Abstract
Centrosomes play a critical role in formation of bipolar mitotic spindles, an essential event for accurate chromosome segregation into daughter cells. Numeral abnormalities of centrosomes (centrosome amplification) occur frequently in cancers, and are considered to be the major cause of chromosome instability, which accelerates acquisition of malignant phenotypes during tumor progression. Loss or mutational inactivation of p53 tumor suppressor protein, one of the most common mutations found in cancers, results in a high frequency of centrosome amplification in part via allowing the activation of the cyclin-dependent kinase (CDK) 2-cyclin E (as well as CDK2-cyclin A) which is a key factor for the initiation of centrosome duplication. In this review, the role of centrosome amplification in tumor progression, and mechanistic view of how centrosomes are amplified in cells through focusing on loss of p53 and aberrant activities of CDK2-cyclins will be discussed.
Basic biology of the centrosome, and its relevance to cancer
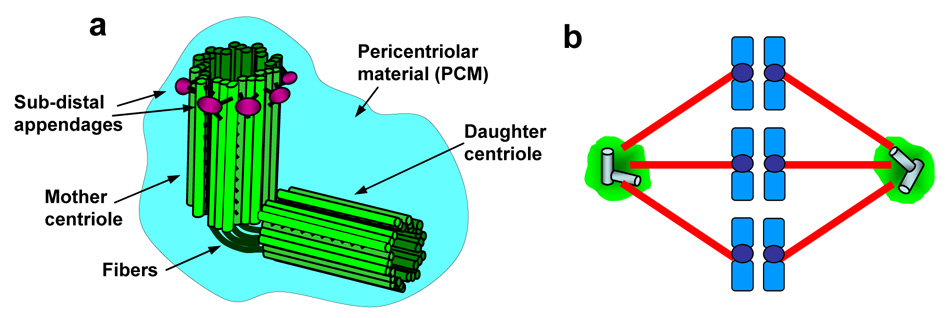
The centrosome is a small non-membranous organelle (1–2 µm in diameter) usually found at the periphery of nucleus during interphase, and its primary function is to nucleate and anchor microtubules. The centrosome in animal cells consists of paired centrioles, and surrounding electron dense materials known as pericentriolar material (PCM) (Fig. 1a). The centrioles in the pair are structurally different from each other; one with a set of appendages at the distal ends (mother centriole) and another without them (daughter centriole), and these appendages are believed to play a role in the microtubule anchoring activity [1]. The PCM is composed of a number of different proteins, and the protein composition of the PCM is highly dynamic: some PCM components reside at the centrosome permanently, while some transiently localize to the centrosome during the cell cycle.
Figure 1. Structure and function of centrosomes.
(a) The basic structure of the centrosome. (b) During mitosis, two centrosomes become the spindle poles, directing the formation of bipolar mitotic spindles.
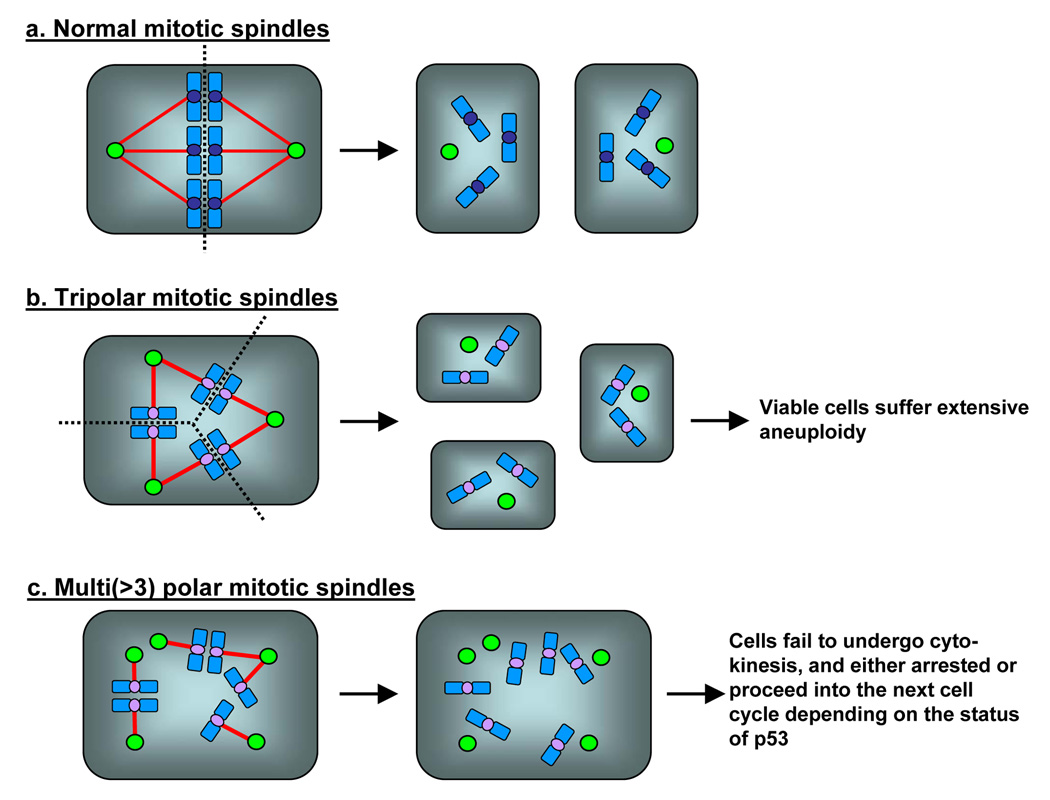
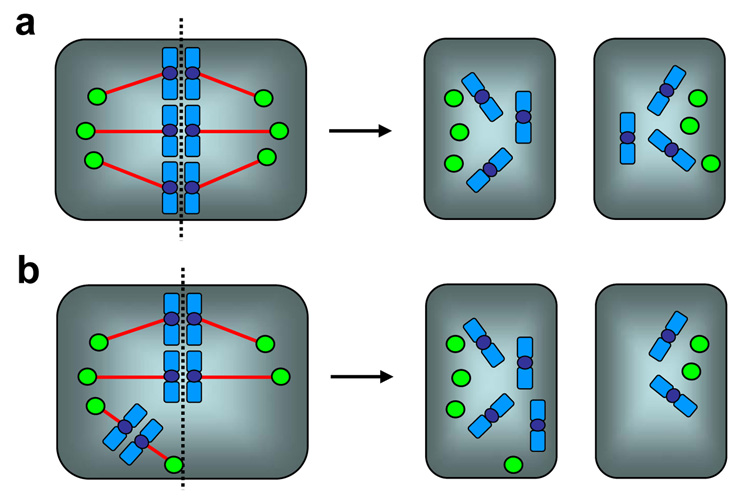
In mitosis, two centrosomes form spindle poles, and direct the formation of bipolar mitotic spindles (Fig.1b). Because formation of proper mitotic spindles is essential for the accurate chromosome segregation into two daughter cells during cytokinesis, two, and only two centrosomes are needed in mitosis. Thus, numeral integrity of centrosomes is carefully controlled, and abrogation of this control results in abnormal amplification of centrosomes (presence of >2 centrosomes). Centrosome amplification leads to aberrant mitotic spindle formation with more than two spindle poles, and subsequent chromosome segregation errors. Cells with amplified centrosomes often form tripolar mitotic spindles, and these cells can undergo cytokinesis (Fig. 2b). Some daughter cells from the tripolar division are viable, yet suffer severe aneuploidy [2]. When the mitotic spindles with more than three poles are formed, cells fail to undergo cytokinesis [2], and become either bi-nucleated or large mono-nucleated cells (Fig. 2c). Since failure to undergo cytokinesis triggers the checkpoint response involving the p53 tumor suppressor protein via a mechanism that is poorly understood [3], the cells become arrested in the presence of p53, and eventually undergo cell death. In contrast, in the absence of p53 or the p53-depependent checkpoint function, those bi- or large mono-nucleated cells continue to cycle, and many of them experience repeated cytokinesis block, become very large multi(>2)-nucleated polyploid cells, and eventually undergo cell cycle arrest/cell death [4]. However, some cells resume cytokinesis likely through the formation of pseudo-bipolar spindles (see below). Since the presence of polyploid chromosomes is known to destabilize chromosomes [5], polyploidy resulting from cytokinesis block due to centrosome amplification further promotes the chromosome instability. It is important to note that centrosome amplification does not always results in formation of multi-polar spindles. Amplified centrosomes frequently form “pseudo-bipolar” spindles by positioning on a bipolar axis (Fig. 3a), resulting in mitotic spindles which structurally resemble the “true” bipolar spindles organized by two centrosomes. Although the mechanism underlying this phenomenon known as “centrosome clustering” is not fully understood, the microtubule motor protein dynein has been shown to play an important role [6]. Cells with “pseudo-bipolar” spindles appear to undergo normal cytokinesis without any chromosome segregation errors. However, even these “pseudo-bipolar” spindles often encounter a risk of chromosome segregation errors (Fig. 3b): one or a few centrosomes fail to line up on the bipolar axis, yet they are functionally intact, nucleating microtubules which capture chromosomes. Depending on which daughter cell receives those chromosomes, aneuploid daughter cells can be generated [7].
Figure 2. Mitotic defects associated with numeral abnormalities of centrosomes.
In normal mitosis, two centrosomes direct the formation of bipolar mitotic spindles (a). In the presence of amplified centrosomes, cells frequently form multiple (>2) spindle poles. (b) Tripolar spindles can undergo cytokinesis, and some daughter cells are viable, yet suffer severe aneuploidy. (c) Cells with spindles with >3 poles fail to undergo cytokinesis in most cases, becoming either bi-nucleated or large mono-nucleated cells. Because of the p53-dependent checkpoint response to cytokinesis failure, the cells become arrested in the presence of p53, and eventually undergo cell death. In contrast, in the absence of p53, cells continue to cycle, and many of them become very large multi(>2)-nucleated cells and often undergo senescence-like arrest and cell death. However, some cells escape from continuous cytokinesis block, and become polyploid cells.
Figure 3. Pseudo-bipolar spindles and the risk of chromosome segregation errors.
Amplified centrosomes frequently form pseudo-bipolar spindles by positioning on a bipolar axis (centrosome clustering), resulting in mitotic spindles which structurally resemble the true bipolar spindles organized by two centrosomes. Cells with pseudo-bipolar spindles undergo normal cytokinesis without any chromosome segregation errors (a). However, one or a few centrosomes often fail to position on the bipolar axis. Those mal-positioned centrosomes are still functionally intact, nucleating microtubules which capture chromosomes. Depending on whether those particular chromosomes are segregated into one or the other daughter cell, aneuploid daughter cells can be generated (b).
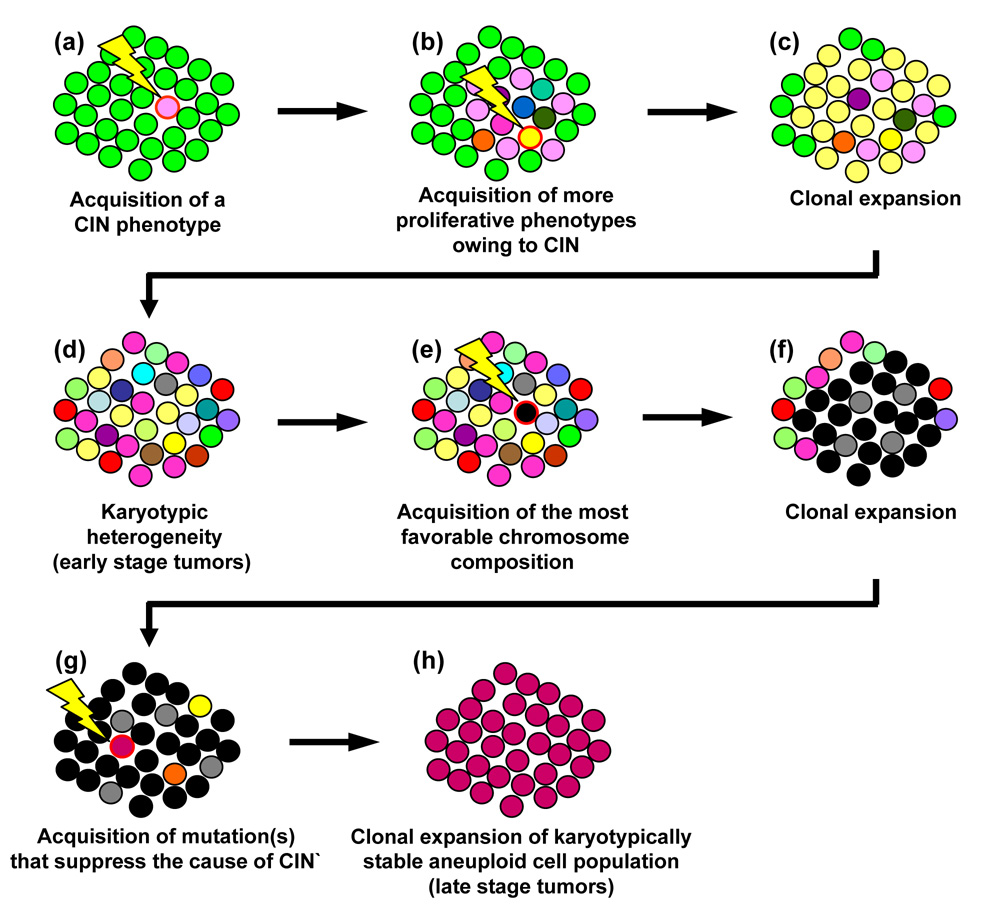
Chromosome segregation errors resulting from centrosome amplification profoundly influences the rate of tumor progression, since loss or gain of even a single chromosome can introduce multiple genetic alterations required for acquisition of more malignant phenotypes. Indeed, centrosome amplification frequently occurs in almost all types of solid tumors and several cases of leukemia and lymphoma [4, 8]. Moreover, many studies have shown a strong association between the occurrence of centrosome amplification and a high degree of aneuploidy. However, it should be noted that correlative studies of aneuploidy and centrosome amplification in tumor tissues, which also apply to other chromosome destabilizing events, need to be approached cautiously. During the tumor progression, tumor cells often undergo a phenomenon called “karyotypic convergence” (Fig. 4) [9], in which tumors in early stages are karyotypically heterogeneous due to unstable chromosomes, while those in later stages are highly aneuploid, yet karyotypically homogenous and their chromosomes are stable. It is believed that tumor cells in early stages continuously re-establish the chromosome compositions that render better growth properties (i.e., faster growth rate, anchorage independent growth, mitogenic stimuli independent growth, etc.). Once the tumor cells acquire a superlative chromosome composition for growth under a given environment, chromosome instability itself becomes a target of the selection pressure, prioritizing the maintenance of the acquired superior chromosome composition. To this end, the cells, which have acquired mutation(s) that either eliminate or counteract the cause of chromosome instability, restore the stability of chromosomes and eventually dominate the tumor population because of their higher proliferative capacities. Thus, if the centrosome amplification is the cause of chromosome instability in the early stage tumors, it will likely be suppressed in the late stage tumors that are chromosomally homogenous. In the study using the cultured p53-deficient cells (loss of p53 leads to centrosome amplification, which will be discussed in details later), it has been shown that cells suffer extensive chromosome instability associated with centrosome amplification and form a karyotypically heterogeneous population. However, after prolonged passaging, cells with a particular chromosome composition dominate the population, and those cells no longer show centrosome amplification [10], supporting the karyotypic convergence theory of tumor progression. Further analysis of the p53-deficient cells after prolonged passages revealed that the cells acquired a mutation that results in a high level expression of BubR1 spindle checkpoint kinase [11]. The spindle checkpoint is a surveillance mechanism that restrains cells from entering anaphase by preventing the activation of Anaphase-promoting complex (APC), an E3 ubiquitin ligase that marks target proteins for degradation by the 26S proteasome, until all chromosomes are properly attached to the bipolar mitotic spindles originated from centrosomes. This checkpoint function is activated by chromosomes (kinetochores) unattached or inadequately attached to the spindles as well as by improper spindle tension exerted on kinetochores [12, 13]. Overexpression of BubR1 appears to increase the potency of the spindle checkpoint function, which efficiently responds to the mitotic defects associated with centrosome amplification. Thus, the cells with defective mitoses associated with amplified centrosomes are effectively targeted by the “super-activated” spindle checkpoint function, and are eventually disposed to cell death. The p53-deficient cells described in the above study achieved the karyotypic convergence by selectively and continuously killing the cells with amplified centrosomes by BubR1 overexpression, hence the favorable chromosome composition could be maintained in the surviving cells. Apparently, BubR1 overexpression is one of the many potential mutations that cells could acquire to suppress the cause of chromosome instability in those cells. However, these findings imply that the efficacy of mitotic checkpoint can profoundly influences the extent of chromosome instability associated with centrosome amplification: abnormally amplified centrosomes can destabilize chromosome more effectively in cells whose spindle checkpoint function is compromised. Taken together, the correlative examination of tumors for aneuploidy and centrosome amplification requires the consideration of many variables such as whether the tumors of interest have undergone karyotypic convergence, and whether the mitotic checkpoint function is compromised.
Figure 4. Karyotypic convergence during tumor progression.
The early stage tumors are often karyotypically heterogeneous, while the late stage tumors are karyotypically homogenous. The karyotypic convergence theory provides the mechanism underlying this phenomenon. A cell acquires a mutation that renders growth advantage over the others, and clonally expands (a). Among this particular population of cells, one or a few cells acquire the mutations that destabilize chromosomes (CIN phenotype, i.e., centrosome amplification) (b). Because of the CIN phenotype, the population gradually becomes karyotypically heterogeneous (c), representing the early stage tumor. A cell within this population eventually acquires the favorable chromosome composition that renders maximal growth/carcinogenic potentials (d), and clonally expands (f). For these cells, maintenance of this particular chromosome composition becomes the priority, selecting the cell(s) that have acquired the mutation(s) that either counteract or eliminate the cause of CIN (g). Such a cell will eventually dominate the population by retaining the favorable karyotypic composition, and thus the population becomes karyotypically homogenous, representing late stage tumors.
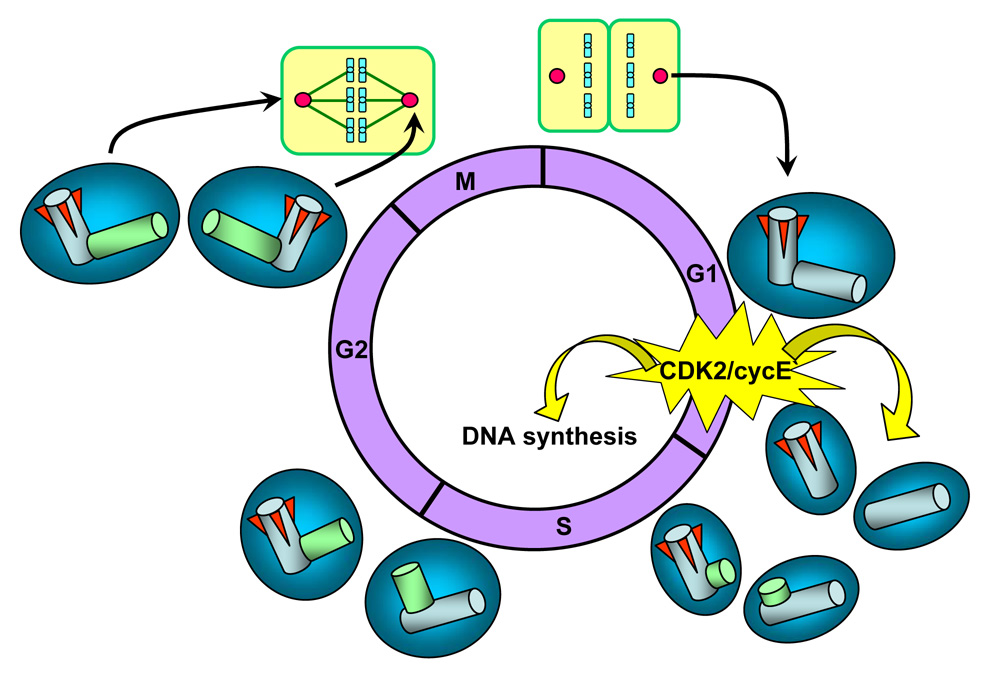
The centrosome duplication cycle and the role of CDK2-cyclin E
Upon cytokinesis, each daughter cell receives only one centrosome along with one half of the replicated DNA. Thus, the centrosome, like DNA, must duplicate once before the next mitosis. In other words, cells have either one unduplicated or two duplicated centrosomes at any given time point during the cell cycle. Since DNA and centrosome are the only two organelles that undergo one duplication in each cell cycle, animal cells are equipped with a mechanism that coordinates these two events, ensuring these two organelles to duplicate only once [14] (Fig. 5). The coupling of the initiation of centrosome and DNA duplication is at least in part achieved by the cyclin-dependent kinase (CDK) 2-cyclin E kinase complex [15]. CDK2-cyclin E, which is a known inducer of S-phase entry in part via phosphorylating pRB retinoblastoma protein [16, 17], has been found to be also a key initiator of centrosome duplication [18, 19]. Thus, the late G1 specific activation of CDK2/cyclin E primarily induced by the temporal increase in the cyclin E expression [20, 21] leads to coordinated initiation of centrosome and DNA duplication. Some of its target centrosomal proteins have been identified, including Mps1 kinase, CP110 and nucleophosmin [22–24]. Centrosome duplication starts with physical separation of the paired centrioles, followed by the formation of the procentriole in the proximity of each pre-existing centriole (Fig. 5). During S and G2, procentrioles elongate and two centrosomes undergo a maturation process by progressively recruiting critical PCM components. At late G2, the daughter centriole of the original pair acquires the sub-distal appendages, and two mature centrosomes are generated.
Figure 5. The centrosome duplication cycle.
CDK2/cyclin E, a known inducer of S-phase entry, is also a key initiator of centrosome duplication. Thus, the late G1 specific activation of CDK2/cyclin E is at least in part responsible for the coordinated initiation of centrosome and DNA duplication. Centrosome duplication begins with the physical splitting of the paired centrioles, followed by formation of procentrioles near the proximal end of each pre-existing centriole. During S and G2, procentrioles elongate, and two centrosomes progressively recruit PCM, an event known as centrosome maturation. In late G2, the daughter centriole of the parental pair acquires sub-distal appendages (red wedges), and two mature centrosomes are generated. At late G2 prior to mitosis, two duplicated centrosomes separate and migrate to opposite ends of the cell. During mitosis, duplicated centrosomes form spindle poles to direct the formation of bipolar mitotic spindles. Upon cytokinesis, each daughter cell receives one centrosome along with one half of the duplicated DNA.
Recent studies examining the CDK2-deficient mice have raised a question for the requirement of CDK2 in the initiation of centrosome duplication. CDK2-deficient mice develop rather normally [25, 26], suggesting that CDK2 is not essential for the induction of DNA as well as centrosome duplication [25–27]. Indeed, cells derived from these mice can proliferate and undergo centrosome duplication [25–27], indicating that the function of CDK2 for the initiation of centrosome duplication can be readily and functionally replaced by other proteins. The limited number of studies has shown that over-activation of CDK4/CDK6, both of which are G1 CDKs that form complexes with cyclin D, can promote centrosome duplication [28, 29]. Although it is not known whether CDK4,6-cyclin D-mediated promotion of centrosome duplication still depends on the presence of CDK2, these studies raise the possibility that CDK4/6-cyclin D may be the proteins that compensates for the loss of CDK2 in the initiation of centrosome duplication. Indeed, CDK2−/−CDK4−/− double knock-out mice are embryonically lethal, and cells prepared from those mice are defective in G1/S transition [30]. It remains to be seen whether initiation of centrosome duplication is also affected in the CDK2−/−CDK4−/− mouse cells. Alternatively, CDK1, a mitotic CDK, may be able to compensate for the loss of CDK2 for the initiation of centrosome duplication by forming a complex with cyclin E, as is the case for induction of the G1/S transition [30]. More recently, it has been shown that mouse embryos lacking all interphase CDKs (CDK2, 3, 4 and 6) undergo cell division, and in those embryos, CDK1 binds to all cyclins, resulting in the pRb phosphorylation and the expression of the E2F-regulated genes [31]. However, cyclin E-deficient cells can initiate centrosome duplication rather normally (see below), indicating that the involvement of CDK1-cyclin E in the initiation of centrosome duplication is unlikely, yet the involvement of CDK1 complexed with other interphase cyclins (i.e., cyclin A) remains highly possible (see below). It remains to be tested whether CDK4/6-cyclin D (as well as CDK1-cyclin E/A) can act on the previously identified centrosomal targets of CDK2-cyclin E. Alternatively, the kinases other than the CDK family may play a role in the initiation of centrosome duplication in the CDK2-deficient cells, including Polo-like Kinase (PLK) 2, PLK4 and ROCKII, all of which have been shown to possess the activities to promote centrosome duplication [32–34].
Similar to CDK2-deficient mice, cyclin E-deficient mice also develop rather normally, and the initiation of centrosome duplication is not significantly affected in cells derived from these mice [35], suggesting that the function of cyclin E can also be readily replaced by other proteins. We have recently found that initiation of centrosome duplication is significantly inhibited by the transient depletion of cyclin A in cyclin E-deficient cells, suggesting that cyclin A is a primary candidate protein that compensates for the loss of cyclin E for initiation of centrosome duplication [35]. Indeed, CDK2-cyclin A has been previously implicated in the initiation of centrosome duplication [36]. However, as long as cyclin E is present, the role of cyclin A in the initiation of centrosome duplication appears to be minimal [35], and similar argument will probably apply to CDK2: as long as CDK2 is present, other CDKs may likely play minimal roles for the initiation of centrosome duplication.
DNA replication requires several critical biochemical events before being finally initiated by CDK2-cyclin E [20, 21]. In contrast, centrosomes rapidly respond to active CDK2/cyclin E, and begin the duplication process. For instance, overexpression of cyclin E, which results in constitutive activation of CDK2/cyclin E, induces centrosome duplication in very early G1 phase [37]. In contrast, newly duplicated centrosomes do not re-duplicate immediately even in the presence of active CDK2-cyclin E [37]. The analysis of centrosomes in CHO cells that are inhibited for S-phase entry/progression by exposure to DNA synthesis inhibitors shows that duplicated centrosomes continue to re-duplicate without DNA synthesis, but do so only after a substantial length of time (~20 h) [38], suggesting that duplicated centrosomes need to regain the duplication competency before undergoing re-duplication. The study by Wong & Stearn further supported this possibility, showing that fusion of G1 phase cells (containing unduplicated centrosomes) and S phase cells (containing newly duplicated centrosomes) results in duplication of only one centrosome presumably of the G1 centrosome [39]. Thus, even in the presence of active CDK2/cyclin E, newly duplicated centrosomes can re-duplicate only after acquisition of the duplication competency. More recently, it has been shown in vitro [40] that centriole disengagement (loss of the strict orthogonal configuration of the paired centrioles) catalyzed by separase, a mitotic protease that acts on the cohesion complex at anaphase to facilitate the separation of sister chromatids [41], may be the mechanistic nature of the duplication competency. Separase is known to be activated by APC [11,12], and thus its activation is likely restricted to mitotic phase. Considering that the duplicated centrosomes in cells arrested in S-phase by exposure to DNA synthesis inhibitors can still acquire the duplication competency and re-duplicate without undergoing mitosis [38], there seems to be more to the story for the molecular mechanism underlying the duplication competency, and further studies are clearly needed for the mechanism of the centrosome duplication competency in vivo. Nevertheless, at least partly because of the requirement of the acquisition of the duplication competency for duplicated centrosomes to re-duplicate, cyclin E overexpression (constitutive activation of CDK2-cyclin E) alone does not efficiently induce centrosome amplification [37, 42]. However, cyclin E overexpression, which is frequently observed in cancer cells [43], will certainly lay a favorable ground for generation of amplified centrosomes.
Loss of p53 and centrosome amplification
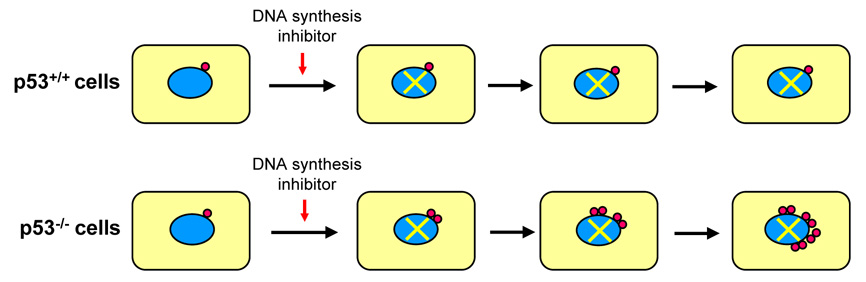
Since CDK2-cyclin E plays a key role in the initiation of centrosome duplication, the proteins that control the CDK2/cyclin E activity are also expected to be involved in the regulation of centrosome duplication, including p53. The involvement of p53 in the regulation of centrosome duplication was initially suggested by the observations that cells and tissues from p53-deficient mice show a high frequency of centrosome amplification [7, 44]. The subsequent studies further supported the role of p53 in the regulation of centrosome duplication. As described briefly above, the cells arrested by exposure to the ribonucleotide reductase inhibitors [i.e., hydroxyurea (HU), mimosine] as well as a DNA polymerase inhibitor [i.e., aphidicolin (Aph)] undergo multiple rounds of centrosome re-duplication without DNA synthesis, resulting in generation of amplified centrosomes. However, efficient centrosome re-duplication in the cell cycle-arrested cells occurs when p53 is either mutated or lost [45] (Fig. 6). In normal cells, p53 is stabilized by the physiological stress associated with the prolonged exposure to the inhibitors such as Aph or HU by the ARF-mediated inhibition of Mdm2 [46], leading to up-regulation of p21Waf1/Cip1, a potent CDK inhibitor and major transactivation target of p53 [47]. This results in continuous inhibition of CDK2, hence blocking initiation of centrosome re-duplication. In contrast, in cells lacking p53, CDK2 activation is unchecked, leading to centrosome re-duplication/centrosome amplification. Even under a normal condition/environment, cells are subjected to internal as well as external stresses that temporarily force cells to halt cell cycling irrespective of the p53 status (i.e., imbalance or deprivation of critical molecules such as dNTPs similar to the situation experimentally induced by HU or mimosine treatment). In such cases, centrosomes re-duplicate if cells lack functional p53, leading to centrosome amplification. Once the stress causing problems are eliminated, cells resume cell cycling, and these cells will likely suffer chromosome loss/gain during mitosis due to the presence of amplified centrosomes [48].
Figure 6. Centrosome re-duplication in cells arrested by exposure to DNA synthesis inhibitors in the absence of p53.
When cells are exposed to DNA synthesis inhibitors such as hydroxyurea, aphidicolin and mimosine, cells become arrested at the G1/S transition and in S-phase in a p53-independent manner. In the presence of the functionally intact p53, p53 is stabilized/activated in response to the stress associated with prolonged arrest by the ARF-dependent mechanism, leading to up-regulation of p21, which in turn continuously inhibits CDK2, hence blocking centrosome duplication (a). In contrast, if p53 is either lost or mutated, CDK2 activity is unchecked and activated, which triggers centrosome re-duplication in the absence of DNA synthesis, resulting in generation of amplified centrosomes (b).
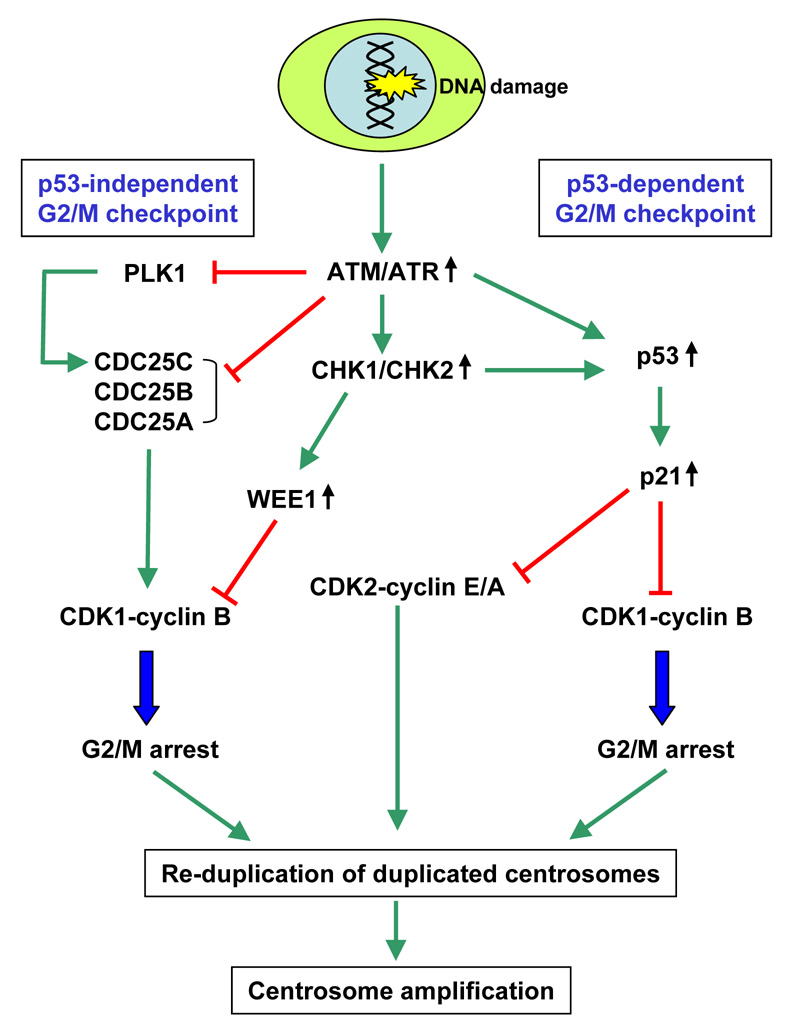
The molecular mechanism of centrosome amplification associated with loss of p53 has recently become clearer primarily from the studies of centrosome amplification associated with DNA damage as well as mutations/losses of DNA damage repair proteins. BRCA1 and BRCA2, both of which are products of familial breast/ovarian cancer susceptibility genes [49] and the Rad51 family proteins, including Rad51 and five Rad51 paralogs, Rad51B, Rad51C, Rad51D, XRCC2 and XRCC3 [50] play major roles in DNA damage repair in vertebrate cells. Expression of the dominant negative Rad51 as well as conditional repression of Rad51 leads to centrosome amplification [51, 52]. Similarly, reduced expression or loss of Rad51B, Rad51C, Rad51D, XRCC2, XRCC3 as well as BRCA1 and BRCA2 all result in high frequencies of centrosome amplification and severe chromosome instability [53–58]. It has recently been shown that, using the inducible Rad51 system in Rad51-deficient cells, centrosome amplification occurs during prolonged G2-arrest due to activation of the G2/M checkpoint triggered by the presence of damaged DNA [59]. Thus, DNA damages, which are normally corrected by the repair system, will accumulate if the repair system is defective, triggering the G2/M checkpoint and cell cycle arrest. The G2/M checkpoint is the surveillance mechanism for ensuring the readiness of cells to commit to mitosis, especially of the integrity of replicated DNA [60], and targets CDK1-cyclin B, a key kinase complex that triggers mitotic entry [61], is a primary target of the G2/M checkpoint. The G2/M checkpoint is exerted via both p53-dependent and p53-independent mechanisms, both of which are triggered by ATM and ATR kinases that act as a sensor of damaged DNA and coordinates the DNA damage response signaling pathways, including Chk1 and Chk2 kinases (Fig. 7). p53 is stabilized by ATM/ATR- as well as Chk1/Chk2-mediated phosphorylation, leading to up-regulation of p21 CDK inhibitor, which prevents activation of CDK1/cyclin B by direct inhibition of the kinase activity as well as nuclear exclusion [62, 63], resulting in late G2 arrest (p53-dependent G2/M checkpoint). CHK1/CHK2 also blocks CDK1-cyclin B activation by directly phosphorylating CDC25 phosphatases (CDC25A, B and C), all of which can act as activators of CDK1/cyclin B [64]. CHK1 has also been shown to block activation of CDC25C by inhibiting Polo-like kinase 1 (PLK1) [65], one of the known activators of CDC25C [66]. CHK1 also up-regulates WEE1, a kinase that catalyzes inhibitory phosphorylation of CDK1 [67]. All these events lead to inhibition of the activation of CDK1-cyclin B, resulting in late G2 arrest (p53-independent G2/M checkpoint). The G2 arrest by the accumulation of damaged DNA caused by mutations/losses of DNA repair proteins provides an opportunity for duplicated centrosomes to acquire the duplication competency and to re-duplicate, leading to generation of amplified centrosomes. Similar scenario can be drawn for cells exposed to high doses of genotoxic agents. Cells inflicted with extensive DNA damage by exposure to γ-irradiation, UV-irradiation or genotoxic drugs become arrested due to activation of the G2/M checkpoint, and undergo centrosome amplification [68–71]. However, initiation of centrosome re-duplication in these G2-arrested cells still requires the CDK2 activity (mostly of those bound by cyclin A, ref. 35), which can be effectively inhibited by activation of the p53-dependent G2/M checkpoint pathway and consequential up-regulation of p21. Thus, centrosome amplification in the G2-arrested cells in response to DNA damage occurs efficiently when the p53-dependent G2/M checkpoint is compromised (Fig. 7). Indeed, it has been shown that centrosome amplification does not occur in the irradiated normal human cells despite of being arrested in G2. Only when p53 expression is silenced in those cells, radiation exposure induced a high frequency of centrosome amplification [71].
Figure 7. Centrosome amplification in cells arrested by the G2/M checkpoint in response to DNA damage occurs efficiently in the absence of p53.
The G2/M checkpoints in response to DNA damage are exerted through the p53-dependent and p53-independent pathways, both of which target CDK1-cyclin B. Both p53-dependent and - independent G2/M checkpoint responses are triggered by activation of ATM/ATR and CHK1/CHK2 kinases. In the p53-independent pathway, CHK1 and CHK2 phosphorylate and inhibit CDC25A, B and C. CHK1 also inhibits the activity of PLK1, which is known to activate CDC25C. CHK1 also appears to suppress CDC25B by blocking the access of CDC25B to centrosomally localized CDK1-cyclin B (initial activation of CDK1-cyclin B is thought to occur at centrosomes) [85, 86]. CHK1 also phosphorylates and up-regulates the activity of WEE1 kinase that catalyzes the inhibitory phosphorylation of CDK1. All these events block the activation of CDK1-cyclin B in a concerted fashion, resulting in late G2 arrest. In the p53-dependent pathway, ATM/ATR and CHK1/CHK2 phosphorylate and stabilize p53, which in turn up-regulates p21. p21 then suppresses the activity of CDK1-cyclin B, leading to late G2 arrest. This G2 arrest provides time for centrosomes to regain duplication competency and re-duplicate. However, CDK2 activity is still required for centrosomes to re-duplicate. In late G2 phase, high levels of already active form of CDK2 (mostly complexed with cyclin A) are present. In cells arrested by the G2/M checkpoint responses, those CDK2 kinases remain active even efficient inhibition of CDC25s. However, if the intact p53-dependent pathway is present, centrosome re-duplication is blocked by the p21-mediated suppression of the CDK2 activity. In contrast, in the absence of the p53-dependent pathway, centrosomes re-duplication is continuously triggered by active CDK2-cyclin E/A.
In sum, loss or mutational inactivation of p53 allows uncontrolled activation of CDK2 in cells arrested for cell cycling due to physiological stress as well as damaged DNA, and active CDK2, mostly those in complex with cyclin A, triggers centrosome re-duplication, leading to generation of amplified centrosomes. Once the cause of the cell cycle arrest is either removed or repaired, cells will resume cell cycling in the presence of amplified centrosomes, leading to mitotic defects and chromosome segregation errors.
Other CDK2 and p53 modulators and centrosome amplification
Because of the involvement of p53 in the regulation of centrosome duplication, the proteins that control the stability of p53 is also involved in the regulation of centrosome duplication, and aberrant expression/activity of those proteins results in centrosome amplification. For instance, MDM2 (HDM2 in human), an E3 ubiquitin ligase that promotes degradation of p53 [72,73], is frequently overexpressed in various types of cancers, especially in those which retain wild-type p53 [74]. When MDM2 is overexpressed in cells harboring wild type p53, the level of p53 is reduced, and centrosome amplification is efficiently induced [75]. Similarly, expression of human papilloma virus E6 protein, which also promotes degradation of p53 [76], leads to centrosome amplification [77]. More recently, p53 has been shown to be primed for MDM2-mediated degradation by phosphorylation mediated by Aurora A (STK15/BTAK) kinase [78]. Indeed, forced expression of Aurora A has been shown to induce centrosome amplification [79], and priming of p53 for MDM2-mediated degradation at least partly contributes to the ability of Aurora A to induce centrosome amplification.
Besides the p53-p21 pathway, CDK2 activity is also controlled by other CDK inhibitors, including p27Kip1 and p16(INK4a) [80]. Both p27Kip1 and p16(INK4a) have been implicated in the regulation of centrosome duplication. For instance, centrosome amplification associated with DNA damage requires down-regulation of p27Kip1 in certain cell types [70]. Loss of p16(INK4a) has also been shown to induce centrosome amplification [81]. In both cases, uncontrolled activation of CDK2/cyclin E was detected, likely contributing to generation of amplified centrosomes.
CDK2 activity is also positively controlled, and CDC25 phosphatase is the major activator of CDK2. In mammals, there are three CDC25 isoforms (CDC25A, B and C). Although early studies suggested a cell cycle stage-specific role for each CDC25 isoform, it has recently been shown that all CDC25 isoforms participate in the control of both early and late cell cycle transitions targeting all types of CDK/cyclin complexes [64]. CDK/cyclin complexes first accumulate in an inactive form in which the CDK subunits are phosphorylated, and dephosphorylation by CDC25 leads to activation of CDKs. Among these isoforms, only CDC25B has been shown to localize to centrosomes [82]. However, the centrosomal localization may not be a critical event for CDC25 to regulate centrosome duplication. Although CDK2-cyclin E is known to localize to centrosomes [18, 83], it is possible that CDK2/cyclin E may be targeted by CDC25 elsewhere, and then recruited to the centrosome. Therefore, the role of CDC25A and C in the regulation of centrosome duplication cannot be ruled out simply because they do not localize to centrosomes. It remains to be shown which CDC25 isoform(s) are more critically involved in the regulation of centrosome duplication, and whether aberrant CDC25 expression/activity contributes to centrosome amplification.
Conclusion
In this review, recent advance in the mechanisms of the numeral abnormality of centrosomes and its role in chromosome instability in cancer cells are discussed through focusing on p53 and CDK2-cyclins. However, many other proteins that are frequently mutated in cancers are found to be involved in the regulation of centrosome duplication, hence mutations of those proteins result in centrosome amplification [84]. Some proteins control centrosome duplication through modulating the stability/activity of p53 as well as CDK2-cyclins, while the others do so independently from p53 and CDK2-cyclins. It is also important to note that functional/structural defects of centrosomes are equally important for its role in destabilization of chromosomes in cancer cells. For instance, failure of the duplicated centrosomes to undergo a proper maturation process (i.e., failure to recruit key PCM components, etc.) results in the functional impairment of centrosomes, leading to formation of defective mitotic spindles and chromosome segregation errors. Many oncogenic and tumor suppressor proteins have been found to be involved in the regulation of the centrosome function as well, and mutations of those proteins results in the functional impairment of centrosomes. The key questions remained to be answered for the role of centrosomes in carcinogensis is how those oncogenes and tumor suppressors interplay to control the numeral and functional integrity of the centrosome, and how concurrent mutations of those proteins exacerbate the numeral and functional abnormalities of the centrosome. Together with further understanding of the molecular biology of centrosomes, these studies may prove the centrosome a superb target of the cancer management.
Acknowledgements
Preparation of this article is in part supported by the grants from National Institute of Health (Bethesda, MD, USA) and Moffitt Cancer Center Institutional fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 2.Sattler CA, Sawada N, Sattler GL, Pitot HC. Electron microscopic and time lapse studies of mitosis in cultured rat hepatocytes. Hepatology. 1988;8:1540–1549. doi: 10.1002/hep.1840080612. [DOI] [PubMed] [Google Scholar]
- 3.Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a "tetraploidy checkpoint". J. Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 5.Levine DS, Sanchez CA, Rabinovitch PS, Reid BJ. Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer. Proc. Natl. Acad. Sci. USA. 1991;88:6427–6431. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 7.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 8.D'Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 9.Heim S, Mandahl N, Mitelman F. Genetic convergence and divergence in tumor progression. Cancer Res. 1988;48:5911–5916. [PubMed] [Google Scholar]
- 10.Chiba S, Okuda M, Mussman JG, Fukasawa K. Genomic convergence and suppression of centrosome hyperamplification in primary p53−/− cells in prolonged culture. Exp. Cell Res. 2000;258:310–321. doi: 10.1006/excr.2000.4916. [DOI] [PubMed] [Google Scholar]
- 11.Oikawa T, Okuda M, Ma Z, Goorha R, Tsujimoto H, Inokuma H, Fukasawa K. Transcriptional control of BubR1 by p53 and suppression of centrosome amplification by BubR1. Mol. Cell. Biol. 2005;25:4046–4061. doi: 10.1128/MCB.25.10.4046-4061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musacchio A, Hardwick KG. The spindle checkpoint: Structural insights into dynamic signaling. Nat. Rev. Mol. Cell. Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- 13.Chan GK, Yen TJ. The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit. Prog. Cell Cycle Res. 2003;5:431–439. [PubMed] [Google Scholar]
- 14.Mazia D. The chromosome cycle and the centrosome cycle in the mitotic cycle. Int. Rev. Cytology. 1987;100:49–92. doi: 10.1016/s0074-7696(08)61698-8. [DOI] [PubMed] [Google Scholar]
- 15.Hinchcliffe EH, Sluder G. Two for two: Cdk2 and its role in centrosome doubling. Oncogene. 2002;21:6154–6160. doi: 10.1038/sj.onc.1205826. [DOI] [PubMed] [Google Scholar]
- 16.Pines J. The cell cycle kinases. Semin. Cancer Biol. 1994;5:305–313. [PubMed] [Google Scholar]
- 17.Heichman KA, Roberts JM. Rules to replicate by. Cell. 1994;79:557–562. doi: 10.1016/0092-8674(94)90541-x. [DOI] [PubMed] [Google Scholar]
- 18.Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- 19.Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dulic V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 21.Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 22.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2-cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 23.Fisk HA, Winey A. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 25.Ortega S, Prieto I, Odajima J, Martín A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 26.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr. Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 27.Duensing A, Liu Y, Tseng M, Malumbres M, Barbacid M, Duensing S. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene. 2006;25:2943–2949. doi: 10.1038/sj.onc.1209310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ussar S, Voss T. MEK1 and MEK2, different regulators of the G1/S transition. J. Biol. Chem. 2004;279:43861–43869. doi: 10.1074/jbc.M406240200. [DOI] [PubMed] [Google Scholar]
- 29.Nelsen CJ, Kuriyama R, Hirsch B, Negron VC, Lingle WL, Goggin MM, Stanley MW, Albrecht JH. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J. Biol. Chem. 2005;280:768–776. doi: 10.1074/jbc.M407105200. [DOI] [PubMed] [Google Scholar]
- 30.Berthet C, Klarmann KD, Hilton MB, Suh HC, Keller JR, Kiyokawa H, Kaldis P. Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Dev. Cell. 2006;10:563–573. doi: 10.1016/j.devcel.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 2005;7:831–836. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 32.Warnke S, Kemmler S, Hames RS, Tsai HL, Hoffmann-Rohrer U, Fry AM, Hoffmann I. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr. Biol. 2004;14:1200–1207. doi: 10.1016/j.cub.2004.06.059. [DOI] [PubMed] [Google Scholar]
- 33.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 34.Ma Z, Kanai M, Kawamura K, Kaibuchi K, Ye K, Fukasawa K. Interaction between ROCK II and nucleophosmin/B23 in the regulation of centrosome duplication. Mol. Cell. Biol. 2006;26:9016–9034. doi: 10.1128/MCB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanashiro K, Kanai M, Geng Y, Sisinski P, Fukasawa K. Roles of cyclin A and E in induction of centrosome amplification in p53-compromised cells. Oncogene. 2008 doi: 10.1038/onc.2008.161. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- 37.Mussman JG, Horn HF, Carroll PE, Okuda M, Donehower LA, Fukasawa K. Synergistic induction of centrosome hyperamplification by loss of p53 and cyclin E overexpression. Oncogene. 2000;19:1635–1946. doi: 10.1038/sj.onc.1203460. [DOI] [PubMed] [Google Scholar]
- 38.Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat. Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- 40.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 41.Natsmyth K, Peters JM, Uhlmann F. Splitting the chromosome:cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 42.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 43.Keyomarsi K, Herliczek TW. The role of cyclin E in cell proliferation, development and cancer. Prog. Cell Cycle Res. 1997;3:171–191. doi: 10.1007/978-1-4615-5371-7_14. [DOI] [PubMed] [Google Scholar]
- 44.Fukasawa K, Wiener F, Vande Woude GF, Mai S. Genomic instability and apoptosis are frequent in p53 deficient mice. Oncogene. 1997;15:1295–1302. doi: 10.1038/sj.onc.1201482. [DOI] [PubMed] [Google Scholar]
- 45.Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. Direct regulation of the centrosome duplication cycle by the p53-p21 (Waf1/Cip1) pathway. Oncogene. 2001;20:3173–3184. doi: 10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- 46.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 47.Harper JW. Cyclin dependent kinase inhibitors. Cancer Surv. 1997;29:91–107. [PubMed] [Google Scholar]
- 48.Kawamura K, Izumi H, Ma Z, Ikeda R, Moriyama M, Tanaka T, Nojima T, Levin LS, Fujikawa-Yamamoto K, Suzuki K, Fukasawa K. Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res. 2004;64:4800–4809. doi: 10.1158/0008-5472.CAN-03-3908. [DOI] [PubMed] [Google Scholar]
- 49.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 50.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 51.Bertrand P, Lambert S, Joubert C, Lopez BS. Overexpression of mammalian Rad51 does not stimulate tumorigenesis while a dominant-negative Rad51 affects centrosome fragmentation, ploidy and stimulates tumorigenesis, in p53-defective CHO cells. Oncogene. 2003;22:7587–7592. doi: 10.1038/sj.onc.1206998. [DOI] [PubMed] [Google Scholar]
- 52.Daboussi F, Thacker J, Lopez BS. Genetic interactions between RAD51 and its paralogues for centrosome fragmentation and ploidy control, independently of the sensitivity to genotoxic stresses. Oncogene. 2005;24:3691–3696. doi: 10.1038/sj.onc.1208438. [DOI] [PubMed] [Google Scholar]
- 53.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 54.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 55.Date O, Katsura M, Ishida M, Yoshihara T, Kinomura A, Sueda T, Miyagawa K. Haploinsufficiency of RAD51B causes centrosome fragmentation and aneuploidy in human cells. Cancer Res. 2006;66:6018–6024. doi: 10.1158/0008-5472.CAN-05-2803. [DOI] [PubMed] [Google Scholar]
- 56.Renglin Lindh A, Schultz N, Saleh-Gohari N, Helleday T. RAD51C (RAD51L2) is involved in maintaining centrosome number in mitosis. Cytogenet. Genome Res. 2007;116:38–45. doi: 10.1159/000097416. [DOI] [PubMed] [Google Scholar]
- 57.Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 58.Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, Ross G, Ashworth A. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 59.Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 61.Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 62.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 63.Charrier-Savournin FB, Chateau MT, Gire V, Sedivy J, Piette J, Dulic V. p21-Mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Mol. Biol. Cell. 2004;15:3965–3976. doi: 10.1091/mbc.E03-12-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boutros R, Lobjois V, Ducommun B. CDC25 phosphatases in cancer cells: key players? Good targets? Nat. Rev. Cancer. 2007;7:495–507. doi: 10.1038/nrc2169. [DOI] [PubMed] [Google Scholar]
- 65.Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc. Natl. Acad. Sci. USA. 2006;103:11964–11969. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roshak AK, Capper EA, Imburgia C, Fornwald J, Scott G, Marshall LA. The human polo-like kinase, PLK, regulates cdc2/cyclin B through phosphorylation and activation of the cdc25C phosphatase. Cell Signal. 2000;12:405–411. doi: 10.1016/s0898-6568(00)00080-2. [DOI] [PubMed] [Google Scholar]
- 67.Lee J, Kumagai A, Dunphy WG. Positive regulation of wee1 by chk1 and 14-3-3 proteins. Mol. Biol. Cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato N, Mizumoto K, Nakamura M, Tanaka M. Radiation-induced centrosome overduplication and multiple mitotic spindles in human tumor cells. Exp. Cell Res. 2000;255:321–326. doi: 10.1006/excr.1999.4797. [DOI] [PubMed] [Google Scholar]
- 69.D'Assoro AB, Busby R, Suino K, Delva E, Almodovar-Mercado GJ, Johnson H, Folk C, Farrugia DJ, Vasile V, Stivala F, Salisbury JL. Genotoxic stress leads to centrosome amplification in breast cancer cell lines that have an inactive G1/S cell cycle checkpoint. Oncogene. 2004;23:4068–4075. doi: 10.1038/sj.onc.1207568. [DOI] [PubMed] [Google Scholar]
- 70.Sugihara E, Kanai M, Saito S, Nitta T, Toyoshima H, Nakayama K, Nakayama KI, Fukasawa K, Schwab M, Saya H, Miwa M. Suppression of centrosome amplification after DNA damage depends on p27 accumulation. Cancer Res. 2006;66:4020–4029. doi: 10.1158/0008-5472.CAN-05-3250. [DOI] [PubMed] [Google Scholar]
- 71.Kawamura K, Morita N, Domiki C, Fujikawa-Yamamoto K, Hashimoto M, Iwabuchi K, Suzuki K. Induction of centrosome amplification in p53 siRNA-treated human fibroblast cells by radiation exposure. Cancer Sci. 2006;97:252–258. doi: 10.1111/j.1349-7006.2006.00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haupt Y, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 73.Kubbutat MH, Jones SN, Vousden K. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 74.Momand J, Zambetti G. Mdm-2: "big brother" of p53. J. Cell. Biochem. 1997;64:343–352. [PubMed] [Google Scholar]
- 75.Carroll PE, Okuda M, Horn HF, Biddinger P, Stambrook PJ, Gleich LL, Li YQ, Tarapore P, Fukasawa K. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene. 1999;18:1935–1944. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- 76.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 77.Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum CP, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA. 2000;97:10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat. Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 79.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 80.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 81.McDermott KM, Zhang J, Holst CR, Kozakiewicz BK, Singla V, Tlsty TD. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4:e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dutertre S, Cazales M, Quaranta M, Froment C, Trabut V, Dozier C, Mirey G, Bouché JP, Theis-Febvre N, Schmitt E, Monsarrat B, Prigent C, Ducommun B. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J. Cell Sci. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 83.Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- 84.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 85.Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 86.Krämer A, Mailand N, Lukas C, Syljuåsen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]