Summary
Despite the critical importance of protein ubiquitination in regulation of diverse cellular processes, the molecular mechanisms by which cells recognize and transmit ubiquitin signals remain poorly understood. The endosomal sorting machinery component hepatocyte growth factor regulated tyrosine kinase substrate (Hrs) contains an ubiquitin-interacting motif (UIM), which is believed to bind ubiquitinated membrane cargo proteins and mediate their sorting to the lysosomal degradation pathway. To gain insight into the role of Hrs UIM-mediated ubiquitin signaling in cells, we performed a proteomic screen for Hrs UIM-interacting ubiquitinated proteins in human brain by using an in vitro expression cloning (IVEC) screening approach. We have identified 48 ubiquitinated proteins that are specifically recognized by the UIM domain of Hrs. Among them, 12 are membrane proteins which are likely to be Hrs cargo proteins, and 4 are membrane protein-associated adaptor proteins whose ubiquitination may act as a signal to target their associated membrane cargo for Hrs-mediated endosomal sorting. Other classes of the identified proteins include components of the vesicular trafficking machinery, cell signaling molecules, proteins associated with the cytoskeleton and cytoskeleton-dependent transport, and enzymes involved in ubiquitination and metabolism, suggesting the involvement of Hrs UIM-mediated ubiquitin signaling in regulation of multiple cellular processes. We have characterized the ubiquitination of two identified proteins, Munc18-1 and Hsc70, and their interaction with Hrs UIM and provided functional evidence supporting a role for Hsc70 in regulation of Hrs-mediated endosome-to-lysosome trafficking.
Keywords: Ubiquitination, ubiquitin-interacting motif, Hrs, endocytic trafficking, in vitro expression cloning
Introduction
Ubiquitination is a post-translational modification in which the 76-amino-acid polypeptide ubiquitin is covalently attached to a lysine residue(s) of substrate proteins [1]. Proteins can be either monoubiquitinated or polyubiquitinated by attachment of a multi-ubiquitin chain linked through one of the internal lysine residues in ubiquitin [2]. K48-linked polyubiquitination is the canonical signal that targets proteins for degradation by the 26S proteasome, whereas monoubiquitination and K63-linked polyubiquitination serve as regulatory signals to modulate protein activity, localization, and interactions [3, 4]. Increasing evidence points to the critical importance of protein ubiquitination in the control of diverse cellular processes, from DNA repair and transcription regulation to vesicular trafficking and virus budding [4–6]. Moreover, dysregulated ubiquitination has been implicated in the pathogenesis of many human diseases, including cancer and neurodegenerative disorders [7]. Elucidation of the molecular mechanisms by which cells recognize and sort ubiquitinated proteins is thus essential for understanding ubiquitin signaling in both normal physiology and diseases.
The ubiquitin-interacting motif (UIM) is a conserved ubiquitin recognition module initially identified based on the sequence homology to the ubiquitin-binding region of the S5a subunit of the 26S proteasome [8]. The UIM has a 20-amino-acid consensus sequence: X-Ac-Ac-Ac-Ac-Φ-X-X-Ala-X-X-X-Ser-X-X-Ac-X-X-X-X, where Φ represents a large hydrophobic residue and Ac represents an acidic residue. UIMs are found in many proteins implicated in a variety of cellular processes, including endocytosis, endosome-to-lysosome trafficking, DNA repair, mRNA splicing, and neurodegeneration [8]. In vitro studies indicate that the UIM binds monoubiquitin and polyubiquitin chains [9–12]. Furthermore, UIM domains from different proteins bind polyubiquitin chains of varying lengths with different affinities [12], suggesting that different UIM domains may recognize distinct subsets of ubiquitinated proteins.
Hepatocyte growth factor regulated tyrosine kinase substrate (Hrs) is an early endosome-associated UIM-containing protein that plays a central role in controlling endosome-to-lysosome trafficking [13–16]. A major sorting decision in the endocytic pathway occurs at the early endosome, where membrane cargo proteins can be sorted to the recycling pathway for delivery to the cell surface or to lumenal vesicles of multivesicular bodies (MVBs) for eventual degradation in the lysosome [6, 16]. A sorting signal for cargo trafficking to the lysosomal pathway is the ubiquitination of cargo proteins. The UIM domain of Hrs has been shown to bind ubiquitin in vitro [9, 11, 14] and to facilitate the sorting of several ubiquitinated cargo proteins to the lysosomal pathway in mammalian cells [17] and yeast [10, 18]. The Hrs UIM domain may also interact with ubiquitinated components of the endosomal trafficking machinery to regulate endosome-to-lysosome trafficking [6]. Recently, Hrs has been shown to preferentially bind K63-linked polyubiquitin chains [19]. Interestingly, the UIM domain is indispensable for monoubiquitination as well as phosphorylation of Hrs [9, 12, 20], raising the possibility that the Hrs UIM domain may bind E3 ubiquitin-protein ligase(s) and/or kinase(s). The identities of ubiquitinated cargo and other cellular proteins that are recognized by the Hrs UIM domain remain largely unknown.
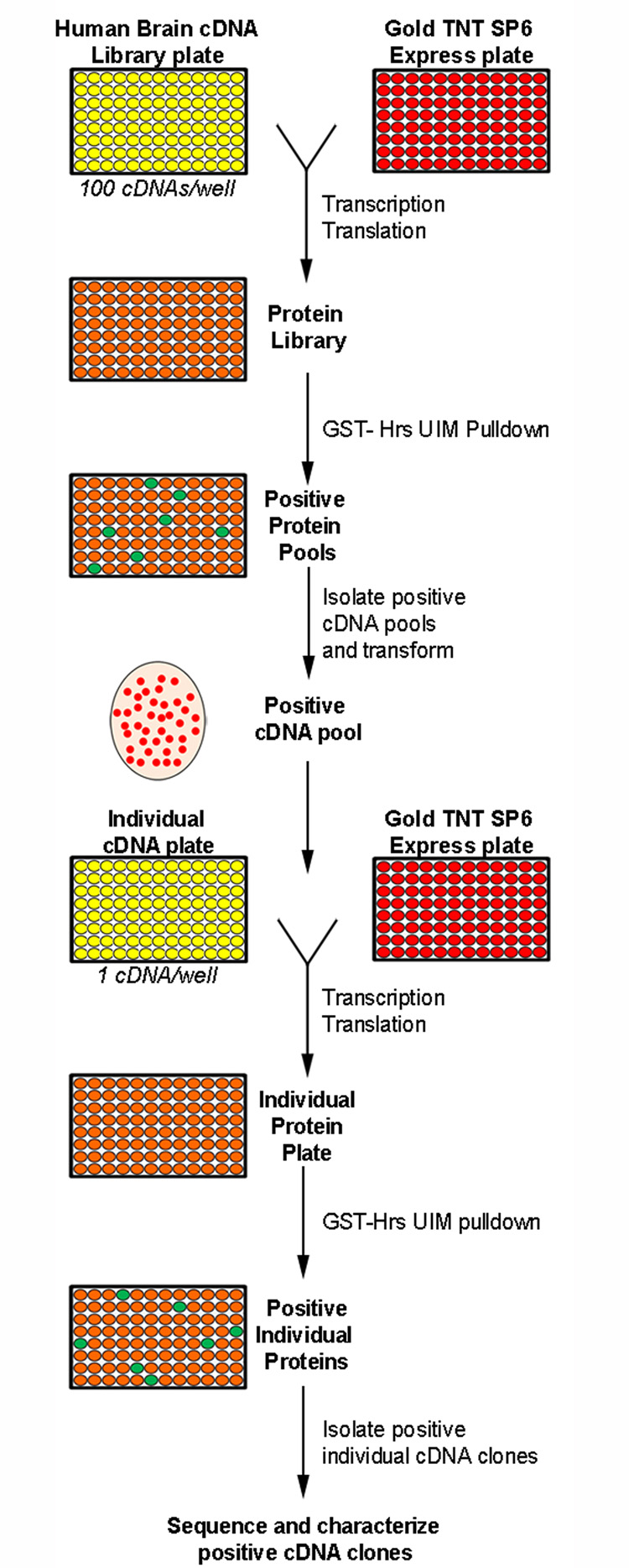
In order to gain insight into the role of Hrs UIM-mediated ubiquitin signaling in cells, we performed a proteomic screen for Hrs UIM-interacting ubiquitinated proteins in human brain by using a combined in vitro expression cloning (IVEC) and GST-pull down approach (Fig. 1). IVEC is a powerful screening method that combines biochemical analysis of radioactively labeled proteins with the ability to quickly isolate the corresponding cDNAs [21, 22]. Compared to yeast two-hybrid screening, IVEC screening offers the advantage of studying direct interaction between two proteins in vitro [23], rather than indirect analysis of the interaction between fusion proteins inside the yeast nucleus. Moreover, our IVEC screening approach complements other proteomic screening strategies [24, 25], which are often contaminated with secondary, non-specific binding proteins.
Fig. 1.
Schematic illustrating the in vitro expression cloning (IVEC) system used to identify and isolate cDNAs from human adult brain library that encode Hrs UIM-interacting proteins.
Here, we report the identification of a set of proteins that are specifically recognized by the UIM domain of Hrs. Our results reveal the involvement of Hrs UIM-mediated protein interactions in the coordination of multiple steps in endosomal trafficking as well as in the regulation of cell signaling, cytoskeleton and membrane dynamics and other cellular processes.
Results
IVEC screen for proteins that are specifically recognized by the UIM domain of Hrs
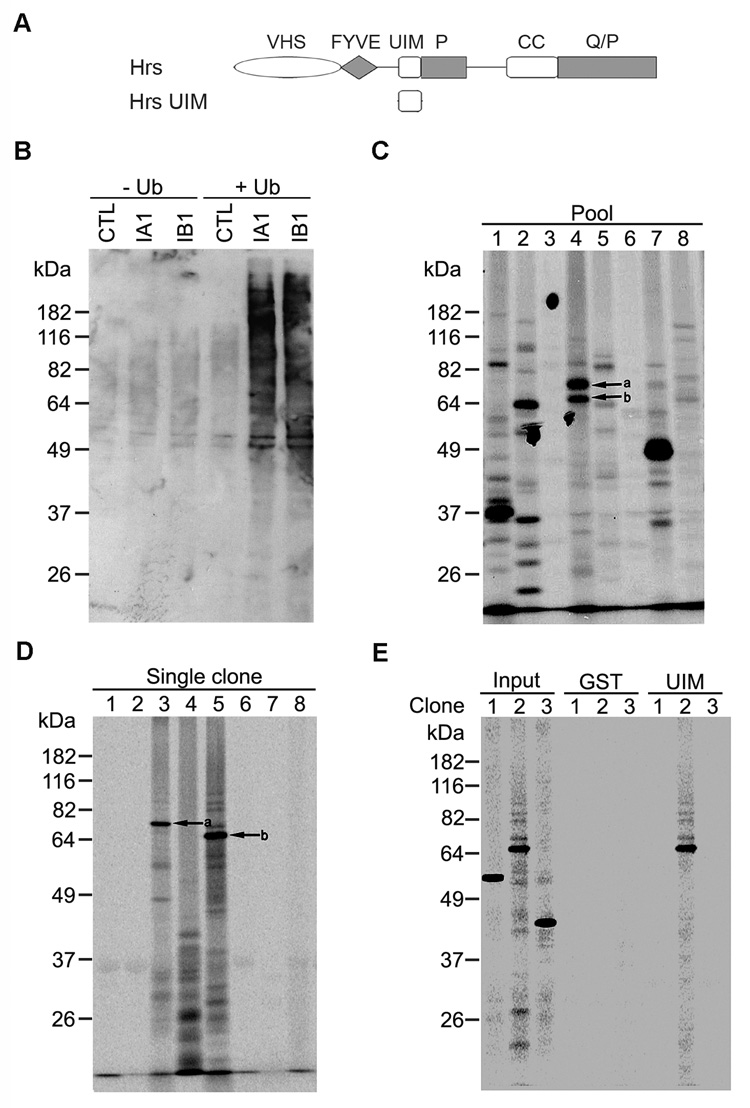
To identify cellular targets of the Hrs UIM domain, we screened a human adult brain cDNA library for Hrs UIM-interacting proteins using an IVEC approach [21, 22], which is summarized in Fig. 1. Pools of cDNAs (100 independent cDNA clones per pool) from the human brain library were in vitro transcribed and translated in the TNT Quick coupled transcription-translation reticulocyte lysate system in the presence of [35S]methionine and ubiquitin to generate 35S-labeled protein pools [23]. It has been well established that such a transcription-translation reticulocyte lysate system is capable of carrying out ubiquitination of in vitro translated proteins [23, 26, 27]. To determine whether the protein pools synthesized in our IVEC system are ubiquitinated, we performed immunoblot analysis with anti-ubiquitin antibody to examine the ubiquitination status of protein pools generated from the in vitro transcription-translation of human brain cDNA pools in the presence or absence of ubiquitin (Fig. 2B). We found that addition of ubiquitin to the in vitro transcription-translation reaction mixture dramatically increased the ubiquitination levels of in vitro translated protein pools, confirming that protein pools synthesized in our IVEC system are indeed ubiquitinated. The 35S-labeled ubiquitinated protein pools were then tested for their ability to bind to a GST-fused UIM domain of Hrs protein (Fig. 2A) in an in vitro binding assay. Fig. 2C shows an example of six positive pools (Pool # 1, 2, 4, 5, 7, and 8) containing Hrs UIM-binding proteins isolated from the primary screen. From each of the positive pools, individual cDNA clones were isolated and subjected to a secondary screen in the same manner to identify positive cDNA clones encoding Hrs UIM-binding proteins (Fig. 2D). The specificity of the observed interactions was confirmed by the specific binding of the identified proteins to GST-Hrs UIM but not to GST control (Fig. 2E).
Fig. 2. IVEC screen for proteins that bind to the UIM domain of Hrs.
(A) Domain structure of full-length Hrs (top) and the GST-fused Hrs UIM domain used in the IVEC screen (bottom). (B) Two cDNA pools, IA1 and IB1, containing 100 independent cDNA clones per pool from a human adult brain cDNA library, were in vitro transcribed and translated in the presence of cold methionine with or without ubiquitin. The control (CTL) reactions were carried out under the same conditions with no cDNAs added. The synthesized protein pools were analyzed by immunoblotting with anti-ubiquitin antibody. (C) Primary screen for positive pools containing Hrs UIM-binding proteins. Pools of cDNAs (100 independent cDNA clones per pool) from a human adult brain cDNA library were in vitro transcribed and translated in the presence of [35S]methionine and ubiquitin and then subjected to a GST-Hrs UIM pull down assay. Bound proteins were analyzed by SDS-PAGE. Autoradiography of gel samples was performed using a phosphoimager. Example of positive pools (Pool # 1, 2, 4, 5, 7, and 8) selected for secondary screen. The two bands labeled a and b in Pool # 4 represent distinct Hrs UIM-binding proteins, which would be individually isolated by secondary screen. (D) Secondary screen for isolation of individual positive cDNA clones encoding Hrs UIM-binding proteins. In vitro translated products from individual cDNA clones isolated from each of the positive pools were analyzed as described above for their ability to bind GST-Hrs UIM. Example of 8 single clones isolated from Pool # 4, of which Clone # 3 and 5 are individual positive cDNA clones encoding Hrs UIM-binding proteins a and b indicated in (C). (E) Specificity of Hrs UIM binding. In vitro translated products from 3 isolated individual cDNA clones (Input) were incubated with immobilized GST-Hrs UIM fusion protein or GST control. Bound proteins were analyzed by SDS-PAGE and autoradiography. Clone # 2 encodes a protein that specifically binds to GST-Hrs UIM but not to GST control, whereas Clone # 1 and 3 are negative interactors that binds neither to GST-Hrs UIM nor to GST control.
From the IVEC screen, we isolated 64 positive clones, which encode 48 proteins that are specifically recognized by the UIM domain of Hrs (Fig. 3 and Table 1). The specific binding of the identified proteins to the Hrs UIM domain suggests that these proteins may be ubiquitinated. In support of this notion, four of the identified proteins, APP [28], β tubulin [29], Hsc70 [30, 31], and MARK4 [32] have been previously shown to be ubiquitinated. However the interaction of these proteins with the Hrs UIM domain has never been reported. The remaining 43 proteins are not previously known to be ubiquitinated or to bind to the Hrs UIM domain.
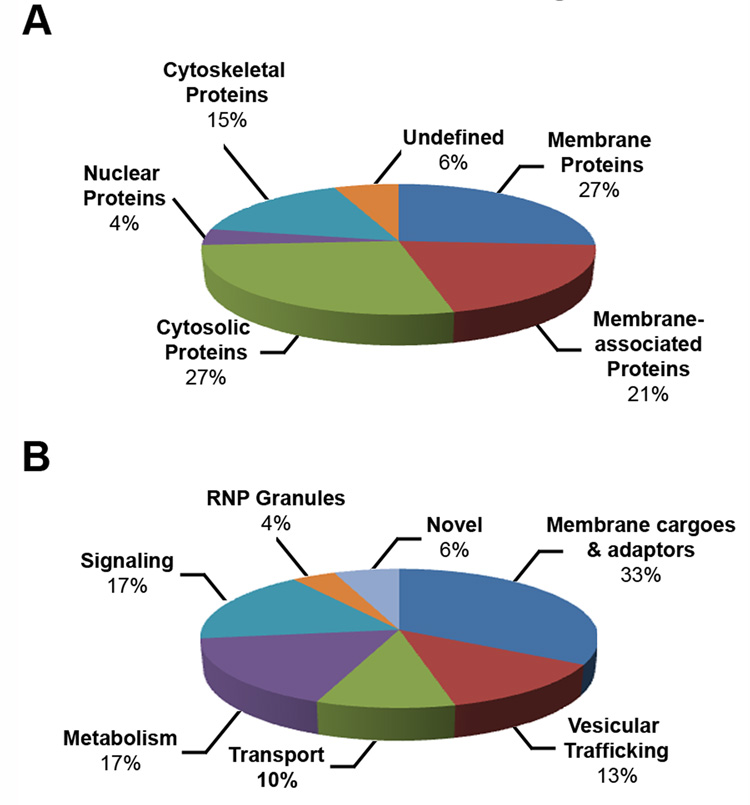
Fig. 3. Classification of the identified proteins according to cellular localization (A) and molecular function (B).
The number of proteins in each category is expressed as the percentage of the total number of different proteins identified from the screen.
Table 1.
Hrs UIM-interacting proteins identified from the IVEC screen
| Gene | Protein Name (Alternate name) | Accession Number |
|---|---|---|
| Membrane proteins | ||
| APPa | amyloid beta (A4) protein | NP_958816 |
| APLP2 | amyloid beta (A4) precursor-like protein 2 | NP_001633 |
| ATP1A1 | Na+/K+ ATPase alpha 1 subunit | NP_000692 |
| ATP2A2 | ATPase, Ca2+ transporting, slow twitch 2 | NP_001672 |
| ARL6IP1 | ADP-ribosylation factor-like 6 interacting protein | NP_055976 |
| BSG | basigin | NP_940991 |
| C3orf1 | hypothetical protein LOC51300 | NP_057673 |
| FAM5C | family with sequence similarity 5, member C | NP_950252 |
| MEST | mesoderm specific transcript | NP_002393 |
| TMCC2 | transmembrane and coiled-coil domain family 2 | NP_055673 |
| TMEM49 | transmembrane protein 49 (VMP1) | NP_112200 |
| UNC84B | unc-84 homolog B (Rab5IP) | NP_056189 |
| Membrane protein-associated adaptor proteins | ||
| AHCYL1 | s-adenosylhomocysteine hydrolase-like 1(IRBIT) | NP_006612 |
| CASKc | calcium/calmodulin-dependent serine protein kinase | NP_003679 |
| TJP2 | tight junction protein 2 (ZO-2) | NP_963923 |
| TRAP1 | TNF receptor-associated protein 1 | NP_057376 |
| Vesicular trafficking | ||
| GGA2b | ADP-ribosylation factor binding protein 2 | NP_055859 |
| HSPA8a | heat shock 70 kDa protein 8 (Hsc70) | NP_006588 |
| MAP3K10c | mitogen-activated protein kinase 10 (MLK2) | NP_002437 |
| STXBP1 | syntaxin binding protein 1(Munc18-1) | NP_003156 |
| SCRN1 | secernin 1 | NP_055581 |
| Cytoskeleton and cytoskeleton-dependent transport | ||
| CRMP1 | collapsin response mediator protein 1 | NP_001304 |
| DCTN2 | dynactin 2 (dynamitin) | NP_006391 |
| GFAPa | glial fibrillary acidic protein | NP_002046 |
| MARK4a | MAP/microtubule affinity-regulating kinase 4 | NP_113605 |
| PPP1R16A | protein phosphatase 1, regulator (inhibitor) subunit 16A (MYPT3) | NP_116291 |
| RHOBTB3 | rho-related BTB domain containing 3 | NP_055714 |
| TUBBa | tubulin, beta | NP_821133 |
| TUBB2A | tubulin, beta 2 | NP_001060 |
| Cell signaling | ||
| ESRRG | estrogen-related receptor gamma (ERR3) | NP_996317 |
| ILKAP | integrin-linked kinase-associated protein phosphatase 2C | NP_789769 |
| PPP1R7 | protein phosphatase 1 regulatory subunit 7 | NP_002703 |
| RSU1 | ras suppressor protein 1 | NP_036557 |
| UBA1 | ubiquitin-activating enzyme E1 | NP_003325 |
| Metabolism | ||
| ACLY | ATP citrate lyase | NP_942127 |
| CBS | cystathionine-beta-synthase | NP_000062 |
| DECR | 2,4-dienoyl CoA reductase 1 | NP_001350 |
| MAT2A | methionine adenosyltransferase II alpha | NP_005902 |
| MTHFD1L | C1 tetrahydrofolate synthase | NP_056255 |
| OSBPL5 | oxysterol-binding protein-like protein 5 | NP_065947 |
| PLD3 | phospholipase D family, member 3 | NP_036400 |
| PFKM | phosphofructokinase, muscle | NP_000280 |
| Ribonucleoprotein granules | ||
| HNRPDL | heterogeneous nuclear ribonucleoprotein D-like | NP_112740 |
| RPS3A | ribosomal protein S3a | NP_000997 |
| SF3B3 | splicing factor 3b, subunit 3 | NP_036558 |
| Novel proteins | ||
| LOC349114 | hypothetical protein LOC349114 | Q8N836 |
| PTCD3 | Pentatricopeptide repeat domain 3 | NP_060422 |
| ZNF302 | zinc finger protein 302 | NP_060913 |
known to be ubiquitinated
thought to be ubiquitinated based on similarity
interacts with an E2 or E3, but is not known to be ubiquitinated
Classification of the identified Hrs UIM-interacting proteins
To shed light on the role of Hrs UIM-mediated protein interactions, we categorized the 48 proteins isolated from the IVEC screen according to functional predictions based on the available literature, Gene Ontology, and homology searches (Table 1). Classification of the identified Hrs UIM-interacting proteins according to their cellular localization (Fig. 3A) reveals that the majority of these proteins are integral membrane proteins (27%), membrane-associated proteins (21%), or cytosolic proteins (27%). We and others have shown that Hrs is associated with both early endosomal membrane and cytosolic fractions [13, 33, 34]. The localization of the majority of the identified Hrs UIM-binding proteins to the membrane and cytosol suggests that they are appropriately positioned to interact with Hrs in cells. In addition, we identified a number of proteins that could be classified as cytoskeletal (15%), which suggests that Hrs UIM-mediated ubiquitin signaling may have a role in regulation of cytoskeleton dynamics. The localization of only 4% and 6% of proteins could be classified as nuclear or unknown, respectively.
Dividing the identified Hrs UIM-interacting proteins based on their functional classes (Fig. 3B) further suggests that the screen largely identified putative membrane cargo proteins and membrane cargo adaptor proteins (33%), consistent with the proposed function of Hrs in endosomal sorting and trafficking. The other major functional groups to which the Hrs UIM-interacting proteins belong include: cell signaling (17%), metabolism (17%), vesicular trafficking (13%), and transport (10%), suggesting an interconnection between Hrs UIM-mediated ubiquitin signaling and these cellular processes.
Characterization of Munc18-1 and Hsc70 as Hrs UIM-interacting ubiquitinated proteins
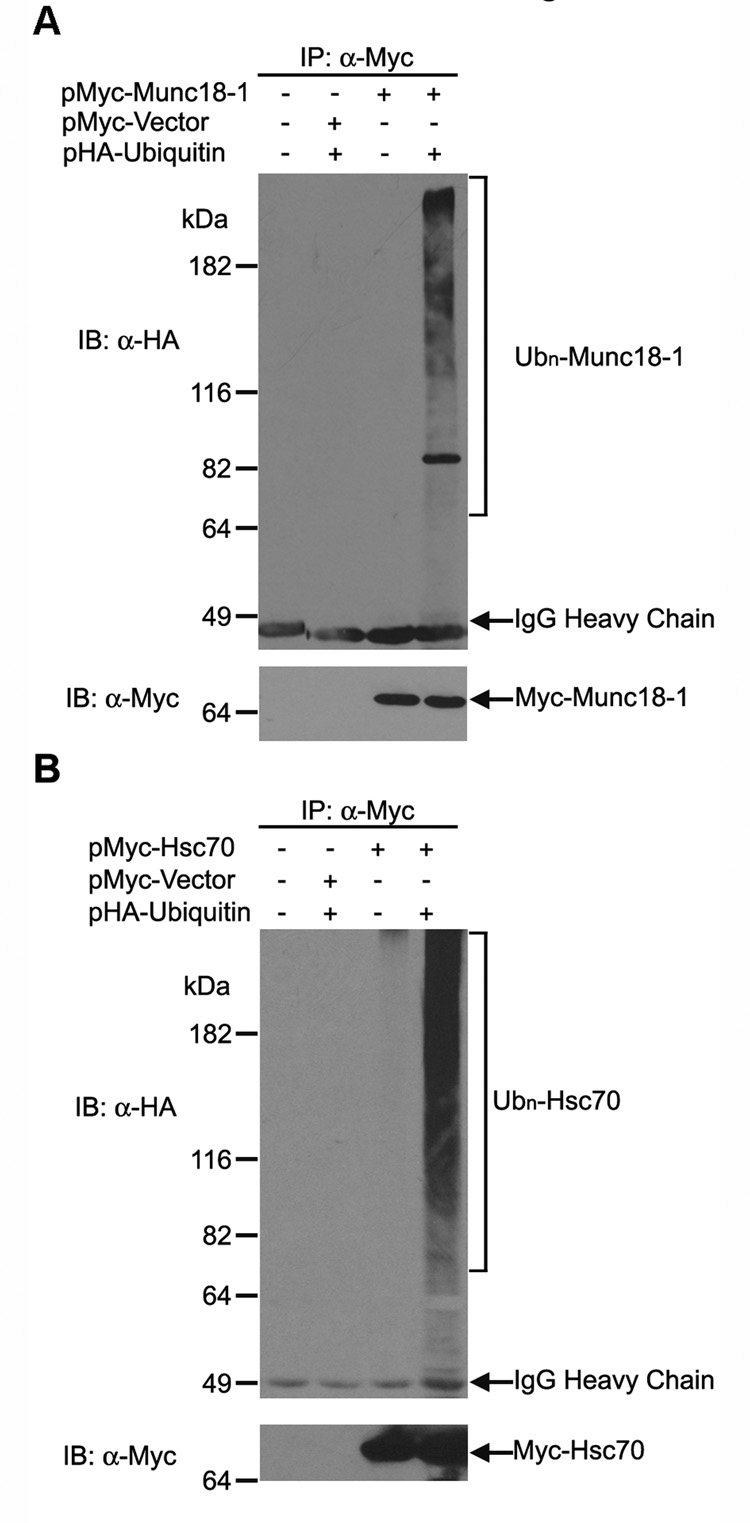
The ability of the Hrs UIM domain to bind ubiquitin and ubiquitinated proteins has been well established [9, 11, 14, 17, 19]. Thus, the direct interaction between the Hrs UIM domain and each of the 48 proteins identified from our IVEC screen (Table 1) raises the possibility that these proteins are ubiquitinated in cells. To test this possibility, we used a well-established in vivo ubiquitination assay [35, 36] to determine the ubiquitination status of the identified Hrs UIM-interacting protein Munc18-1, a key regulator of Ca2+-dependent exocytosis [37] which is previously unrecognized as a ubiquitinated protein. Lysates from HeLa cells expressing HA-tagged ubiquitin and Myc-tagged Munc18-1 were subjected to immunoprecipitation with anti-Myc antibodies, followed by immunoblotting with anti-HA antibodies to detect HA-ubiquitin-conjugated Munc18-1 protein (Fig. 4A). We observed a prominent band around 82 kDa that may represent a di-ubiquitinated species of Munc18-1 as well as higher molecular weight smear that may represent polyubiquitinated forms of Munc18-1. These results provide the first evidence that Munc18-1 is ubiquitinated in vivo and support the notion that Hrs UIM-binding proteins isolated from our IVEC screen likely represent ubiquitinated proteins.
Fig. 4. Munc18-1 and Hsc70 are ubiquitinated in cell-based assays.
(A) HeLa cells were transfected with the indicated plasmids and treated with proteasome inhibitor MG132 for 8 hours before harvest. Cell lysates were subjected to immunoprecipitation with anti-Myc antibody, followed by immunoblotting with anti-HA antibody to detect HA-tagged ubiquitin conjugated to Munc18-1 (upper panel). The blot was then re-probed with anti-Myc antibody to detect Myc-tagged Munc18-1 protein (lower panel). (B) In vivo ubiquitination of Hsc70 was analyzed using the same assay as described above. Data are representative of at least 3 independent experiments.
Next, we sought to determine whether ubiquitinated Munc18-1 is specifically recognized by Hrs UIM. In vitro binding assays were performed by incubating immobilized GST-Hrs UIM, GST-Hrs full-length, GST-HrsΔUIM, or GST control proteins (Fig. 5A) with soluble Myc-tagged Munc18-1 immunopurified from transfected HeLa cells. Bound proteins were probed with an anti-ubiquitin antibody and anti-Myc antibody to detect ubiquitinated Munc18-1 and non-ubiquitinated Munc18-1, respectively (Fig. 5B). We found that both GST-fused Hrs UIM domain and full-length Hrs selectively interact with ubiquitinated Munc18-1 but not with non-ubiquitinated Munc18-1 protein. The ability of Hrs to bind ubiquitinated Munc18-1 was dramatically reduced by the deletion of the UIM domain. Furthermore, the GST control did not pull down any detectable level of ubiquitinated or non-ubiquitinated Munc18-1. Together, these results indicate that Hrs UIM domain is both necessary and sufficient for binding Munc18-1 in an ubiquitin-dependent manner and support the validity and specificity of our IVEC screen.
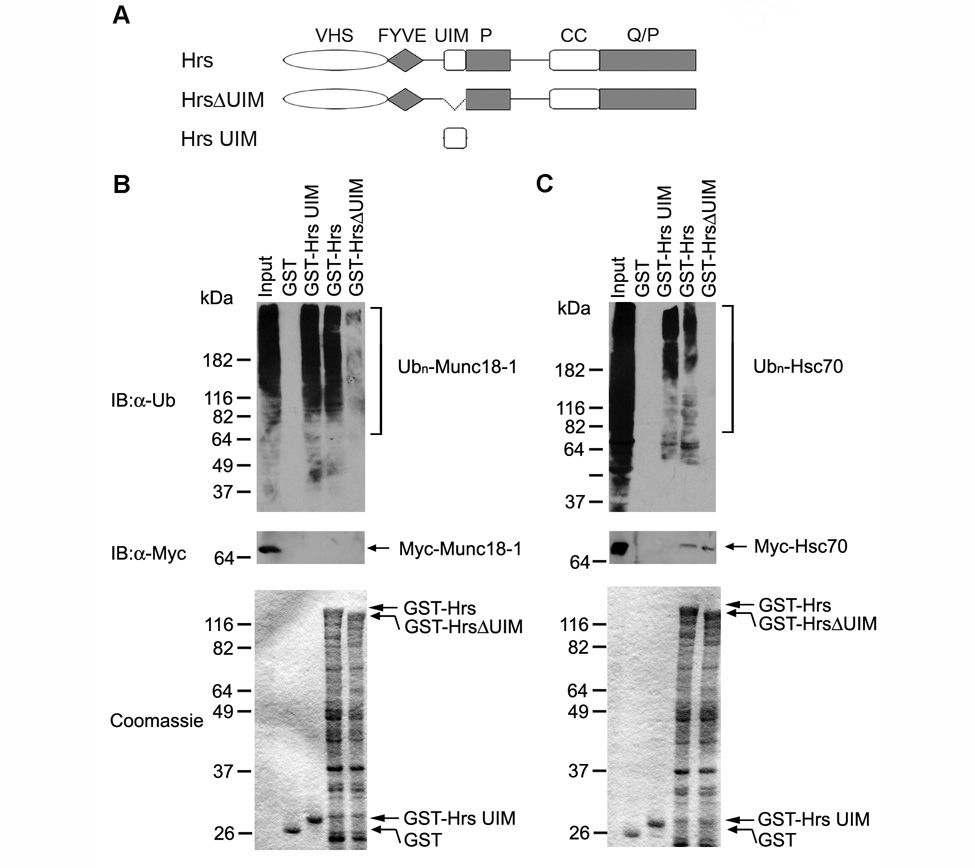
Fig. 5. Hrs directly binds ubiquitinated Munc18-1 or ubiquitinated Hsc70 in an UIM-dependent manner.
(A) Domain structure of GST-Hrs fusion proteins. (B) Soluble immunopurified Myc-tagged Munc18-1 (input) was incubated with similar amounts of immobilized GST or GST-Hrs fusion proteins (lower panel). Immunoblot analysis of bound proteins with anti-ubiquitin antibody (upper panel) and anti-Myc antibody (middle panel) reveals an UIM-dependent interaction of Hrs with ubiquitinated Munc18-1 but not with non-ubiquitinated Munc18-1 protein. (C) Soluble immunopurified Myc-tagged Hsc70 (input) was incubated with similar amounts of immobilized GST or GST-Hrs fusion proteins (lower panel). Immunoblot analysis of bound proteins with anti-ubiquitin antibody (upper panel) and anti-Myc antibody (middle panel) reveals that Hrs binds ubiquitinated Hsc70 and non-ubiquitinated Hsc70 protein through different domains. The immunopurified Myc-tagged, ubiquitinated and non-ubiquitinated forms of Munc18-1 or Hsc70 in the input lane were detected by immunoblotting with anti-ubiquitin antibody and anti-Myc antibody, respectively, but their amounts were too low for detection by the Coomassie stain.
Our identification of 48 proteins as novel binding partners for the Hrs UIM domain has led to a number of interesting hypotheses. For example, previous studies have shown that Hrs is enriched with ubiquitinated cargo proteins in flat clathrin-coated microdomains of early endosomes [17, 38, 39]. These clathrin-coated microdomains have been proposed to play a role in endosomal sorting and retention of ubiquitinated cargo proteins [17, 39]. The flat clathrin coat has to be dissociated prior to endosomal invagination and budding of the MVB lumenal vesicles [17, 39], but the molecular machinery for the disassembly of the endosomal clathrin coat remains unknown. Hsc70 is a constitutively expressed member of the Hsp70 molecular chaperone family and has been shown to regulate clathrin uncoating processes [40, 41]. Although Hsc70 is known to be ubiquitinated [30, 31], it is previously unrecognized as an Hrs-binding protein. Our identification of Hsc70 as an Hrs UIM-interacting protein raises an intriguing hypothesis that the Hrs UIM-mediated interaction recruits Hsc70 to endosomes for clathrin uncoating prior to the budding of MVB lumenal vesicles. As a first step to test this hypothesis, we performed in vivo ubiquitination analysis to confirm that Hsc70 is indeed ubiquitinated in cells (Fig. 4B). Furthermore, we performed binding experiments and found that ubiquitinated Hsc70 specifically bound to GST-Hrs UIM and GST-Hrs, but not to GST-HrsΔUIM or the GST control (Fig. 5C), indicating that the Hrs UIM domain is both necessary and sufficient for binding ubiquitinated Hsc70. Our results showed that the Hrs UIM domain is unable to bind Hsc70 in the absence of ubiquitination, as GST-Hrs UIM did not pull down any detectable level of non-ubiquitinated Hsc70 (Fig. 5C). Interestingly, our analysis revealed that the full-length Hrs was capable of interacting with non-ubiquitinated Hsc70 and this interaction was not affected by the deletion of the UIM domain (Fig. 5C), suggesting that the interaction of Hrs with non-ubiquitinated Hsc70 is mediated by a binding site on Hrs which is located outside of its UIM domain.
Hsc70 is essential for ligand-induced epidermal growth factor receptor degradation
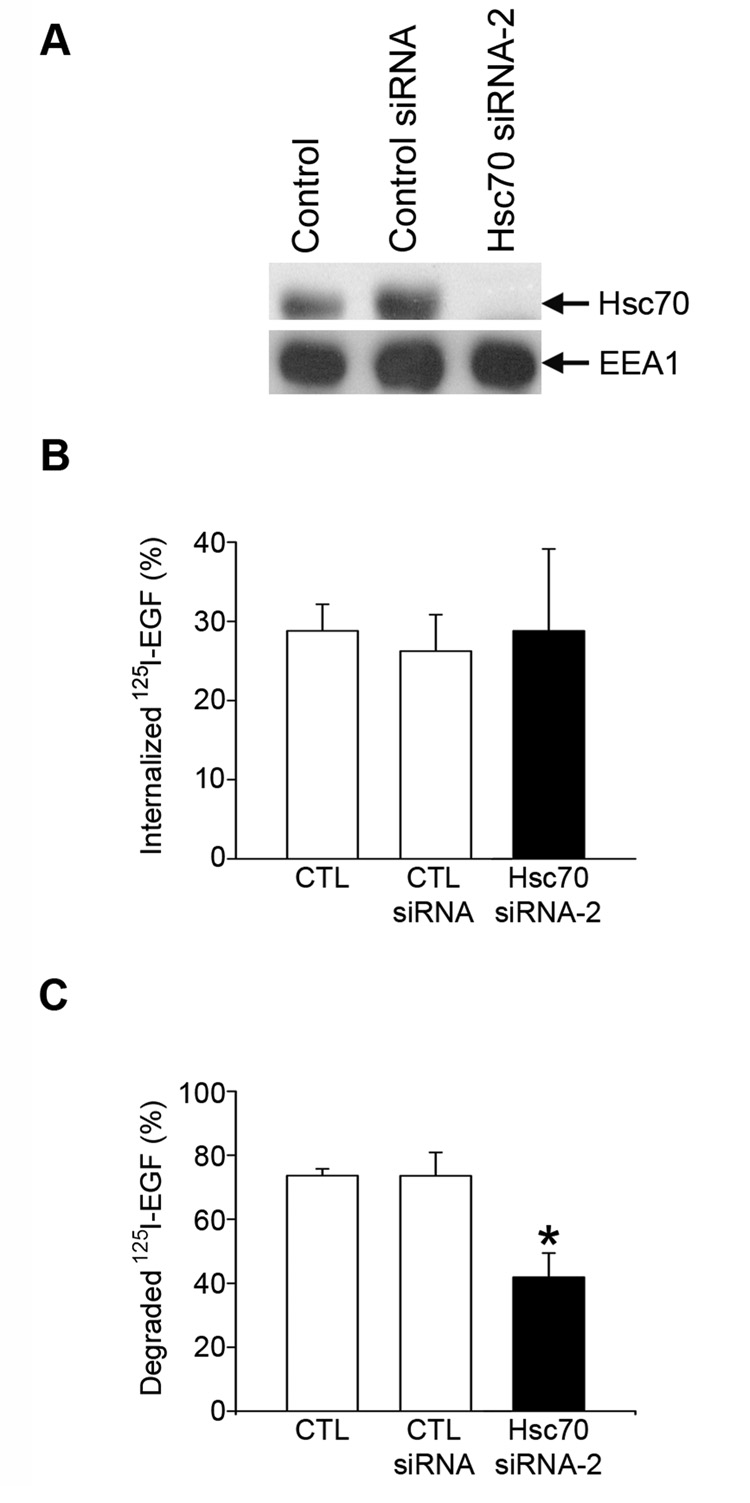
Next, we assessed the role of Hsc70 in regulation of Hrs-mediated endosomal trafficking by using the epidermal growth factor (EGF) receptor (EGFR) as a cargo protein. Previous studies have shown that binding of EGF to the EGFR at the plasma membrane causes rapid internalization of the EGF-EGFR complex and subsequent sorting at the early endosome for delivery to the lysosome for degradation [42–44]. The role of Hrs-mediated early endosomal sorting in the regulation of EGF-induced EGFR degradation is well-established; both the overexpression and the depletion of Hrs inhibit ligand-induced degradation of the EGFR [45, 46]. Our identification of the interaction between Hsc70 and Hrs raises the possibility that Hsc70 may participate in the regulation of ligand-induced endocytic trafficking of the EGF-EGFR complex to the lysosome for degradation. To test this possibility, we examined the effect of depleting Hsc70 through small-interfering RNA (siRNA) on EGF-induced EGFR degradation. For selective depletion of endogenous Hsc70, we used two distinct siRNA duplexes, Hsc70 siRNA-1 and Hsc70 siRNA-2, which specifically target different regions of the Hsc70 mRNA. Immunoblot analysis confirmed that Hsc70 siRNA-1 (data not shown) and Hsc70 siRNA-2 (Fig. 6A) both specifically inhibited the expression of endogenous Hsc70, but not EEA1.
Fig. 6. Hsc70 knockdown inhibits EGF-induced EGFR degradation.
(A) Equal amounts of proteins from HeLa cell lysates transfected with the indicated siRNA were analyzed by immunoblotting with antibodies against Hsc70 and EEA1. (B) HeLa cells transfected with the indicated siRNAs were incubated with 125I-EGF for 10 minutes at 37°C. The internalized 125I-EGF is expressed as a percentage of the initially bound 125I-EGF. (C) HeLa cells transfected with the indicated siRNAs were allowed to internalize 125I-EGF for 10 minutes and then chased for 1 hour at 37°C. The degraded 125I-EGF is expressed as a percentage of the initially internalized 125I-EGF. Data represent mean ± s.e.m. from 3 independent experiments. The asterisks indicate a statistically significant difference (p < 0.05) from the control siRNA-transfected cells.
Next, we examined the effect of siRNA-mediated knockdown of Hsc70 expression on the uptake and degradation of 125I-EGF in HeLa cells. We found that depletion of Hsc70 by Hsc70 siRNA-2 (Fig. 6B) had no statistically significant effect on 125I-EGF internalization. As shown in Fig. 6C, we observed a statistically significant (p < 0.05) decrease in 125I-EGF degradation in Hsc70 siRNA-2 (41.9 ± 7.5%, n = 4) transfected HeLa cells compared to the untransfected controls (73.6 ± 2.2%, n = 4) and control siRNA transfected cells (73.6 ± 7.3%, n = 4). Similar effects were observed when using Hsc70 siRNA-1. Together, these data provide strong evidence supporting a functional role for Hsc70 in regulation of the trafficking of internalized EGF-EGFR complexes to the lysosome for degradation.
Discussion
The present study represents the first large-scale unbiased screen for candidate proteins that are specifically recognized by the UIM domain of Hrs. Our screening results demonstrate that the IVEC screen for identification of Hrs UIM-interacting proteins is highly specific, as out of 48,000 independent human brain cDNA clones screened, we only isolated 64 positive clones corresponding to 48 distinct proteins. Furthermore, among the identified proteins, we did not find any proteins that are exclusively localized to the extracellular matrix. The validity of our IVEC screen is supported by our in vivo ubiquitination assays showing that two identified Hrs UIM-interacting proteins, Munc18-1 and Hsc70, are indeed ubiquitinated in cells. Furthermore, the results of our deletion mutagenesis and binding experiments clearly demonstrate that the Hrs UIM domain is both necessary and sufficient for selective interaction with the ubiquitinated forms of Munc18-1 and Hsc70 but not with the non-ubiquitinated form of these proteins. Together, these data strongly suggest that the Hrs UIM-interacting proteins identified in our IVEC screen (Table 1) are likely to be ubiquitinated proteins.
The current model for Hrs UIM function is that the Hrs UIM binds ubiquitinated membrane cargo proteins at early endosomes, thereby facilitating the sorting of these proteins to the lysosomal pathway [6, 15, 16]. In support of this model, we identified 9 known and 3 novel membrane proteins as Hrs UIM-interacting proteins (Table 1), which likely represent endosomal cargo proteins that undergo ubiquitination-dependent sorting by Hrs. Among the Hrs UIM-interacting membrane proteins, we identified amyloid beta A4 protein (APP) and the related APP-like protein 2 (APLP2). Mutations in the APP gene are associated with Alzheimer’s disease (AD) [47]. Previous studies have shown that APP localizes to endosomes [48] and that APP is ubiquitinated [28]. Our finding that the Hrs UIM binds to APP is of particular interest given the increasing evidence that endosomal abnormalities, specifically enlarged early endosomes, precede the appearance of symptoms in AD [49]. Our study provides the first report of an interaction between a component of the endosomal sorting machinery and APP and suggests an aberrant Hrs-mediated endosomal sorting of APP may be involved in AD pathogenesis.
Our IVEC screen results support an additional role for Hrs UIM in the sorting of non-ubiquitinated membrane cargo proteins to the lysosomal pathway. Recent studies reveal that not all membrane cargo proteins require ubiquitination for trafficking to lysosomes [50] and that "ubiquitination-independent" cargo trafficking also requires Hrs for sorting to lysosomes [51]. The mechanism underlying Hrs-dependent endosome-to-lysosome trafficking of non-ubiquitinated membrane cargo is not understood. Interestingly, our identification of 4 membrane protein-associated adaptor proteins, CASK [52], ZO-2 [53], IRBIT [54], and TRAP1 [55], as putative ubiquitinated proteins recognized by the Hrs UIM raises an intriguing possibility that the ubiquitination of adaptor proteins may act as a sorting signal for targeting their associated membrane proteins to the lysosomal pathway.
In addition to membrane cargo and adaptor proteins, we identified five proteins that function in vesicular trafficking (Table 1), including GGA2 and MLK2. GGA2 belongs to a family of Arf-dependent adaptors that bind clathrin and mediate the sorting of cargo proteins at the trans-Golgi network for delivery to endosomes [56]. Recent evidence indicates that GGA proteins not only function at the trans-Golgi network, but also at early endosomes to facilitate the transport of endosomal cargo proteins into the MVB [57]. MLK2 is a protein kinase that functions in the stress-activated JNK signaling pathway and has been shown to bind clathrin via its C-terminal clathrin box motif and regulates clathrin-coated vesicle trafficking [58]. Interestingly, Hrs also contains a C-terminal clathrin box motif that binds clathrin, and the ability of Hrs to bind clathrin is essential for the formation of Hrs-clathrin sorting microdomains on early endosomes [17, 38, 39, 59]. The identification of GGA2 and MLK2 as Hrs UIM-interacting ubiquitinated proteins suggests that these two proteins may work in concert with Hrs in the clathrin-dependent endosomal sorting and retention process.
Since clathrin is not incorporated into MVB lumenal vesicles, the flat clathrin coat on the early endosome has to be dissociated prior to the budding of the lumenal vesicles [17, 39]. The molecular machinery for the dissociation of the endosomal clathrin coat remains undefined. In this study, we identified the clathrin uncoating ATPase Hsc70 as an Hrs UIM-interacting ubiquitinated protein and provided evidence that Hsc70 is an essential component of the machinery that regulates Hrs-mediated endosome-to-lysosome trafficking of internalized EGF-EGFR complexes. Our findings support the idea that Hsc70 is part of the clathrin uncoating machinery at early endosomes and that loss of Hsc70 inhibits this uncoating process and subsequently delivery of cargo proteins to the MVB pathway for degradation in the lysosome.
The other two identified proteins in the vesicular trafficking category are Munc18-1, an essential component of the molecular machinery for synaptic vesicle exocytosis [37, 60], and secernin 1, a cytosolic protein involved in regulation of exocytosis from mast cells [61]. Our identification of these two proteins as Hrs UIM-binding partners suggests a role for Hrs in regulation of Ca2+-dependent exocytosis. Consistent with this role, we and others have previously reported a functional interaction between Hrs and SNAP-25, a vesicular SNARE protein involved in synaptic vesicle exocytosis [62–65]. Our results obtained from the present study provides the first evidence that Munc18-1 is ubiquitinated in cells and suggests that Munc18-1 ubiquitination and Hrs UIM-mediated ubiquitin signaling may regulate the exocytosis process.
Our IVEC screen also resulted in the isolation of 8 proteins that function in the regulation of microtubule, actin, and intermediate filament cytoskeletal networks and their associated motors (Table 1). Microtubules are dynamic protein filaments that serve as tracks for regulated movement and intracellular positioning of organelles, including endosomes [66]. The identification of β-tubulins, MARK4 [32, 67, 68], and dynactin 2 [69] as Hrs UIM-interacting proteins suggests a previously unrecognized role of Hrs in regulating microtubule dynamics and microtubule-based transport of endosomes. The interaction of the Hrs UIM with dynactin 2 is of particular interest because it provides a mechanism for loading endosomes onto microtubules and converting them to a motile pool. In addition to microtubules, the dynamics of actin cytoskeleton and intermediate filaments have also been implicated in regulation of endosomal trafficking [66, 70, 71]. Our identification of RhoBTB3 [72], CRMP-1 [73], MYPT3 [74, 75], and GFAP [76, 77] as Hrs UIM-interacting proteins suggests a role for Hrs in the coordinated regulation of actin dynamics, intermediate filament dynamics, and endosomal trafficking.
We and other laboratories have shown that Hrs exists in both cytosolic and endosomal membrane-associated pools [13, 33, 34]. Our screening results (Table 1) raise the possibility that, in addition to endosome-associated Hrs UIM-mediated ubiquitin signaling, cytosolic Hrs UIM may play a role in regulation of multiple cellular processes, including exocytosis, signal transduction, transport of RNP granules, and various metabolic processes. The wealth of data and interesting hypotheses generated from this study provide a basis for further studies to elucidate the molecular mechanisms underlying Hrs UIM-mediated ubiquitin signaling in cells.
Experimental procedures
Expression constructs and antibodies
Standard molecular biological techniques were used to generate pGST-Hrs UIM, which directs the expression of an N-terminal GST-tagged Hrs UIM domain corresponding to amino acid residues 251–286 of rat Hrs [33]. The pGST-Hrs UIM expression construct was sequenced to ensure that the fusion was in the correct reading frame and there were no unwanted changes in the codons. The pHA-ubiquitin [35], pGST-Hrs, and pGST-HrsΔUIM [78] constructs have been described previously. The pMyc-Hsc70 and pMyc-Munc18-1 plasmids were obtained as generous gifts from Drs. Cam Patterson and Tom Südhof, respectively. Antibodies used in this study include the following: anti-HA (3F10, Boehringer Mannheim, Mannheim, Germany; HA.11, Covance, Princeton, NJ, USA), anti-Hsc70 (Stressgen, Ann Arbor, MI, USA), anti-Myc (9E10.3, Neomarkers, Fremont, CA, USA), anti-ubiquitin (P4G7 and FL76, Covance), anti-EEA1 (BD Transduction Laboratories, San Jose, CA, USA), and secondary antibodies conjugated to horseradish peroxidase (Jackson Immunoresearch Labs, Inc., West Grove, PA, USA).
IVEC screen for Hrs UIM-interacting proteins
For identification of ubiquitinated proteins that bind to the UIM domain of Hrs, IVEC screen (Fig. 1) of a human adult brain cDNA library was performed using the ProteoLink IVEC system (Promega Corporation, Madison, WI, USA). The brain library cDNAs in a 96-well format with 100 cDNAs per well were in vitro transcribed and translated in the Gold TNT Quick coupled transcription-translation reticulocyte lysate system (Gold TNT SP6 Express 96-well plate) in the presence of [35S]methionine and ubiquitin as described previously [23]. The obtained protein pools were incubated at 4°C for 2 hours in binding buffer with GST-Hrs UIM fusion protein (Fig. 2A, bottom) or GST control immobilized on glutathione-agarose beads. After extensive washes with washing buffer, bound proteins were eluted by boiling in the Laemmli sample buffer, and analyzed by SDS-PAGE. Autoradiography of gel samples was performed using a phosphoimager. For each positive protein pool, the corresponding cDNA pool was progressively subdivided and re-examined in the same manner until individual positive cDNA clones were isolated [22]. Positive clones were then analyzed by DNA sequencing and by BLAST searches for sequence homology in the NCBI database. Putative transmembrane proteins were identified using both the predictive HMMTOP servers [79, 80].
Classification of Hrs UIM-interacting proteins
The identified Hrs UIM-interacting proteins were classified according to their subcellular localization and molecular function as determined based on the available literature, Gene Ontology, and homology searches. The percentage of proteins in each category was calculated by normalizing the number of proteins in each group to the total number of different proteins identified from the IVEC screen.
In vivo ubiquitination assays
In vivo ubiquitination assays were performed as described previously [35, 36]. Briefly, HeLa cells were transfected with pHA-ubiquitin in combination with pMyc-Munc18-1 or pMyc-Hsc70 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Twenty-four hours after transfection, the cells were incubated for 8 hours with proteasome inhibitor MG132 (20 µM in DMSO). The cells were then lysed and an equal amount of proteins from each lysate was subjected to a denaturing immunoprecipitation using anti-Myc antibodies. Immunoprecipitates were analyzed by SDS-PAGE, followed by immunoblotting with an anti-HA antibody to detect HA-ubiquitin conjugated to Munc18-1 or Hsc70.
Ubiquitin binding assays
GST-Hrs fusion proteins (~200 pmol) or GST control immobilized on glutathione-agarose beads were incubated at 4°C for 2 hours in binding buffer (25 mM Tris pH 7.5, 125 mM NaCl, 0.1% IGEPAL CA630) with ubiquitinated Munc18-1 or Hsc70 immunopurified from transfected HeLa cells [36, 81]. After extensive washes, bound proteins were eluted by boiling in the Laemmli sample buffer, and analyzed by SDS-PAGE and immunoblotting [82].
Small interfering RNAs (siRNAs) transfection
Two siRNAs (Dharmacon, Lafayette, CO, USA) were generated against the following human Hsc70 mRNA sequences: 3′-GGAGGUGUCUUCUAUGGUUUU-5′ and 3′-GAACAAGAGAGCUGUAAGAUU-5′, called Hsc70 siRNA-1 and siRNA-2, respectively. In addition, a control siRNA with no known mammalian homology (siCONTROL Non-Targeting siRNA #1, Dharmacon) was used as a negative control. HeLa cells were transfected with the indicated siRNA (50 nM) using the TransIT siQUEST (Mirus, Madison, WI, USA) reagent according to the manufacturer's instructions. At 72 hours post-transfection, cells were lysed and an equal amount of protein from each lysate was subjected to SDS-PAGE and immunoblotting with anti-Hsc70 and anti-EEA1 antibodies.
125I-EGF internalization and degradation assays
For measurement of 125I-EGF internalization, cells were serum starved for 2 hours, then incubated on ice with ~20 ng/ml 125I-EGF (MP Biochemicals, Solon, OH, USA) in binding buffer (1% BSA in serum-free DMEM). Cells were then washed with cold binding buffer and either lysed immediately to measure the initially bound 125I-EGF or transferred to 37°C for 10 minutes. After washing cells with acid wash (0.5 M NaCl, 0.2 M acetic acid, pH 2.8) on ice, the internalized 125I-EGF was measured as described [83, 84] and expressed as a percentage of the initially bound 125I-EGF. For measurement of 125I-EGF degradation after internalization, cells were chased in serum-free DMEM containing 1.5 µg/ml EGF and 1% BSA at 37°C for 60 minutes. Degraded 125I-EGF was measured as described [83, 84] and expressed as a percentage of the initially internalized 125I-EGF. Data is presented as the mean (± s.e.m.) and is representative of at least 3 independent experiments.
Acknowledgements
We thank Drs. Tom Südhof and Cam Patterson for providing the expression constructs for Munc18-1 and Hsc70, respectively. J.W.P was supported by National Institute of Neurological Disorders and Stroke Training Grant T32NS007480. This work was supported by grants from National Institutes of Health (NS047575 and GM082828 to L.L. and NS050650 to L.-S.C.).
Abbreviations
- AD
Alzheimer’s disease
- EEA1
early endosome antigen 1
- EGF
epidermal growth factor
- EGFR
EGF receptor
- GST
glutathione S-transferase
- Hrs
Hepatocyte growth factor regulated tyrosine kinase substrate
- IP3
inositol triphosphate
- IVEC
in vitro expression cloning
- JNK
Jun amino-terminal kinase
- MAP
microtubule-associated protein
- MVB
multivesicular body
- NMDA
N-methyl-D-aspartic acid
- PIP2
phosphatidylinositol bisphosphate
- siRNA
small-interfering RNA
- TNF
tumor necrosis factor
- Ub
ubiquitin
- UIM
ubiquitin-interacting motif
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 4.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM. Mechanisms Underlying Ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover A, Schwartz AL. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim Biophys Acta. 2004;1695:3–17. doi: 10.1016/j.bbamcr.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 9.Polo S, Sigismund S, Faretta M, Guidi M, Capua MR, Bossi G, Chen H, De Camilli P, Di Fiore PP. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 10.Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- 11.Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- 12.Miller SL, Malotky E, O'Bryan JP. Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J Biol Chem. 2004;279:33528–33537. doi: 10.1074/jbc.M313097200. [DOI] [PubMed] [Google Scholar]
- 13.Chin LS, Raynor MC, Wei X, Chen H, Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J Biol Chem. 2001;276:7069–7078. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, Bellen HJ. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 15.Clague MJ, Urbe S. Hrs function: viruses provide the clue. Trends Cell Biol. 2003;13:603–606. doi: 10.1016/j.tcb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 17.Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 18.Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- 19.Barriere H, Nemes C, Du K, Lukacs GL. Plasticity of Poly-Ubiquitin Recognition as Lysosomal Targeting Signals by the Endosomal Sorting Machinery. Mol Biol Cell. 2007 doi: 10.1091/mbc.E07-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbe S, Sachse M, Row PE, Preisinger C, Barr FA, Strous G, Klumperman J, Clague MJ. The UIM domain of Hrs couples receptor sorting to vesicle formation. J Cell Sci. 2003;116:4169–4179. doi: 10.1242/jcs.00723. [DOI] [PubMed] [Google Scholar]
- 21.King RW, Lustig KD, Stukenberg PT, McGarry TJ, Kirschner MW. Expression cloning in the test tube. Science. 1997;277:973–974. doi: 10.1126/science.277.5328.973. [DOI] [PubMed] [Google Scholar]
- 22.Lustig KD, Stukenberg PT, McGarry TJ, King RW, Cryns VL, Mead PE, Zon LI, Yuan J, Kirschner MW. Small pool expression screening: identification of genes involved in cell cycle control, apoptosis, and early development. Methods Enzymol. 1997;283:83–99. doi: 10.1016/s0076-6879(97)83009-1. [DOI] [PubMed] [Google Scholar]
- 23.Pridgeon JW, Geetha T, Wooten MW. A Method to Identify p62's UBA Domain Interacting Proteins. Biol Proced Online. 2003;5:228–237. doi: 10.1251/bpo66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 25.Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- 26.Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- 27.Tibbles KW, Brierley I, Cavanagh D, Brown TD. A region of the coronavirus infectious bronchitis virus 1a polyprotein encoding the 3C-like protease domain is subject to rapid turnover when expressed in rabbit reticulocyte lysate. The Journal of general virology. 1995;76(Pt 12):3059–3070. doi: 10.1099/0022-1317-76-12-3059. [DOI] [PubMed] [Google Scholar]
- 28.d'Abramo C, Massone S, Zingg JM, Pizzuti A, Marambaud P, Dalla Piccola B, Azzi A, Marinari UM, Pronzato MA, Ricciarelli R. Role of peroxisome proliferator-activated receptor gamma in amyloid precursor protein processing and amyloid beta-mediated cell death. Biochem J. 2005;391:693–698. doi: 10.1042/BJ20050560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren Y, Zhao J, Feng J. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J Neurosci. 2003;23:3316–3324. doi: 10.1523/JNEUROSCI.23-08-03316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM. Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 32.Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem J. 2008;411:249–260. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- 33.Kwong J, Roundabush FL, Moore PH, Montague M, Oldham W, Li Y, Chin LS, Li L. Hrs interacts with SNAP-25 and regulates Ca(2+)-dependent exocytosis. J Cell Sci. 2000;113:2273–2284. doi: 10.1242/jcs.113.12.2273. [DOI] [PubMed] [Google Scholar]
- 34.Urbe S, Mills IG, Stenmark H, Kitamura N, Clague MJ. Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate. Mol Cell Biol. 2000;20:7685–7692. doi: 10.1128/mcb.20.20.7685-7692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler TC, Chin LS, Li Y, Roudabush FL, Li L. Regulation of synaptophysin degradation by Mammalian homologues of seven in absentia. J Biol Chem. 2002;277:10273–10282. doi: 10.1074/jbc.M107857200. [DOI] [PubMed] [Google Scholar]
- 36.Chin LS, P VJ, Li L. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem. 2002;277:35071–35079. doi: 10.1074/jbc.M203300200. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Chin LS. The molecular machinery of synaptic vesicle exocytosis. Cell Mol Life Sci. 2003;60:942–960. doi: 10.1007/s00018-003-2240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raiborg C, Bache KG, Mehlum A, Stang E, Stenmark H. Hrs recruits clathrin to early endosomes. Embo J. 2001;20:5008–5021. doi: 10.1093/emboj/20.17.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sachse M, Urbe S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newmyer SL, Schmid SL. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J Cell Biol. 2001;152:607–620. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang HC, Newmyer SL, Hull MJ, Ebersold M, Schmid SL, Mellman I. Hsc70 is required for endocytosis and clathrin function in Drosophila. J Cell Biol. 2002;159:477–487. doi: 10.1083/jcb.200205086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morino C, Kato M, Yamamoto A, Mizuno E, Hayakawa A, Komada M, Kitamura N. A role for Hrs in endosomal sorting of ligand-stimulated and unstimulated epidermal growth factor receptor. Exp Cell Res. 2004;297:380–391. doi: 10.1016/j.yexcr.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 43.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 44.Mellman I. Membranes and sorting. Curr Opin Cell Biol. 1996;8:497–498. doi: 10.1016/s0955-0674(96)80026-3. [DOI] [PubMed] [Google Scholar]
- 45.Chin LS, Raynor MC, Wei X, Chen HQ, Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J Biol Chem. 2001;276:7069–7078. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- 46.Kanazawa C, Morita E, Yamada M, Ishii N, Miura S, Asao H, Yoshimori T, Sugamura K. Effects of deficiencies of STAMs and Hrs, mammalian class E Vps proteins, on receptor downregulation. Biochem Biophys Res Commun. 2003;309:848–856. doi: 10.1016/j.bbrc.2003.08.078. [DOI] [PubMed] [Google Scholar]
- 47.Wolfe MS, Guenette SY. APP at a glance. J Cell Sci. 2007;120:3157–3161. doi: 10.1242/jcs.03481. [DOI] [PubMed] [Google Scholar]
- 48.Nixon RA. Niemann-Pick Type C disease and Alzheimer's disease: the APP-endosome connection fattens up. The American journal of pathology. 2004;164:757–761. doi: 10.1016/S0002-9440(10)63163-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. The American journal of pathology. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanowitz M, Von Zastrow M. Ubiquitination-independent trafficking of G protein-coupled receptors to lysosomes. J Biol Chem. 2002;277:50219–50222. doi: 10.1074/jbc.C200536200. [DOI] [PubMed] [Google Scholar]
- 51.Hislop JN, Marley A, Von Zastrow M. Role of mammalian vacuolar protein-sorting proteins in endocytic trafficking of a non-ubiquitinated G protein-coupled receptor to lysosomes. J Biol Chem. 2004;279:22522–22531. doi: 10.1074/jbc.M311062200. [DOI] [PubMed] [Google Scholar]
- 52.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 53.Kiener TK, Sleptsova-Friedrich I, Hunziker W. Identification, tissue distribution and developmental expression of tjp1/zo-1, tjp2/zo-2 and tjp3/zo-3 in the zebrafish, Danio rerio. Gene Expr Patterns. 2007;7:767–776. doi: 10.1016/j.modgep.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 54.Ando H, Mizutani A, Matsu-ura T, Mikoshiba K. IRBIT, a novel inositol 1,4,5-trisphosphate (IP3) receptor-binding protein, is released from the IP3 receptor upon IP3 binding to the receptor. J Biol Chem. 2003;278:10602–10612. doi: 10.1074/jbc.M210119200. [DOI] [PubMed] [Google Scholar]
- 55.Song HY, Dunbar JD, Zhang YX, Guo D, Donner DB. Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J Biol Chem. 1995;270:3574–3581. [PubMed] [Google Scholar]
- 56.Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 57.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 58.Akbarzadeh S, Ji H, Frecklington D, Marmy-Conus N, Mok YF, Bowes L, Devereux L, Linsenmeyer M, Simpson RJ, Dorow DS. Mixed lineage kinase 2 interacts with clathrin and influences clathrin-coated vesicle trafficking. J Biol Chem. 2002;277:36280–36287. doi: 10.1074/jbc.M204626200. [DOI] [PubMed] [Google Scholar]
- 59.Raiborg C, Wesche J, Malerod L, Stenmark H. Flat clathrin coats on endosomes mediate degradative protein sorting by scaffolding Hrs in dynamic microdomains. J Cell Sci. 2006;119:2414–2424. doi: 10.1242/jcs.02978. [DOI] [PubMed] [Google Scholar]
- 60.Toonen RF, Verhage M. Munc18-1 in secretion: lonely Munc joins SNARE team and takes control. Trends in neurosciences. 2007;30:564–572. doi: 10.1016/j.tins.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 61.Way G, Morrice N, Smythe C, O'Sullivan AJ. Purification and identification of secernin, a novel cytosolic protein that regulates exocytosis in mast cells. Mol Biol Cell. 2002;13:3344–3354. doi: 10.1091/mbc.E01-10-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwong J, Roundabush FL, Hutton Moore P, Montague M, Oldham W, Li Y, Chin LS, Li L. Hrs interacts with SNAP-25 and regulates Ca(2+)-dependent exocytosis. J Cell Sci. 2000;113(Pt 12):2273–2284. doi: 10.1242/jcs.113.12.2273. [DOI] [PubMed] [Google Scholar]
- 63.Komada M, Kitamura N. Hrs and hbp: possible regulators of endocytosis and exocytosis. Biochem Biophys Res Commun. 2001;281:1065–1069. doi: 10.1006/bbrc.2001.4441. [DOI] [PubMed] [Google Scholar]
- 64.Tsujimoto S, Bean AJ. Distinct protein domains are responsible for the interaction of Hrs-2 with SNAP-25. The role of Hrs-2 in 7 S complex formation. J Biol Chem. 2000;275:2938–2942. doi: 10.1074/jbc.275.4.2938. [DOI] [PubMed] [Google Scholar]
- 65.Sun W, Yan Q, Vida TA, Bean AJ. Hrs regulates early endosome fusion by inhibiting formation of an endosomal SNARE complex. J Cell Biol. 2003;162:125–137. doi: 10.1083/jcb.200302083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murray JW, Wolkoff AW. Roles of the cytoskeleton and motor proteins in endocytic sorting. Adv Drug Deliv Rev. 2003;55:1385–1403. doi: 10.1016/j.addr.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 67.Trinczek B, Brajenovic M, Ebneth A, Drewes G. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J Biol Chem. 2004;279:5915–5923. doi: 10.1074/jbc.M304528200. [DOI] [PubMed] [Google Scholar]
- 68.Mandelkow EM, Thies E, Trinczek B, Biernat J, Mandelkow E. MARK/PAR1 kinase is a regulator of microtubule-dependent transport in axons. J Cell Biol. 2004;167:99–110. doi: 10.1083/jcb.200401085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qualmann B, Mellor H. Regulation of endocytic traffic by Rho GTPases. Biochem J. 2003;371:233–241. doi: 10.1042/BJ20030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Styers ML, Kowalczyk AP, Faundez V. Intermediate filaments and vesicular membrane traffic: the odd couple's first dance? Traffic. 2005;6:359–365. doi: 10.1111/j.1600-0854.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 72.Ramos S, Khademi F, Somesh BP, Rivero F. Genomic organization and expression profile of the small GTPases of the RhoBTB family in human and mouse. Gene. 2002;298:147–157. doi: 10.1016/s0378-1119(02)00980-0. [DOI] [PubMed] [Google Scholar]
- 73.Leung T, Ng Y, Cheong A, Ng CH, Tan I, Hall C, Lim L. p80 ROKalpha binding protein is a novel splice variant of CRMP-1 which associates with CRMP-2 and modulates RhoA-induced neuronal morphology. FEBS Lett. 2002;532:445–449. doi: 10.1016/s0014-5793(02)03736-5. [DOI] [PubMed] [Google Scholar]
- 74.Skinner JA, Saltiel AR. Cloning and identification of MYPT3: a prenylatable myosin targetting subunit of protein phosphatase 1. Biochem J. 2001;356:257–267. doi: 10.1042/0264-6021:3560257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vereshchagina N, Bennett D, Szoor B, Kirchner J, Gross S, Vissi E, White-Cooper H, Alphey L. the essential role of PP1beta in Drosophila is to regulate nonmuscle myosin. Mol Biol Cell. 2004;15:4395–4405. doi: 10.1091/mbc.E04-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hughes EG, Maguire JL, McMinn MT, Scholz RE, Sutherland ML. Loss of glial fibrillary acidic protein results in decreased glutamate transport and inhibition of PKA-induced EAAT2 cell surface trafficking. Brain Res Mol Brain Res. 2004;124:114–123. doi: 10.1016/j.molbrainres.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 77.Tang G, Xu Z, Goldman JE. Synergistic effects of the SAPK/JNK and the proteasome pathway on glial fibrillary acidic protein (GFAP) accumulation in Alexander disease. J Biol Chem. 2006;281:38634–38643. doi: 10.1074/jbc.M604942200. [DOI] [PubMed] [Google Scholar]
- 78.Kirk E, Chin LS, Li L. GRIF1 binds Hrs and is a new regulator of endosomal trafficking. J Cell Sci. 2006;119:4689–4701. doi: 10.1242/jcs.03249. [DOI] [PubMed] [Google Scholar]
- 79.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics (Oxford, England) 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 80.Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. Journal of molecular biology. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 81.Li Y, Chin LS, Weigel C, Li L. Spring, a Novel RING Finger Protein That Regulates Synaptic Vesicle Exocytosis. J Biol Chem. 2001;276:40824–40833. doi: 10.1074/jbc.M106141200. [DOI] [PubMed] [Google Scholar]
- 82.Chin LS, Nugent RD, Raynor MC, Vavalle JP, Li L. SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J Biol Chem. 2000;275:1191–1200. doi: 10.1074/jbc.275.2.1191. [DOI] [PubMed] [Google Scholar]
- 83.Valiathan RR, Resh MD. Expression of human immunodeficiency virus type 1 gag modulates ligand-induced downregulation of EGF receptor. J Virol. 2004;78:12386–12394. doi: 10.1128/JVI.78.22.12386-12394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]