Abstract
Constitutive NF-κB activation is among the many deregulated signaling pathways that are proposed to drive pancreatic cancer cell growth and survival. Recent reports suggest that glycogen synthase kinase-3β (GSK-3β) plays a key role in maintaining basal NF-κB target gene expression and cell survival in pancreatic cancer cell lines. However, the mechanism by which GSK-3β facilitates constitutive NF-κB signaling in pancreatic cancer remains unclear. In this report, we analyze the contributions of both GSK-3 isoforms (GSK-3α, GSK-3β) in regulating NF-κB activation and cell proliferation in pancreatic cancer cell lines (Panc-1 and MiaPaCa-2). We demonstrate that GSK-3 isoforms are differentially required to maintain basal NF-κB DNA binding activity, transcriptional activity, and cell proliferation in Panc-1 and MiaPaCa-2 cells. Our data also indicate that IKK subunits are not equally required to regulate pancreatic cancer-associated NF-κB activity and cell growth. Importantly, we provide the first evidence that GSK-3 maintains constitutive NF-κB signaling in pancreatic cancer by regulating IKK activity. These data provide new insight into GSK-3-dependent NF-κB regulation, and further establishes GSK-3 and IKK as potential therapeutic targets for pancreatic cancer.
Keywords: Pancreatic cancer, glycogen synthase kinase-3, IκB kinase, NF-κB
Introduction
Pancreatic cancer represents the fourth leading cause of cancer-related death in the United States with a five-year patient survival rate of 5% (1). Moreover, an estimated 33,370 patients were expected to die from this disease in 2007 (1). The dismal mortality rate from pancreatic cancer stems from its aggressive metastatic nature and its ability to resist conventional chemotherapies (2). Therefore, expanding our knowledge of the complex molecular pathways responsible for the development of pancreatic cancer is critical for the discovery of new therapeutic strategies. Deregulated NF-κB and glycogen synthase kinase-3 signaling are among the many pathways that have been implicated in the pathogenesis of pancreatic cancer.
NF-κB represents a family of evolutionarily conserved transcription factors consisting of five members: c-Rel, RelA (p65), RelB, p50 (NF-κB1/p105 precursor), and p52 (NF-κB2/p100 precursor) (3). The most studied NF-κB complex consists of the p65/p50 heterodimer. In resting cells, NF-κB is rendered inactive within the cytoplasm through association with inhibitory IκB proteins. Various inflammatory stimuli can trigger the activation of the IκB kinase (IKK) complex which consists of a regulatory subunit (IKKγ) and two catalytic subunits (IKKα and IKKβ) (4). Upon IKK activation, IκB is phosphorylated and subsequently targeted for rapid proteosomal degradation thus liberating NF-κB for nuclear translocation, enhanced DNA binding and transcriptional regulation (5).
Constitutive activation of NF-κB has been characterized in numerous human cancers and is associated with regulating genes that control cell survival, proliferation, metastasis, and angiogenesis (6). Importantly, constitutive NF-κB activation has been observed in 70% of human pancreatic cancers as well as in human pancreatic cancer cell lines and animal models (7–9). Studies have also demonstrated constitutive IKK activity to play a key role in regulating cell survival and cell cycle progression in multiple in vitro pancreatic cancer models (8, 10). Thus, there has been growing interest in utilizing IKK as a chemotherapeutic target for pancreatic cancer.
Glycogen synthase kinase-3 (GSK-3) is a serine/theronine kinase that exists as two highly similar mammalian isoforms (GSK-3α and GSK-3β) (11, 12). GSK-3 is recognized for its role in downregulating β-catenin, thus suppressing the transcriptional activity of T-cell-specific transcription factor (TCF)/lymphoid enhancer factor (LEF) complexes within the Wnt/β-catenin pathway (13). Numerous reports have subsequently demonstrated involvement of this multifunctional kinase in regulating a variety of transcription factors involved in cancer progression, including NF-κB (13–18). A GSK-3β deficient mouse model provided the first evidence of GSK-3-dependent NF-κB regulation (19). These data show that the loss of GSK-3β results in defective NF-κB signaling in response to TNF-α. Furthermore, we previously reported that GSK-3β specifies promoter-specific recruitment of p65/RelA to NF-κB-dependent genes in response to TNF-α (20). A previous report has also implicated GSK-3β in playing a critical role in regulating constitutive NF-κB reporter activity and target gene expression within in vitro pancreatic cancer models (21). However, the mechanism by which GSK-3β drives constitutive or inducible NF-κB has not been characterized.
Despite the structural similarity between GSK-3α and GSK-3β evidence suggests that these isoforms are not functionally redundant in regulating NF-κB (19, 22). In this report, we characterize the individual roles that each GSK-3 isoform play in maintaining constitutive NF-κB activity and cell proliferation in pancreatic cancer cell lines (Panc-1 and MiaPaCa-2). We show that both GSK-3 isoforms can function to regulate basal NF-κB DNA binding and transcriptional activity, whereas GSK-3α predominantly controls cell growth and survival. Our data also demonstrate that IKKα and IKKβ exhibit different requirements to drive constitutive NF-κB activity in a pancreatic cancer cell type-dependent manner. Additionally, we provide the first evidence that links GSK-3 to constitutive IKK activity in pancreatic cancer cells.
Materials and Methods
Cell Culture and Reagents
Panc-1 (CRL-1496) and MiaPaCa-2 (CRL-1420) pancreatic cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA). Panc-1 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum and 100 units/ml penicillin/streptomycin. MiaPaCa-2 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 2.5% horse serum. Cells were cultured in DMEM supplemented with 0.5% fetal bovine serum for 24 hour prior to experimentation. All cell culture reagents were obtained from Invitrogen (Carlsbad, CA). The following antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA): p65 (SC-109), p50 (SC-7178), GSK-3α/β (SC-7291), β-tubulin (SC-9104), and GST (SC-33613). IKKα clone 14A231 and IKKβ clone10AG2 antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). The following antibodies were obtained from Cell Signaling Technology (Beverly, MA): phospho-p65 (serine 536), phospho-glycogen synthase (serine 641), glycogen synthase, cleaved caspase-3 (Asp 175), and caspase-3. TNF-α was purchased from Promega (Madison, WI). GSK-3 inhibitors (AR-A014418 and SB216763) were obtained from Sigma-Aldrich (St. Louis, MO). The IKKβ inhibitor (Compound A) was provided by Bayer Healthcare (Wuppertal, Germany).
Small RNA interference
The following siRNA (siGenome SMARTpool) was obtained from Dharmacon (Layfayette, CO) as a pool of four annealed double-stranded RNA oligonucleotides: IKKα (M-003473-02), IKKβ (M-003503-03), GSK-3α (M-003009-01), GSK-3β (M-003010-03), and non-targeting control #3 (D001201-03). In brief, cells were cultured to 70% confluency in six-well plates. Dharmafect 1 transfection reagent (Layfayette, CO) was used to transfect 100nM siRNA according to manufacturer’s instruction.
Electrophoretic mobility shift assay (EMSA) and NF-κB DNA binding ELISA
EMSA and NF-κB super-shift analysis was performed on nuclear extracts as previously described (20) using 32P-labeled oligonucleotide probe corresponding to an NF-κB site within the MHC class I promoter region. Relative p65 DNA binding activity was quantified using the TransAM NF-κB p65 transcription factor assay kit (Active Motif, Carlsbad, CA) according to manufacturer’s instructions. DNA binding activity was measured in triplicate at 450nm wavelength on a Versamax Microplate Reader (Molecular Devices Corp., Sunnyvale, CA)
Western blot analysis
Whole-cell lysates were prepared on ice using Mammalian Protein Extraction Reagent (M-PER) (Pierce Biotechnology, Rockford, IL) according to manufacturer’s instructions. Cytoplasmic extracts were prepared as previously described (23). Protein extracts were quantified by Bradford assay (Bio-Rad Laboratories, Hercules, CA) and analyzed by SDS-PAGE as previously described (20).
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS tetrazolium) cellular proliferation assay
Cells were seeded in triplicate at 3 × 103 cells per well (96-well plate) and cultured in the presence or absence of GSK-3 or IKKβ inhibitors at the indicated time course. Alternatively, cells were transiently transfected with appropriate siRNA and cultured at the indicated time points post-transfection. At the end of each time point, MTS tetrazolium compound (Promega, Madison, WI) was added and absorbance was read at 490nm on a Versamax Microplate Reader (Molecular Devices Corp., Sunnyvale, CA)
Dual-luciferase reporter assay
Cells were seeded in triplicate at 2 × 104 cells per well (24-well plate), transfected with the appropriate siRNA as described above, and cultured for 24 hours. After siRNA transfection, cells were co-transfected with 200ng of luciferase reporter construct containing tandem NF-κB binding sites from the MHC class I promoter region and 5ng pRL-TK Renilla luciferase construct (Promega, Madison, WI) using Fugene6 transfection reagent (Roche Applied Science, Indianapolis, IN). Cells were cultured for an additional 24 hours, harvested in passive lysis buffer, and analyzed according to Dual-Luciferase Assay System protocol (Promega, Madison, WI). Relative light units were measured on an Lmax Microplate Luminometer (Molecular Devices Corp., Sunnyvale, CA) and normalized to pRL-TK Renilla luciferase light units.
Real-time PCR
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. Two micrograms of RNA was reverse transcribed into cDNA using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA). Real-time PCR was performed and analyzed as previously described (20) using TaqMan Gene Expression Assay primer-probe sets (Applied Biosystems, Foster City, CA) for IκBα (Hs00153283_m1) and Bcl-xL (Hs00236329_m1).
IKK kinase assay
Whole-cell lysate were prepared on ice for 45 minutes in the following lysis buffer containing 20mM Tris (pH 8.0), 500mM NaCl, 0.25% Triton-X100, 1mM EDTA, 1mM EGTA, 1mM dithiothreitol (DTT), 1X protease inhibitor (Roche Applied Science, Indianapolis, IN), and 1X phosphatase inhibitor cocktail 1 (Sigma-Aldrich, St. Louis, MO). IKK complexes were immunoprecipitated from 500μg protein extract using an IKKα antibody (Upstate Biotechnology). An in-vitro kinase assay was performed and analyzed as previously described (20). GST-IκBα substrate phosphorylation was visualized by autoradiography and quantitated using Image Quant Version 5.2 software (Molecular Dynamics, Sunnyvale, CA).
Luminescent-based caspase-3/7 activity assay
Cells were plated in triplicate at 2 × 103 cells per well in white-walled 96-well plates (Becton Dickinson, Franklin Lakes, NJ). Cells were transiently transfected with siRNA as described above. Caspase-3/7 activity was measured at 48, 72, and 96 hours post-transfection using the Caspase-Glo 3/7 assay (Promega, Madison, WI) according to manufacturer’s instructions. Caspase-Glo 3/7 assay utilizes a caspase-3/7 tetrapeptide DEVD substrate which produces a luminescent signal upon cleavage. Relative light units were measured on an Lmax Microplate Luminometer (Molecular Devices Corp., Sunnyvale, CA).
Statistical analysis
Prism software (GraphPad Software, Inc. San Diego, CA) was used for statistical analysis of data. An unpaired t test was used to evaluate differences between group means. P values less than 0.05 were considered to be statistically significant.
Results
GSK-3 isoforms regulate constitutive NF-κB activity in pancreatic cancer cells
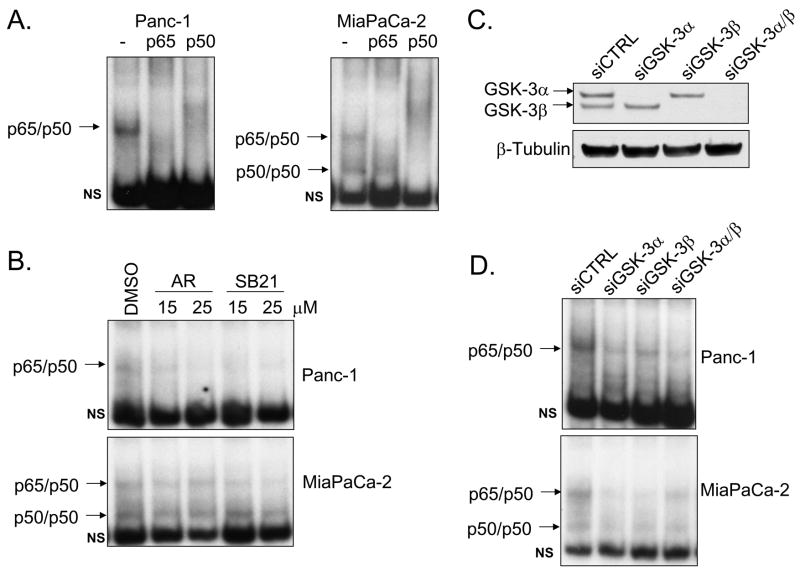
A recent study emphasized the role GSK-3β plays in maintaining constitutive NF-κB reporter and target gene expression in pancreatic cancer cells (21). However, this report did not address the individual requirements of both GSK-3 isoforms in regulating NF-κB. Furthermore, the mechanism by which GSK-3 regulates NF-κB activity requires understanding. Here we have analyzed constitutive NF-κB DNA binding and transcriptional activity in two well-known in vitro pancreatic carcinoma models (Panc-1 and MiaPaCa-2). As shown in Fig. 1A, constitutive NF-κB DNA binding activity is detected in Panc-1 cells through EMSA analysis of nuclear extracts. This form of NF-κB activity is shown to be the p65/p50 heterodimer through supershift analysis (Fig. 1A). EMSA analysis of MiaPaca-2 cells demonstrated constitutive DNA binding of p65/p50 heterodimers as well as p50 homodimers (Fig. 1A). Cells were treated with two structurally distinct pharmacological inhibitors of both GSK-3 isoforms (AR-A014418 and SB-216763) to determine a potential requirement of GSK-3 in controlling constitutive NF-κB DNA binding activity. Previously reported treatment duration and inhibitor concentrations (21) were used in our studies. Treatment with either GSK-3 inhibitor for 24 hours at 15 and 25μM reduces p65/50 DNA binding in both cell lines (Fig. 1B). DNA binding activity of p50 homodimers was largely unaffected by GSK-3 inhibition in MiaPaCa-2 cells. To ask whether direct inhibition of GSK-3 isoforms would affect NF-κB constitutive DNA binding activity in pancreatic cancer cells, RNA interference of GSK-3α and GSK-3β was utilized. Western blot analysis confirms that GSK-3 RNA interference was isoform-specific and significantly reduced GSK-3 protein levels relative to non-targeting siRNA control (Fig. 1C). Loss of either GSK-3 isoform reduced constitutive NF-κB DNA binding in both cell lines (Fig. 1D). Moreover, an ELISA-based DNA binding assay demonstrated a significant reduction of p65 binding activity following the knock-down of either GSK-3 isoform (Supplemental Fig. 1)
Figure 1. GSK-3 is required for constitutive NF-κB DNA binding.
EMSA was performed on Panc-1 and MiaPaCa-2 nuclear extracts using 32P-labeled NF-κB-specific probe. (A) Super-shift analysis was performed using antibodies against p65 and p50. Arrows indicate p65/50 and p50/p50 NF-κB complexes. Non-specific binding is specified by NS. (B) Cells were treated with vehicle control (DMSO) or GSK-3 inhibitors (AR-A014418 and SB216763) at 15 and 25μM for 24 hours. (C) Panc-1 cells were transiently transfected with 100nM siRNA targeted against GSK-3α, GSK-3β, combined GSK-3α/β, and non-targeting control (siCTRL) for 48 hours. Cytoplasmic extracts were harvested and separated by SDS-PAGE. Immunoblots were performed using the specified antibodies. (D) Nuclear extracts were harvested from Panc-1 and MiaPaCa-2 cells transfected with siRNA as described above and EMSA was performed.
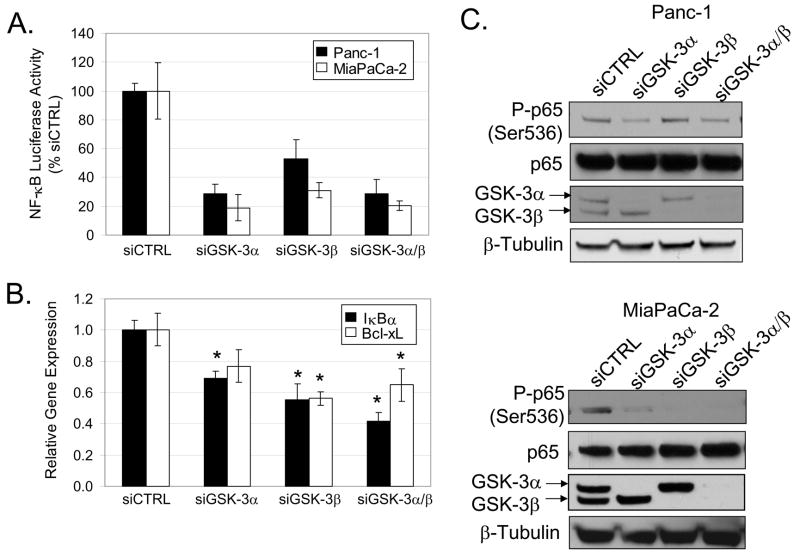
To determine whether GSK-3 isoforms play roles in regulating NF-κB transcriptional activity, we measured the effect of GSK-3 RNA interference on basal NF-κB-luciferase reporter activity. Our data show that knock-down of either GSK-3 isoform resulted in a significant reduction of constitutive NF-κB-luciferase reporter activity in Panc-1 and MiaPaCa-2 cells (Fig. 2A). Next, we examined whether GSK-3-dependent regulation of reporter activity corresponded with the expression of two NF-κB-regulated genes (IκBα and Bcl-xL). Loss of GSK-3β alone and both GSK-3α/β significantly suppressed expression of Bcl-xL (Fig. 2B). Furthermore, knock-down of either GSK-3 isoform alone or combined GSK-3α/β resulted in reduced expression of IκBα (Fig. 2B). Serine 536 phosphorylation of the NF-κB subunit, p65/RelA, correlates with its transcriptional activity (24). To determine whether the GSK-3 isoforms regulate p65 phosphorylation, RNA interference was used. Extracts were prepared from cells treated with control siRNA or siRNA for either GSK-3 isoform. Consistent with the elevated NF-κB activity in Panc-1 in MiaPaCa-2 cells, we also observe constitutive serine 536 phosphorylation of p65 (Fig. 2C). RNA interference of GSK-3α or combined GSK-3α/β reduced p65 phosphorylation in Panc-1 cells (Fig. 2C). Conversely, knock-down of GSK-3β had a greater effect on basal p65 phosphorylation in MiaPaCa-2 cells (Fig. 2C). Taken together, these data suggest that both GSK-3 isoforms play individual roles in maintaining constitutive NF-κB DNA binding and transcriptional activity in Panc-1 and MiaPaCa-2 cells.
Figure 2. GSK-3 regulates NF-κB transcriptional activity.
(A) Panc-1 and MiaPaCa-2 cells were transiently transfected with 100nM siRNA targeted against GSK-3α, GSK-3β, combined GSK-3α/β, and non-targeting control (siCTRL) for 24 hours. Cells were subsequently transfected with 3X-NF-κB and renilla luciferase reporter constructs for 24 hours. Luciferase activity was measured in triplicate and indicated as % activity relative to siCTRL. Reporter activity from cells transfected with GSK-3 siRNA were all found to be significantly different relative to siCTRL (P < 0.05). (B) Panc-1 cells were transfected with siRNA as described above for 48 hours. RNA was harvested and expression of IκBα and Bcl-xL was analyzed by real-time PCR in triplicate. Astericks indicates statistical significance relative to siCTRL (P < 0.05). (C) Panc-1 and MiaPaCa-2 were transiently transfected with siRNA as described above for 48 hours. Whole cell extracts were harvested and separated by SDS-PAGE. Immunoblots were performed using the specified antibodies.
GSK-3α promotes pancreatic cancer cell growth and survival
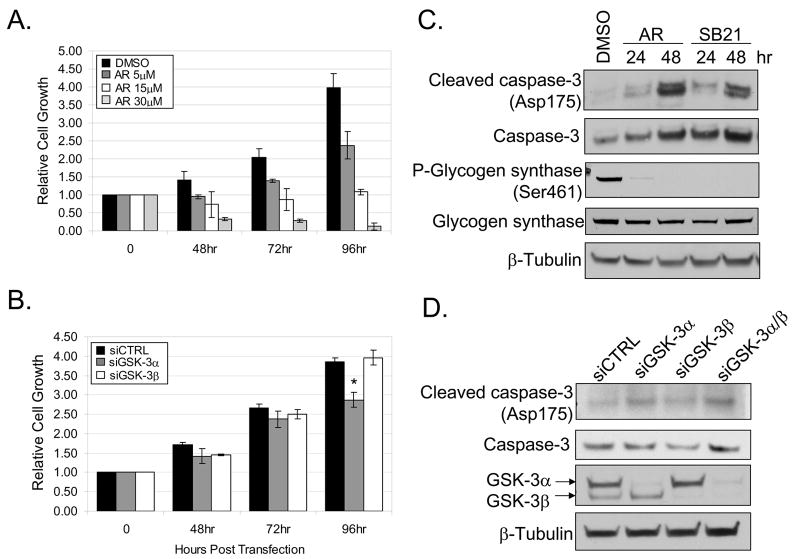
Constitutive NF-κB activity has been reported to play mitogenic and anti-apoptotic roles in pancreatic cancer cell lines (8, 9). Pharmacological GSK-3 inhibition and GSK-3 RNA interference were used to determine a role for GSK-3 in controlling proliferation and survival of Panc-1 and MiaPaCa-2 cells. Consistent with a previous report (21), GSK-3 inhibition (AR-A014418) decreased growth of Panc-1 cells in a dose-dependent manner (5, 15, 30μM) over a 96 hour time course (Fig. 3A). Similar results were observed in MiaPaCa-2 cells (data not shown). Next, we used RNA interference to determine the individual requirements for GSK-3α and GSK-3β on cell viability. We observed a significant reduction in Panc-1 cell growth following GSK-3α RNA interference at 96 hours post siRNA transfection when compared to non-targeting control and GSK-3β siRNA (Fig. 3B). Similar results were observed in MiaPaCa-2 cells (data not shown). Moreover, the knock-down of GSK-3α in Panc-1 cells resulted in a significant reduction in the total number of cells at 72 and 96 hours post siRNA transfection (Supplemental Fig. 2A). Western blot analysis confirmed knock-down of GSK-3 isoforms at each time-point post siRNA transfection (Supplemental Fig. 2B). Importantly, we did not observe complete GSK-3 knock-down until 72 and 96 hours post siRNA transfection (Supplemental Fig. 2B). These data could explain why the effects of GSK-3α knock-down were only seen at later time-points post siRNA transfection.
Figure 3. Blockade of GSK-3 suppresses cell proliferation.
(A) Panc-1 cells were treated with DMSO or 5, 15, and 30μM of the GSK-3 inhibitor (AR-A014418) for 48, 72, and 96 hours. Cell growth was measured in triplicate at each time-point using a colormetric MTS tetrazolium assay. (B) Panc-1 cells were transiently transfected with siRNA targeted against GSK-3α, GSK-3β and siCTRL. Cell growth was measured as described above at 48, 72, and 96 hours post-transfection. Data was normalized to the initial cell density prior to GSK-3 inhibitor treatment or siRNA transfection respectively. Astericks indicates statistical significance relative to siCTRL (P < 0.05). (C) Panc-1 cells were treated with DMSO or 30μM of GSK-3 inhibitors (AR-A014418 and SB216764) for 24 and 48 hours. Whole-cell extracts were harvested and separated by SDS-PAGE. (D) Panc-1 cells were transiently transfected with 100nM siRNA targeted against GSK-3α, GSK-3β, combined GSK-3α/β, and siCTRL for 48 hours. Whole-cell extracts were harvested and separated by SDS-PAGE. Immunoblots were performed using the specified antibodies.
The induction of caspase-3 cleavage was also measured to determine whether the loss of cell viability following the disruption of GSK-3 was due to apoptosis. An increase in caspase-3 cleavage following 30μM pharmacologic GSK-3 inhibition was observed over 24 and 48 hours in Panc-1 (Fig. 3C). The efficiency of GSK-3 inhibition was indicated by the reduced basal phosphorylation of a GSK-3 substrate, glycogen synthase at serine 461 (Fig. 3C). Moreover, GSK-3α knock-down induced caspase-3 cleavage 48 hours post-transfection of siRNA (Fig. 3D). Similar results were observed in MiaPaCa-2 cells (data not shown). A luminescent caspase-3/7 assay was used to confirm a significant increase in caspase activity following the knock-down of GSK-3α or combined GSK-3α/β in Panc-1 cells (Supplemental Fig. 3). Notably, pharmacologic GSK-3 inhibition was more effective than GSK-3 knock-down in suppressing cell growth and inducing caspase-3 cleavage. These observations could indicate unknown off-target effects of pharmacologic GSK-3 inhibition (Fig. 3, see discussion). Taken together, we show that the loss of GSK-3α is more efficient than GSK-3β in suppressing cell growth and survival in Panc-1 and MiaPaCa-2 cells. Importantly, these observations correlate with the ability of GSK-3α to regulate constitutive NF-κB activity.
Constitutive NF-κB activity in pancreatic cancer cells is IKK-dependent
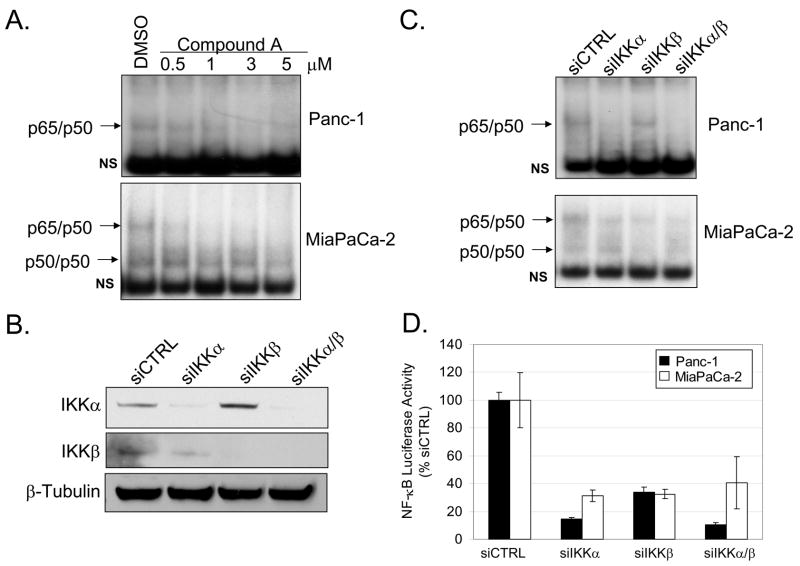
In addition to the studies shown above relative to GSK-3, studies have also shown IKK to be essential in regulating constitutive NF-κB signaling in pancreatic cancer cells (8–10). To confirm these findings, NF-κB DNA binding activity was analyzed in the presence of an established small molecule IKKβ inhibitor (Compound A) (25). Compound A treatment for 24 hours showed dose-dependent suppression of constitutive NF-κB DNA binding activity in Panc-1 and MiaPaCa-2 cells (Fig. 4A). We note that IKKβ inhibition is more effective in blocking constitutive NF-κB DNA binding activity in MiaPaCa-2 cells. Next, RNA interference to IKKα and IKKβ was utilized to determine the individual requirements of IKK subunits for constitutive NF-κB DNA binding activity and transcriptional activation. The knock-down of IKK subunits was specific and significantly reduced protein levels relative to non-targeting siRNA control (Figure 4B). We observed a decrease in p65/p50 DNA binding activity following the knock-down of either IKK subunit in MiaPaCa-2 cells (Fig. 4C). Interestingly, knock-down of IKKα alone or combined IKKα/β knock-down caused significant loss of NF-κB DNA binding activity in Panc-1 cells (Fig. 4C). The individual role that each IKK subunit plays in regulating basal NF-κB transcriptional activity was measured by an NF-κB reporter assay. RNA interference against either IKK subunit caused substantial suppression of reporter activity in both Panc-1 and MiaPaCa-2 cells (Fig. 4D). However, the suppressive effect of IKKα and combined IKKα/β knock-down were stronger than IKKβ knock-down alone (Fig. 4D). Thus, our data suggests that both IKK subunits play a role in regulating constitutive NF-κB activity in pancreatic cancer cells, but IKKα may play a more significant role in Panc-1 cells.
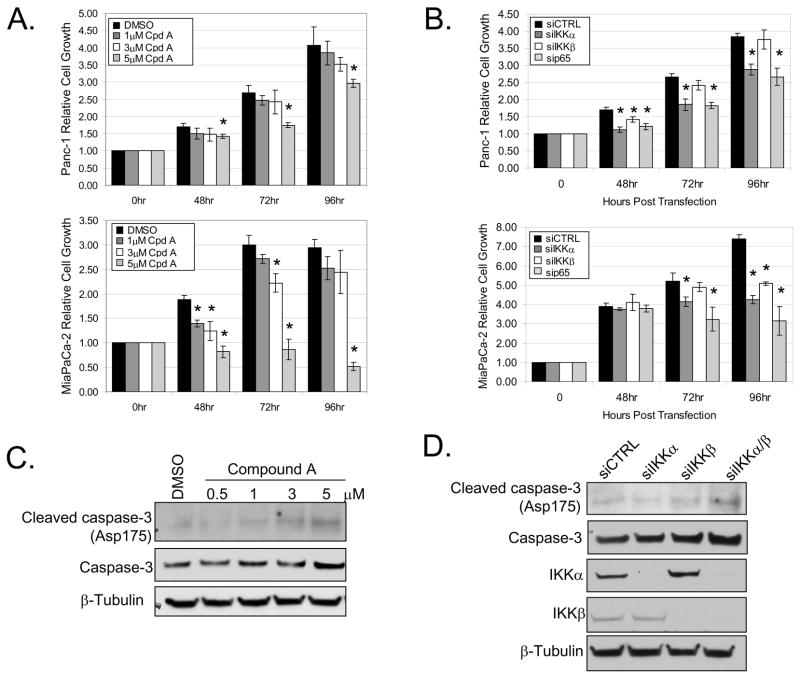
Figure 4. IKK is required for constitutive NF-κB activation.
(A) Panc-1 and MiaPaCa-2 cells were treated with DMSO or 0.5, 1, 3, and 5μM of IKKβ inhibitor (Compound A) for 24 hours. Nuclear extracts were harvested and EMSA was performed using 32P-labeled NF-κB-specific probe. Arrows indicate NF-κB complexes as determined by supershift analysis in Fig. 1A. Non-specific binding is indicated with NS. (B) Panc-1 cells were transiently transfected with 100nM siRNA targeted against IKKα, IKKβ, combined IKKα/β, and siCTRL for 48 hours. Cytoplasmic extracts were harvested from cells transfected with siRNA as described above. Extracts were separated by SDS-PAGE and immunoblots were performed using the specified antibodies. (C) EMSA was performed on nuclear extracts harvested from cells transfected with siRNA as described above. (D) Panc-1 and MiaPaCa-2 cells were transfected with siRNA as described above for 24 hours. Cells were subsequently transfected with 3X-NF-κB and renilla luciferase reporter constructs for 24 hours. Luciferase activity was measured in triplicate and indicated as % activity relative to siCTRL. Reporter activity from cells transfected with IKK siRNA were all found to be significantly different relative to siCTRL.
IKK regulates pancreatic cancer cell growth and survival
To determine the functional significance of NF-κB regulation by IKK in pancreatic cancer cells, growth and survival was measured following IKKβ inhibition (Compound A) or IKK RNA interference. We showed that a higher concentration of Compound A (5μM) was required to significantly reduce Panc-1 cell growth (Fig. 5A). However, MiaPaCa-2 cells were more sensitive to increasing concentration of Compound A, as shown by significant loss of cell growth after 48 hour treatment at 1, 3, and 5μM (Fig. 5A). These results are consistent with our previous observations of Compound A being more effective in suppressing constitutive NF-κB DNA binding in MiaPaCa-2 cells (Fig. 4A). Cell growth was also measured in cells at 48, 72, and 96 hours post-transfection of IKKα and IKKβ siRNA. Furthermore, we used p65/RelA RNA interference to test the dependency of NF-κB for cell growth. In Panc-1 cells, loss of either IKK subunit or p65/RelA significantly reduced cell growth 48 hours post siRNA transfection (Fig. 5B). However, knock-down of IKKα and p65/RelA was more effective in suppressing cell growth at the 72 and 96 hour time-points (Fig. 5B). In MiaPaCa-2 cells, knockdown of IKKα, IKKβ, and p65/RelA each resulted in a significant reduction in cell growth 96 hours post siRNA transfection (Fig. 5B). We note that MiaPaCa-2 cell growth is more sensitive to the loss of both IKK subunits, while Panc-1 cell growth is more dependent on IKKα. While siRNA effects are not complete in their ability to knockdown individual proteins, these experiments demonstrate that the IKK subunits and p65/RelA are involved in growth/survival of pancreatic cancer cells.
Figure 5. Blockade of IKK suppresses cell proliferation.
(A) Panc-1 and MiaPaCa-2 cells were treated with DMSO or 1, 3, and 5μM of IKKβ inhibitor (Compound A) for 48, 72, and 96 hours. Cell growth was measured in triplicate at each time-point using a colormetric MTS tetrazolium assay. (B) Panc-1 and MiaPaCa-2 cells were transiently transfected with 100nM siRNA targeted against IKKα, IKKβ, p65, and siCTRL. Cell growth was measured as described above at 48, 72, and 96 hours post-transfection. Data was normalized to the initial cell density prior to Compound A treatment or siRNA transfection respectively. Asterisks indicate statistical significance relative to DMSO or siCTRL (P < 0.05). (C) Panc-1 cells were treated with Compound A as described above for 24 hours. Whole-cell extracts were harvested and separated by SDS-PAGE. (D) Panc-1 cells were transiently transfected with 100nM siRNA targeted against IKKα, IKKβ, combined IKKα/β, and siCTRL. Whole-cell extracts were harvested, separated by SDS-PAGE, and immunoblotted using the specified antibodies.
The induction of caspase-3 cleavage was also measured to determine whether the reduction of cell growth following the disruption of IKK was due to apoptosis. We observe a dose-dependent increase in caspase-3 cleavage upon 24 hour treatment with Compound A in Panc-1 cells (Fig. 5C). Similar results were obtained with MiaPaCa-2 cells (data not shown). Additionally, maximal caspase-3 cleavage was induced in Panc-1 cells following knock-down of both IKKα and IKKβ subunits (Fig. 5D). Furthermore, we observe a significant increase in caspase-3/7 activity following RNA interference against either IKKα or IKKβ (Supplemental Fig. 4). Overall, our data suggest that differential NF-κB regulation by IKK subunits correlates with cell growth and survival in Panc-1 and MiaPaCa-2 cells.
GSK-3 is required for constitutive IKK activity
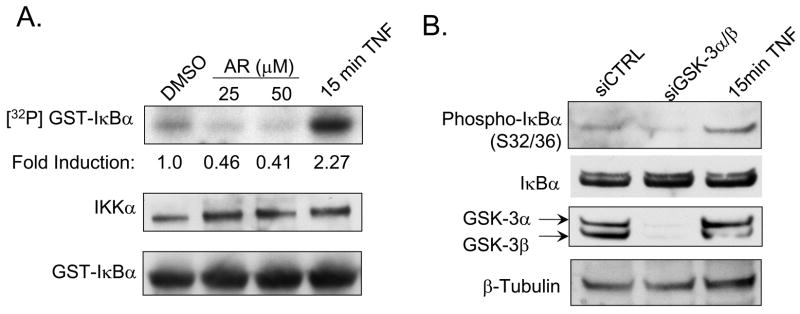
The mechanism by which GSK-3 regulates constitutive NF-κB signaling in pancreatic cancer is poorly understood. Our data thus far has shown that loss of GSK-3 isoforms and IKK subunits suppresses constitutive NF-κB activity, cell growth and survival in Panc-1 and MiaPaCa-2 cells. To determine whether GSK-3 regulates constitutive IKK activity in pancreatic cancer cells, the effects of pharmacological GSK-3 inhibition on IKK-dependent IκBα phosphorylation was measured via an in vitro IKK kinase assay. Panc-1 cells were treated for 24 hours with GSK-3 inhibitor (AR-A014418) at 25 and 50μM. Endogenous IKK complexes were immunoprecipitated using an IKKα-specific antibody. Immunoprecipitates were incubated with GST-IκBα and an in vitro kinase reaction was performed. GSK-3 inhibition reduced phosphorylation of GST-IκBα in a dose-dependent manner (Fig. 6A). Moreover, the basal phosphorylation status of IκBα at serine 32/36 was measured in Panc-1 cells following the knock-down of both GSK-3 isoforms. Consistent with our in vitro kinase assay data, the loss of GSK-3 also resulted in a reduction of endogenous IκBα phosphorylation (Fig. 6B), and is consistent with an inhibition of IKK activity. Notably, these data provide the first evidence that GSK-3 regulates cancer cell-associated NF-κB by maintaining IKK activity.
Figure 6. GSK-3 inhibition suppresses constitutive IKK kinase activity.
(A) Panc-1 cells were treated with DMSO or GSK-3 inhibitor (AR-A014418) at 25 and 50μM for 24 hours. As control, cells were treated with 10ng/ml TNF-α for 15min. Whole cells extracts were harvested. IKKα antibody was used to immunoprecipitate IKK complex. Kinase activity within immunocomplexes were assayed following incubation with wild-type GST-IκBα substrate in the presence of γ[32P]-ATP. Autoradiography was quantified and indicated as fold induction relative to DMSO. Immunoblots were performed against IKKα and GST to confirm equal loading. (B) Panc-1 cells were treated with 10ng/ml TNF-α for 15 minutes or transiently transfected with 100nM siRNA targeted against GSK-3α/β and siCTRL for 48 hours. Whole-cell extracts were harvested, separated by SDS-PAGE, and immunoblotted using the specified antibodies.
Discussion
GSK-3β has been previously reported to play an essential role in maintaining constitutive NF-κB reporter activity and expression of NF-κB target genes in pancreatic cancer cells (21). In this study, we show that both GSK-3 isoforms function to regulate constitutive NF-κB activity in Panc-1 and MiaPaCa-2 cells. Although both GSK-3 isoforms contribute to regulating NF-κB activity, GSK-3α was shown to be primarily required for cell growth (Fig. 3B and Supplemental Fig. 2A). Moreover, the suppression in cell growth following the loss GSK-3α may be due to increased apoptotic signaling, as shown by an induction of caspase-3 activity (Fig. 3D and Supplemental Fig. 3). We note that pharmacological inhibition of GSK-3 was more effective than GSK-3α knock-down in suppressing Panc-1 cell growth (Fig. 3A and 3B). Although the GSK-3 inhibitor (AR-A014418) was not shown to affect a panel of closely related protein kinases (26), unknown off-target effects cannot be ruled out when interpreting growth inhibition in pancreatic cancer cells. Importantly, we show that complete knock-down of GSK-3 and significant growth suppression was not observed until later time-points post siRNA transfection (Supplemental Fig. 2). These data may also explain why GSK-3α siRNA was less effective in suppressing cell growth relative to GSK-3 pharmacologic inhibition. Overall, our data suggest that GSK-3 isoforms are not functionally redundant in regulating NF-κB activity and cell growth in pancreatic cancer cells.
IκB kinases play a central role in regulating NF-κB signal transduction. Therefore, efforts have been made in utilizing IKK as a therapeutic target to block constitutive NF-κB activity. In this report, we use an IKKβ-specific small molecule inhibitor (Compound A) that has been shown to be effective in blocking constitutive (27) and inducible (25) NF-κB activity in various cell lines. Our data shows that Compound A is also efficient in inhibiting constitutive NF-κB activity (Fig. 4) and cell proliferation (Fig. 5) in pancreatic cancer cells. In addition to IKKβ inhibition, we show that IKKα is the predominant regulator of constitutive NF-κB activity and cell growth in Panc-1 (Fig. 4, 5). Notably, higher concentrations of the IKKβ inhibitor (Compound A) were required to diminish Panc-1 cell growth. These data raises the possibility that Compound A may target IKKα at higher doses. Indeed, Comound A was shown to inhibit recombinant IKKα activity at higher concentrations (Ki for ATP: 135nM) (25). Overall, we show that the individual requirements for IKKα and IKKβ for driving NF-κB activity vary between Panc-1 and MiaPaCa-2 cells. These data underscore the heterogeneity between pancreatic carcinoma cell lines and prompts the need to understand the variety of oncogenic mutations that potentiates NF-κB activity. Moreover, our results emphasize the need for IKKα-specific small molecule inhibitors to target constitutive NF-κB activity in select pancreatic carcinoma cell types.
The mechanism by which GSK-3 regulates constitutive NF-κB activity in pancreatic cancer is not fully understood. Our data suggests that GSK-3 and IKK may function together to regulate constitutive NF-κB activity. Previous reports have speculated on whether IKK is required for GSK-3-dependent NF-κB activity. Analysis from our group and others provide evidence that GSK-3β functions independent of the IKK complex during TNF-α-induced NF-κB signaling (19, 20, 28). Moreover, GSK-3 was also suggested to function independent of IKKβ in pancreatic cancer cell lines (21). In this regard, our current data demonstrates GSK-3-dependent regulation of constitutive IKK activity in certain pancreatic cancer cell lines. We show that inhibition (AR-A014418) or knock-down of both GSK-3 isoforms suppressed constitutive IKK activity in Panc-1 cells (Fig. 6). Notably, this abrogation in constitutive IKK activity correlated with reduced NF-κB activity, cell growth, and survival. Thus, we provide the first evidence that links GSK-3 and IKK in constitutive NF-κB signaling in pancreatic cancer cells.
Pancreatic cancer is a highly aggressive, metastatic disease known for its dismal mortality rate. Despite current chemotherapeutic efforts to improve clinical prognosis, pancreatic cancer still remains to be among the most drug-resistant tumors. Consequently, there is growing interest on specifically targeting deregulated signaling pathways that drive the molecular pathogenesis of pancreatic cancer. We emphasize in this report the critical roles both GSK-3 and IKK play in maintaining constitutive NF-κB signaling. Collectively, our data provide new insight into GSK-3-dependent NF-κB regulation, and further establishes GSK-3 and IKK as potential therapeutic targets for pancreatic cancer.
Supplementary Material
Acknowledgments
Research support was provided by NIH grants: CA73756, CA75080, and AI35098; by GI SPORE 5P50CA106991; and by Waxman Cancer Research Foundation
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar FH, Banerjee S, Li Y. Pancreatic cancer: Pathogenesis, prevention and treatment. Toxicol Appl Pharmacol. 2006 doi: 10.1016/j.taap.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 4.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–52. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–63. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer Cell. 2004;6:203–8. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Rigas B. NF-kappaB, inflammation and pancreatic carcinogenesis: NF-kappaB as a chemoprevention target (review) Int J Oncol. 2006;29:185–92. [PubMed] [Google Scholar]
- 8.Liptay S, Weber CK, Ludwig L, Wagner M, Adler G, Schmid RM. Mitogenic and antiapoptotic role of constitutive NF-kappaB/Rel activity in pancreatic cancer. Int J Cancer. 2003;105:735–46. doi: 10.1002/ijc.11081. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Aggarwal BB, Shishodia S, Abbruzzese J, Kurzrock R. Nuclear factor-kappaB and IkappaB kinase are constitutively active in human pancreatic cells, and their down-regulation by curcumin (diferuloylmethane) is associated with the suppression of proliferation and the induction of apoptosis. Cancer. 2004;101:2351–62. doi: 10.1002/cncr.20605. [DOI] [PubMed] [Google Scholar]
- 10.Schneider G, Saur D, Siveke JT, Fritsch R, Greten FR, Schmid RM. IKKalpha controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. Embo J. 2006;25:3801–12. doi: 10.1038/sj.emboj.7601259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. Embo J. 1990;9:2431–8. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodgett JR. cDNA cloning and properties of glycogen synthase kinase-3. Methods Enzymol. 1991;200:564–77. doi: 10.1016/0076-6879(91)00172-s. [DOI] [PubMed] [Google Scholar]
- 13.Ali A, Hoeflich KP, Woodgett JR. Glycogen synthase kinase-3: properties, functions, and regulation. Chem Rev. 2001;101:2527–40. doi: 10.1021/cr000110o. [DOI] [PubMed] [Google Scholar]
- 14.Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–4. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 15.Ross SE, Erickson RL, Hemati N, MacDougald OA. Glycogen synthase kinase 3 is an insulin-regulated C/EBPalpha kinase. Mol Cell Biol. 1999;19:8433–41. doi: 10.1128/mcb.19.12.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyle WJ, Smeal T, Defize LH, et al. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell. 1991;64:573–84. doi: 10.1016/0092-8674(91)90241-p. [DOI] [PubMed] [Google Scholar]
- 17.Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 18.Fiol CJ, Mahrenholz AM, Wang Y, Roeske RW, Roach PJ. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J Biol Chem. 1987;262:14042–8. [PubMed] [Google Scholar]
- 19.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 20.Steinbrecher KA, Wilson W, 3rd, Cogswell PC, Baldwin AS. Glycogen synthase kinase 3beta functions to specify gene-specific, NF-kappaB-dependent transcription. Mol Cell Biol. 2005;25:8444–55. doi: 10.1128/MCB.25.19.8444-8455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ougolkov AV, Fernandez-Zapico ME, Savoy DN, Urrutia RA, Billadeau DD. Glycogen synthase kinase-3beta participates in nuclear factor kappaB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005;65:2076–81. doi: 10.1158/0008-5472.CAN-04-3642. [DOI] [PubMed] [Google Scholar]
- 22.Liang MH, Chuang DM. Differential roles of glycogen synthase kinase-3 isoforms in the regulation of transcriptional activation. J Biol Chem. 2006;281:30479–84. doi: 10.1074/jbc.M607468200. [DOI] [PubMed] [Google Scholar]
- 23.Mayo MW, Wang CY, Cogswell PC, et al. Requirement of NF-kappaB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–5. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 24.Mattioli I, Sebald A, Bucher C, et al. Transient and selective NF-kappa B p65 serine 536 phosphorylation induced by T cell costimulation is mediated by I kappa B kinase beta and controls the kinetics of p65 nuclear import. J Immunol. 2004;172:6336–44. doi: 10.4049/jimmunol.172.10.6336. [DOI] [PubMed] [Google Scholar]
- 25.Ziegelbauer K, Gantner F, Lukacs NW, et al. A selective novel low-molecular-weight inhibitor of IkappaB kinase-beta (IKK-beta) prevents pulmonary inflammation and shows broad anti-inflammatory activity. Br J Pharmacol. 2005;145:178–92. doi: 10.1038/sj.bjp.0706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhat R, Xue Y, Berg S, et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J Biol Chem. 2003;278:45937–45. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- 27.Duncan EA, Goetz CA, Stein SJ, et al. IkappaB kinase beta inhibition induces cell death in Imatinib-resistant and T315I Dasatinib-resistant BCR-ABL+ cells. Mol Cancer Ther. 2008;7:391–7. doi: 10.1158/1535-7163.MCT-07-0305. [DOI] [PubMed] [Google Scholar]
- 28.Schwabe RF, Brenner DA. Role of glycogen synthase kinase-3 in TNF-alpha-induced NF-kappaB activation and apoptosis in hepatocytes. Am J Physiol Gastrointest Liver Physiol. 2002;283:G204–11. doi: 10.1152/ajpgi.00016.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.