Abstract
Summary: We present a tool designed for visualization of large-scale genetic and genomic data exemplified by results from genome-wide association studies. This software provides an integrated framework to facilitate the interpretation of SNP association studies in genomic context. Gene annotations can be retrieved from Ensembl, linkage disequilibrium data downloaded from HapMap and custom data imported in BED or WIG format. AssociationViewer integrates functionalities that enable the aggregation or intersection of data tracks. It implements an efficient cache system and allows the display of several, very large-scale genomic datasets.
Availability: The Java code for AssociationViewer is distributed under the GNU General Public Licence and has been tested on Microsoft Windows XP, MacOSX and GNU/Linux operating systems. It is available from the SourceForge repository. This also includes Java webstart, documentation and example datafiles.
Contact: brian.stevenson@licr.org
Supplementary information: Supplementary data are available at http://sourceforge.net/projects/associationview/ online.
1 INTRODUCTION
Advances in genotyping platforms have enabled the identification of millions of single nucleotide polymorphism (SNPs) in the human genome, which are intensively used to study the impact of genomic variation on phenotype. Dedicated software like WGAViewer (Ge et al., 2008) was developed to facilitate the interpretation of results from early genome-wide association (GWA) studies. Recent dramatic increases in array resolution—the latest Affymetrix and Illumina arrays offer more than 1.8 M features—have created a novel and immediate need for efficient and scalable visualization tools. Scientists and clinicians strongly rely on such tools to interpret their results, while bioinformaticians need scalable applications to check the results from their high-throughput analyses. In this context, we have developed AssociationViewer, a software tool for visualization of GWA studies in genomic context. The program can efficiently handle large genomic datasets, is extensible to any genomic data represented in BED or WIG format and implements aggregation (union) or intersection of data tracks.
2 PROGRAM OVERVIEW
2.1 Cache and memory management
With increasing data volumes, efficient resource management is essential. One approach is to store the data in a cache with fast indexing mechanisms to retrieve the data, and to keep in memory only the information that is visualized. We implemented such a system in AssociationViewer. For comparison, loading a single dataset with 500 K SNPs in WGAViewer needs about 224 MB of RAM, whereas loading 10 different datasets (a total of 10 M data points) and displaying all genes on chromosome 1 needs only 50 MB in AssociationViewer.
2.2 Data import and export
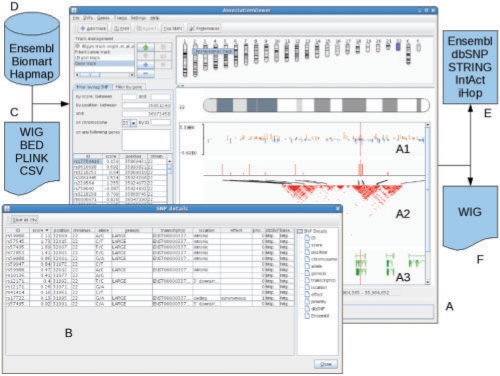
A typical GWA dataset consists of a list of SNPs with P-values derived from an association analysis. In AssociationViewer, such data can be imported from PLINK (Purcell et al., 2007) output or other text files. Import of data in BED and WIG format is also possible (Fig. 1C). These formats are extensively used by the bioinformatics community and in the UCSC genome browser (Kent et al., 2002) to describe genomic and transcriptomic data. BED describes gene features, whereas WIG allows representation of any single position associated with a score (Fig. 1A1). AssociationViewer allows export in WIG format (Fig. 1F). Window images can also be exported in many popular formats.
Fig. 1.
General view of AssociationViewer (A and B). Also displayed are the input files (C), annotation data downloaded (D), cross-references (E) and export format (F).
2.3 Annotation retrieval
Gene and transcript data (Fig. 1A3) can be downloaded from Ensembl (Hubbard et al., 2007) and Biomart (Kasprzyk et al., 2004). Tag SNPs can be retrieved from the Hapmap website (The International HapMap Consortium, 2007) (Fig. 1D). The user can choose to connect to Ensembl or HapMap releases for NCBI Builds 35 or 36.
2.4 Genome navigation and data interaction
Navigation in AssociationViewer is intuitive (Fig. 1A). The user selects a chromosome either by clicking on the appropriate ideogram or via genomic coordinates. Scrolling or zooming is done via a mouse or the appropriate icons. One can search for SNPs, either by providing IDs, a coordinate range, a score cut-off or a list of neighbouring genes. Genes are found using similar options except that there is no score-based filter. Retrieved data (position, function description) are displayed in a table (Fig. 1B) which includes cross-references to Ensembl (Hubbard et al., 2007), IntAct (Kerien et al., 2007), iHop (Fernández et al., 2007), dbSNP (http://www.ncbi.nlm.nih.gov/), STRING (Von Mering et al., 2007) (Fig. 1E). The sequence surrounding a SNP and any associated SNPs can be downloaded and displayed in a table summary and in a linkage disequilibrium (LD) plot (Fig. 1A2).
2.5 GWA specialized functions
To better understand the distribution of GWA P-values, AssociationViewer can produce QQ plots to identify where a SNP's P-value strongly deviates from random expectation. To compare SNP P-values between different data tracks, it can generate a Manhattan plot. To rank SNPs with highly significant P-values and obtain information for possible gene candidates, it can generate a ‘top hit’ report.
2.6 Track merging—aggregation and intersection
When browsing multiple tracks, it can become tedious to visualize a region of interest. Merging two or more tracks can help this situation. In AssociationViewer WIG (score) tracks are aggregated in two steps: (i) within each track, set all values to 1 if they are greater than the mean score for that track, otherwise set them to 0; (ii) sum the discretized values at each position over all tracks. BED (gene) tracks are aggregated by merging features together and providing a colour code representing the overlap density.
Intersection between WIG tracks is also possible, generating a tabulated report of common positions and scores. This is useful when comparing GWA results from different studies on the same phenotype. For example, intersecting SNPs with significant P-values from different GWAs and deriving a top hit report will sort these SNPs by the number of times they were replicated in the different GWAs. This is a useful functionality to integrate different studies, to reduce the data complexity and to facilitate interpretation of the results.
3 CONCLUSION AND DISCUSSION
AssociationViewer is a flexible software tool that permits visualization of GWA data. It implements essential features such as a ‘top hits’ report, SNP annotation retrieval, QQ and LD plots. Any genomic or transcriptomic data represented in BED or WIG format can be imported. Genomic annotation can be downloaded from Ensembl, BioMart and Hapmap.
The ability to handle very large datasets is often limited in visualization software. We optimized resource management by using an efficient cache system and limiting the amount of information held in memory. As a result, our software performs remarkably well when simultaneously visualizing several large-scale GWA datasets.
The aggregation and intersection of data tracks are useful functionalities to reduce data complexity. The intersection feature report offers the possibility to integrate and visualize results from different studies. As a proof of concept, simple aggregation methods were implemented in the current version of AssociationViewer, but more elaborate algorithms will be developed in future versions.
Dedicated resources for SNP and copy number variant datasets are being set up [e.g. Ensembl Variation, European Genotype Archive (http://www.ebi.ac.uk/ega/), Database of Genomic Variants (Iafrate et al., 2004)]. Once connection to these resources is possible, we plan to enable queries via the API to visualize results within AssociationViewer.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nicolas Guex for helpful input on the features and usability of AssociationViewer, Sébastien Moretti and Laurent Falquet for testing the software and Victor Jongeneel for encouragement and support. We also thank Dongliang Ge and David Goldstein (Duke University) for discussions during the early stages of this work.
Funding: Swiss National Science Foundation; Infectigen; University of Lausanne; Swiss Institute of Bioinformatics; Ludwig Institute for Cancer Research.
Conflict of Interest: none declared
REFERENCES
- Fernández JM, et al. iHOP Web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJP, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kasprzyk A, et al. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerien S, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mering C, et al. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.