6F; Supplementary Figure S9C). This was accompanied by the induction of the heat-shock-related genes Hsp70 and Hsp40, which remained unaltered in young cells (Supplementary Figure S9C). These data suggest that, in particular, old cells depend on BAG3 to maintain proteostasis, and that BAG3 knockdown provoked a general stress response, including Hsp and SQSTM1 induction, exclusively in old cells.
Next, we were interested in whether BAG3 overexpression in young cells activates the autophagy pathway for the removal of polyUb-proteins. BAG3 overexpression led to an increase of autophagy activity as determined by the lysosomal flux of SQSTM1 (Figure 6G) and LC3-II (Figure 6H; Supplementary Figure S8B). Strikingly, in BAG3-transfected cells, polyUb-proteins accumulated in the presence of NH4Cl/Leu (Figure 6G). This could be efficiently suppressed by knockdown of SQSTM1 (Figure 6G), indicating that BAG3 acts in concert with SQSTM1 to stimulate lysosomal polyUb-protein degradation. To investigate whether the increased lysosomal polyUb-turnover involves the autophagy pathway, we analysed this effect in Atg7 knockdown cells. Atg7 depletion, which led to a clear suppression of autophagy (see LC3 blot, Figure 6H), counteracted the BAG3-dependent accumulation of polyUb-protein in the presence of NH4Cl/Leu (Figure 6H), corroborating that BAG3 stimulates degradation of polyUb-proteins by autophagy. Interestingly, Atg7-depletion resulted in increased BAG3 levels (see Supplementary data for further discussion).
In BAG3-overexpressing young cells, the basal level of polyUb-proteins was increased (Figure 6G and H), indicating that BAG3 interfered with the PQC system of young cells. Young and old cells showed a different ratio of soluble/insoluble polyUb-proteins (Figure 6A). Thus, we conclude that a proper ratio of BAG1/BAG3, adjusted to the ratio of soluble to insoluble (aggregated) quality control substrates, might be more important to maintain proteostasis than the abundance of BAG3.
BAG3 to BAG1 ratio is increased in the aged rodent brain
Many age-related neurodegenerative diseases including Alzheimer, Parkinson and Huntington disease are associated with the aberrant accumulation of SQSTM1-positive inclusion bodies (Pankiv et al, 2007), suggesting alterations of PQC during neuronal aging. Hence, we investigated whether the age-related change of PQC observed in the cellular aging models could be confirmed in models of CNS aging. When comparing brain homogenates from young (3 months) and old (24 months) mice, we found increased BAG3 and decreased BAG1L levels in various brain regions of old mice (Figure 7A and C; Supplementary Figure S11A for quantification). Gene expression analyses in the cerebellum revealed increased BAG3 and decreased BAG2 mRNA levels during aging (Figure 7B). BAG1 mRNA levels were slightly decreased, but this effect was not statistically significant. SQSTM1 mRNA levels were significantly increased (Figure 7B), and immunoblot analysis revealed increased steady-state SQSTM1 levels in all brain regions during aging (Figure 7D). Moreover, similar to the in vitro aging model, we found elevated levels of SDS-stable SQSTM1 polymers (Figure 7D). The highest levels of SQSTM1 polymers were found in the aged cerebellum correlating well with the highest BAG3 levels detected in this area (Supplementary Figure S11B). The increased BAG3 to BAG1 ratio in the aged brain was also accompanied by increased levels of LC3-II (Figure 7D) and increased activity of cathepsins (shown in Figure 7E for hippocampus and cerebellum). Together, these data suggest that during brain aging, a shift from BAG1 to BAG3 occurs, which is possibly associated with an increase of autophagy activity.
Figure 7.
(A) Protein extracts from cerebellum (CER) of young (Y, 3 months) and old (O, 24 months) mice were analysed for BAG1 and BAG3 expression by immunoblot analysis. (B) Real-time PCR analysis of indicated mRNA levels in cerebellum of young and old mice. Depicted is the log2 expression ratio of target genes in old mice relative to young mice. Values are expressed as mean±s.e.m. *P<0.05 and **P<0.01 versus young, n=3. (C) Immunoblot analysis as in (A) but in cortex (CTX), hippocampus (HIP) and mid-brain (MB). (D) Western-blot analysis of indicated proteins in different brain regions of young and old mice. (E) Total cathepsin and specific cathepsin B activity in brain extracts from young and old mice was determined using fluorescence probes Z-FR-AMC and Z-RR-AMC, respectively, as described in the Material and methods section. Values are expressed as mean±s.e.m. *P<0.05 versus young, n=3. (F) Expression analysis of indicated proteins in primary hippocampal astrocytic and neuronal cell cultures from young (2 months) and old (24 months) rats. Detection of GLT1 and NeuN served as astrocyte and neuron markers, respectively. Note the NeuN signals in astrocytic cultures. We attributed these signals to the presence of resting nuclei of dead neurons, which we observed under the microscope.
To identify the cell population in the brain responsible for the altered BAG levels during aging, we used cultured hippocampal neurons as well as astrocytes from young (2 months) and old (24 months) rats. We found strongly elevated levels of BAG3 in neurons isolated from old animals compared with their young counterparts (Figure 7F). Furthermore, the levels of BAG1L and BAG1 were down regulated. In contrast, astrocytes showed no age-associated alterations of BAG levels (Figure 7F). These results suggest that in the aged rodent brain, an increase of the BAG3 to BAG1 ratio occurs in neuronal cell populations.
Discussion
This study identifies BAG1 and BAG3 as key modulators of the proteasomal and autophagic pathways for the degradation of polyUb-proteins. Moreover, we show that BAG1 and BAG3 are reciprocally regulated during aging leading to an increased BAG3 to BAG1 ratio in aged cells. Triggered by increased BAG3 expression, proteostasis in aged cells is maintained by recruitment of the autophagy pathway involving the polyUb- and LC3-binding protein SQSTM1.
How do BAG1 and BAG3 function in differentially regulating the UPS and autophagy? With respect to the basic role of BAG1 in the UPS, Höhfeld and colleagues have reported convincing mechanistic insights showing that BAG1 can act as a physical link between the chaperone system and the proteasome (Luders et al, 2000). Moreover, BAG1 can accept substrates from Hsc/Hsp70 and direct them to proteasomal degradation in cooperation with the E3 ubiquitin ligase CHIP (Demand et al, 2001). CHIP and BAG1 can simultaneously bind to Hsc/Hsp70. Thus, a ternary complex of Hsc/Hsp70 together with CHIP and BAG1 is thought to function in protein degradation by the UPS (Demand et al, 2001). BAG3 binds to the same domain of Hsc/Hsp70 as BAG1, presumably in a competitive manner. One might therefore speculate that a ternary complex composed of Hsc/Hsp70, CHIP and BAG3 handles substrates for autophagic degradation. In line with this view, CHIP can target substrates for both degradation pathways (Shin et al, 2005). Our finding that BAG3 can stimulate autophagy processes is in agreement with a previous report showing enhanced autophagic degradation of polyQ aggregates by BAG3 (Carra et al, 2008). These authors hypothesised that BAG3 might recruit the autophagy machinery in close vicinity to the chaperone-loaded substrates. Our finding that BAG3 interacts, directly or indirectly, with SQSTM1 could be a new piece in the puzzle of how the chaperone network is linked to the autophagy machinery. Owing to its ability to bind both, ubiquitin and LC3, SQSTM1 combines two features important for selective degradation of proteins by autophagy: substrate sequestration and recruitment of the autophagosome membrane (Pankiv et al, 2007). Sequestration of substrates by SQSTM1 involves the build-up of protein aggregates or inclusion bodies. Our studies indicate that BAG3 might function in sequestration of chaperone substrates and the formation of inclusion bodies along with SQSTM1. In accordance with this view, BAG3-GFP co-localised with the large SQSTM1-positive aggregates. However, unlike SQSTM1, BAG3 is not subject to autophagic degradation. Thus, BAG3 is either not captured within inclusion bodies and autophagosomes or resistant to lysosomal hydrolysis. Although this aspect remains to be addressed, we can conclude that BAG3 stimulates the degradation of polyUb-proteins by a SQSTM1-dependent autophagy pathway.
Our findings further suggest that BAG1 and BAG3 control a molecular switch between proteasomal and autophagic degradation pathways, respectively. Several reports indicate a cross-talk between the two degradation systems, as autophagy is induced upon proteasome inhibition (Ding and Yin, 2008). In line with a role of BAG3 in this adaptive process, BAG3 expression has been shown to be induced upon proteasome inhibition (Wang et al, 2008). Moreover, as previously shown, BAG3 gene expression is controlled by heat-shock-factor-1 (Franceschelli et al, 2008). Consistent with this finding, several studies showed increased BAG3 expression under protein denaturating conditions (Pagliuca et al, 2003). We conclude that under physiological conditions, PQC is mainly achieved by BAG1-dependent proteasomal degradation. However, under acute stress conditions, when misfolded proteins accumulate and the aggregation potential increases, the UPS might be insufficient for complete clearance of defective proteins. In addition, the proteasome cannot degrade insoluble proteins and nondissociable aggregates (Ding and Yin, 2008). Moreover, aggregates might even impair proteasome function which in turn enhances protein aggregation—a vicious cycle (Bence et al, 2001). In contrast, such protein aggregates can be effectively degraded by autophagy (Ding and Yin, 2008). Thus, in conditions where increased protein misfolding and aggregation occurs, the cell has to increasingly rely on the autophagic degradation system to maintain proteostasis. We suggest that BAG3 has a critical role in mediating this adaptation.
Importantly, during aging, a persistent shift from BAG1 to BAG3 determines the constitutive activation of the autophagic system for degradation of defective proteins. The finding that in aged cells PQC is achieved by a shift towards autophagy is plausible considering the elevated levels of oxidatively modified proteins that accumulate in aged cells (Breusing and Grune, 2008). The occurrence of oxidised proteins is a hallmark of aging in a large number of cell types and tissues from a variety of organisms. These proteins are prone to form cross-linked aggregates, which are poor proteasome substrates (Ding and Yin, 2008). Thus, we conclude that old cells need to adapt to chronic oxidative stress and recruit constitutively the autophagy pathway to maintain proteostasis. On the other hand, it has been suggested that autophagic activity declines with age (Cuervo, 2003). It should be noted, however, that the term autophagy encompasses three different degradation pathways involving lysosomal activity: (macro)autophagy, microautophagy and chaperone-mediated autophagy (CMA). To our knowledge, microautophagy has so far not been investigated in the context of aging. It is well acknowledged that rates of CMA, a selective form of autophagy involving the lysosomal receptor LAMP2A, decrease with age because of a decrease in the levels of the receptor (Kiffin et al, 2007). Although it is widely assumed that autophagy declines with age, original literature addressing this issue is sparse. We found only one study describing decreased autophagic activity in the liver of aged rats in response to an anti-lipolytic agent (Del Roso et al, 2003). However, basal autophagy in heart and liver was not found to decrease with age (Wohlgemuth et al, 2007). The perceived decrease in the autophagic potential during aging is probably explained by the progressive, age-dependent accumulation of biological ‘garbage', such as lipofuscin (age pigment), an intralysosomal, polymeric, undegradable material (Brunk and Terman, 2002). Apparently, the accumulation of such ‘waste' material is indicative of a catabolic insufficiency with respect to this material, but not of a general decline in autophagy. Rather, it reflects an imbalance between the production of this material and the degradation capacity. Our studies were performed on aged but pre-senescent cells before the massive accumulation of lipofuscin-like material has occurred. Hence, we understand the integration of the autophagy pathway into the cellular PQC system as an important adaptation to the heightened pro-oxidant milieu developing during aging. This view is supported by the observed life-span extensions of model organisms by promoting either antioxidant defense systems or autophagy (Melendez et al, 2003; Balaban et al, 2005).
In conclusion, our findings suggest that the co-chaperones BAG1 and BAG3 represent key players of cellular PQC by stimulating the turnover of polyUb-proteins by proteasomal and autophagic degradation pathways, respectively. Moreover, our study shows that during aging, an increased BAG3/BAG1 ratio triggers the recruitment of the autophagy pathway for PQC. This is, at least in part, mediated by BAG3 in association with the polyUb- and LC3-binding protein SQSTM1. We propose that protein turnover by autophagy is gaining importance during the aging process, because of elevated protein oxidation and protein cross-linking leading to protein aggregates that cannot be degraded by the proteasome. Future studies will have to show whether an impairment of this adaptation process may contribute to age-related proteinopathies.
Materials and methods
Inhibitors
Stock solutions of all inhibitors used throughout this study were prepared in DMSO, except NH4Cl and leupeptin, which were solved in H2O. Leupeptin, pepstatinA and E64d were purchased from Sigma, E64, Bafilomycin A1 and lactacystin were from Alexis, MG132 was from Calbiochem and NH4Cl was from Roth.
Cell culture
Human embryonic kidney cells 293 (HEK, 293) were purchased from the American Type Culture Collection. Primary human fibroblasts IMR90 (I90) and WI38 cells were purchased from Coriell Institute for Medical Research. All cell culture products were purchased from Invitrogen. Cells were cultured in Dulbecco modified Eagle medium supplemented with 1 mM sodium pyruvate, 10% (v/v) fetal bovine serum (FBS), 1 × nonessential amino acids, 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in a 5% CO2-humidified atmosphere. Medium was refreshed every 3 days during cultivation and every 24 h in an experimental setting.
Ex vivo cell culture
Mixed primary hippocampal cultures from young (2 months) and aged (24 months) animals were prepared essentially as described previously (Brewer, 1997). In detail, Sprague–Dawley rats were anesthetised with halothane and decapitated by guillotine. The hippocampi were rapidly dissected from the brain in 3 ml HibernateA (Life technologies) and the menniges and excess white matter were removed under the stereomicroscope. Hippocampi were placed on a sterile prewet filter paper and cut into 0.5-mm slices perpendicular to the long axes of the hippocampi using a tissue chopper. Slices were transferred to 5 ml HibernateA supplemented with B27 (HibernateA/B27). After shaking 8 min at 30°C, slices were transferred to 5 ml HibernateA/B27 containing 12 mg papain and incubated for 30 min in a 30°C water bath with a rotating platform. Digested slices were transferred to 2 ml HibernateA/B27 and triturated 10 times with a 1-ml blue polypropylene pipet tip with a 0.9-mm opening. Supernatants were collected and subjected to subcellular fractionation using Optiprep/Hibernate 1:1. Neuron enriched fractions were resuspended in Neurobasal medium (Invitrogen) supplemented with B27, 5 ng/ml basal fibroblast growth factor (Sigma) and cultured in a humidified atmosphere with 5% CO2 at 37°C. Protein samples were prepared after 7 days in vitro (7DIV). In a parallel set of experiment, cultures were treated with 100 μM NMDA for 24 h to specifically eliminate neurons.
Plasmids, siRNAs and transfection method
Expression plasmids for human BAG3 (pBAG3-N1) and human BAG3 fused to EGFP (pBAG3.EGFP-N1) were constructed by cloning partial human BAG3 cDNA containing the whole CDS into pEGFP-N1 (Clontech). Primer sequences used to clone BAG3 plasmids are listed in Supplementary Table S1. Following vector linearisation with BamH1 and Not1 (for pBAG3-N1) or BamH1 only (for pBAG3.EGFP-N1) PCR products were inserted using the In-Fusion reaction according to manufacturer's protocol (Clontech). p103QHtt.EGFP-N1 and 25Q.Htt.EGFP-N1 expression plasmids were obtained by subcloning the coding sequences of p426-103Q-GPD and p426-25Q-GPD (Addgene plasmids 1184 and 1181, respectively; Krobitsch and Lindquist, 2000) into pEGFP-N1 (Clontech). Expression plasmids for GFP-LC3 (Jackson et al, 2005), d2GFP (Matsuda and Cepko, 2007), Ub-R-GFP and Ub-G76V-GFP (Dantuma et al, 2000) were provided by Addgene. Construction of human BAG1L and BAG1S expression plasmid is described elsewhere (Froesch et al, 1998; Schmidt et al, 2003).
siRNAs were purchased from Eurofins MWG Operon as duplexes with 3′-dTdT overhangs. To verify specificity of knockdown effects, two independent sets of siRNA duplexes were used for each target gene. Sequences of used siRNAs are listed in Supplementary Table S1. Generally, cells were transfected with 20 μg of siRNA. In all knockdown experiments, same amounts of siRNA targeting a nonsense sequence were transfected as control.
Cells were transfected by electroporation using the Amaxa Nucleofector I (program U-24) and standard electroporation cuvettes (Sigma). Electroporation buffer contained 135 mM KCl, 0.2 mM CaCl2, 2 mM MgCl2, 5 mM EGTA, 10 mM HEPES (pH 7.5) and 25% heat-inactivated FBS. To achieve 293 cells stably expressing d2GFP, cells were seeded after transfection in 96-well plates with one cell per well. After 2 weeks, cell clones with weak green fluorescence were isolated and analysed for d2GFP expression.
Immunoblotting and Immunocytochemistry
Immunoblot analyses were performed as described previously (Gamerdinger et al, 2006). Generally, 15 μg of total protein were subjected to SDS–PAGE using precast NuPAGE 4–12% Bis-Tris gels with MES running buffer (Invitrogen) or hand-cast gels together with the Laemmli buffer-driven Mini Protean III system (Bio-Rad). Proteins were detected by chemiluminescence using either SuperSignal (Pierce) or, for weak signals, Immobilon (Millipore) substrates and visualised with the Fuji LAS-3000 dark box (Fujifilm).
For immunocytochemical staining, I90 cells were grown on glass cover slips in 6-well plates. Cells were fixed with 3.5% (w/v) PFA for 5 min at 4°C and 10 min at RT. After a second methanol fixation step for 6 min at −20°C, unspecific antibody binding sites were blocked with 3% (v/v) serum. After incubation of cells with primary antibodies, cells were incubated with Cyanine- (Cy2-, Cy3- or Cy5-) conjugated secondary antibodies (Jackson). Microscopic analysis was performed with an inverted Axiovert 200 microscope (Zeiss) equipped with a SPOT RT CCD-camera from Diagnostic Instruments (Visitron).
The cBAG antibody was raised in rabbits against human BAG domain in BAG1M (aa 151–263). Antibodies used throughout this study are listed in Supplementary Table S1.
Co-immunoprecipitation
Cells were washed twice with ice cold PBS and lysed on ice in IP-buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 0.5% NP40) supplemented with 1% (v/v) protease inhibitor mix and 1% phosphatase inhibitor mix (both from Sigma) for 20 min. After centrifugation (30 min, 15 000 g, 4°C), supernatants were collected and normalised to the protein content. For Co-IP studies, generally 2 μg of antibody were added to an input volume of 300 μl with 1.5–2 μg/μl protein. As control IPs were done with same amounts of purified rabbit or mouse IgG (Sigma). After shaking for 1 h at 4°C, protein G sepharose beads (GE Healthcare) were added and the samples incubated for 1 h at 4°C with constant rotation. Immunocomplexes were washed three times with IP-buffer and solved by heating for 10 min at 99°C in SDS–PAGE loading buffer (10% SDS, 20% glycerine, 125 mM Tris, 1 mM EDTA, 0.002% bromphenol blue and 10% β-mercaptoethanol) for subsequent immunoblot analysis.
Quantitative real-time reverse transcription–PCR analysis
RNA extraction, cDNA synthesis and quantitative real-time PCR were performed as described previously (Gamerdinger et al, 2006). Quantitative real-time PCR data were applied to REST (Pfaffl et al, 2002) for calculation and to test for significance by a randomisation test. Statistical significance was accepted at a level of P<0.05. Actin was used as a reference gene in the I90 aging cell model, in all other experiments the ribosomal protein gene L19. Primer sequences are listed in Supplementary Table S1.
Measurement of proteasome and cathepsin activity
For preparation of cell extracts, young and old IMR90 cells were washed twice with ice cold PBS and collected by trypsinisation. Cells were resuspended and incubated for 10 min in hypotonic buffer (10 mM HEPES pH 7.6, 10 mM K-acetate, 1.5 mM Mg-acetate, 2 mM DTT) at 4°C and then passed 10 times through a 25-gauge needle. The homogenate was spun at 640 g at 4°C for 5 min, and then, after the supernatant was brought to 90 mM K-acetate, centrifuged again at 10 000 g for 20 min at 4°C. Supernatants were collected, normalised to protein content and cryo-freezed in liquid nitrogen. Enzymatic reaction was started by mixing active cell extracts (6–8 μg protein in 25 μl) from young and old I90 cells (see Supplementary data) with 25 μl of assay buffer (15 mM HEPES pH 7.6, 130 mM K-acetate, 1.5 mM Mg-acetate, 1.5 mM CaCl2, 1.6 mM DTT, 8 mM ATP), supplemented either with 70 μM Suc-LLVY-AMC (Sigma; for proteasome activity) or 70 μM Z-FR-AMC (Calbiochem; for total cathepsin activity). AMC fluorescence was recorded in a black 96-well plate at 37°C in 2-min intervals for a total time period of 30 min using the Victor3V Multilabel counter (Perkin Elmer). Specific proteasomal and cathepsin L activity was determined by subtracting unspecific AMC fluorescence obtained in the presence of proteasome inhibitor MG132 (20 μM) and lysosomal inhibitors E64 and pepstatinA (both 10 μg/ml), respectively.
Brain samples from young and old mice were resuspended in hypotonic buffer, sonicated on ice and then passed 20 times through a 25-gauge needle followed by the same purification as described for I90 cells. Specific cathepsin B activity was measured with the cathepsin B-specific fluorescent substrate Z-RR-AMC (Biomol).
Transmission electron microscopy
Transmission electron microscopy (TEM) was performed as described previously (Mersseman et al, 2008). In brief, cells were prefixed in 2.5% glutaraldehyde, 0.1 M sucrose, 0.1 M cacodylate buffer (pH 7.3) for 1 h at room temperature. After three washes for 10 min in 0.1 M cacodylate buffer, cells were post-fixed for 1 h in 1 ml 2% OsO4, 0.1 M sucrose and 0.1 M cacodylate buffer, dehydrated and embedded in araldite resin. Ultrathin sections were cut with a Leica Ultracut S microtome and were counterstained with 2% aqueous uranyl acetate. Sections were analysed in a FEI Tecnai 12 BioTwin transmission electron microscope and imaged with an SCCD SIS MegaView III camera.
Statistical methods
Statistical significance was determined by Student's t-test using SIGMA STAT software (SPSS Science). Statistical significance was accepted at a level of P<0.05. The results are expressed as mean±s.e.m.
Supplementary Material
Supplementary data
Acknowledgments
We are grateful to Dr Theo Rein for providing the BAG1S and BAG1L expression plasmid. We thank Elisabeth Sehn for her excellent technical assistance in the TEM studies and appreciate Drs Karla Kirkegaard, Connie Cepko and Nico Dantuma for providing us their expression plasmids via Addgene. This work was supported by grants from the Fritz und Hildegard Berg-Stiftung (to CB).
References
- Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J (2005) BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell 16: 5891–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T (2005) Mitochondria, oxidants, and aging. Cell 120: 483–495 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292: 1552–1555 [DOI] [PubMed] [Google Scholar]
- Bimston D, Song J, Winchester D, Takayama S, Reed JC, Morimoto RI (1998) BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J 17: 6871–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breusing N, Grune T (2008) Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem 389: 203–209 [DOI] [PubMed] [Google Scholar]
- Brewer GJ (1997) Isolation and culture of adult rat hippocampal neurons. J Neurosci Methods 71: 143–155 [DOI] [PubMed] [Google Scholar]
- Brunk UT, Terman A (2002) Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med 33: 611–619 [DOI] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Lambert H, Landry J (2008) HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem 283: 1437–1444 [DOI] [PubMed] [Google Scholar]
- Cuervo AM (2003) Autophagy and aging—when ‘all you can eat' is yourself. Sci Aging Knowledge Environ 2003: pe25. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG (2000) Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol 18: 538–543 [DOI] [PubMed] [Google Scholar]
- Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, Pollera M, Bergamini E (2003) Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol 38: 519–527 [DOI] [PubMed] [Google Scholar]
- Demand J, Alberti S, Patterson C, Hohfeld J (2001) Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol 11: 1569–1577 [DOI] [PubMed] [Google Scholar]
- Ding WX, Yin XM (2008) Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4: 141–150 [DOI] [PubMed] [Google Scholar]
- Doong H, Rizzo K, Fang S, Kulpa V, Weissman AM, Kohn EC (2003) CAIR-1/BAG-3 abrogates heat shock protein-70 chaperone complex-mediated protein degradation: accumulation of poly-ubiquitinated Hsp90 client proteins. J Biol Chem 278: 28490–28500 [DOI] [PubMed] [Google Scholar]
- Franceschelli S, Rosati A, Lerose R, De Nicola S, Turco MC, Pascale M (2008) Bag3 gene expression is regulated by heat shock factor 1. J Cell Physiol 215: 575–577 [DOI] [PubMed] [Google Scholar]
- Froesch BA, Takayama S, Reed JC (1998) BAG-1L protein enhances androgen receptor function. J Biol Chem 273: 11660–11666 [DOI] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Manthey D, Behl C (2006) Oestrogen receptor subtype-specific repression of calpain expression and calpain enzymatic activity in neuronal cells—implications for neuroprotection against Ca-mediated excitotoxicity. J Neurochem 97: 57–68 [DOI] [PubMed] [Google Scholar]
- Hohfeld J, Jentsch S (1997) GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J 16: 6209–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K (2005) PLoS Biol 3: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia SK, Lee S, Smith PD, Liu L, Crocker SJ, Thorarinsdottir TE, Glover JR, Fon EA, Park DS, Lozano AM (2004) BAG5 inhibits parkin and enhances dopaminergic neuron degeneration. Neuron 44: 931–945 [DOI] [PubMed] [Google Scholar]
- Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM (2007) Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci 120: 782–791 [DOI] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S (2000) Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA 97: 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto E, Suuronen T, Salminen A (2001) Ubiquitin-binding protein p62 expression is induced during apoptosis and proteasomal inhibition in neuronal cells. Biochem Biophys Res Commun 280: 223–228 [DOI] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem 275: 4613–4617 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL (2007) Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA 104: 1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391 [DOI] [PubMed] [Google Scholar]
- Mersseman V, Besold K, Reddehase MJ, Wolfrum U, Strand D, Plachter B, Reyda S (2008) Exogenous introduction of an immunodominant peptide from the non-structural IE1 protein of human cytomegalovirus into the MHC class I presentation pathway by recombinant dense bodies. J Gen Virol 89: 369–379 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3: 542–545 [DOI] [PubMed] [Google Scholar]
- Nichols WW, Murphy DG, Cristofalo VJ, Toji LH, Greene AE, Dwight SA (1977) Characterization of a new human diploid cell strain, IMR-90. Science 196: 60–63 [DOI] [PubMed] [Google Scholar]
- Pagliuca MG, Lerose R, Cigliano S, Leone A (2003) Regulation by heavy metals and temperature of the human BAG-3 gene, a modulator of Hsp70 activity. FEBS Lett 541: 11–15 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443: 780–786 [DOI] [PubMed] [Google Scholar]
- Schmidt U, Wochnik GM, Rosenhagen MC, Young JC, Hartl FU, Holsboer F, Rein T (2003) Essential role of the unusual DNA-binding motif of BAG-1 for inhibition of the glucocorticoid receptor. J Biol Chem 278: 4926–4931 [DOI] [PubMed] [Google Scholar]
- Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ (2005) The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem 280: 23727–23734 [DOI] [PubMed] [Google Scholar]
- Takayama S, Reed JC (2001) Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol 3: E237–E241 [DOI] [PubMed] [Google Scholar]
- Wang HQ, Liu HM, Zhang HY, Guan Y, Du ZX (2008) Transcriptional upregulation of BAG3 upon proteasome inhibition. Biochem Biophys Res Commun 365: 381–385 [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, Dunn WA Jr (2007) Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation Res 10: 281–292 [DOI] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5: 781–791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data