Abstract
Huntington's disease (HD) is an inherited neurodegenerative disorder caused by polyglutamine (polyQ) expansions in the huntingtin (Ht) protein. A hallmark of HD is the proteolytic production of an N-terminal fragment of Ht, containing the polyQ repeat, that forms aggregates in the nucleus and cytoplasm of affected neurons. Proteins with longer polyQ repeats aggregate more rapidly and cause disease at an earlier age, but the mechanism of aggregation and its relationship to disease remain unclear. To provide a new, genetically tractable model system for the study of Ht, we engineered yeast cells to express an N-terminal fragment of Ht with different polyQ repeat lengths of 25, 47, 72, or 103 residues, fused to green fluorescent protein. The extent of aggregation varied with the length of the polyQ repeat: at the two extremes, most HtQ103 protein coalesced into a single large cytoplasmic aggregate, whereas HtQ25 exhibited no sign of aggregation. Mutations that inhibit the ubiquitin/proteasome pathway at three different steps had no effect on the aggregation of Ht fragments in yeast, suggesting that the ubiquitination of Ht previously noted in mammalian cells may not inherently be required for polyQ length-dependent aggregation. Changing the expression levels of a wide variety of chaperone proteins in yeast neither increased nor decreased Ht aggregation. However, Sis1, Hsp70, and Hsp104 overexpression modulated aggregation of HtQ72 and HtQ103 fragments. More dramatically, the deletion of Hsp104 virtually eliminated it. These observations establish yeast as a system for studying the causes and consequences of polyQ-dependent Ht aggregation.
Polyglutamine (polyQ) expansions in several unrelated proteins are responsible for at least eight inherited neurodegenerative diseases. These include Huntington's disease (HD), spinobulbar muscular atrophy, dentatorubral pallidoluysian atrophy, and spinocerebellar ataxia types 1, 2, 3, 6, and 7 (1–3). Perhaps the most baffling aspect of these diseases is that the proteins are expressed widely in brain and other tissues, yet each is toxic in a different, highly specific group of neurons and produces a distinct pathology (2).
The major characteristic of HD is a selective loss of neurons in the striatum and cortex leading to movement disorders, dementia, and eventually, death (4, 5). The causative agent is a 350-kDa protein, huntingtin (Ht), with glutamine expansions in the N-terminal region (6). The toxicity of Ht in specific neurons correlates with the length of the glutamine expansion, but the mechanism of toxicity is unknown (7, 8).
A central event in HD is the production of an N-terminal fragment of Ht that aggregates in affected neurons during the natural progression of the disease in humans (9). In transgenic animal models an N-terminal fragment is sufficient to produce an HD-like phenotype, which also depends on the length of the Q repeat (10, 11). Aggregates are found in both the nucleus and/or the cytoplasm of affected neurons in human patients, transgenic animals, and cell lines (12). It is by no means clear, however, whether the aggregates are themselves pathogenic, simply benign byproducts (and thereby markers) of other pathogenic polyQ misfolding events, or a defense mechanism whose purpose is to reduce the interaction of toxic misfolded polyQ proteins with other proteins. Indeed, even the mechanisms underlying the aggregation of these fragments are unknown. A further complexity is that several other proteins interact with Ht. These include HAP1, HIP1, Hsp35, WW domain-containing proteins, the ubiquitin-conjugating enzyme hE2–25k, the SH3GL3 protein, cystathione β-synthase, and calmodulin (12–14).
These problems are inherently difficult to study. Although several mammalian cell-line and transgenic-animal models exist for studying Ht, none is as readily amenable to genetic analysis as yeast. We demonstrate that the aggregation of N-terminal fragments of Ht in yeast cells depends on the length of its polyQ repeat and that this aggregation depends on the balance of chaperone protein activities in the cell. Furthermore, N-terminal fragments with up to 103 glutamines have little or no toxicity in either the aggregated or the soluble state. Thus, yeast cells should provide a valuable system for investigating general factors that affect aggregation and for determining what neuronal cell-type specific factors might influence toxicity.
Materials and Methods
Plasmid Construction.
Plasmids encoding fusions between the N-terminal region of Ht and green fluorescent protein (GFP) were the kind gift of G. Lawless (Univ. of California, Los Angeles).
To create yeast expression plasmids for HtQ25 or HtQ103, DNAs were digested with XhoI and XbaI, and the resulting XhoI–XbaI fragments were ligated into the vector pYES (Invitrogen) to obtain plasmids pYES/PQ25 or pYES/PQ103, respectively. These DNAs were digested with SalI, and ends were filled with Klenow enzyme. Afterward, DNAs were digested with EcoRI, and the resulting fragments were subcloned into a high-copy (2 μ) expression vector p426 for constitutive expression or p426GAL for galactose induction, respectively (15, 16).
To create low-copy expression plasmids with either constitutive (glycerol-3-phosphate dehydrogenase; GPD) or galactose (GAL) inducible promotor DNAs p426/PQ25 or p426/PQ103 were digested with XhoI. The resulting XhoI fragments were subcloned into p416 or p416GAL, respectively (15, 16).
To generate the same set of yeast expression plasmids for HtQ47 or HtQ72, DNAs were double-digested with Acc65I and XbaI, and fragments were blunted with Klenow enzyme and subcloned into a ClaI-blunted vector p426 for constitutive expression. To generate low-copy expression plasmids with constitutive expression (GPD), DNA p426/PQ47, or p426/PQ72 were digested with SpeI and XhoI, and the resulting fragments were subcloned into p416.
The expression plasmids used in this study are listed in Table 1 (17–19).
Table 1.
Plasmids used in this study
| Plasmid | Promotor | Copy number | Ref. |
|---|---|---|---|
| p416 | GPD | CEN, low | 16 |
| p416/PQ25 | GPD | CEN, low | This study |
| p416/PQ47 | GPD | CEN, low | This study |
| p416/PQ72 | GPD | CEN, low | This study |
| p416/PQ103 | GPD | CEN, low | This study |
| p426 | GPD | 2μ, high | 16 |
| p426/PQ25 | GPD | 2μ, high | This study |
| p426/PQ47 | GPD | 2μ, high | This study |
| p426/PQ72 | GPD | 2μ, high | This study |
| p426/PQ103 | GPD | 2μ, high | This study |
| p416GAL | GAL | CEN, low | 15 |
| p416Gal/PQ25 | GAL | CEN, low | This study |
| p416Gal/PQ103 | GAL | CEN, low | This study |
| p426GAL | GAL | 2μ, high | 15 |
| p426Gal/PQ25 | GAL | 2μ, high | This study |
| p426Gal/PQ103 | GAL | 2μ, high | This study |
| pTVSIS1 | GPD | 2μ, high | Unpublished data |
| pRSYDJ1 | GPD | CEN, low | 17 |
| pLH101 | GPD | 2μ, high | Unpublished data |
| pTGpd/P82 | GPD | CEN, low | 18 |
| p2HG(104) | GPD | 2μ, high | 19 |
Yeast Strains, Transformation, and Cultivation.
In this study we used five isogenic series of yeast strains, in the backgrounds: W303 (MATa can1–100 ade2–1 his3–11, 15 trp1–1 ura3–1 leu2–3, 112), YPH499 (MATa ade2–101ochre his3-Δ200 leu2-Δ1 lys2–801amber trp-Δ63 ura3–52), MHY810 (MATa his3-Δ200 leu2-Δ1 lys1–1 met14 ura3-Δ1∷TRP1 trp1-Δ1), MHY501 (MATa his3-Δ200 leu2–3, 112 ura3–52 lys2–801 trp1–1) and MHY803 [MHY501 derivative: MATa his3-Δ200 leu2–3, 112 ura3–52 lys2–801 trp1–1 (doa3∷HIS3+) (Ycplac22-Doa3-His6)]. The MHY strains were kind gifts from Mark Hochstrasser (Univ. of Chicago). Yeast strains used are listed in Table 2.
Table 2.
Aggregation of mutant huntingtin in different yeast strains
| 2μ plasmids Aggregation of
|
CEN plasmids Aggregation of
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Q25 | Q47 | Q72 | Q103 | Q25 | Q47 | Q72 | Q103 | ||
| Strain | |||||||||
| MHY810 | wild-type | − | −/+ | + | ++ | ||||
| MHY898 | sen3-1 | − | −/+ | + | ++ | − | + | ++ | |
| MHY803 | wild-type | − | −/+ | + | ++ | ||||
| MHY792 | doa3-1 | − | −/+ | + | ++ | − | + | ++ | |
| MHY501 | wild-type | − | −/+ | + | ++ | ||||
| MHY1408 | uba1 | − | −/+ | + | ++ | ||||
| YPH499 | wild-type | − | −/+ | + | ++ | ||||
| DYJ1 | Δydj1 | − | −/+ | + | ++ | ||||
| W303 | wild-type | − | −/+ | + | ++ | − | −/+ | + | ++ |
| LP6-2 | Δhsp26 | − | −/+ | + | ++ | ||||
| LP8-1 | Δhsp35 | − | −/+ | + | ++ | ||||
| SL314-A1 | Δssa1ssa2 | − | −/+ | + | ++ | − | + | ++ | |
| SL318-2A | Δssa3ssa4 | − | −/+ | + | ++ | ||||
| CLD82a | Δhsc82 | − | −/+ | + | ++ | − | + | ++ | |
| iLEP1α | Δhsc82 | − | −/+ | + | ++ | ||||
| SL304A | Δhsp104 | − | − | − | − | − | − | − | − |
| Overexpression | |||||||||
| Wild-type | YDJ1 | − | + | ++ | |||||
| Wild-type | SIS1 | − | + | ++ | |||||
| Wild-type | SSA1 | − | + | + | |||||
| Wild-type | HSP82 | − | + | ++ | |||||
| Wild-type | HSP104 | − | + | + | |||||
| Δhsp104 | YDJ1 | − | − | − | |||||
| Δhsp104 | SIS1 | − | − | − | |||||
Empty space, no transformation made; −, no foci fluorescence; −/+, a minority of cells have one small focus; +, one or more foci, with considerable background fluorescence; ++, one or two intense fluorescence foci, with lower background fluorescence.
Transformation of yeast was performed using a standard lithium/polyethylene glycol method (20).
Yeast cells were grown in rich media (YPD) or in minimal glucose/raffinose/galactose medium (21) deficient for the required amino acids for plasmid selection. For experimental purposes, cells were grown overnight at 25°C into logarithmic, late logarithmic, or early stationary phase.
Sedimentation Analysis.
Yeast cells were harvested by centrifugation at 1,500 × g for 5 min at room temperature and washed once in 10 mM EDTA. Cells were resuspended in spheroplasting buffer [1 M sorbitol/0.1 M EDTA/0.5 mg/ml zymolyase 100T (Seikagaku Corporation)/50 mM DTT, pH 7.5] and incubated for 2 hr at 30°C. Afterward, spheroplasts were harvested by mild centrifugation at 325 × g for 5 min at 4°C and lysed in 1× TNE (50 mM Tris, pH 7.5/150 mM NaCl/2 mM EDTA) containing a protease-inhibitor mixture (complete Mini-tablets, Boehringer Mannheim). After incubation in 1× TNE + 2% Sarkosyl for 5 min on ice, samples were loaded onto a 5% (wt/vol) sucrose cushion (1 M sucrose/100 mM NaCl/0.5% sulfobetaine), and centrifugation was performed at 315,000 × g for 1 hr at 4°C. Afterward, supernatant and pellet fractions were subjected to SDS/8% PAGE (Novex) and transferred to a poly(vinylidene difluoride) membrane (Millipore). Membranes were blocked with 5% nonfat dehydrated milk powder in PBS for 1 hr. Incubation with the primary antibody was performed overnight at 4°C. After incubation with protein A-peroxidase (1:5,000, Boehringer Mannheim), the immune complexes were visualized by treating membranes with ECL reagent (Amersham Pharmacia). Antibody α-GFP was used at 1:100 (Clontech).
Microscopy.
Yeast cells were allowed to adhere onto polylysine-treated slides for 10 min. For nucleus staining, cells were fixed with 1% formaldehyde for 5 min and washed 3 times with PBS. After treatment with 4′,6-diamidine-2-phenylindole·dihydrochloride (DAPI, Sigma) for 5 min, cells were washed 3 times with PBS. Microscopy was performed with an Axioplan 2 microscope (Zeiss), and micrographs were taken at a magnification of ×100.
Results
Coalescence of Mutant Ht in Yeast.
To investigate Ht in yeast, the N-terminal region (amino acids 1–68 of the wild-type protein) with a wild-type polyQ repeat length of 25 residues or with a mutant repeat length of 47, 72, or 103 residues was fused to GFP. Each was placed under the control of GPD, a strong constitutive yeast promoter, on a single-copy plasmid (Fig. 1, Top Left). Homopolymeric tracts of CAG, the naturally occurring glutamine codon in Ht, are inherently unstable, and particularly so in yeast (22, 23). To reduce this problem, we took advantage of the facts that glutamine is encoded by both CAG and CAA and that mixed-codon repeats are considerably more stable (24). To minimize instability problems, all experiments reported herein were performed with mixed-codon polyQ repeats and all work was performed with fresh transformants, using at least two independent colonies in each case, and repeated at least two times.
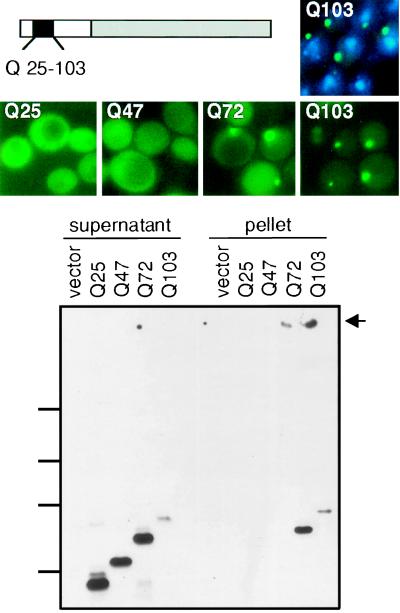
Figure 1.
Expression of Ht fragments in yeast. (Top) Left, Schematic representation of Ht–GFP fragments used in this study. Gray box, GFP; white boxes, amino acids 1–68 of the N-terminal region of human Ht protein containing a stretch of 25, 47, 72, or 103 glutamines (black box). (Middle) Visualization of Ht aggregates with GFP. For wild-type cells expressing HtQ103, costaining with DAPI is also shown (Top Right). (Bottom) Sedimentation analysis. Spheroplasts were lysed in 1× TNE and Sarkosyl was added to a final concentration of 2%. After centrifugation, supernatant and pellet fractions were analyzed by SDS/8% PAGE and immunoblotting with antibodies against GFP. Equal loading was demonstrated by Coomassie staining (data not shown). Marker protein positions are indicated on the left (212 kDa, 122 kDa, 83 kDa, 51.8 kDa). Arrow marks the top of the gel.
Fluorescence from GFP-fusion proteins containing wild-type polyQ tracts (25 residues; HtQ25) was always distributed diffusely throughout the cell (Fig. 1, Middle). HtQ47 fluorescence was also diffusely distributed, although coalescent foci were observed in a small percentage of cells (<2%). More than half of cells expressing HtQ72 exhibited a single intense spot of fluorescence against a diffuse fluorescent background (Fig. 1, Middle). Virtually all cells expressing HtQ103 exhibited a single intense spot of fluorescence, with much less background fluorescence than seen with other variants (Fig. 1, Middle). When the same constructs were expressed from high-copy plasmids (p426 series, Table 1), fluorescence intensity was much greater, but the pattern of fluorescence was very similar (data not shown). Immunoblotting of total cellular protein indicated that all four variants were expressed at similar levels (data not shown). Thus, the degree of coalescence exhibited by the N-terminal fragment of Ht depends more on the length of the polyQ tract than the level of protein expressed.
Newly Induced Mutant Ht Can Aggregate in Any Cell.
Cells expressing different Q repeat variants exhibited the same frequency of plasmid loss (determined by plating cells to nonselective and selective media) and grew at similar rates, with only a slight deficit in cells expressing HtQ103. Final densities were typically 0.7–0.8 for cells expressing HtQ25, HtQ47, or HtQ72 and 0.5–0.7 for cells expressing HtQ103. Thus, the long polyQ Ht fragments were not overtly toxic in yeast. However, because the proteins were expressed from a constitutive promoter, it was possible that a subset of cells competent to grow in the presence of polyQ proteins had been selected during transformation. If so, selection might also have influenced the aggregation state of the proteins. To determine whether the coalescence of expanded glutamine reflected an inherent property of the protein or was the result of a selective process, the Ht–GFP constructs were transferred to the control of a galactose-inducible promoter (Table 1). Transformants were selected on glucose plates to keep the construct tightly repressed. To initiate induction, cells were first grown in raffinose medium overnight, to eliminate glucose repression, and then transferred to galactose medium, to induce Ht expression.
Bright GFP fluorescence was observed after 4 hr, but for all three variants tested, HtQ25, HtQ72, and HtQ103, fluorescence was diffusely distributed (data not shown). With continued expression HtQ72 and HtQ103 coalescence began to appear in some cells after 9 hr (2 doublings). After 24 hr, coalescence was indistinguishable from that observed in cultures expressing the Ht variants constitutively and all cultures had reached similar densities (data not shown). Thus, coalescence of expanded glutamine repeats occurs in most, if not all, cells in the culture, but many hours of expression are required for it to occur.
Mutant Ht Forms Cytoplasmic Aggregates in Yeast.
Costaining cells with DAPI, a DNA-binding dye that fluoresces blue, demonstrated that foci of Ht coalesence were in the cytoplasmic compartment not in the nucleus (Fig. 1, Top Right). To determine whether these foci reflected the sequestration of Ht–GFP fusions into a membrane-bounded compartment or the formation of higher order protein complexes, cell walls were removed and cells were lysed in the presence of the detergent Sarkosyl (2%). After sedimentation, supernatant and pellet fractions were boiled in sample buffer containing 5% SDS for 10 minutes and analyzed by immunoblotting (Fig. 1, Bottom).
HtQ25 and HtQ47 were detected only in supernatant fractions. HtQ72 was distributed between supernatant and pellet fractions, whereas virtually all HtQ103 protein was found in the pellet fraction. Note that after electrophoresis, a major fraction of HtQ103 remained at the top of the gel. Apparently, the coalescence detected through GFP fluorescence was due to the formation of higher order complexes. For HtQ103, and less so for HtQ72, these complexes resisted solubilization by boiling in 5% SDS.
Aggregates Are Unaltered in Proteasome-Deficient Cells.
Because aggregates of Ht (25) and other glutamine-repeat proteins associated with disease (26–28) are ubiquitinated in mammalian cells and are associated with components of the proteasome, it has been suggested that the ubiquitin/proteasome pathway might be involved in aggregate formation. We were unable to detect ubiquitinated Ht protein in yeast cells (data not shown). However, even for proteins known to be degraded by this pathway, ubiquitin conjugates can be difficult to detect. To investigate this question more rigorously, we took a genetic approach. We used three strains, each containing a lesion in a different component of the ubiquitin/proteasome degradation pathway: (i) uba1, the ubiquitin activating enzyme (M. Hochstrasser, personal communication), (ii) doa3, a catalytic subunit of the 20S proteasome (29), and (iii) sen3, a subunit of the 19S proteasome regulatory complex (30). Because each of these genes is essential, we used partial loss-of-function mutations, which severely impair this pathway. In each of the strains, the Ht variants behaved in the same manner as they did in wild-type cells. There were no changes in the number of cells containing coalescent foci, nor in the size or intracellular distribution of those foci (Table 2, and data not shown).
Molecular Chaperones Affect Aggregation of Ht.
Chaperone proteins are a highly conserved but diverse group of proteins that control the folding of other proteins by interacting with different types of folding intermediates and off-pathway folding products (31). They have profound effects on the aggregation of abnormal proteins. To determine how changes in the levels of chaperone proteins would affect the coalescence of the Ht polyQ variants, we generated an isogenic series of strains containing deletion mutations or overexpression plasmids for various chaperone proteins that produced wild-type or polyQ expanded Ht fragments. Note that some chaperone deletions could not be tested because they are lethal.
Most of the tested alterations in chaperone proteins had no noticeable effects on the intracellular distribution of Ht variants as determined by GFP fluorescence (Table 2) and no significant effect on the manner in which the Ht fragments were partitioned between the supernatant and pellet fractions after sedimentation (data not shown). This category included mutations that (i) eliminated the expression of the major small Hsp (in yeast, Hsp26) (32), (ii) increased the expression of Hsp90 (in yeast, Hsc/p82) several fold (33) or reduced the expression of Hsp90 by 10- to 15-fold (34), (iii) eliminated the expression of various members of the essential cytosolic Hsp70 family [constitutive members Ssa1 and Ssa2 (35) and stress inducible members Ssa3 and Ssa4 (J. L. Vogel and S.L., unpublished data)], and (iv) increased or eliminated expression of Ydj1 (a member of the Hsp40 family) (17). We also examined a deletion of Hsp35. This heat-inducible protein is a member of the glyceraldehyde-3-phosphate dehydrogenase family and is postulated to be a chaperone because it is both heat inducible and one of the most abundant proteins in yeast (36). Mammalian glyceraldehyde-3-phosphate dehydrogenase exhibits a glutamine length-dependent association with Ht (37).
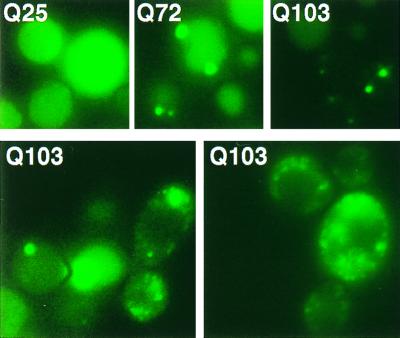
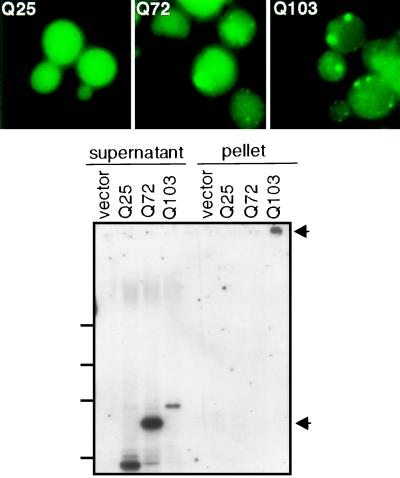
Overexpression of three chaperones had significant effects. Sis1, a member of the Hsp40 family, caused two intense foci of aggregation to appear in most cells with HtQ72 and HtQ103, rather than the single focus of coalescence observed in virtually all wild-type cells (Fig. 2, Upper). In cells overexpressing Hsp70 (Ssa1), HtQ72 and HtQ103 fluorescence was much more variable than in wild-type cells. Multiple foci of fluorescence were observed in many cells, and many also contained a higher background of diffuse fluorescence (Fig. 2, Lower). This variability likely reflects differences in plasmid copy number, which is commonly observed with Hsp70 expression plasmids (38). Overexpression of Hsp104 also increased the number of fluorescent foci and the background fluorescence observed with the Ht variants HtQ72 and HtQ103 (Fig. 3, Upper; Table 2). It also increased the relative quantities of HtQ72 and HtQ103 found in the supernatant fractions after centrifugation (compare Fig. 1, Bottom and Fig. 3, Lower). Curiously, although HtQ72 protein appeared at least partially aggregated in these cells, little protein fractionated in the pellet in three of three experiments. The protein may be more loosely packed or in a Sarkosyl-soluble state.
Figure 2.
Overexpression of Sis1 and Hsp70 affect Ht aggregation. (Upper) Wild-type cells expressing Ht–GFP fusion proteins with 25, 72, or 103 Q residues were transformed with pTVSIS I to overexpress Sis1. (Lower) Wild-type cells containing expression-plasmid p416/PQ103 were transformed with plasmid pLH101 to generate Hsp70 overexpression. Two pictures of cells expressing HtQ103 are shown for adequate representation. Focal planes that are best for visualizing large aggregates often leave multiple small aggregates out of focus.
Figure 3.
Overexpression of Hsp104 reduces Ht aggregation. (Upper) Visualization of Ht–GFP fusion proteins. Yeast wild-type cells producing Ht variants were transformed with plasmid p2HG(104) to overexpress Hsp104. (Lower) Sedimentation analysis was performed as for Fig. 1. Upper arrow marks the top of the gel, lower arrow indicates migration position of SDS-soluble HtQ72.
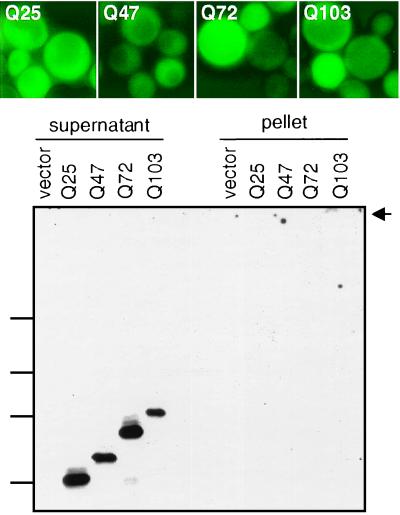
Of all the chaperone alterations tested, a deletion of the HSP104 gene had the most dramatic effect. In these strains, all of the Ht variant fragments exhibited diffuse fluorescence (Fig. 4, Upper). The same results were obtained with both the high and low copy Ht expression constructs (Table 2). Moreover, by sedimentation, all of the proteins were only detected in supernatant fractions (Fig. 4, Lower).
Figure 4.
The loss of Hsp104 affects Ht aggregation. (Upper) Visualization of GFP fusion proteins in a Δhsp104 strain. (Lower) Sedimentation analysis was performed as for Fig. 1.
Discussion
Our work establishes the genetically tractable and readily manipulated yeast Saccharomyces cerevisiae as a model system for investigating the misfolding of a critical, polyQ-containing fragment of Ht, the protein that is responsible for HD. The number of progressive neurodegenerative diseases determined to be caused by the expansion of CAG (polyQ) codons has increased dramatically in recent years, and likely will continue to do so (3). There are currently no effective therapies for these devastating illnesses, and a better understanding of the pathogenic processes underlying them is urgently needed. Overwhelming evidence indicates that the misfolding of proteins that contain polyQ expansions, but are otherwise unrelated, is a central element in the pathology of each disease. The polyQ expansion is the common determinative factor in their misfolding (39). By extension, then, our studies in yeast hold promise for investigating the mechanisms underlying all of these pathologies.
In these disorders, disease is generally accompanied by aggregation of the polyQ protein (or a fragment thereof) in the nucleus and/or cytoplasm of affected neurons (2, 12, 40). However, aggregation and pathogenicity are each closely linked to the length of the polyQ expansion. In every case (except that of spinocerebellar ataxia type 6), polyQ expansions of up to ≈35 residues are benign, whereas expansions of >40 residues are pathogenic, with earlier disease onsets associated with longer repeats (12, 41). We have found a similar length dependence for polyQ misfolding and aggregation when Ht fragments are expressed in yeast. Fragments with 25 Q residues are fully soluble. Fragments with 47, 72, and 103 Qs show progressively greater tendencies to aggregate.
Taking advantage of genetic manipulations in yeast, we examined the effects of two major cellular mechanisms for protein homeostasis on polyQ-dependent aggregation. First, we examined the role of the ubiquitin/proteasome pathway. Aggregates of several polyQ disease proteins in human neurons are ubiquitinated and contain proteasome components (26–28, 40). We used three different mutant yeast strains that affect the ubiquitin/proteasome pathway at different steps. Each mutant has a severe effect on the ubiquitin/proteasome pathway in yeast. None had a noticeable effect on the aggregation state of Ht fragments. This observation could suggest that the ubiquitination and proteasome associations of polyQ proteins in mammalian neurons are not directly germane to their aggregation or solubilization. More likely, they are the remnants of futile attempts to degrade the proteins.
Second, we used a set of isogenic strains with a wide variety of HSP deletion mutations and overexpression plasmids to investigate the influence of protein chaperones on the aggregation of Ht. Deletion or overexpression of most chaperones, including Ydj1 (the yeast Hsp40 homologue of human HDJ2) (42) had little effect on the distribution of Ht fragments containing polyQ expansions. We were unable to test a deletion of Sis1 (the yeast Hsp40 homologue of human HDJ1) (42) because it is lethal. However, overexpression of Sis1 altered the distribution of HtQ72 and HtQ103 in yeast. In mammalian cells, both members of the Hsp40 family are associated with aggregates of glutamine-expansion proteins, and their overexpression suppresses polyQ aggregation (26, 28). Interestingly, in the only study that directly compared the effects of HDJ1 and HDJ2, HDJ1 (the Sis1 homologue) had a much stronger effect than HDJ2 (the Ydj1 homologue) (43). Overexpression of Hsp70 in yeast had a stronger effect on HtQ72 and HtQ103 localization than any of the Hsp40 manipulations we tested. Hsp70 localizes to the aggregates of polyQ expansion mutants in mammalian cells, but the effects of its overexpression on aggregation have not yet been reported. However, overexpression of Hsp70 has just been reported to reduce toxicity of an ataxin-3 mutant in transgenic Drosophila (44). Overexpression of Hsp104 also reduced the aggregation of Ht fragments containing the longer polyQ repeats in yeast. Similar results were obtained when yeast Hsp104 was coexpressed with polyQ–GFP fusion proteins in nematodes (S. Satyal and R. Morimoto, personal communication). Although this work is still in a preliminary stage, it begins to suggest that the effects of chaperones on polyQ mutant proteins may be universal.
Of all of the chaperone alterations we tested, by far the most dramatic effect was observed with a deletion of Hsp104. Eliminating Hsp104 expression virtually eliminated the aggregation of Ht fragments, even of those containing 103 Q residues. In previous in vitro studies, purified N-terminal Ht fragments with polyQ tracts of 20 or 30 residues remained soluble, whereas those with 51 or more residues aggregated (45). The effect of the Hsp104 deletion on Ht fragments containing polyQ, however, demonstrates that no matter what their intrinsic tendency to aggregate may be, their fate in the context of a living cell is completely subject to the protein folding milieu.
Remarkably, Hsp104 has unexpectedly similar effects on the propagation of a protein-based genetic element (prion) in yeast. This element, [PSI+], propagates through self-perpetuating coalescence of the glutamine-rich domain of its prion determinant, Sup35. Both HSP104 deletion and overexpression plasmids cure cells of the [PSI+] element and return Sup35 to the soluble state (46, 47). These highly unusual patterns of genetic interaction with Hsp104 suggest that a common mechanism controls the aggregation state of Ht and Sup35. Two models have been proposed for [PSI+] inheritance (47, 48), which differ chiefly in their explanation for the effects of HSP104 deletions. The effects of Hsp104 deletions on Ht fragments support one of these: Hsp104 is required to place the glutamine-rich, prion domain of Sup35 into an aggregation competent state and, therefore, for prion formation. They further suggest that glutamine residues can be a determining factor in Hsp104 interactions, either through direct recognition of glutamines or the altered structures they impose. Paralogs of Hsp104 exist in mammalian cells, but an orthologous protein has not yet been identified. It seems likely, that mammalian cells contain proteins with similar functions and, if so, polyQ proteins may provide ideal substrates for their identification.
The polyQ fragments of Ht exhibited minimal toxicity in yeast, whether they were present in the aggregated or the soluble state. Even the longest expansion (Q103) caused only a slight reduction in final culture density. In humans, Ht proteins with the longest repeat expansions not only produce disease earlier, but also expand the cell-type distribution of toxicity (9). Engineering longer polyQ expansions in Ht, with perhaps the addition of a nuclear targeting signal, might produce a toxicity in yeast that could be used to study generalized features of the pathogenic mechanism.
Lack of toxicity for the aggregated and soluble Ht fragments in yeast is advantageous for three reasons. First, it provides an opportunity to study natural cellular factors that control the fate of misfolded polyQ proteins and to search for potential therapeutic agents that affect misfolding, aggregation, and degradation, without the complication of deciphering the contributions of toxicity to the outcome of the assay. We have developed two methods for screening for such factors: visual assays of GFP fluorescence patterns and sedimentation assays. Based on assays we have established for the prion-determining domain of Sup35 (49), fusing the polyQ Ht fragment to a reporter protein whose function depends on solubility should provide an easy, high-throughput genetic screen for aggregation.
Second, polyQ-expanded proteins may perturb the distribution of other cellular proteins by providing novel interaction surfaces (37, 50). The nature of these interactions is most readily and rapidly tested in an environment where the aggregation state of the polyQ protein can readily be manipulated and in which neither the aggregated nor the soluble form is toxic.
Finally, perhaps the greatest conundrum presented by the polyQ diseases is that most of the proteins are ubiquitously expressed, yet each manifests its toxicity in a unique and distinct set of neurons. Yeast screens with different polyQ proteins may provide an opportunity to identify the cell type-specific factors that contribute to the unique spectrum of toxicities and subsequently to search for factors that ameliorate their effects.
Acknowledgments
We are grateful to G. Lawless for providing the Ht–GFP constructs and to M. Hochstrasser and R. Swanson for providing yeast strains. We thank members of the Lindquist lab for helpful comments. S.K. was supported by a Deutsche Forschungsgemeinschaft postdoctoral fellowship.
Abbreviations
- GFP
green fluorescent protein
- Ht
huntingtin
- HD
Huntington's disease
- polyQ
polyglutamine
- Hsp
heat shock protein
References
- 1.Reddy P S, Housman D E. Curr Opin Cell Biol. 1997;9:364–372. doi: 10.1016/s0955-0674(97)80009-9. [DOI] [PubMed] [Google Scholar]
- 2.Ross C A. Neuron. 1997;19:1147–1150. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 3.Davies S W, Beardsall K, Turmaine M, DiFiglia M, Aronin N, Bates G P. Lancet. 1998;351:131–133. doi: 10.1016/S0140-6736(97)08360-8. [DOI] [PubMed] [Google Scholar]
- 4.Martin J B, Gusella J F. N Engl J Med. 1986;315:1267–1276. doi: 10.1056/NEJM198611133152006. [DOI] [PubMed] [Google Scholar]
- 5.Vonsattel J P, Myers R H, Stevens T J, Ferrante R J, Bird E D, Richardson E P., Jr J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 6.The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 7.Gusella J F, Persichetti F, MacDonald M E. Mol Med. 1997;3:238–246. [PMC free article] [PubMed] [Google Scholar]
- 8.Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, McCutcheon K, Singaraja R, Kazemi-Esfarjani P, Devon R, Kim S U, Bredesen D E, Tufaro F, Hayden M R. Nat Genet. 1998;18:150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- 9.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 10.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 11.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies S W, Bates G P. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 12.Reddy P H, Williams M, Tagle D A. Trends Neurosci. 1999;22:248–255. doi: 10.1016/s0166-2236(99)01415-0. [DOI] [PubMed] [Google Scholar]
- 13.Boutell J M, Wood J D, Harper P S, Jones A L. Hum Mol Genet. 1998;7:371–378. doi: 10.1093/hmg/7.3.371. [DOI] [PubMed] [Google Scholar]
- 14.Bao J, Sharp A H, Wagster M V, Becher M, Schilling G, Ross C A, Dawson V L, Dawson T M. Proc Natl Acad Sci USA. 1996;93:5037–5042. doi: 10.1073/pnas.93.10.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mumberg D, Muller R, Funk M. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 17.Kimura Y, Yahara I, Lindquist S. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 18.Nathan D F, Lindquist S. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel J L, Parsell D A, Lindquist S. Curr Biol. 1995;5:306–317. doi: 10.1016/s0960-9822(95)00061-3. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams A, Gottschling D E, Kaiser C, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 22.Moore H, Greenwell P W, Liu C P, Arnheim N, Petes T D. Proc Natl Acad Sci USA. 1999;96:1504–1509. doi: 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweitzer J K, Livingston D M. Hum Mol Genet. 1997;6:349–355. doi: 10.1093/hmg/6.3.349. [DOI] [PubMed] [Google Scholar]
- 24.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 26.Stenoien D L, Cummings C J, Adams H P, Mancini M G, Patel K, DeMartino G N, Marcelli M, Weigel N L, Mancini M A. Hum Mol Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 27.Chai Y, Koppenhafer S L, Shoesmith S J, Perez M K, Paulson H L. Hum Mol Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 28.Cummings C J, Mancini M A, Antalffy B, DeFranco D B, Orr H T, Zoghbi H Y. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 29.Chen P, Hochstrasser M. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeMarini D J, Papa F R, Swaminathan S, Ursic D, Rasmussen T P, Culbertson M R, Hochstrasser M. Mol Cell Biol. 1995;15:6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gething M-J. Guidebook to Molecular Chaperones and Protein Folding Catalysts. Oford, U.K.: Oxford Univ. Press; 1997. [Google Scholar]
- 32.Petko L, Lindquist S. Cell. 1986;45:885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- 33.Borkovich K A, Farrelly F W, Finkelstein D B, Taulien J, Lindquist S. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathan D F, Vos M H, Lindquist S. Proc Natl Acad Sci USA. 1999;96:1409–1414. doi: 10.1073/pnas.96.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 36.Boucherie H, Bataille N, Fitch I T, Perrot M, Tuite M F. FEMS Microbiol Lett. 1995;125:127–133. doi: 10.1111/j.1574-6968.1995.tb07348.x. [DOI] [PubMed] [Google Scholar]
- 37.Burke J R, Enghild J J, Martin M E, Jou Y S, Myers R M, Roses A D, Vance J M, Strittmatter W J. Nat Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- 38.Stone D E, Craig E A. Mol Cell Biol. 1990;10:1622–1632. doi: 10.1128/mcb.10.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ordway J M, Tallaksen-Greene S, Gutekunst C A, Bernstein E M, Cearley J A, Wiener H W, Dure L S t, Lindsey R, Hersch S M, Jope R S, et al. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 40.Zoghbi H Y, Orr H T. Curr Opin Neurobiol. 1999;9:566–570. doi: 10.1016/S0959-4388(99)00013-6. [DOI] [PubMed] [Google Scholar]
- 41.Perutz M F. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 42.Cheetham M E, Caplan A J. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chai Y, Koppenhafer S L, Bonini N M, Paulson H L. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 45.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 46.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 47.Patino M M, Liu J J, Glover J R, Lindquist S. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 48.Kushnirov V V, Ter-Avanesyan M D. Cell. 1998;94:13–16. doi: 10.1016/s0092-8674(00)81216-7. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Lindquist S. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 50.Li S H, Hosseini S H, Gutekunst C A, Hersch S M, Ferrante R J, Li X J. J Biol Chem. 1998;273:19220–19227. doi: 10.1074/jbc.273.30.19220. [DOI] [PubMed] [Google Scholar]