Abstract
DNA damage induced by ionizing radiation (IR) activates p53, leading to the regulation of downstream pathways that control cell-cycle progression and apoptosis. However, the mechanisms for the IR-induced p53 activation and the differential activation of pathways downstream of p53 are unclear. Here we provide evidence that the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) serves as an upstream effector for p53 activation in response to IR, linking DNA damage to apoptosis. DNA-PKcs knockout (DNA-PKcs−/−) mice were exposed to whole-body IR, and the cell-cycle and apoptotic responses were examined in their thymuses. Our data show that IR induction of apoptosis and Bax expression, both mediated via p53, was significantly suppressed in the thymocytes of DNA-PKcs−/− mice. In contrast, IR-induced cell-cycle arrest and p21 expression were normal. Thus, DNA-PKcs deficiency selectively disrupts p53-dependent apoptosis but not cell-cycle arrest. We also confirmed previous findings that p21 induction was attenuated and cell-cycle arrest was defective in the thymoctyes of whole body-irradiated Atm−/− mice, but the apoptotic response was unperturbed. Taken together, our results support a model in which the upstream effectors DNA-PKcs and Atm selectively activate p53 to differentially regulate cell-cycle and apoptotic responses. Whereas Atm selects for cell-cycle arrest but not apoptosis, DNA-PKcs selects for apoptosis but not cell-cycle arrest.
In response to DNA damage induced by ionizing radiation (IR), the tumor suppressor p53 becomes activated as a transcription factor, leading to the regulation of distinct downstream pathways controlling cell cycle progression and apoptosis (1, 2). However, the mechanisms for p53 activation by IR and the differential activation of downstream pathways by p53 are unclear. Candidates for the upstream activators of p53 include two members of the phosphatidylinositol 3-kinase family, Ataxia telangiectusia mutated (ATM) and DNA-PKcs (the catalytic subunit of DNA-dependent protein kinase). Studies of the thymus in Atm−/− mice (3) showed a lack of IR-induced cell-cycle arrest and abnormal p21 induction. However, IR induction of apoptosis and Bax were normal. Based on these data, it was postulated that the effect of Atm on p53-dependent function is selective (3).
DNA-PKcs is another candidate as an upstream activator of p53 in response to IR, linking DNA damage to cell-cycle arrest and apoptosis (4–6). DNA-PK is a serine/threonine kinase that consists of a 465-kDa catalytic subunit (DNA-PKcs), and a DNA-targeting component Ku, which itself is a heterodimer of a 70-kDa and an 86-kDa polypeptide (Ku70 and Ku80) (7). When assembled on a suitable DNA molecule in vitro, DNA-PK becomes activated and phosphorylates many transcription factors, including p53 (8). It recently has been reported that a SCID (severe combined immunodeficiency) cell line SCGR11 accumulates a transcriptionally inactive form of p53 upon IR (4). As SCID cells are defective in DNA-PKcs (9), it would appear that DNA-PK acts upstream of p53 in response to DNA damage. However, the role of DNA-PK in p53 activation has been questioned by reports of normal IR-induced p53 activation and cell-cycle arrest in primary cells from SCID mice (10–13). Primary SCID cells, however, have been shown to retain residual DNA-PK activity (4), possibly because the SCID mutation resides downstream of the conserved kinase motifs of the DNA-PKcs gene, resulting in a truncated product missing only 83 amino acid residues from the carboxyl-terminal end (9). To avoid the ambiguity associated with SCID cells, we have generated DNA-PKcs−/− mice in which the DNA-PKcs gene was disrupted via homologous recombination and derived DNA-PKcs−/− primary mouse embryo fibroblasts (MEFs) (14).
We previously have reported that the DNA-PKcs−/− mice are hypersensitive to radiation and are severely immunodeficient (14), and that DNA-PKcs transcript, DNA-PKcs protein or kinase activity are not detected in them (14, 15). Using these reagents, we now present evidence on the role of DNA-PKcs in distinct radiation-induced p53-dependent signal transduction pathways in vivo. Specifically, we exposed DNA-PKcs−/− mice to whole-body IR and examined the cell-cycle and apoptotic responses of their thymuses. This approach minimizes potentially confounding artifacts associated with cultured primary and immortalized cells. Furthermore, it has been demonstrated that IR-induced apoptosis in mouse thymocytes is p53 dependent (16, 17); whereas other cell types that exhibit apoptosis upon IR exposure, such as endothelial cells, activate alternative apoptotic pathways that are p53 independent (17). Our data show that IR induction of apoptosis and Bax, both mediated by p53, is significantly suppressed in the absence of DNA-PKcs. In contrast, IR-induced cell-cycle arrest and p21 induction are normal. Thus, DNA-PKcs deficiency selectively disrupts p53-dependent apoptosis but not cell-cycle arrest. In this regard, DNA-PKcs and Atm (3) are similar in the selective activation of pathways downstream of p53, but dissimilar in that DNA-PKcs selects for apoptosis but not cell-cycle arrest, and Atm for cell-cycle arrest but not apoptosis.
Materials and Methods
Generation and Genotyping of DNA-PKcs−/− Mice.
The generation of the DNA-PKcs−/− mice was as described (14). Mice from heterozygous crosses were genotyped by PCR that distinguishes endogenous from the targeted DNA-PKcs allele (14). PCR contains 1 μg genomic DNA, 0.6 μM (each) of primers MD-20 (TATCCGGAAGTCGCTTAGCATTG), MD-21 (CCTGAAGACTGAAGTTGGCAGAA), and POL-8 (TTCACATACACCTTGTCTCCGACG), 0.2 mM (each) dNTP, 1.5 mM MgCl2, and 2.5 units of Taq polymerase. Primers MD-20 and MD-21 give a product of wild-type allele that is 264 bp; primers MD-20 and Pol-8 yield a product of the targeted allele that is 360 bp. Both DNA-PKcs−/− male and female mice were fertile and good breeders and were used to generate DNA-PKcs−/− mice for our experiments. p53−/− and Atm−/− mice were obtained from Jackson Laboratories and genotyped by PCR (18, 19).
Cell-Cycle Analysis in Vivo.
To assess cell-cycle regulation after IR, mice were irradiated with 10 Gy, then immediately given an i.p. injection of BrdUrd/5-fluorodeoxyuridine (FdUrd) aqueous solution (RPN 201, Amersham Pharmacia; 1 ml). Unirradiated control mice also were injected with 1 ml of BrdUrd/FdUrd aqueous solution. The mice were sacrificed 2 hr later by asphyxiation, and their thymuses were removed for thymocytes isolation. The cells were fixed sequentially with 70% ethanol and 2 M hydrochloric acid, centrifuged, and resuspended in 1 ml of 0.1 M Na2B4O7. Cells were incubated with anti-BrdUrd (0.2 μg/106 cells) for 30 min on ice, washed, and incubated with FITC-conjugated F(ab′)2 goat anti-mouse IgG (1 μg) for 30 min at room temperature. All samples were counterstained with propidium iodine (5 μg/ml) and analyzed by using a FACstar system.
Apoptosis Assays.
Wild type, DNA-PKcs−/−, p53−/−, and Atm−/− mice (2–4 months old) were given graded doses of whole-body irradiation (10 or 20 Gy) and sacrificed by asphyxiation 10 hr afterward. Apoptosis in vivo was assessed in the thymus by the terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) assay, as described (20). In brief, tissue specimens were fixed overnight in 10% formalin and embedded in paraffin blocks. Tissue sections (5 μm thick), adherent to polylysine-treated slides, were deparaffinized by heating at 70°C for 10 min and then at 60°C for 5 min. Tissue-mounted slides first were washed with xylene and rehydrated in 100% ethanol and then 95% ethanol. The slides were incubated in 10 mM Tris⋅HCl (pH 8) for 5 min, digested with 20 μg/ml proteinase K, rinsed in distilled water, and treated with 3% H2O2 in PBS for 5 min at 22°C to inactivate endogenous peroxidase. After three washes in PBS, the slides were incubated for 15 min at 22°C in buffer (140 mM Na-cacodylate, pH 7.2/30 mM Trizma base/1 mM CoCl2) and then for 30 min at 37°C in reaction mixture (0.1 units/μl terminal deoxynucleotidyl transferase/0.1 nM biotin-16-dUTP/100 mM Na-cacodylate, pH 7.0/0.1 mM DTT/0.05 mg/ml BSA/2.5 mM CoCl2). The reaction was stopped by transferring the slices to a bath of 300 mM NaCl, 30 mM Na citrate for 15 min at 22°C. The slides were washed in PBS, blocked with 2% human serum albumin in PBS for 10 min, rewashed, and incubated with avidin-biotin peroxidase. After 30 min at 22°C, cells were stained with the chromogen 3,3′-diaminobenzidine tetrachloride and counterstained with hematoxylin. Nuclei of apoptotic cells appear brown and granular, whereas normal nuclei stain blue.
Immunoblot Analysis.
Immunoblotting was performed by standard techniques (21) directly on crude thymic tissue samples or isolated thymocytes. Protein concentrations were determined with the BCA assay (Pierce). Immunoreactive bands were visualized with Supersignal CR-horseradish peroxidase (HRP) (Pierce) chemiluminescent substrate and recorded on Kodak XAR x-ray film. The primary antibodies used in this study were mouse monoclonal anti-p53 antibody (p240, Santa Cruz Biotechnology), rabbit anti-p21 antisera (Santa Cruz Biotechnology), rabbit anti-Bax antisera (Upstate Biotechnology, Lake Placid, NY), and monoclonal anti-actin antibody (Sigma). HRP-conjugated secondary antibodies (Boehringer Mannheim) were used to detect primary antibody signals.
Results
DNA-PKcs Deficiency Suppresses IR-Induced Thymic Apoptosis.
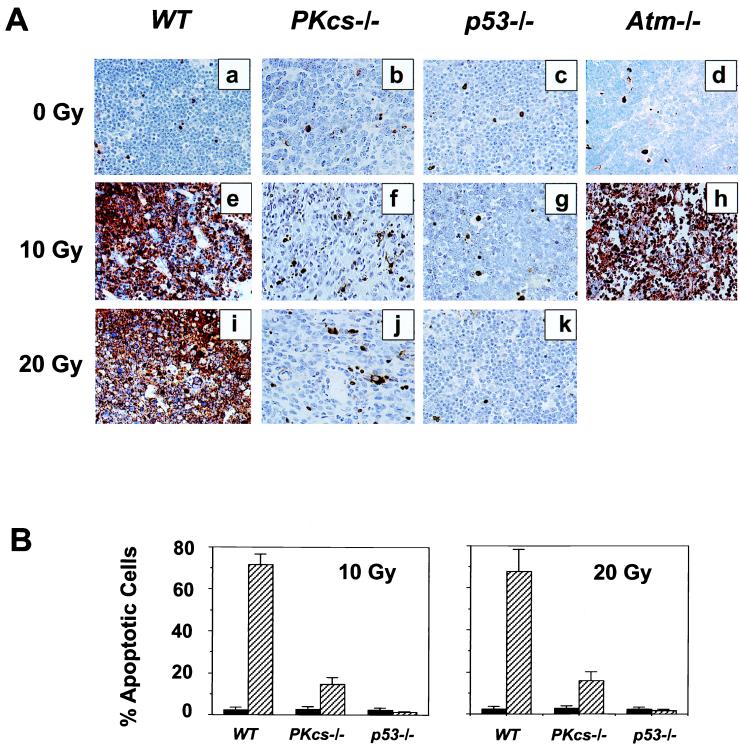
One consequence of radiation-induced DNA damage is cell death by apoptosis. Given the importance of DNA-PKcs in DNA damage repair, we studied its effect on radiation-induced apoptosis in vivo. Wild-type, DNA-PKcs−/−, p53−/−, and ATM−/− mice were given whole-body irradiation, and their thymic tissues were evaluated for apoptosis by using the TUNEL in situ assay. Unirradiated thymic tissue from wild-type and knockout animals showed a 2–3% incidence of apoptosis (Fig. 1A a–d). At 10 hr after irradiation, a substantial amount of apoptosis was observed in the thymus of wild-type and ATM−/− mice (Fig. 1A e, h, and i). In contrast, in DNA-PKcs−/− thymuses the proportion of apoptotic cells was significantly less and more evenly distributed (Fig. 1A f and j). This decrease of apoptotic response in the DNA-PKcs−/− thymus was probably not caused by the structure abnormality associated with immunodeficiency, because IR-induced apoptosis in the thymic tissues from immunodeficient Ku70−/− (22), Ku80−/− (23), and Atm−/− (24) mice was comparable to the wild-type controls (data for Ku70−/− and Ku80−/− mice not shown, for Atm−/− mice see Fig. 1Ah). As expected, there were rare TUNEL-positive cells in the p53−/− thymus (Fig. 1A g and k). Quantitatively, of the 3,000 cells counted for each genotype of animals at 10 hr after a dose of 10 Gy, >70% of cells were apoptotic in the wild-type and Atm−/− thymus, but only 15% (±3%) in DNA-PKcs−/− thymus and very few in the thymus of p53−/− mice (≈2%) (Fig. 1B). Similar results were obtained at 10 hr after 20 Gy of IR (Fig. 1B). Double staining with anti-CD34 antibody and TUNEL demonstrated that the apoptosis observed in DNA-PKcs−/− thymus represents mostly endothelial cells (data not shown). Thus, DNA-PKcs deficiency significantly suppresses IR-induced p53-dependent apoptosis in the thymocytes of whole body-irradiated DNA-PKcs−/− mice.
Figure 1.
Radiation-induced apoptosis in thymic tissues of wild-type (WT) , DNA-PKcs−/−, p53−/−, and Atm−/− mice. (A) Animals were mock-irradiated (a–d), irradiated with 10 Gy (e–h), or 20 Gy (i–k), and apoptosis was evaluated 10 hr postirradiation (e–k) by using the TUNEL in situ assay. (B) Graphic representation of IR-induced apoptosis in thymus. At least 3,000 cells were counted in a minimum of three animals for each genotype either before irradiation, or 10 hr after 10 Gy and 20 Gy, respectively. Each value represents the mean ± SD in percentage of apoptotic cells. Black bars represent values obtained in unirradiated animals, and hatched bars those from irradiated animals. Double staining with anti-CD34 antibody and TUNEL demonstrated that the apoptosis observed in DNA-PKcs−/− thymus represents mostly endothelial cells (data not shown).
DNA-PKcs Deficiency Does Not Disrupt Cell-Cycle Arrest After IR.
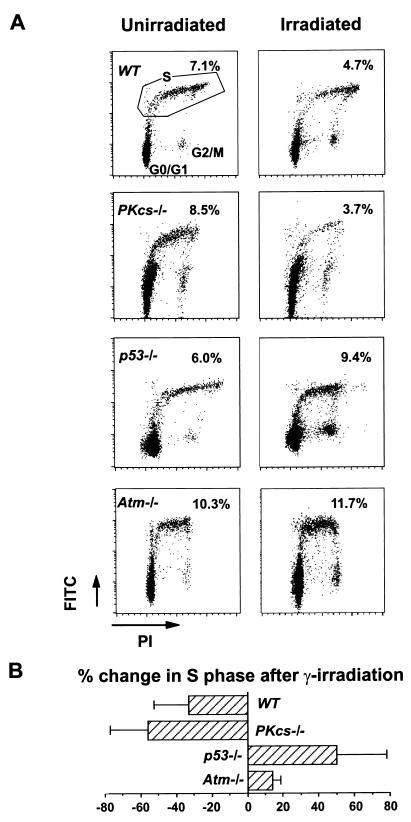
Radiation-induced G1/S checkpoint delay was studied in vivo by flow cytometric analysis of cells extracted from the thymus of irradiated and mock-irradiated animals (Fig. 2). In wild-type animals, IR induced a G1/S checkpoint delay, resulting in a decrease in the number of thymic BrdUrd-positive S-phase cells, from 7.1% in control mice to 4.7% in irradiated mice. The G1/S checkpoint delay also was observed in DNA-PKcs−/− mice as evidenced by a reduction in BrdUrd-positive S-phase cells, from 8.5% in control mice to 3.7% in irradiated mice. In contrast, the G1/S checkpoint in p53−/− and Atm−/− mice was defective, in agreement with previous reports (3, 25–27). In similar in vitro experiments using primary MEFs (data not shown), IR resulted in a decrease in the number of BrdUrd-positive S-phase cells in both wild-type and DNA-PKcs−/− MEFs, indicative of cell-cycle arrest at the G1/S transition. On the other hand, IR did not induce G1/S cell-cycle arrest in p53−/− MEFs (3, 25–27).
Figure 2.
IR-induced cell-cycle checkpoints in vivo in wild-type (WT), DNA-PKcs−/−, p53−/−, and Atm−/− mice. (A) Representative flow-cytometry scatter plots of thymocytes isolated from unirradiated or irradiated mice 2 hr after injection with BrdUrd, plotted as increasing fluorescence of propidium iodine (PI, x axis) versus increasing FITC fluorescence obtained with an anti-BrdUrd antibody and FITC-conjugated second antibody to detect BrdUrd incorporation (y axis). Gating for S-phase cells is shown from unirradiated wild-type mice (Top Left). The percentage of S-phase cells before and after irradiation for each genotype is shown in the corresponding panels. (B) Graph representing the mean ± SD change in the percentage of S-phase cells after irradiation.
p21 But Not Bax Was Significantly Induced by IR in DNA-PKcs−/− Thymus.
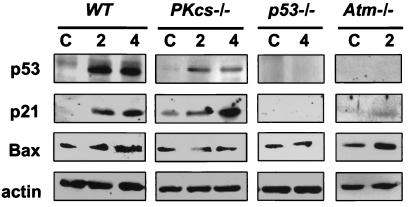
p53 transactivates p21 and Bax, mediating cell-cycle arrest and apoptosis (1, 2, 28–30). To determine whether DNA-PKcs was required for the p53-dependent transactivation of p21 and Bax, we performed immunoblot analysis (Fig. 3). We found that p53 protein could not be detected in the unirradiated wild-type and DNA-PKcs−/− mice, but was markedly induced in both wild-type and DNA-PKcs−/− thymuses at 2 and 4 hr after 10 Gy of IR (Fig. 3 Top). As expected, p53 was absent in the p53−/− mice (18). There was very low level of p21 and Bax in the thymuses of unirradiated wild-type animals and substantial induction of p21 and Bax protein at 4 hr after 10 Gy (Fig. 3 Middle). The constitutive p21 level in the thymus of unirradiated DNA-PKcs−/− mice was higher than that of the wild-type mice, perhaps because of the presence of DNA double-strand breaks in the thymus caused by their deficiency in V(D)J recombination (14). Nonetheless, p21 was significantly induced by IR in DNA-PKcs−/− thymus, with the level of increase comparable to that in the wild-type mice. Although p21 was induced by IR in both genotypes, Bax was not induced in DNA-PKcs−/− thymus (Fig. 3 Middle). Neither p21 nor Bax was induced by IR in the thymus of p53−/− mice (Fig. 3), consistent with the p53 dependency of their induction. Experiments conducted at other doses (e.g., 4, 5, and 7 Gy) yielded similar results (data not shown).
Figure 3.
Immunoblot analysis of p53, p21, and Bax protein expression in irradiated wild-type (WT), DNA-PKcs−/−, p53−/−, and Atm−/− mice. Wild-type, DNA-PKcs−/−, p53−/−, and Atm−/− mice were exposed to 10 Gy of whole-body irradiation. Two and four hours after IR, mice were sacrificed by asphyxiation, the thymuses dissected, and thymocytes prepared. Immunoblotting was performed directly on isolated thymocytes by using standard techniques (21). Immunoblots (50 μg/lane) of thymic extracts isolated from wild-type, DNA-PKcs−/−, p53−/−, and Atm−/− mice were probed with a mouse mAb against p53 and rabbit antisera against p21 and Bax, respectively. Equivalent protein concentrations in extracts from each genotype were demonstrated by separate immunoblot analysis of actin. C, control, unirradiated; 2 and 4: 2 hr and 4 hr after a dose of 10 Gy (whole-body irradiation). Experiments were repeated 2–3 times, and duplicates or triplicates of immunoblots were analyzed, which yielded consistent results.
We also confirmed previous findings on the IR-induced responses of Atm−/− mice (3). In contrast to DNA-PKcs−/− mice, p53 protein levels were not induced by IR in thymus of Atm−/− mice (Fig. 3). IR-induced cell-cycle (G1/S) checkpoint function was also defective (Fig. 2), and induction of p21 was attenuated in thymus from Atm−/− mice (Fig. 3). However, IR-induced apoptosis (Fig. 1) and Bax induction (Fig. 3) were normal compared with the wild-type irradiated controls.
Discussion
The above results suggest that, in response to IR, DNA-PKcs actives p53 in such a way to selectively induce certain p53-dependent function, i.e., apoptosis, but not others, i.e., the p21 induction and cell-cycle arrest. Consistent with the current in vivo results are our previous in vitro findings using primary DNA-PKcs −/− MEFs (15). Specifically, our data showed that IR induced normal p53 accumulation and serine-18 (equivalent to serine-15 of human p53) phosphorylation, sequence-specific DNA binding by p53, transactivation of p21 gene expression, and normal G1/S cell-cycle arrest, even in the absence of DNA-PKcs (15).
Given the above, that DNA-PKcs is not essential for p53-mediated p21 induction and cell-cycle arrest in response to IR, other proteins must be involved in the signal transduction pathway. In this regard, Atm appears to be a likely candidate. It has been shown by Barlow et al. (3) that Atm deficiency is related to defective induction of p53 by IR and associated with a lack of G1/S cell-cycle arrest and p21 induction, although normal apoptotic responses and Bax induction are retained. Those authors suggest that the IR-induced p21 increase and cell-cycle arrest are Atm mediated, but that the apoptotic response and Bax induction are independent of Atm function. Our data on the IR response of Atm−/− mice (Figs. 1–3) are in good agreement with Barlow et al.'s findings and further support this notion. A summary of different responses to IR in the wild-type, DNA-PKcs−/−, Atm−/−, and p53−/− mice is presented in Table 1. Based on these results, we suggest that DNA-PKcs and Atm complement each other to activate p53, but lead to distinct downstream pathways, resulting in either cell-cycle arrest or apoptosis. In this model, DNA-PKcs is required for normal apoptotic responses to IR, but not for cell-cycle arrest. Conversely, Atm is required for normal cell-cycle arrest but not for apoptosis (3). However, we cannot rule out the possibility that DNA-PKcs may have some effects on p53-dependent cell-cycle arrest that cannot be detected because of redundancy in pathways that activates p53 to stimulate cell-cycle checkpoint function. Conversely, Atm may have some effects on p53-dependent apoptosis that cannot be detected because of redundant pathways that activate p53 to trigger apoptosis.
Table 1.
Responses of the wild-type, DNA-PKcs−/−, Atm−/−, and p53−/− mice after 10 Gy of IR
| Wild type | DNA-PKcs−/− | Atm−/− | p53−/− | |
|---|---|---|---|---|
| p53* | ++++ | ++ | Not detectable | Not detectable |
| p21* | ++++ | ++/+++ | + | Not detectable |
| Bax* | + | − | + | − |
| G1/S arrest† | Normal | Normal | Deficient | Deficient |
| Apoptosis‡ | Normal | Deficient | Normal | Deficient |
Wild-type, DNA-PKcs−/−, Atm−/−, and p53−/− mice were exposed to 10 Gy of whole-body γ-irradiation. Two and/or four hours after IR, mice were sacrificed by asphyxiation, the thymuses dissected and thymocytes prepared. Immunoblotting was performed by using standard techniques and antibodies specific to p53, p21, and Bax, respectively. Signals of p53, p21, and Bax expression were quantified using AMBIS scanning densitometer and software. Relative increase of p53, p21, and Bax expression after IR is expressed as follows: ++++: greater than 4-fold increase; +++: 3- to 4-fold increase, ++: 2- to 3-fold increase, +: less than 2-fold increase; −: no change or decrease in protein expression after IR.
†See Fig. 2.
‡See Fig. 1.
As shown in Fig. 3, there is significantly less p53 in the DNA-PKcs−/− thymic tissue than in wild-type tissue. It has been shown previously that the level of p53 can dictate whether a cell arrests (low p53 level) or undergoes apoptosis (high p53 level) (31). Therefore, it is possible that DNA-PKcs regulates apoptosis through controlling the level of p53 as well as its state of modification. The induction of Bax expression might require a higher threshold of p53 than that needed for p21 expression as suggested by the IR responses of DNA-PKcs−/− thymus (Table 1). However, it appears that apoptotic response and Bax induction, both being normal in irradiated Atm−/− thymus, do not require increased p53 level (ref. 3 and Table 1). It is possible that phosphorylation of distinct sites of p53 (e.g., serine 20) results in differential activity of p53. Thus, distinct, differentially activated p53 may lead to disparate downstream pathways, mediating the cellular choice between cell-cycle arrest and apoptosis. Furthermore, one cannot exclude the possibility that apoptosis in irradiated thymic tissue requires both p53 function and other as-yet-unidentified factor(s). DNA-PKcs could conceivably regulate this hypothetical factor rather than p53, and, because regulation of this factor would be intact in Atm−/− thymic tissue, such cells would be able to undergo apoptosis. This possibility would, of course, still leave open the question as to why Atm regulates cell-cycle arrest mediated by p53 (3).
The differential activation of p53 has been suggested as a common mechanism to mediate diverse responses to different types of DNA damage. Recently, p53 has been shown to be modified, upon DNA damage, at a number of N- and C-terminal phosphorylation sites including serines 15, 20, and 33 (32). Even though it has not yet been established which kinases are responsible for phosphorylating p53 in vivo, a number of candidates have been suggested. It has been shown that the serine-15 site can be phosphorylated by DNA-PKcs (33), ATM (34–36), and ATR (ATM + Rad3-related) (35, 37). Much less is known about regulation of serine 33 and serine 20. It is possible that IR triggers different downstream target genes by inducing combinations of phosphorylation via Atm and/or DNA-PKcs, thus selectively regulating distinct p53-dependent cell-cycle arrest and apoptosis. There has been much recent discussion as to whether there is any connection between DNA-PKcs and p53 (4, 15, 38, 39). Our study provides a biological framework strongly suggesting a link between DNA-PKcs and p53 in response to IR.
Acknowledgments
We thank Carol Prives for stimulating discussions, Tom Delohery for flow cytometric analysis, Maria S. Jiao for technical assistance, and P. Krechmer for word processing. The work was supported in part by National Institutes of Health Grants CA-31397, CA-56909, and CA-78497 (G.C.L.), and the Department of Energy Office of Health and Environmental Research (D.J.C.). H.O. is a postdoctoral fellow supported in part by National Institutes of Health Training Grant CA-61801.
Abbreviations
- ATM
Ataxia telangiectusia mutated
- IR
ionizing radiation
- DNA-PKcs
catalytic subunit of DNA-dependent protein kinase
- SCID
severe combined immunodeficiency
- MEF
mouse embryo fibroblast
- TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
References
- 1.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Barlow C, Brown K D, Deng C X, Tagle D A, Wynshaw-Boris A. Nat Genet. 1997;17:453–456. doi: 10.1038/ng1297-453. [DOI] [PubMed] [Google Scholar]
- 4.Woo R A, McLure K G, Lees-Miller S P, Rancourt D E, Lee P W K. Nature (London) 1998;394:700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- 5.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt C A, Lowe S W. J Pathol. 1999;187:127–137. doi: 10.1002/(SICI)1096-9896(199901)187:1<127::AID-PATH251>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 7.Smith G C M, Jackson S P. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 8.Anderson C W. Trends Biochem Sci. 1993;18:433–437. doi: 10.1016/0968-0004(93)90144-c. [DOI] [PubMed] [Google Scholar]
- 9.Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, Fukumura R, Morimyo M, Muto M, Itoh M, et al. Proc Natl Acad Sci USA. 1997;94:2438–2443. doi: 10.1073/pnas.94.6.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L C, Clarkin K C, Wahl G M. Cancer Res. 1996;56:2940–2944. [PubMed] [Google Scholar]
- 11.Guidos C J, Williams C J, Grandal I, Knowles G, Huang M T F, Danska J S. Genes Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 12.Gurley K E, Kemp C J. Carcinogenesis. 1996;17:2537–2542. doi: 10.1093/carcin/17.12.2537. [DOI] [PubMed] [Google Scholar]
- 13.Rathmell W K, Kaufmann W K, Hurt J C, Byrd L L, Chu G. Cancer Res. 1997;57:68–74. [PubMed] [Google Scholar]
- 14.Kurimasa A, Ouyang H, Dong L-J, Wang S, Li X, Cordon-Cardo C, Chen D J, Li G C. Proc Natl Acad Sci USA. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burma S, Kurimasa A, Xie G, Taya Y, Araki R, Abe M, Crissman H A, Li G C, Chen D. J Biol Chem. 1999;274:17139–17143. doi: 10.1074/jbc.274.24.17139. [DOI] [PubMed] [Google Scholar]
- 16.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 17.Santana P, Pena L A, Haimovitz-Friedman A, Martin S, Green D, McLoughlin M, Cordon-Cardo C, Schuchman E H, Fuks Z, Kolesnick R. Cell. 1996;86:189–199. doi: 10.1016/s0092-8674(00)80091-4. [DOI] [PubMed] [Google Scholar]
- 18.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 19.Liao M-L, Yin C, Barlow C, Wynshaw-Boris A, Van Dyke T. Mol Cell Biol. 1999;19:3095–3102. doi: 10.1128/mcb.19.4.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuks Z, Alfieri A, Haimovitz-Friedman A, Seddon A, Cordon-Cardo C. Cancer J. 1995;1:62–72. [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 22.Ouyang H, Nussenzweig A, Kurimasa A, da Costa Soares V, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M, Iliakis G, et al. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 24.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley J N, Ried T, Tagle D, Wynshaw-Boris A. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 25.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 26.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 27.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Nature (London) 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 28.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 29.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 30.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Ko L J, Jayaraman L, Prives C. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 32.Shieh S-Y, Taya Y, Prives C. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lees-Miller S P, Chen Y-R, Anderson C W. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 35.Canman C, Lim D, Cimprich K, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M, Siliciano J. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 36.Khanna K, Keating K K, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller S P, Lavin M F. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 37.Tibbetts R, Brunbaugh K, Williams J, Sarkaria J, Cliby W, Shieh S-Y, Prives C, Abraham R. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Araki R, Fukumura R, Fujimori A, Taya Y, Shiloh Y, Kurimasa A, Burma S, Li G C, Chen D J, Sato K, et al. Cancer Res. 1999;59:3543–3546. [PubMed] [Google Scholar]
- 39.Jimenez G S, Bryntesson F, Torres-Arayus M I, Priestly A, Beeche M, Saito S, Sakaguchi K, Appella E, Jeggo P A, Taccioli G E, et al. Nature (London) 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]