Abstract
Summary: The modeling tool ProMoT facilitates the efficient and comprehensible setup and editing of modular models coupled with customizable visual representations. Since its last major publication in 2003, ProMoT has gained new functionality in particular support of logical models, efficient editing, visual exploration, model validation and support for SBML.
Availability: ProMoT is an open source project and freely available at http://www.mpi-magdeburg.mpg.de/projects/promot/.
Contact: mirschel@mpi-magdeburg.mpg.de; mirschel@mpi-magdeburg.mpg.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Research in systems biology requires efficient and easy-to-use software tools for modeling and visualization. This gains even more importance as size and complexity of models continue to increase. A suitable way to efficiently handle such models is the decomposition into smaller, more manageable parts resulting in modular models. Such models are characterized by a tree of modules which provides a hierarchical structure for the content. The modular description is based on network theory which was adapted for application to model biological systems (Kremling et al., 2000). Network theory allows to specify elements (e.g. species) and coupling elements (e.g. reactions). Modular networks additionally consist of modules and interfaces between modules. Modules, interface elements, variables and equations are called modeling entities and constitute the modular modeling concept. ProMoT implements the modular modeling concept (Ginkel et al., 2003). Modeling entities can be extended by the user. Additionally, libraries containing modeling entities for specific application areas, such as biochemical reaction networks, logic-based signaling networks or chemical engineering are provided. Although different other modeling tools such as CellDesigner (Funahashi et al., 2008), JDesigner (Sauro et al., 2003) and Edinburgh Pathway Editor (Sorokin et al., 2006) also facilitate visual editing of models, they do not provide a modular approach for model setup.
ProMoT is implemented in Common Lisp and Java. It runs on Windows and Linux. Further system requirements, installation instructions, several tutorials and up-to-date versions can be found at http://www.mpi-magdeburg.mpg.de/projects/promot/.
2 DESCRIPTION
ProMoT supports models based on DAE (differential algebraic equation) systems and models based on a logical (Boolean) modeling formalism (Klamt et al., 2006; Saez-Rodriguez et al., 2006). Modeling in ProMoT is subdivided into several steps which are associated to different components. In the following the functionality of the ProMoT Browser, the ProMoT Visual Editor and the ProMoT Visual Explorer are introduced.
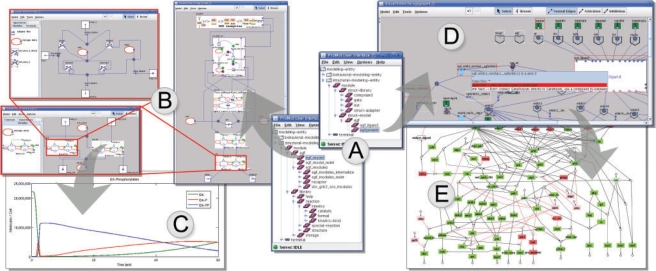
The ProMoT Browser facilitates loading and saving of models defined in ProMoT's own MDL format. It displays all loaded modeling entities (Fig. 1A), allows to copy or delete them and to create new ones. All other GUI components (e.g. for graphical editing and exploring) can be started from the ProMoT Browser. Models defined in SBML (Hucka et al., 2003) or CELLNETANALYZER (Klamt et al., 2007) format can be imported and exported. Furthermore, ProMoT models can be exported to the simulation environments Matlab (Fig. 1C), DIVA or Diana (Krasnyk et al., 2007). Export in formats which do not support modularity preserves the complete contents and discards all information about the original partition in modules. Prior to model export ProMoT validates and optimizes the model. Documentation for models can be generated in LaTeX or HTML format. For detailed information about different aspects of the export see Supplementary Material.
Fig. 1.
GUI components of ProMoT and associated modeling steps for two different modeling types—the dynamic model of the Epidermal Growth Factor Receptor (EGFR) by Saez-Rodriguez et al. (2005) (left) and the logical, signaling EGFR model taken from Samaga et al. (2009) The logic of EGFR/ErbB signaling: theoretical properties and analysis of high-throughput data, submitted (right). Arrows indicate a feasible sequence of modeling steps. The ProMoT Browser shows loaded modeling entities (A). Models can be edited using the ProMoT Visual Editor (B, D). While editing a background syntax checking marks structural modeling errors (D). Models can be exported to different simulation environments (C) or analysis tools. Subsequently the analysis results from CELLNETANALYZER can be visualized on top of the network structure (E).
The ProMoT Visual Editor aims to setup and edit the model holistically. Hence, the topological structure of a module based on a graph metaphor as well as behavioral properties (variables and equations) should be graphically editable. Modeling entities can be placed and connected by links (Fig. 1B and D). Afterwards their variables can be altered using a variable editor. In previous versions, all properties could only be edited in the MDL code of the model. For models (e.g. imported SBML models without an initial layout) different layout algorithms are available which take advantage of ProMoT's inherent modularity. To provide model validation while editing, a background syntax check validates the model against structural modeling errors (see Supplementary Material and Fig. 1D). This is particularly elaborated for the logical modeling formalism. Additionally, editing shortcuts reduce the amount of user interactions and the probability to introduce errors in the modeling process.
The ProMoT Visual Explorer provides basically two functionalities—exploring and visualizing a modular model. The former uses context sensitive zooming through the different levels of modules and helps to get a quick view of edits or to search for specific components in context of the whole model. The latter allows to tailor visualizations specifically to users needs. The goal is twofold: first, it aims to reduce visual complexity by presenting only a relevant subset of the model. This results in a less cluttered and more descriptive visualization. Second, it provides adequate visual representations for the logical modeling formalisms supported by ProMoT. Among others it visually aids the user in specific modeling tasks, e.g. the visual interpretation of analysis results (Fig. 1E). The ProMoT Visual Explorer is also used to export the model representation into common bitmap, vector and graph formats.
3 CONCLUSIONS
ProMoT is suitable for different application areas, e.g. dynamic models (Bettenbrock et al., 2006), logical models (Samaga et al., 2009, The logic of EGFR/ErbB signaling: theoretical properties and analysis of high-throughput data, submitted) and synthetic biology (Marchisio et al., 2008).
ProMoT, a tool for efficient and comprehensible setup and editing of modular models is presented. A sophisticated graphical user interface assists the modeler in setup and management of modules and additionally includes a component for graphical zooming and flexible visual presentations. The described software is well integrated with external tools (e.g. CELLNETANALYZER, DIVA, Diana, MATLAB) for model analysis and simulation. Moreover, ProMoT can be used as a command line tool which is helpful to start from external tools such as SYCAMORE (Weidemann et al., 2008). ProMoT is suitable for different application areas, e.g. dynamic models (Bettenbrock et al., 2006), logical models (?., ? The logic of EGFR/ErbB signaling: theoretical properties and analysis of high-throughput data, submitted) and synthetic biology (Marchisio et al., 2008).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank R. Samaga and J. Saez-Rodriguez who provided models. The authors are also thankful to anonymous reviewers for constructive comments.
Funding: German Ministry of Research and Education [HepatoSys grant 0313077 (to S.M. and M.R.), ForSys grant 0313922 (to K.S.)].
Conflict of Interest: None declared.
REFERENCES
- Bettenbrock K, et al. A quantitative approach to catabolite repression in Escherichia coli. J. Biol. Chem. 2006;281:2578–2584. doi: 10.1074/jbc.M508090200. [DOI] [PubMed] [Google Scholar]
- Funahashi A, et al. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE. 2008;96:1254–1265. [Google Scholar]
- Ginkel M, et al. Modular modeling of cellular systems with ProMoT/Diva. Bioinformatics. 2003;19:1169–1176. doi: 10.1093/bioinformatics/btg128. [DOI] [PubMed] [Google Scholar]
- Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Klamt S, et al. A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinformatics. 2006 doi: 10.1186/1471-2105-7-56. doi: 10.1186/1471-2105-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt S, et al. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Sys. Biol. 2007 doi: 10.1186/1752-0509-1-2. doi:10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnyk M, et al. Proceeding of the 16th European Symposium on Computer Aided Process Engineering. Garmisch-Partenkirchen: Elsevier; 2006. The ProMoT/Diana simulation environment; pp. 445–450. [Google Scholar]
- Kremling A, et al. The organization of metabolic reaction networks: a signal-oriented approach to cellular models. Metab. Eng. 2000;2:190–200. doi: 10.1006/mben.2000.0159. [DOI] [PubMed] [Google Scholar]
- Marchisio MA, Stelling J. Computational design of synthetic gene circuits with composable parts. Bioinformatics. 2008;24:1903–1910. doi: 10.1093/bioinformatics/btn330. [DOI] [PubMed] [Google Scholar]
- Saez-Rodriguez J, et al. Dissecting the puzzle of life: modularization of signal transduction networks. Comput. Chem. Eng. 2005;29:619–629. [Google Scholar]
- Saez-Rodriguez J, et al. Visual setup of logical models of signaling and regulatory networks with ProMoT. BMC BioInformatics. 2006;7 doi: 10.1186/1471-2105-7-506. doi: 10.1186/1471-2105-7-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauro HM, et al. Next generation simulation tools: the systems biology workbench and BioSPICE integration. OMICS. 2003;7:355–372. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- Sorokin A, et al. The pathway editor: a tool for managing complex biological networks. IBM J. Res. Dev. 2006;50:561–573. [Google Scholar]
- Weidemann A, et al. SYCAMORE - a systems biology computational analysis and modeling research environment. Bioinformatics. 2008;24:1463–1464. doi: 10.1093/bioinformatics/btn207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.