Abstract
P450 enzymes comprising the human CYP4F gene subfamily are catalysts of eicosanoid (e.g., 20-HETE and leukotriene B4) formation and degradation, although the role that individual CYP4F proteins play in these metabolic processes is not well defined. Thus, we developed antibodies to assess the tissue-specific expression and function of CYP4F2, one of four CYP4F P450s found in human liver and kidney. Peptide antibodies elicited in rabbits to CYP4F2 amino acid residues 61–74 (WGHQGMVNPTEEG) and 65–77 (GMVNPTEEGMRVL) recognized on immunoblots only CYP4F2 and not CYP4F3b, CYP4F11 or CYP4F12. Immunoquantitation with anti-CYP4F2 peptide IgG showed highly-variable CYP4F2 expression in liver (16.4 ± 18.6 pmol/mg microsomal protein; n = 29) and kidney cortex (3.9 ± 3.8 pmol/mg; n = 10), with two subjects lacking the hepatic or renal enzyme entirely. CYP4F2 content in liver microsomes was significantly correlated (r ≥ 0.63; p < 0.05) with leukotriene B4 and arachidonate ω-hydroxylase activities, which are both CYP4F2-catalyzed. Our study provides the first example of a peptide antibody that recognizes a single CYP4F P450 expressed in human liver and kidney, namely CYP4F2. Immunoquantitation and correlation analyses performed with this antibody suggest that CYP4F2 functions as a predominant LTB4 and arachidonate ω-hydroxylase in human liver.
Keywords: CYP450, CYP4F2, fatty acids, eicosanoids, ω-hydroxylation, peptide antibodies
INTRODUCTION
P450 enzymes comprise a superfamily of hemoproteins that oxidize both xenobiotics and endobiotics, including drugs, environmental contaminants, steroid hormones, and fatty acids. While most research on P450 enzymes has focused on their capacity to metabolize therapeutic agents [1, 2], more attention is being paid to the endobiotic-metabolizing properties of these enzymes, of which many belong to the CYP4A and CYP4F gene subfamilies [3, 4]. Indeed, the human CYP4F/A enzymes have been shown to: a) function as catalysts of medium-, long-, and very long-chain fatty acid ω-hydroxylation [5–7], which facilitates the lowering of excessive free fatty acid levels; b) convert one particular fatty acid, AA, into the potent eicosanoid 20-HETE [8–10], a powerful vasoconstrictor, inhibitor of ion transport and cellular proliferation agent; c) catabolize various AA oxygenated derivatives, including leukotrienes (e.g., LTB4), prostaglandins, and lipoxins, thereby inactivating these bioactive lipid mediators [11–17]. CYP4F-mediated metabolism of another endogenous substrate, namely vitamin E, represents the initial step in this compound’s biotransformation [18, 19]. In contrast to other P450s, the CYP4F/A proteins possess the unique ability to oxidize, or ω-hydroxylate, the thermodynamically-unfavorable terminal methyl group present on saturated and unsaturated fatty acids of different chain lengths, AA-derived eicosanoids, and tocopherol phytyl side chains.
Considering their catalytic properties, it is not surprising that the CYP4F/A enzymes are thought to play an important role in the regulation of certain physiological and/or pathological phenomena. Indeed, potential relationships have been described between CYP4A and CYP4F protein levels and/or associated metabolic activities and the amplitude of LTB4-dependent inflammatory processes [20 and references therein], blood pressure regulation [21], and cellular proliferation [22]. However, the assignment of a specific CYP4F gene subfamily P450 to a particular endobiotic-metabolizing activity occuring in intact tissue has proven problematic. While CYP4A11 constitutes the single, major CYP4A P450 expressed in human liver and kidney, at least 4 distinct CYP4F P450s are abundantly expressed in these same tissues. In fact, the human CYP4F gene subfamily is comprised of 6 different members, including CYP4F2, CYP4F3a, CYP4F3b, CYP4F8, CYP4F11 and CYP4F12, which possess 74% overall amino acid sequence homology. CYP4F2, CYP4F3b, CYP4F11 and CYP4F12 are found in liver and kidney [10, 16, 23–25] whereas CYP4F3a is found only in myeloid tissue [26, 27] and CYP4F8 is expressed exclusively in seminal vesicles [28]. CYP4F3a and CYP4F3b have identical amino acid sequences except at residues 67–114, where alternative exon splicing has led to the integration of exon 3 into CYP4F3b and exon 4 into CYP4F3a. Such alternative exon splicing gives two P450 enzymes with markedly different substrate specificity and tissue distribution [10, 16].
Several of the above findings, including CYP4F2 involvement in hepatic and renal 20-HETE formation and LTB4 catabolism [8, 9, 15], were arrived at using CYP4F2-reactive polyclonal antibodies as metabolic probes2. The extensive sequence homology found among the CYP4F P450s predicts, however, that our original polyclonal CYP4F2 antibodies reacted not only with their immunogen but also with the other CYP4F subfamily members now known to be expressed in human liver and kidney. A similar phenomenon, namely antibody cross-reactivity among the members of a P450 gene subfamily, was observed with antibodies elicited against human liver CYP2C9 [29]. As such, it is possible that CYP4F enzymes other than CYP4F2, such as CYP4F3b, CYP4F11 or CYP4F12, also function as catalysts of fatty acid and LTB4 ω-hydroxylation. Indeed, CYP4F3b heterologous expression systems (e.g., Sf9 insect cells infected with a CYP4F3b-cDNA containing baculovirus) ω-hydroxylate AA as well as LTB4 at extensive rates [10], although it is not known whether CYP4F3b promotes the same functions in native hepatic or renal tissues. To further address this issue, we utilized another immunochemical approach, namely that using peptide antibodies specifically targeted against an individual CYP4F enzyme. This methodology has been used successfully by other investigators to develop targeted antibodies against either CYP450 COOH-terminal peptides [30, 31] or internal peptides [32–35]. Herein, we describe the development of antibodies against two overlapping peptides comprising CYP4F2 amino acid residues 61–73 and 66–77. The resultant CYP4F2 peptide IgG preparation, which reacted with only CYP4F2 and no other CYP4F P450 on Western blots, was used to demonstrate that CYP4F2 is expressed at highly variable levels in human liver and kidney. The targeted antibody was also utilized to define a role for CYP4F2 in catabolism of LTB4 and the conversion of AA to 20-HETE, a powerful vasoactive eicosanoid.
MATERIALS AND METHODS
Human Liver and Kidney Microsomes
Human liver tissue was obtained from organ donors through the Liver Tissue Procurement and Distribution System (Minneapolis, Minnesota), which was funded by NIH Contract #N01-DK-9-2310. The donors included 29 male and female subjects ranging in age from 3 – 63 years old. With the exception of one donor with primary sclerotic cholangitis (subject N), these subjects were without obvious liver pathology. Livers were removed within 30 min of death, frozen in liquid nitrogen, and stored at −80°C until microsomes were prepared [5, 36]. Microsomes from human renal cortical samples were obtained from sources described elsewhere [9]. The donors entailed 10 subjects, of whom 5 were males and 5 were females, with ages ranging from 15 – 69 years old. P450 content was determined from CO-reduced difference spectra [37] while protein concentration was measured using the BCA procedure [38].
Microsomal Enzymes
Recombinant human CYP4F2, CYP4F3b and CYP4F12 were derived from hemin-fortified suspension cultures of Spodoptera frugiperda (Sf9) insect cells that had been infected with the corresponding CYP4F/A cDNA-containing baculovirus constructs. After a 72 h infection period, Sf9 cells were harvested, lysed by sonication, and the recombinant CYP4F/A P450s purified to near homogeneity from the lysates using a combination of hydrophobic, adsorption and/or metal ion affinity chromatography. Additional details regarding human CYP4F P450 cDNA cloning, baculovirus construct preparation, and expressed enzyme purification will be published elsewhere. P450 reductase and b5 were purified to near electrophoretic homogeneity from human liver microsomes as previously reported [39, 40]. The specific contents of b5 and P450 reductase were 25.7 nmol/mg protein and 33.5 nmol/mg protein (113,000 units/mg); one nmol P450 reductase activity was defined as that amount catalyzing reduction of 3,400 µmol ferricytochrome c/min at 22°C in 300 mM potassium phosph ate buffer (pH 7.7). CYP4F2, CYP4F3b and CYP4F12 supersomes®, which are microsomes derived from Trichoplusia ni insect cells co-infected with the corresponding human P450 cDNA, P450 reductase cDNA and b5 cDNA baculovirus constructs, were obtained from BD Biosciences (Bedford, MA).
Oleate, AA, and LTB4 ω-Hydroxylation Assays
Oleate oxidation to its ω- and ω-1 hydroxylated metabolites was determined under conditions where product formation was linear with respect to time and to P450 concentration as described previously [41, 42]. Reaction mixtures (0.25 ml) contained: a) purified reconstituted systems consisting of 25–50 pmol CYP4F enzyme, 75–150 pmol P450 reductase, 7.5–15 µg DLPC and 100–200 pmol b5; b) supersomes® derived from insect cells expressing human CYP4F2, CYP4F3b or CYP4F11 (protein equivalent to 25 pmol P450) or; c) human liver microsomes (protein equivalent to 50–150 pmol P450). Other incubation components included 100 mM KPO4 buffer (pH 7.4),100 µM oleate, and 1 mM NADPH. For antibody inhibition studies, the reconstituted recombinant P450s or liver microsomes were first incubated with either immune-specific or preimmune IgG for 3 min at 37°C and then for 10 min at room temperature, followed by addition of the remaining reaction components. Reactions were initiated with the cofactor, and were terminated after 10 min at 37°C by placing the tubes on ice. Incubation mixtures were extracted with 4 volumes of ethyl acetate, the organics extracts were evaporated using nitrogen gas, and the residues resolubilized in 80 µl of methanol to which 20 µl of a freshly-prepared solution of PDAM (1 mg/ml in ethyl acetate) was added [43]. Oleate-PDAM derivatives were allowed to form overnight at room temperature in the dark, and the samples (30 µl) were then injected onto a Waters Alliance HPLC comprised of a 2690 separation module and 2487 UV/visible detector. Oleate and its hydroxylated metabolites (mainly 18-OH oleate; see Results) were resolved on a Waters Novapak C18 column (3.9 × 150 mm) using a concave gradient (curve 10 at 0.3% per min) of 90% to 99% acetonitrile in water at a flow rate of 1.0 ml/min. At 27 min post-injection, the column was washed isocratically with acetonitrile:methanol (50:50, v/v) at a flow rate of 1.0 ml/min for 5 min to remove residual PDAM-derivatized material. Column eluants were continuously monitored for UV absorbance at 275 nm. These HPLC conditions gave retention times for 17-OH oleate, 18-OH oleate-PDAM and oleate-PDAM of 13.5, 14.5 min and 28 min, respectively. Rates of 18-OH oleate formation were determined from standard curves prepared by adding varying amounts of ω-hydroxylated fatty acid metabolite3 to incubations conducted without NADPH.
AA conversion to 20-HETE was performed as described previously [8, 9] but with modifications. Reaction mixtures were the same as those given above except that incubations contained 100 µM AA instead of oleate, and the reactions were terminated after 15 min at 37°C by addition of 10 µl of 2N HCl, followed by placing the tubes on ice. The samples were extracted with 1.0 ml of ethyl acetate, the organic extracts were placed into Total Recovery® autosampler vials (Waters Corporation, Milford, MA), and were evaporated to dryness with nitrogen gas. Residues were resolubilized by adding 25 µl of acetonitrile to the vials, of which 15 µl was subjected to HPLC analysis. AA was resolved from its hydroxylated metabolites using a Waters YMC-Pro C18 column (2.0 × 250 mm; 5 µm particle size) with a linear gradient (0.3% per min) of 45% to 55% acetonitrile in 0.1% acetic acid at a flow rate of 0.3 ml/min; at 30 min postinjection, the gradient was increased from 55% acetonitrile to 100% acetonitrile at 6.4%/min. Column eluants were continuously monitored for UV absorbance at 200 nm. Under these conditions, 20-HETE had a retention time of 27.5 min while the parent compound AA had a retention time of 39.5 min. Rates of 20-HETE formation were determined from standard curves prepared by adding varying amounts of the authentic metabolite to incubations conducted without NADPH. LTB4 20-hydroxylation by purified CYP4F enzymes, CYP4F supersomes and human liver microsomes was assessed as described by Jin et al. [15].
Production of CYP4F2 Peptide Antibodies
Two overlapping CYP4F2 peptides, designated peptide A (61-WGHQGMVNPTEEG-74) and peptide B (65-GMVNPTEEGMRVL-77), were synthesized by 21st Century Biochemicals (Marlboro, MA) using a MultiPep synthesizer (Intavis LLC, San Jose, CA) employing Fmoc solid-phase chemistry. The sequences of the peptides and their purity (> 80%) were confirmed using MALDI/CID tandem mass spectrometry. An NH2-terminal aminohexanoic acid moeity and a COOH-terminal cysteine residue were added to peptides A and B in order to permit their conjugation to two different carrier proteins, namely HCN and ovalbumin, using m-maleimido-benzoyl-N-hydroxysuccinimide ester as the coupling agent (Fig. 1A). We also prepared two other CYP4F2 peptides that were designated as peptide C (89-KVWMGPISPLLSL-101) and peptide D (89-KVWMGPISPLLSLC-102). A cysteine residue was added to the NH2-terminal of peptide C to facilitate its conjugation to HCN while the tripeptide lysine-glycine-cysteine was added to peptide D at the amino terminus (Fig. 1B) to enable internal cyclization via cysteine sulfhydryl oxidation. The CYP4F2 peptide conjugates were then used to immunize New Zealand male rabbits, with high-titer antisera (as assessed by ELISA) harvested 10 days after the 3rd through 6th booster injections. IgG fractions were derived from the antisera using caprylic acid/ammonium sulfate precipitation [44]. The preparation and properties of polyspecific human CYP4F+ IgG2 have been described elsewhere [8, 9, 15]. Preimmune (control) IgG was prepared from rabbit sera obtained prior to immunization.
Figure 1. Sequence homology among human CYP4F gene subfamily P450s and the CYP4F2 peptides used for antibody production.
Panel A. Peptides employed to develop CYP4F2 antibodies. Residues are numbered relative to the CYP4F2 NH2-terminal amino acid (see Panel B). The cysteine (C) residues added to peptide A facilitates carrier protein coupling while the KGC residues added to peptide B enables oxidative cyclization of the cysteine SH groups. Ahx denotes 6-aminohexanoic acid, which was added to the NH2-terminal residues of peptides C and D also to facilitate carrier protein coupling while the C residue added to the COOH-terminal end of these peptides served the same function. Panel B. An alignment of the CYP4F2, CYP4F3b, CYP4F11, and CYP4F12 cDNA-deduced amino acid sequences is shown from residues 1 through 520–524. The region demarcated by the solid arrows contains amino acids 67–114, which are encoded by exon 3 in all four CYP4F sequences [Christmas, 1999 #2170]. The CYP4F P450s depicted share 75% overall sequence identity although residues 67–114 share only 46% sequence similarity.
Other Immunochemical Methods
Protein blotting of microsomal proteins and purified enzymes to nitrocellulose and subsequent immunochemical staining with anti-human CYP4F or anti-human CYP4F2 peptide IgG were conducted as described previously [8, 9, 15]. To increase assay sensitivity, Western blots performed with CYP4F2 peptide antibodies were incubated with alkaline phosphatase-conjugated strepavidin (rather than streptavidin-biotinylated horseradish peroxidase), followed by staining with a mixture of NBT/BCIP as chromogens. Total CYP4F and CYP4F2 content were measured by scanning immunoreactive bands with an Epson Expression 1600 flat-bed scanner interfaced to a computer, and then quantitating the staining intensities with UnScanIt V6.1 software (Silk Scientific Inc., Orem, UT). Staining intensity was expressed as absorbance units (AU)/mg protein applied to the gel, and was ranked relative to the total CYP4F and CYP4F2 content in liver microsomes from human subject UC9410.
Reagents
AA, 20-HETE, and oleic acid were purchased from Cayman Chemical Co. (Ann Arbor, MI), PDAM was from Invitrogen (Carlsbad, CA) and synthetic DLPC was obtained from Avanti Polar Lipids, Inc. (Alabaster, AL). LTB4 and 20-OH LTB4 were supplied by Dr. Joshua Rokash, Florida Institute Technology (Melbourne, FL). Streptavidin-alkaline phosphatase conjugate and NBT/BCIP were purchased from Roche Applied Science (Indianapolis, IN). HPLC-grade solvents were purchased from Burdick and Jackson (Muskegon, MI). All other chemicals used were of the highest grade commercially available.
RESULTS
CYP4F2 Peptide Selection and Antibody Production
CYP4F2 peptides A, B, C and D (Fig. 1A) were synthesized using conventional methods, coupled to KLH and/or ovalbumin, and these hapten-carrier protein conjugates then used to elicit in rabbits antibodies with the desired specificity towards human CYP4F2. Our rationale underlying this particular choice of internally-located CYP4F2 peptides was as follows. An alignment of the CYP4F2, CYP4F3b, CYP4F11, and CYP4F12 cDNA-deduced amino acid sequences, which range in length from 520–524 residues, gave 74.8% overall sequence homology (Fig. 1B) whereas the alignment of any two of these CYP4F P450 protein sequences gave 81–93% similarity. The 15 amino acids comprising the COOH-termini of the four CYP4F P450s displayed 72% identity overall, with 10 of these residues exhibiting 100% homology, thereby ruling out their use for peptide antibody derivation [31]. For sake of comparison, CYP4F2 shares only 43% overall sequence identity with CYP4A11, a human CYP4A gene subfamily member, with only 33% homology found at the carboxyl termini.
A closer examination of CYP4F primary structure revealed multiple amino acid differences (only 46% similarity) between CYP4F2 and the other CYP4F P450s amongst residues 67–114 (Fig. 1B), which comprise a region of the protein encoded by exon 3 of the corresponding structural gene [10, 16]. The antigenicity and hydrophobicity of peptides derived from this region were initially screened using standard Kyle-Doolittle and Hopp-Woods scales, which led to the design and synthesis of peptides A and B encompassing CYP4F2 residues 89–102 and 89–101, respectively (Fig. 1A); peptide B was cyclized by oxidation of its two cysteine SH groups [32, 45]. These sequences were chosen since they contained amino acids residues unique to CYP4F2 that were also contiguous (Fig. 1B). However, the antibodies produced against HCN-conjugated CYP4F2 peptides A and B were of limited utility since they failed to bind to CYP4F2 on Western blots (and only weakly recognized the corresponding peptide immunogens in ELISA)(data not shown).
The CYP4F2-CYP2C5 sequence alignment led to the selection of alternative peptides (C and D) comprised of amino acids located closer to the NH2-terminal end of CYP4F2 (Fig. 1A). Residues 61–74 and 65–77 likely correspond to rabbit CYP2C5 residues 43–65 and 50–62 encompassing regions including a loop leading into helix A as well as helix A itself, which are predicted from homology modeling to be surface-exposed [46, 47]. We attempted to foster a stronger immune response by coupling these two overlapping CYP4F2 peptides to dissimilar carrier proteins, namely HCN and ovalbumin. Immunization of rabbits with conjugated CYP4F2 peptides C and D gave rise to targeted antibodies that reacted strongly with their corresponding uncoupled immunogens in ELISA; antibody titers after 4 booster injections ranged between 1:50,000 and 1:100,000 on plates coated with either of the peptides (data not shown).
Characterization of CYP4F2 Peptide Antibodies
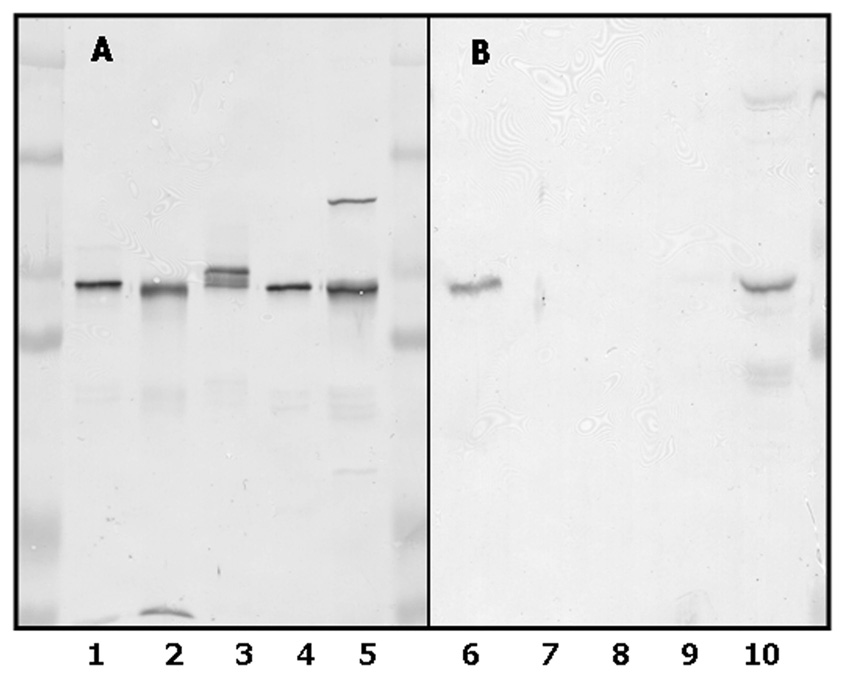
As already mentioned, there are at least 4 distinct human CYP4F gene subfamily members expressed in liver and kidney, namely CYP4F2, CYP4F3b, CYP4F11 and CYP4F12 [10, 16, 23–25]. The extensive sequence homology (75%) among these CYP4F P450s predicts that our original polyclonal CYP4F+ antibody (first termed anti-CYP4F2 IgG)2, which was developed in rabbits against a CYP4F2 enzyme preparation purified from human liver [8, 15], cross-reacted with most, if not all, of these hemeproteins. This extensive cross-reactivity is depicted in Fig. 2A. In contrast, antibodies developed against a mixture of conjugated CYP4F2 peptides C and D were found to react exclusively with CYP4F2 protein in purified form (Fig 2B, lane 6) as well as in native liver microsomes (Fig. 2B, lane 10). Cross-reactivity between the CYP4F2 peptide antibodies and other CYP4F subfamily members was no longer observed, regardless of the amount of these other proteins subjected to immunoblotting. Although not shown here, anti-CYP4F2 peptide IgG also failed to recognize CYP4F3a, the alternatively-spliced CYP4F3 enzyme [16].
Figure 2. Immunochemical properties of CYP4F2 peptide IgG.
Recombinant CYP4F enzymes and human liver microsomes were subjected to SDS-PAGE, electrophoretically transferred to a nitrocellulose membrane, and the membrane immunochemically stained with either anti-CYP4F+ IgG (Panel A) or anti- CYP4F2 peptide IgG (Panel B) as described in Materials and Methods. Lanes 1 and 6, purified CYP4F2 (0.1 µg); lane 2 and 7, purified CYP4F3b (0.1 µg); lanes 3 and 8, CYP4F11-containing Sf9 insect cell lysates (5 µg); lanes 4 and 9, CYP4F12-containing Sf9 insect cell lysates (1 µg); lanes 5 and 10, liver microsomes from subject G (15 µg). The cluster of immunoreactive bands observed in lane 3 stem from the proteolysis of CYP4F11 that occured during preparation of lysates from Sf9 cells expressing this P450.
Expression of CYP4F2 in Human Liver and Kidney
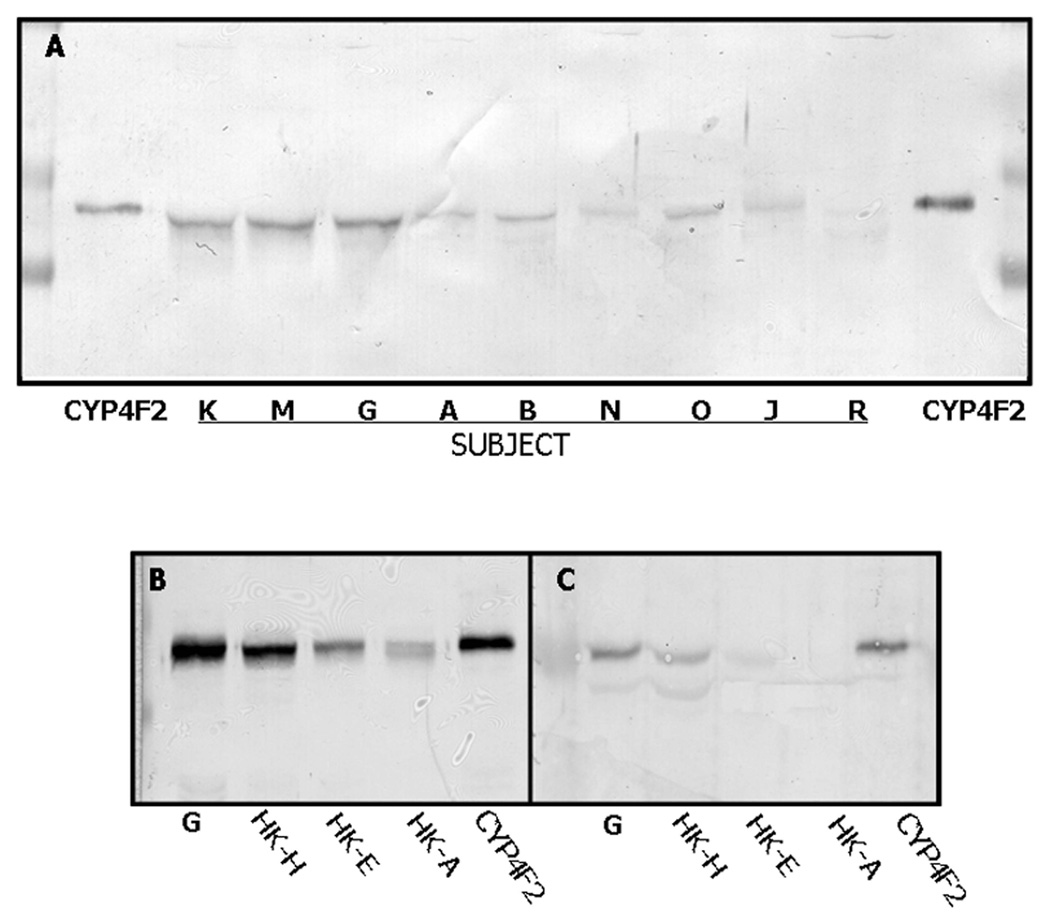
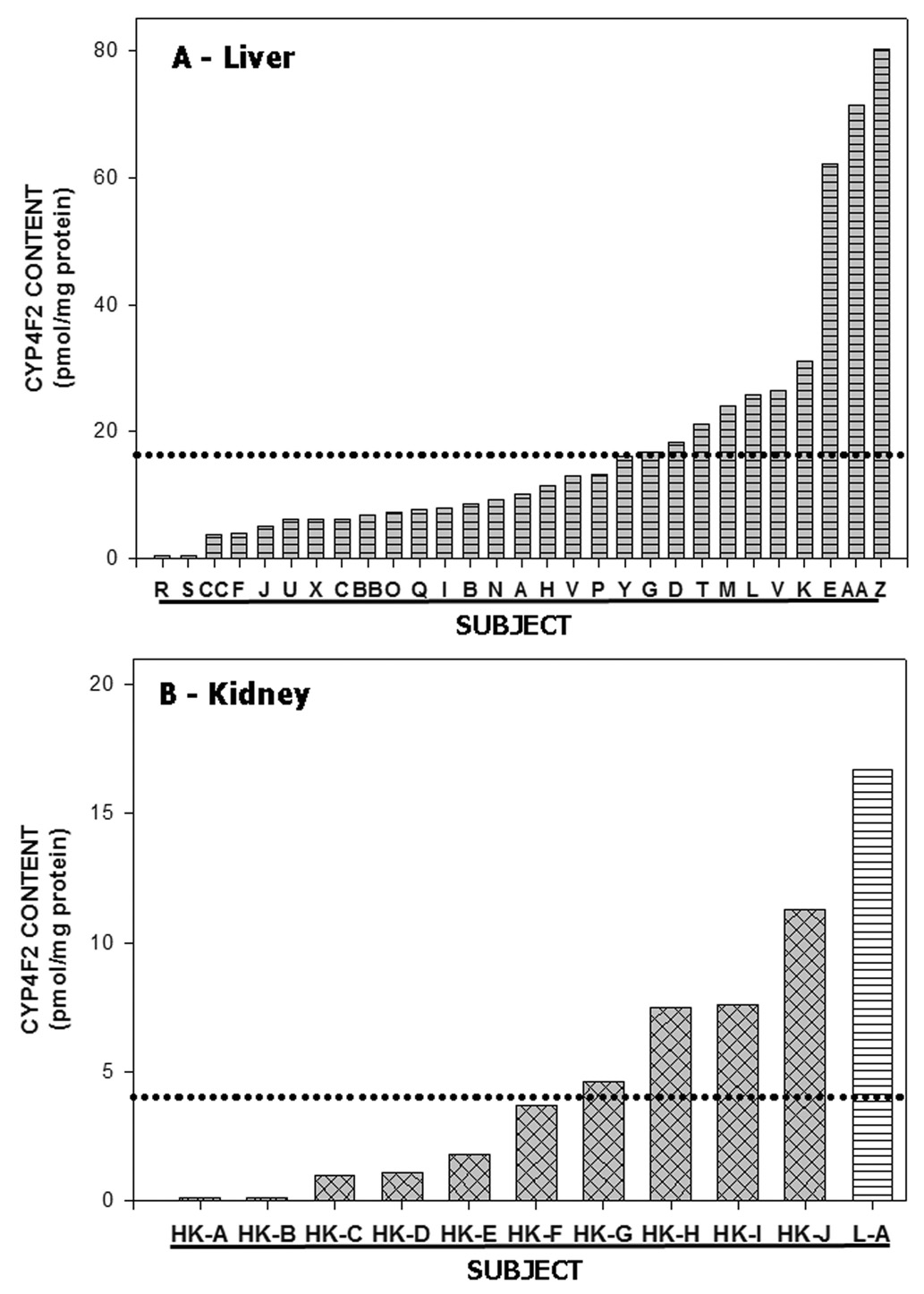
Using anti-CYP4F2 peptide IgG as a specific probe, CYP4F2 protein expression in human liver was found to be highly variable. Hepatic microsomes from 29 different subjects were subjected to Western blotting with the CYP4F2 peptide antibody, and immunoreaction intensities were quantitated by scanning densitometry. As shown in Fig. 3A, which depicts the immunostaining results obtained with 9 of these patients, subjects K, M and G (lane 2–4, respectively) expressed CYP4F2 at high levels while subjects A, B and O (lanes 5, 6 & 8, respectively) expressed CYP4F2 at more moderate levels. This CYP4F enzyme was expressed at very low levels in subjects N and J (lanes 7 and 9, respectively) while CYP4F2 levels in subject R (lane 10) were below detection. Another individual, namely subject S, also failed to express CYP4F2 (data not shown). Immunoreactive CYP4F2 levels varied from 0 – 80.1 pmol/mg microsomal protein among the 29 human subjects examined (Fig. 4A), with a mean of 18.2 ± 19.9 pmol/mg. There was no significant relationship (r = 0.238; p = 0.214; n = 29) between the expression in liver of CYP4F2 and total immunoreactive CYP4F (quantitated using anti- CYP4F+ IgG), suggesting that hepatic CYP4F2 expression is regulated in a manner different than that of CYP4F3b, CYP4F11, and/or CYP4F12.
Figure 3. Expression of CYP4F2 in human liver and kidney.
Panel A. Samples were subjected to Western blotting with anti-CYP4F2 peptide IgG as described in Materials and Methods. The lanes marked CYP4F2 contain purified enzyme (0.1 µg) while the remaining lanes contain liver microsomes (30 µg) from the indicated subjects. The disparity in immunoreactive CYP4F2 migration observed here stems from the large differences in protein amounts applied. Panels B & C. Liver and kidney samples were subjected to immunoblotting with either anti-CYP4F+ IgG (panel B) or anti-CYP4F2 peptide IgG (panel C). The lane marked G contain liver microsomes from subject G while the lanes marked HK-H, HK-E and HK-A contain kidney cortical microsomes from those subjects. Liver microsomes were applied at 15 or 30 µg/lane for immunostaining with anti-CYP4F+ IgG or anti-CYP4F2 peptide IgG, respectively, while renal cortical microsomes were applied at 25 or 45 µg/lane. The lanes marked CYP4F2 containing purified enzyme (0.1 µg). Additional details of the Western blotting procedure are described in Materials and Methods.
Figure 4. Variability of CYP4F2 expression in human liver and kidney cortex.
Panel A. CYP4F2 content was quantitated in liver microsomes obtained from 29 different subjects (A thru CC) using Western blots similar to those shown in Figure 3. The dotted line denotes the mean hepatic CYP4F2 content (16.4 pmol/mg protein) noted among the 29 subjects. Hepatic CYP4F2 levels in subjects R and S were below the level of detection. Panel B. Same as Panel A except that the CYP4F2 content in kidney cortical microsomes from 10 different subjects is shown. The column marked L-A depicts the CYP4F2 content in liver microsomes from subject G above while the dotted line denotes the mean renal CYP4F2 content (3.9 pmol/mg protein; n =10). CYP4F2 levels in kidney cortex from subjects HK-A and HK-B were below the level of detection.
As assessed by immunoblotting with anti-CYP4F2 peptide IgG, CYP4F2 content also varied widely in microsomes prepared from human kidney cortex (Fig. 3C and Fig. 4B). Of the three specimens shown in Figure 3C, subject HK-H (lane 2) contained CYP4F2 levels clearly higher than those of subject HK-E (lane 3) and HK-A (lane 4). This Western blot also shows that renal CYP4F2 levels were considerably less than hepatic CYP4F2 levels (compare lane 6 with lanes 7–9 in Fig. 3C). The same phenomenon applied to total CYP4F levels, as shown upon reaction of kidney versus liver microsomes with polyspecific anti-CYP4F+ IgG (Fig 3B). Data depicting the overall variability of CYP4F2 content in kidney microsomes from 10 different subjects is given in Fig. 4B. Two subjects failed to express the enzyme, 3 subjects exhibited modest CYP4F2 levels, and the remaining 5 subjects had appreciably higher CYP4F2 expression. The mean kidney CYP4F2 content in these subjects was 3.9 ± 3.8 pmol/mg protein (range of 0 – 11.3 pmol/mg protein), a value which is nearly 5-fold less than that observed in liver.
Relationship between CYP4F2 expression and endobiotic metabolism in liver
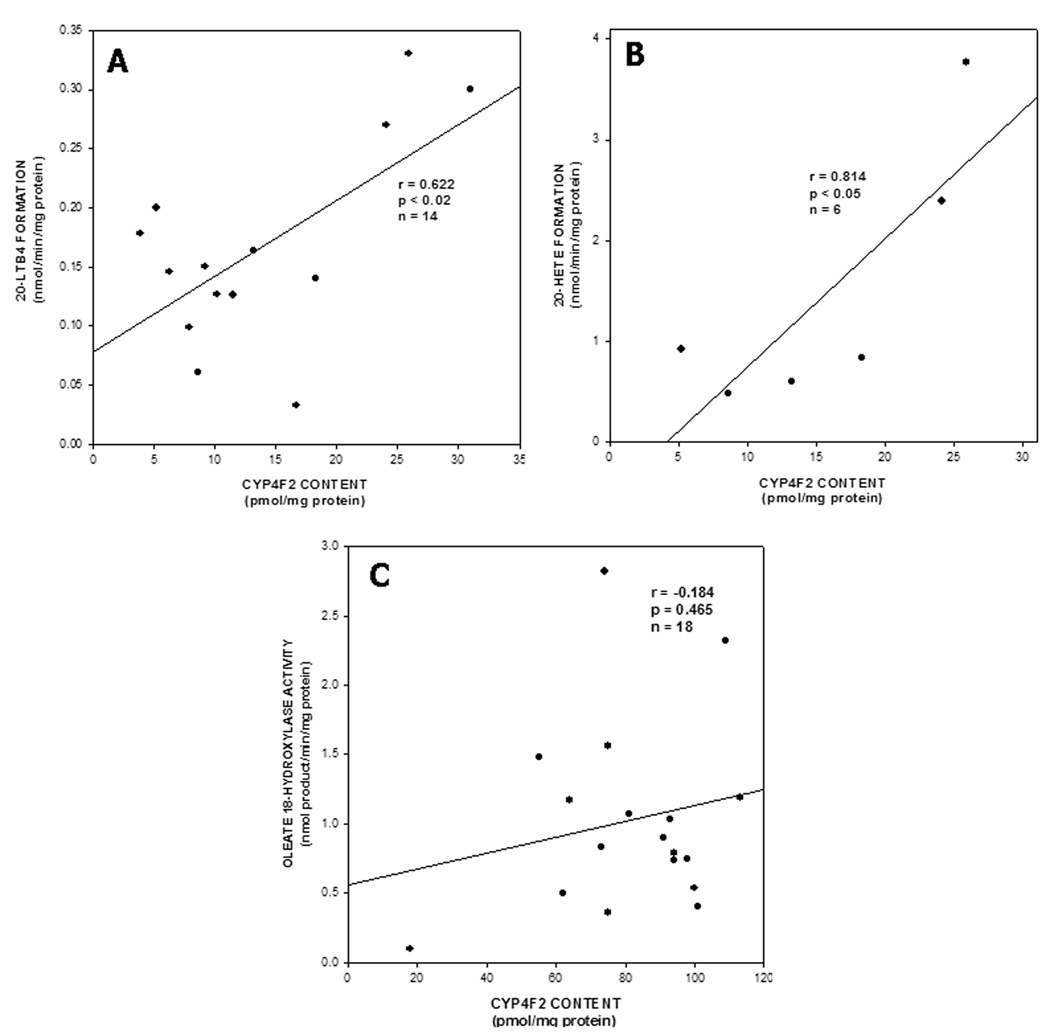
P450 antibodies, particularly those that are inhibitory, are often used to define the role of their corresponding immunogen in a given microsomal drug-metabolizing reaction [8, 9, 29, 31, 48, 49]. However, anti-human CYP4F2 peptide IgG had little effect on CYP4F2-catalyzed reactions, as demonstrated by its limited (< 20%) capacity to inhibit ω-hydroxylation of oleate, an exemplary long-chain fatty acid, by CYP4F2 supersomes® or human liver microsomes at IgG:P450 ratios of up to 30 mg/nmol. In contrast, polyspecific anti-CYP4F+ IgG elicited at least 85% inhibition of oleate ω-hydroxylation by CYP4F2 supersomes® or human liver microsomes at 5–10 mg IgG/nmol P450 (data not shown). It was thus necessary to evaluate the relationship between CYP4F2 expression and endobiotic metabolism in liver using an indirect method, namely correlation analysis. As shown in Fig. 5A, a significant correlation (r = 0.622; p < 0.02) was found between CYP4F2 content and LTB4 20-hydroxylase activity in human liver microsomes. A strong correlation (r = 0.814; p < 0.05) was also observed between microsomal CYP4F2 levels and AA ω- hydroxylation (Fig. 5B), whereas no relationship (r = −0.184; p = 0.465) was found between levels of this CYP4F enzyme and oleate ω-hydroxylase activity in liver microsomes (Fig. 5C). The former results suggest that CYP4F2 plays a prevalent role in the ω-hydroxylation of LTB4 and AA by liver microsomes, at least in the subjects studied here. Indeed, as shown in Table 1, LTB and AA 20-hydroxylation are activities catalyzed by purified, reconstituted CYP4F2 and, in the latter case, by CYP4F2-containing supersomes® as well4. Of interest here is that CYP4F2 (and CYP4F3b) reconstituted systems converted LTB4 to a single metabolite, namely 20-OH LTB4, while liver microsomes and CYP4F3a supersomes converted LTB4 not only to 20-OH LTB4 but also to 20-carboxy LTB4, especially after longer (>15 min) incubation periods6. As discussed below, the capacity of CYP4F3b to also catalyze these ω-hydroxylation reactions may explain, at least in part, why stronger correlations were not obtained between microsomal CYP4F2 content and rates of LTB4 and AA hydroxylation. The lack of correlation between CYP4F2 levels and oleate 18-hydroxylation suggests that a CYP4F enzyme(s) other than CYP4F2 may be an equally efficient catalyst of this reaction in intact liver microsomes. In fact, purified CYP4F3b and CYP4F3b supersomes® converted oleate to its ω- hydroxylated metabolite at rates similar to those observed with CYP4F2 (Table 1). Both CYP4F enzymes hydroxylated oleate at only the ω position whereas human liver microsomes oxidized this monosaturated fatty acid at the ω and ω-1 positions (data not shown). Purified CYP4F12 and CYP4F12 supersomes® failed to oxidize these three substrates, suggesting that this particular CYP4F enzyme did not contribute to microsomal ω-hydroxylation in the correlation analyses.
Figure 5. Correlation between CYP4F2 content and ω-hydroxylation of LTB4, AA and oleate in human liver microsomes.
CYP4F2 expression in hepatic microsomes from the different subjects was determined by quantitative immunoblotting (see Fig. 3) while rates of 20-OH LTB4 (Panel A), 20-HETE (Panel B), and 18-OH oleate formation (Panel C) by the same liver samples were measured as described in Materials and Methods. In each case, the correlation coefficient (r) was derived from the line of best fit by regression analysis, and a test of slope was used to determine the level of significance (p).
TABLE 1.
LTB4, AA and Oleate ω-Hydroxylation by Recombinant Human CYP4F2 and Liver Microsomes
| SUBSTRATE | |||
|---|---|---|---|
| Enzyme | LTB4a | AAb | Oleateb |
| CYP4F2 | 0.20 | 2.6 ± 0.9 | 5.1 |
| CYP4F2 Supersomes | < 0.1 | 3.8 | 21.6 |
| CYP4F3b | 0.22 | 2.4 | 4.7 |
| CYP4F3b Supersomes | 2.13 ± 1.6 | 16.9 | 33.0 |
| CYP4F12 Prep #1 | nd | < 0.1 | < 0.1 |
| CYP4F12 Supersomes | < 0.1 | < 0.1 | < 0.1 |
| Liver Microsomes Subject A | 0.56 | 2.4 ± 0.2 | 2.1 |
| Liver Microsomes Subject B | 0.73 ± 0.1 | 6.9 | 2.0 ± 0.9 |
LTB4, AA and oleate ω-hydroxylation were assessed in incubation mixtures (0.25 ml) containing: a) reconstituted systems [25–50 pmol CYP4F enzyme, 75–150 pmol P450 reductase, 7.5–15 µg DLPC and 100–200 pmol b5; b) supersomes® from T. ni insect cells expressing human CYP4F2, CYP4F3b or CYP4F11 (protein equivalent to 25 pmol P450) or; c) human liver microsomes (protein equivalent to 50–150 pmol P450). Other incubation components included 100 mM potassium phosphate buffer (pH 7.4), 0.5 mM NADPH, and LTB4 (30 µM), AA (100 µM) or oleate (100 µM). Reactions were initiated with NADPH, and were terminated after 10 – 15 min at 37°C. Formati on of 20-OH LTB4, 20-HETE and 18-OH oleate were then assessed by HPLC as described in Materials and Methods.
Data expressed as pmol product formed/min/nmol P450, and denote the average of at least 3 individual determinations. Results given as the mean ± SD represent 9–24 individual determinations.
Data expressed as nmol product formed/min/nmol P450, and denote the average of at least 3 individual determinations. Results given as the mean ± SD represent 6–15 individual determinations.
DISCUSSION
The enzymes comprising the human CYP4F gene subfamily are thought to partake in the regulation of several key physiological and/or pathological processes, including fatty acid oxidation [6, 50], LTB4- dependent inflammation [20 and references therein], cellular proliferation [22, 51] and, perhaps most important, blood pressure control [21, 52]. However, the assignment of a given CYP4F P450 to a particular catalytic function (e.g., conversion of AA to 20-HETE) in intact tissue has proved challenging due to the lack of highly-specific metabolic probes for these enzymes. Without such probes, it is difficult, if not impossible, to gauge how CYP4F enzyme expression in a given cell type or tissue can modulate the above-described processes. To that end, we developed a targeted antibody in order to assess the tissue-specific expression and function of CYP4F2, one of four major CYP4F P450 enzymes found in human liver and kidney. Peptide antibodies were elicited in rabbits to CYP4F2 amino acid residues 61–74 (WGHQGMVNPTEEG) and 65–77 (GMVNPTEEGMRVL) upon their conjugation to the carrier proteins BSA and ovalbumin. On Western blots, these CYP4F2 peptide antibodies recognized only CYP4F2 and not CYP4F3b, CYP4F11 or CYP4F12. Subsequent immunoquantitation studies revealed that the expression of CYP4F2 in liver varied eighty-fold among the 29 human subjects examined. CYP4F2 expression in kidney cortical tissue, which was nearly five-fold less than in liver, exhibited a similar degree of variability (Fig. 4). Moreover, CYP4F2 content in liver microsomes was found to be significantly correlated (r ≥ 0.63; p < 0.05) with microsomal leukotriene B4 ω-hydroxylation as well as arachidonate ω-hydroxylation, both of which are CYP4F2-catalyzed reactions (Table 1).
The anti-CYP4F2 peptide IgG described herein represents the first instance of an antibody which recognizes only a single member of the human CYP4F gene subfamily5. An antibody we designated as anti-CYP4F2 IgG2 was employed in our prior studies on CYP4F2 involvement in hepatic and renal 20- HETE formation and LTB4 catabolism [8, 9, 15], although it is now known that this antibody reacts not only with CYP4F2 but also with CYP4F3b, CYP4F11, CYP4F12 and CYP4F3a [53]. Indeed, the extensive degree of amino acid sequence homology found among these 5 individual CYP4F P450s likely rules out their use as immunogens for deriving naïve yet highly-specific polyclonal antibodies. We therefore modified our usual immunochemical approach to one that involved the production of antibodies against CYP4F2-derived peptides. The sequence relatedness of CYP4F2 with CYP4F3b at their COOH-termini (Fig. 1B) precluded our utilization of peptides derived from this region as antigens, thereby obviating the targeted antibody strategy successfully employed by Edwards and co-workers [31, 54]. In fact, an antibody that we produced against the CYP4F2 carboxyl terminal hexapeptide recognized neither CYP4F11 nor CYP4F12 but cross-reacted strongly with CYP4F3b6. The use of amino acid sequences localized near the CYP4F2 loop region between the G and H helices, which appear particularly well-suited for deriving specific, inhibitory peptide antibodies against various CYP2 and CYP3 gene family P450s including CYP2D6, CYP2C19, and CYP3A4 [33–35, 45, 55] was also not a viable option here due to CYP4F overlapping structure. Thus, we initially focused on CYP4F2 amino acids 67–114, which comprise that region of the hemoprotein encoded by exon 3 of the cognate structural gene [10, 16]. Several sequence differences between CYP4F2 and the other CYP4F P450s are found within this particular region, especially at residues 96–100 (Fig. 1B), which is why CYP4F2 residues 89–102 and 89–101 (peptides A and B, respectively; Fig. 1A) were employed first as peptide immunogens. However, the antibodies elicited against CYP4F2 peptides A and B failed to bind to CYP4F2 on immunoblots and were only weakly reactive with the peptides themselves in ELISA (data not shown). Such results likely stemmed from several factors, one of which was the relative lack of antigenicity of these particular amino acid sequences. Another factor is the location of peptides A and B within the CYP4F2 polypeptide chain, as an alignment of the human CYP4F2 amino acid sequence with that of rabbit CYP2C5, the three-dimensional structure of which has been elucidated [47, 56], showed that CYP4F2 residues 89–102 are localized within the B and C helixes of the hemoprotein, and may thus be inaccessible to bulky IgG molecules [57].
Reassessment of the CYP4F2-CYP2C5 sequence alignment led to the choice of amino acid target sequences (peptides C and D; Fig. 1A) that were localized closer to the NH2-terminal end of CYP4F2. Peptides C and D contain CYP4F2 amino acids 61–74 and 65–77, respectively, which are also encoded mainly by CYP4F2 exon 3 [10, 16], and likely correspond to rabbit CYP2C5 residues 43–65 and 50–62. Homology modeling predicted that these residues are at least partially surface-exposed, since they reportedly comprise the helix A region and a loop leading into this helix [46, 47]. Indeed, immunization with CYP4F2 peptides C and D gave rise to targeted antibodies that, in contrast to peptides A and B, specifically recognized only CYP4F2 on Western blots (Fig. 2 and Fig 3) and were strongly reactive with their corresponding antigens in ELISA (titers ranging from 1:50,000 to 1:100,000)(data not shown). These results indicate the relative lack of influence of three-dimensional structure on anti-CYP4F2 peptide IgG immunoreactivity, and confirm that CYP4F2 amino acid residues 61–77 are not deeply buried within the protein but remain accessible to antibody binding.
Once developed, we used these CYP4F2 peptide antibodies to demonstrate that liver CYP4F2 content varied extensively (80-fold) among our human subject population. The lack of detailed, pre-operative drug intake histories with many of these donor subjects precluded determination of whether exposure to CYP4F inducers (e.g., statins)[58] contributed to the marked interindividual variation observed in hepatic CYP4F2 levels. Intriguingly, 2 of these 29 individuals (subjects R and S) failed to express any immunoreactive CYP4F2. This deficiency in hepatic CYP4F2 protein expression was apparently not due to global P450 proteolytic degradation since these same subjects exhibited substantial levels of other CYP4F P450s, as deduced from Western blots immunostained with anti-CYP4F+ IgG (data not shown). Whether subject R, a 45-year old male Caucasian, and subject S, a 20-year old male Caucasian, are also deficient in renal CYP4F2 protein is not known but is a very relevant question since, in the affirmative, such an observation would strongly support a genetic polymorphism affecting control of CYP4F2 expression. In addition, no relationship (r = 0.238; p = 0.214) was found between hepatic CYP4F2 levels and total immunoreactive CYP4F content among the 29 subjects, suggesting that the regulation of CYP4F2 protein expression in liver is not coordinate with that of CYP4F3b, CYP4F11, and/or CYP4F12.
Polyclonal anti-CYP4F2 peptide IgG proved incapable of inhibiting catalysis by its target, a phenomenon that is not unique among P450 peptide antibodies [31, 54]. IgG:P450 ratios as high as 30 mg/nmol failed to inhibit associated ω-hydroxylation by purified CYP4F2, CYP4F2 supersomes®, or intact liver microsomes. In contrast, anti-CYP4F+ IgG, which is directed against multiple antigenic sites on CYP4F2, is a potent inhibitor of reactions catayzed by this enzyme [8, 9, 15, 59, 60]. Such results indicate that while CYP4F2 amino acid residues 61–77 are accessible to targeted antibody binding, this region of the protein comprising helix A and a loop leading into this helix [46, 47] is not critical for substrate oxidation. Since immunoinhibition assays could not be employed to directly assess the role of CYP4F2 in microsomal fatty acid/eicosanoid ω-hydroxylation, we relied instead on another commonly-used, albeit indirect criterion for determining this role, namely correlation analyses [48]. Three prospective CYP4F2 substrates were utilized, namely, LTB4, AA and oleate, the former two of which have been shown by our group and other investigators to undergo ω-hydroxylation by either purified reconstituted CYP4F2 or CYP4F2 supersomes® [8, 9, 61, 62]. Oleate, a long-chain, monounsaturated fatty acid abundant in human liver [63, 64], was employed as a substrate due to evidence for its extensive oxidation by CYP4F enzymes [41]. CYP4F2- catalyzed metabolism of these substrates was verified using systems reconstituted with the purified recombinant enzyme as well as the cognate supersomes®; analogous CYP4F3b and CYP4F12 systems were included for comparison (Table 1). Although none of these substrates exhibited a catalytic preference for CYP4F2 compared to CYP4F3b, the other major CYP4F P450 expressed in human liver and kidney [10, 50], a significant correlation (r = 0.622; p < 0.02) was still observed between hepatic CYP4F2 content and LTB4 20-hydroxylase activity (Fig. 5A). Similarly, CYP4F2 levels and rates of AA ω-hydroxylation in liver microsomes were also strongly correlated (r = 0.814; p < 0.05)(Fig. 5B). Such results imply that in the subjects studied here, CYP4F2 plays a significant role in microsomal ω-hydroxylation of LTB4 and AA. The capacity of CYP4F3b to promote these same reactions may, however, explain why more robust correlations between CYP4F2 content and LTB4 and AA ω-hydroxylation were not obtained. Indeed, the contribution of CYP4F3b to substrate metabolism but not to immunoreactive enzyme levels would effectively weaken any relationship found between the two parameters. Despite extensive oleate metabolism by purified CYP4F2 and CYP4F2-containing supersomes® (Table 1) no correlation was found between CYP4F2 protein expression and oleate 18-hydroxylation (Fig. 5C), an observation which suggests that CYP4F3b and, perhaps, CYP4F11 [65] function as more efficient cataysts of this reaction in intact liver microsomes. Finally, it is apparent from the reconstitution studies presented here that CYP4F12 is not an especially effective fatty acid ω-hydroxylase despite a high degree of structural homology (85%) with CYP4F2 and CYP4F3b.
In conclusion, we have developed a peptide antibody that specifically targets CYP4F2, and does not recognize the other major CYP4F P450 proteins expressed in human liver and kidney. Immunoquantitation and correlation analyses performed with this antibody indicate that CYP4F2 functions as an important LTB4 and arachidonate ω-hydroxylase in human liver. Polymorphisms in the CYP4F2 structural gene [62, 66] and upstream regulatory regions [67] are now being studied to assess relationships between renal CYP4F2 function and/or expression and essential hypertension. Analogous work is being conducted with CYP4A11 [68–70] which, like CYP4F2, is expressed in human kidney and converts AA to the powerful vasoactive metabolite 20-HETE [9]. Unlike CYP4A11, however, there remains a paucity of specific probes permitting unequivocal identification of the CYP4F enzyme(s) that functions as primary catalyst of 20-HETE formation in intact tissue. Further studies of the type described here are required to derive the immunochemical and/or metabolic probes capable of precisely discriminating among the CYP4F P450s in terms of their in vivo function.
ACKNOWLEDGEMENTS
We thank Dr. Jordan Fishman of 21st Century Biochemicals (Marlborough, MA) for his invaluable advice and assistance with regards to CYP4F2 peptide selection and carrier protein conjugation. We also thank Dr. Michael Wester, Pfizer Global R&D, La Jolla Laboratories (La Jolla, CA), for performing the homology modeling and amino acid sequence alignments for rabbit CYP2C5 with human CYP4F2.
Abbreviations Used
- CYP4F/A
P450 enzymes belonging to the CYP4F and/or CYP4A gene subfamilies, which are designated here according to the nomenclature given on Dr. David Nelson’s website for cytochrome P450s (http://drnelson.utmem.edu/cytochromeP450.html)
- P450 reductase
NADPH:P450 oxidoreductase
- b5
cytochrome b5
- DLPC
L-α-dilauroylphosphatidylcholine
- AA
arachidonic acid
- 20-HETE
20-hydroxy-5,8,11,14-eicosatetraenoic acid
- 18-OH oleate
18-hydroxyoleic acid
- LTB4
leukotriene B4
- 20-OH LTB4
20-hydroxy LTB4
- PDAM
1-pyrenyldiazomethane
- IgG
immunoglobulin G.
- HCN
hemocyanin from keyhole limpet
- NBT/BCIP
nitroblue tetrazolium chloride + 5-bromo-4-chloro-3-indolyl-phosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This research was supported by NIH grant AA07842.
In a previous study, we purified a P450 enzyme from human liver that was designated CYP4F2 based upon its unique NH2-terminal-terminal sequence [71], and also produced polyclonal antibodies against this P450 in rabbits. The AA and leukotriene b4 ω-hydroxylase activities of human liver microsomes were attributed mainly to CYP4F2 since, at that time, it was the only CYP4F P450 known to be expressed in human liver [8, 15]. Subsequent investigations identified other CYP4F proteins in human hepatic tissue, including CYP4F3b, CYP4F11 and CYP4F12 [16, 23, 25], the former of which also catalyzes AA and LTB4 20-hydroxylation. Due to the extensive amino acid sequence relatedness (75%) among these CYP4F gene subfamily members, an antibody produced against purified hepatic CYP4F2 recognizes not only its cognate immunogen but also CYP4F3b, CYP4F11, and CYP4F12 on Western blots (see Fig. 2)..
We substituted 12-hydroxylaurate for 18-OH oleate since the latter compound was not commercially available, and since the PDAM derivatives of medium- and long-chain fatty acids such as laurate, oleate, and palmitate exhibit nearly identical extinction coefficients at 275 nm [43].
The capacity of two different preparations of CYP4F2 supersomes® to convert LTB4 to 20-OH LTB4 was examined although, in both cases, these CYP4F2 heterologous expression systems proved to be devoid of LTB4 ω-hydroxylase activity. The reason for this lack of LTB4 metabolism by CYP4F2 supersomes® is unclear at present, especially since LTB4 is recommended as the test substrate by the supplier, and the same supersomes were efficient catalysts of AA and oleate ω-hydroxylation (Table 1).
A CYP4F11 monoclonal antibody and a polyclonal antibody to a CYP4F11 peptide are commercially available although publications describing their immunochemical properties have not yet appeared.
A Kozeska and JM Lasker, unpublished observations.
Contributor Information
Vandana Hirani, Jurist Institute for Research, Hackensack University Medical Center, Hackensack, NJ 07601.
Anton Yarovoy, Jurist Institute for Research, Hackensack University Medical Center, Hackensack, NJ 07601.
Anita Kozeska, Jurist Institute for Research, Hackensack University Medical Center, Hackensack, NJ 07601.
Ronald P. Magnusson, Kinnakeet Biotechnology, Midlothian, VA 23112
Jerome M. Lasker, Jurist Institute for Research, Hackensack University Medical Center, Hackensack, NJ 07601
REFERENCES
- 1.Guengerich FP. Cytochrome P450: Structure, Mechanism, and Biochemistry. In: Ortiz de Montellano PR, editor. 2nd Edition. New York: Plenum Press; 1995. pp. 473–536. [Google Scholar]
- 2.Guengerich FP, Hosea NA, Parikh A, Bell-Parikh LC, Johnson WW, Gillam EM, Shimada T. Drug Metab Dispos. 1998;26:1175–1178. [PubMed] [Google Scholar]
- 3.Nelson DR. Methods Mol Biol. 2006;320:1–10. doi: 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- 4.Okita RT, Okita JR. Curr Drug Metab. 2001;2:265–281. doi: 10.2174/1389200013338423. [DOI] [PubMed] [Google Scholar]
- 5.Powell PK, Wolf I, Lasker JM. Arch Biochem Biophys. 1996;335:219–226. doi: 10.1006/abbi.1996.0501. [DOI] [PubMed] [Google Scholar]
- 6.Sanders RJ, Ofman R, Duran M, Kemp S, Wanders RJ. J Biol Chem. 2006;281:13180–13187. doi: 10.1074/jbc.M513481200. [DOI] [PubMed] [Google Scholar]
- 7.Adas F, Salaun JP, Berthou F, Picart D, Simon B, Amet Y. J Lipid Res. 1999;40:1990–1997. [PubMed] [Google Scholar]
- 8.Powell PK, Wolf I, Jin R, Lasker JM. J Pharmacol Exp Ther. 1998;285:1327–1336. [PubMed] [Google Scholar]
- 9.Lasker JM, Chen WB, Wolf I, Bloswick BP, Wilson PD, Powell PK. J Biol Chem. 2000;275:4118–4126. doi: 10.1074/jbc.275.6.4118. [DOI] [PubMed] [Google Scholar]
- 10.Christmas P, Jones JP, Patten CJ, Rock DA, Zheng Y, Cheng S-M, Weber BM, Carlesso N, Scadden DT, Rettie AE, Soberman RJ. J. Biol. Chem. 2001;276:38166–38172. doi: 10.1074/jbc.M104818200. [DOI] [PubMed] [Google Scholar]
- 11.Hoch U, Zhang Z, Kroetz DL, Ortiz de Montellano PR. Arch Biochem Biophys. 2000;373:63–71. doi: 10.1006/abbi.1999.1504. [DOI] [PubMed] [Google Scholar]
- 12.Roman RJ, Maier KG, Sun CW, Harder DR, Alonso-Galicia M. Clin Exp Pharmacol Physiol. 2000;27:855–865. doi: 10.1046/j.1440-1681.2000.03349.x. [DOI] [PubMed] [Google Scholar]
- 13.McGiff JC, Quilley J. Am J Physiol. 1999;277:R607–R623. doi: 10.1152/ajpregu.1999.277.3.R607. [DOI] [PubMed] [Google Scholar]
- 14.Capdevila JH, Falck JR, Harris RC. J Lipid Res. 2000;41:163–181. [PubMed] [Google Scholar]
- 15.Jin R, Koop DR, Raucy JL, Lasker JM. Arch Biochem Biophys. 1998;359:89–98. doi: 10.1006/abbi.1998.0880. [DOI] [PubMed] [Google Scholar]
- 16.Christmas P, Ursino SR, Fox JW, Soberman RJ. J Biol Chem. 1999;274:21191–21199. doi: 10.1074/jbc.274.30.21191. [DOI] [PubMed] [Google Scholar]
- 17.Kalsotra A, Turman CM, Kikuta Y, Strobel HW. Toxicol Appl Pharmacol. 2004;199:295–304. doi: 10.1016/j.taap.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 18.Sontag TJ, Parker RS. J Biol Chem. 2002;277:25290–25296. doi: 10.1074/jbc.M201466200. [DOI] [PubMed] [Google Scholar]
- 19.Sontag TJ, Parker RS. J Lipid Res. 2007;48:1090–1098. doi: 10.1194/jlr.M600514-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Christmas P, Tolentino K, Primo V, Berry KZ, Murphy RC, Chen M, Lee DM, Soberman RJ. J Biol Chem. 2006;281:7189–7196. doi: 10.1074/jbc.M513101200. [DOI] [PubMed] [Google Scholar]
- 21.Sarkis A, Roman RJ. Curr Drug Metab. 2004;5:245–256. doi: 10.2174/1389200043335603. [DOI] [PubMed] [Google Scholar]
- 22.Muthalif MM, Benter IF, Karzoun N, Fatima S, Harper J, Uddin MR, Malik KU. Proc Natl Acad Sci U S A. 1998;95:12701–12706. doi: 10.1073/pnas.95.21.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashizume T, Imaoka S, Hiroi T, Terauchi Y, Fujii T, Miyazaki H, Kamataki T, Funae Y. Biochem Biophys Res Commun. 2001;280:1135–1141. doi: 10.1006/bbrc.2000.4238. [DOI] [PubMed] [Google Scholar]
- 24.Bylund J, Bylund M, Oliw EH. Biochem Biophys Res Commun. 2001;280:892–897. doi: 10.1006/bbrc.2000.4191. [DOI] [PubMed] [Google Scholar]
- 25.Cui X, Nelson DR, Strobel HW. Genomics. 2000;68:161–166. doi: 10.1006/geno.2000.6276. [DOI] [PubMed] [Google Scholar]
- 26.Kikuta Y, Kusunose E, Endo K, Yamamoto S, Sogawa K, Fujii-Kuriyama Y, Kusunose M. J Biol Chem. 1993;268:9376–9380. [PubMed] [Google Scholar]
- 27.Christmas P, Carlesso N, Shang H, Cheng SM, Weber BM, Preffer FI, Scadden DT, Soberman RJ. J Biol Chem. 2003;278:25133–25142. doi: 10.1074/jbc.M302106200. [DOI] [PubMed] [Google Scholar]
- 28.Bylund J, Hidestrand M, Ingelman-Sundberg M, Oliw EH. J Biol Chem. 2000;275:21844–21849. doi: 10.1074/jbc.M001712200. [DOI] [PubMed] [Google Scholar]
- 29.Wester MR, Lasker JM, Johnson EF, Raucy JL. Drug Metab Dispos. 2000;28:354–359. [PubMed] [Google Scholar]
- 30.Murray BP, Edwards RJ, Murray S, Singleton AM, Davies DS, Boobis AR. Carcinogenesis. 1993;14:585–592. doi: 10.1093/carcin/14.4.585. [DOI] [PubMed] [Google Scholar]
- 31.Edwards RJ, Boobis AR, Davies DS. Drug Metab Dispos. 2003;31:1476–1480. doi: 10.1124/dmd.31.12.1476. [DOI] [PubMed] [Google Scholar]
- 32.Schulz-Utermoehl T, Edwards RJ, Boobis AR. Drug Metab Dispos. 2000;28:544–551. [PubMed] [Google Scholar]
- 33.Schulz-Utermoehl T, Mountfield RJ, Bywater RP, Madsen K, Jorgensen PN, Hansen KT. Drug Metab Dispos. 2000;28:718–725. [PubMed] [Google Scholar]
- 34.Wang RW, Lu AY. Drug Metab Dispos. 1997;25:762–767. [PubMed] [Google Scholar]
- 35.Wang RW, Newton DJ, Liu NY, Shou M, Rushmore T, Lu AY. Drug Metab Dispos. 1999;27:167–172. [PubMed] [Google Scholar]
- 36.Raucy JL, Lasker JM. Methods Enzymol. 1991;206:577–587. doi: 10.1016/0076-6879(91)06127-o. [DOI] [PubMed] [Google Scholar]
- 37.Omura T, Sato R. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 38.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 39.Lasker JM, Wester MR, Aramsombatdee E, Raucy JL. Arch Biochem Biophys. 1998;353:16–28. doi: 10.1006/abbi.1998.0615. [DOI] [PubMed] [Google Scholar]
- 40.Jin HJ. Arch Pharm Res. 1998;21:35–40. doi: 10.1007/BF03216750. [DOI] [PubMed] [Google Scholar]
- 41.Lasker JM, Ozaki K, Duan K. Pharmacologist. 2002;44:A123. [Google Scholar]
- 42.Adas F, Berthou F, Picart D, Lozac'h P, Beauge F, Amet Y. J Lipid Res. 1998;39:1210–1219. [PubMed] [Google Scholar]
- 43.Nimura N, Kinoshita T, Yoshida T, Uetake A, Nakai C. Anal Chem. 1988;60:2067–2070. doi: 10.1021/ac00170a017. [DOI] [PubMed] [Google Scholar]
- 44.McKinney MM, Parkinson A. Journal of Immunological Methods. 1987;96:271–278. doi: 10.1016/0022-1759(87)90324-3. [DOI] [PubMed] [Google Scholar]
- 45.Schulz-Utermoehl T, Mountfield RJ, Madsen K, Jorgensen PN, Hansen KT. Drug Metab Dispos. 2000;28:715–717. [PubMed] [Google Scholar]
- 46.Cheng MK, McGiff JC, Carroll MA. Am J Physiol Renal Physiol. 2003;284:F474–F479. doi: 10.1152/ajprenal.00239.2002. [DOI] [PubMed] [Google Scholar]
- 47.Wester MR, Stout CD, Johnson EF. Methods Enzymol. 2002;357:73–79. doi: 10.1016/s0076-6879(02)57667-9. [DOI] [PubMed] [Google Scholar]
- 48.Wrighton S, Vandenbranden M, Stevens J, Shipley L, Ring B, Rettie A, Cashman J. Drug Metab Rev. 1993;25:453–484. doi: 10.3109/03602539308993982. [DOI] [PubMed] [Google Scholar]
- 49.Gelboin HV, Krausz K. J Clin Pharmacol. 2006;46:353–372. doi: 10.1177/0091270005285200. [DOI] [PubMed] [Google Scholar]
- 50.Hsu MH, Savas U, Griffin KJ, Johnson EF. Drug Metab Rev. 2007;39:515–538. doi: 10.1080/03602530701468573. [DOI] [PubMed] [Google Scholar]
- 51.Liang CJ, Ives HE, Yang CM, Ma YH. J Lipid Res. 2008;49:66–73. doi: 10.1194/jlr.M700155-JLR200. [DOI] [PubMed] [Google Scholar]
- 52.Singh H, Cheng J, Deng H, Kemp R, Ishizuka T, Nasjletti A, Schwartzman ML. Hypertension. 2007;50:123–129. doi: 10.1161/HYPERTENSIONAHA.107.089599. [DOI] [PubMed] [Google Scholar]
- 53.Ito O, Nakamura Y, Tan L, Ishizuka T, Sasaki Y, Minami N, Kanazawa M, Ito S, Sasano H, Kohzuki M. Mol Cell Biochem. 2006;284:141–148. doi: 10.1007/s11010-005-9038-x. [DOI] [PubMed] [Google Scholar]
- 54.Edwards RJ, Adams DA, Watts PS, Davies DS, Boobis AR. Biochem Pharmacol. 1998;56:377–387. doi: 10.1016/s0006-2952(98)00033-1. [DOI] [PubMed] [Google Scholar]
- 55.Cribb A, Nuss C, Wang R. Drug Metab Dispos. 1995;23:671–675. [PubMed] [Google Scholar]
- 56.Lewis DF, Ito Y, Goldfarb PS. Curr Drug Metab. 2006;7:589–598. doi: 10.2174/138920006778017812. [DOI] [PubMed] [Google Scholar]
- 57.Lewis DF. Xenobiotica. 2002;32:305–323. doi: 10.1080/00498250110112015. [DOI] [PubMed] [Google Scholar]
- 58.Hsu M-H, Savas U, Griffin KJ, Johnson EF. J. Biol. Chem. 2007;282:5225–5236. doi: 10.1074/jbc.M608176200. [DOI] [PubMed] [Google Scholar]
- 59.Wang MZ, Saulter JY, Usuki E, Cheung Y-L, Hall M, Bridges AS, Loewen G, Parkinson OT, Stephens CE, Allen JL, Zeldin DC, Boykin DW, Tidwell RR, Parkinson A, Paine MF, Hall JE. Drug Metab Dispos. 2006 doi: 10.1124/dmd.106.010587. dmd.106.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang MZ, Wu JQ, Bridges AS, Zeldin DC, Kornbluth S, Tidwell RR, Hall JE, Paine MF. Drug Metab Dispos. 2007 doi: 10.1124/dmd.107.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kikuta Y, Kusunose E, Kusunose M. J Biochem (Tokyo) 2000;127:1047–1052. doi: 10.1093/oxfordjournals.jbchem.a022696. [DOI] [PubMed] [Google Scholar]
- 62.Stec DE, Roman RJ, Flasch A, Rieder MJ. Physiol Genomics. 2007;30:74–81. doi: 10.1152/physiolgenomics.00003.2007. [DOI] [PubMed] [Google Scholar]
- 63.Gibson GG, Cinti DL, Sligar SG, Schenkman JB. J Biol Chem. 1980;255:1867–1873. [PubMed] [Google Scholar]
- 64.Cullen P. Am J Cardiol. 2000;86:943–949. doi: 10.1016/s0002-9149(00)01127-9. [DOI] [PubMed] [Google Scholar]
- 65.Dhar M, Sepkovic DW, Hirani V, Magnusson RP, Lasker JM. J. Lipid Res. 2008;49:612–624. doi: 10.1194/jlr.M700450-JLR200. [DOI] [PubMed] [Google Scholar]
- 66.Ward NC, Tsai IJ, Barden A, van Bockxmeer FM, Puddey IB, Hodgson JM, Croft KD. Hypertension. 2008;51:1393–1398. doi: 10.1161/HYPERTENSIONAHA.107.104463. [DOI] [PubMed] [Google Scholar]
- 67.Liu H, Zhao Y, Nie D, Shi J, Fu L, Li Y, Yu D, Lu J. J Am Soc Nephrol. 2008;19:714–721. doi: 10.1681/ASN.2007060713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gainer JV, Bellamine A, Dawson EP, Womble KE, Grant SW, Wang Y, Cupples LA, Guo CY, Demissie S, O'Donnell CJ, Brown NJ, Waterman MR, Capdevila JH. Circulation. 2005;111:63–69. doi: 10.1161/01.CIR.0000151309.82473.59. [DOI] [PubMed] [Google Scholar]
- 69.Mayer B, Lieb W, Gotz A, Konig IR, Kauschen LF, Linsel-Nitschke P, Pomarino A, Holmer S, Hengstenberg C, Doering A, Loewel H, Hense HW, Ziegler A, Erdmann J, Schunkert H. J Hypertens. 2006;24:1965–1970. doi: 10.1097/01.hjh.0000244944.34546.8e. [DOI] [PubMed] [Google Scholar]
- 70.Laffer CL, Gainer JV, Waterman MR, Capdevila JH, Laniado-Schwartzman M, Nasjletti A, Brown NJ, Elijovich F. Hypertension. 2008 doi: 10.1161/HYPERTENSIONAHA.107.102921. HYPERTENSIONAHA.107.102921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kikuta Y, Kusunose E, Kondo T, Yamamoto S, Kinoshita H, Kusunose M. FEBS Lett. 1994;348:70–74. doi: 10.1016/0014-5793(94)00587-7. [DOI] [PubMed] [Google Scholar]