Abstract
LT-IIb-B5, a doughnut-shaped oligomeric protein from enterotoxigenic Escherichia coli, is known to activate the TLR2/TLR1 heterodimer (TLR2/1). We investigated the molecular basis of the LT-IIb-B5 interaction with TLR2/1 in order to define the structure-function relationship of LT-IIb-B5 and, moreover, to gain an insight into how TLR2/1 recognizes large, non-acylated protein ligands that cannot fit within its lipid-binding pockets, as previously shown for the Pam3CSK4 lipopeptide. We first identified four critical residues in the upper region of the LT-IIb-B5 pore: Corresponding point mutants (M69E, A70D, L73E, S74D) were defective in binding TLR2 or TLR1 and could not activate antigen-presenting cells, despite retaining full ganglioside-binding capacity. Point mutations in the TLR2/1 dimer interface, as determined in the crystallographic structure of the TLR2/1-Pam3CSK4 complex, resulted in diminished activation by both Pam3CSK4 and LT-IIb-B5. Docking analysis of the LT-IIb-B5 interaction with this apparently “predominant” activation conformation of TLR2/1 revealed that LT-IIb-B5 may primarily contact the convex surface of the TLR2 central domain. Although the TLR1/LT-IIb-B5 interface is relatively smaller, the leucine-rich repeat motifs 9–12 in the central domain of TLR1 were found to be critical for cooperative TLR2-induced cell activation by LT-IIb-B5. Moreover, the putative LT-IIb-B5 binding site overlaps partially with that of Pam3CSK4; consistent with this, Pam3CSK4 suppressed TLR2 binding of LT-IIb-B5, albeit not as potently as self-competitive inhibition. In conclusion, we identified the upper pore region of LT-IIb-B5 as a TLR2/1 interactive domain, which contacts the heterodimeric receptor at a site that is distinct from, though overlaps with, that of Pam3CSK4.
Keywords: cell activation, molecular biology, cell surface molecules, dendritic cells, monocytes/macrophages
Introduction
In common to other heat-labile enterotoxins from Vibrio cholerae or Escherichia coli, the Type IIb enterotoxin of E. coli (LT-IIb) displays AB5 oligomeric structure, in which an enzymatically toxic A subunit is linked to a pentameric ganglioside-binding (B5) subunit (1, 2). The actual catalytic moiety is the A1 polypeptide, whereas the C-terminal A2 segment acts as a noncovalent linker into the central pore of the doughnut-shaped B pentamer (1). Although LT-IIb and related heat-labile enterotoxins are potent mucosal adjuvants, their intrinsic enterotoxicity precludes their use as adjuvants for human vaccines (3, 4). In our efforts to identify immunostimulatory activities mediated exclusively by the non-catalytic B pentameric subunits, we discovered that TLR2 is uniquely activated by the B pentamers of Type II but not Type I enterotoxins such as the cholera toxin (5). Subsequent work focusing on the 60-kDa B pentamer of LT-IIb (designated LT-IIb-B5) revealed that its interaction with and activation of TLR2 in membrane lipid rafts is facilitated by binding to the GD1a ganglioside (6). Strikingly, the LT-IIb holotoxin does not bind or activate TLR2; this is attributable to the presence of the A subunit which, in either wild-type or catalytically inactive version, appears to sterically interfere with TLR2 binding (7). We moreover found that TLR1 serves as a signaling partner of TLR2 in response to LT-IIb-B5 (6, 7).
The TLR family of pattern-recognition receptors fulfill their central role as sensors of infection by recognizing multiple microbial ligands (8, 9), and this is particularly true for TLR2 which, in cooperation with TLR1 or TLR6, interacts with a variety of structurally diverse molecules (10–12). The triacylated lipopeptide Pam3CSK4 is a prototypical TLR2 ligand which interacts with a hydrophobic pocket in the convex region of TLR2, at the border of its central and C-terminal domains (13). The TLR2 pocket accommodates two of the acyl chains of the lipopeptide, whereas the third acyl chain is inserted into a similar hydrophobic channel in the TLR1 component of the TLR2/1 heterodimer (13). In addition to microbial lipopeptides/lipoproteins, TLR2 interacts with several microbial proteins that do not bear acyl chains (14–19) and are unlikely to be accommodated within the TLR2/1 hydrophobic pockets due to size constraints. The issue of how microbial protein ligands interact with TLR2 or other TLRs was raised in a recent review (20) and, in the absence of specific crystallographic data, has remained a matter of conjecture. The objective of this study was to define the molecular basis of the LT-IIb-B5 interaction with the TLR2/1 heterodimer, thereby offering an insight into how non-lipidated ligands may interact with TLR2/1.
Although TLR2/1 employs hydrophobic interactions for ligand binding (13, 21, 22), LT-IIb-B5 can participate in both hydrophilic and hydrophobic interactions mediated by distinct regions of the molecule (1). The so-called “lower region” interacts hydrophilicly with the oligosaccharide moiety of GD1a, whereas the “upper region” of the B pentamer pore contains a large hydrophobic surface (494-Å2) which engages in hydrophobic interactions with the A2 segment of the A subunit (1). In the absence of the A subunit, however, the upper region hydrophobic surface becomes solvent accessible (1) and thus potentially receptive to additional interactions. Since lack of the A subunit is a strict requirement for TLR2/1 activation (6), we reasoned that the solvent-accessible hydrophobic upper region of LT-IIb-B5 presents a potential TLR2/1-interactive site. This region comprises a ring of hydrophobic residues, namely M69, A70, L73, and the Cβ of S74 (the Oγ atom of S74 is involved in a hydrogen bond in a way that leaves the hydrophobic Cβ atom of S74 pointing towards the interior of the pore), of each of the five B subunits (1).
We have, therefore, created point substitution mutations rendering these residues hydrophilic (M69E, A70D, L73E, and S74D), under the hypothesis that the TLR2/1 interaction would be abrogated. These tools were used in functional studies with wild-type or mutated TLR2/1 followed by structural docking analysis. We conclude that the upper pore of LT-IIb-B5 defines an interactive site for TLR2/1 binding and activation through contacts that may primarily involve the convex surface of the central domain of TLR2. This is the first study that describes a possible mode of TLR2/1 recognition of non-lipidated protein ligands, which cannot fit into small hydrophobic pockets and can engage in TLR interactions through specific residues.
Materials and Methods
Microbial molecules
The construction of recombinant plasmids encoding His-tagged LT-IIb holotoxin or its B pentamer (LT-IIb-B5), in wild-type or GD1a-nonbinding version (T13I), was previously described (5, 23). Single-point substitution mutants (M69E, A70D, L73E and S74D) of LT-IIb-B5 were engineered by means of site-directed mutagenesis (QuikChange® kit; Stratagene). The proteins were extracted from the periplasmic space of transformed E. coli DH5αF’Kan using polymyxin B treatment, and were purified using ammonium sulfate precipitation, followed by nickel affinity chromatography and size-exclusion chromatography using a Sephacryl-100 column and an ÄKTA-FPLC system (GE Healthcare) (5, 23). Identity and purity were confirmed by SDS-PAGE, immunoblotting with specific rabbit IgG Abs, and by quantitative Limulus amebocyte lysate assay kits (BioWhittaker or Charles River Endosafe) which determined negligible endotoxic activity (< 0.007 ng/μg protein). Further evidence against contamination with LPS or other heat-stable contaminants was obtained upon LT-IIb-B5 or holotoxin boiling, which destroys their biological activities (5, 23). Control TLR agonists, including E. coli LPS (ultrapure grade), Pam3CSK4 lipopeptide, and FSL-1 lipopeptide were purchased from InvivoGen. The reagents were used at effective concentrations determined in preliminary experiments or in previous publications.
Cell isolation and culture
All animal procedures associated with tissue harvesting were conducted in compliance with established federal guidelines and institutional policies. BMDC were generated as described by Lutz et al (24). Briefly, bone marrow cells from femurs and tibia of 8–12-week-old BALB/c mice (The Jackson Laboratory) were plated at 2×105 cells/ml and cultured at 37°C and 5% CO2 atmosphere, in complete RPMI (RPMI 1640 containing 10% heat-inactivated FBS, 2 mM L-glutamine, 10 mM HEPES, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 0.05 mM 2-ME; InVitrogen) supplemented with 20 ng/ml recombinant murine GM-CSF (Peprotech). The nonadherent cells were harvested on day 8 and were phenotypically characterized by flow cytometry (see below). Thioglycollate-elicited macrophages were isolated from the peritoneal cavity of BALB/c mice, as previously described (5) and were cultured in complete RPMI. Human monocytic THP1-Blue™ cells (InvivoGen) were maintained in complete RPMI. Cell viability was monitored using the CellTiter-Blue™ assay kit (Promega). None of the experimental treatments affected cell viability compared to medium-only control treatments.
FACS analysis and Abs
Flow cytometry was used for phenotypic characterization of the generated BMDC, as well as for determining costimulatory molecule upregulation in activated BMDC. Briefly, BMDC were analyzed using the BD FACSCalibur and the CellQuest software after staining with the following fluorescently-labeled Abs to cell surface markers or with appropriate isotype controls: CD11c (clone HL3); CD11b (M1/70); CD40 (HM40-3); CD54 (YN1/1.7.4); CD80 (16-10A1); CD86 (GL1); I-A/I-E (M5/114.15.2); Gr1 (RB6-8C5) (all reagents from eBioscience except for anti-CD11c from BD Pharmingen). The analysis showed that the harvested BMDC contained < 1% macrophages and < 5% granulocytes.
Binding assays
Binding of ligands to plate-immobilized GD1a or TLRs was assessed as we have previously described (7, 25). Briefly, 96-well microtiter wells were coated overnight at 4°C with GD1a (Matreya LLC), or with recombinant mouse or human TLR2, TLR1, or TLR6 (R&D systems). After blocking non-specific binding sites with 5% (w/v) BSA, wild-type or mutant LT-IIb-B5 was incubated in PBS containing 10 mg/ml BSA. Bound protein was detected colorimetrically using rabbit IgG anti-LT-IIb Ab followed by peroxidase-conjugated goat anti-rabbit IgG (adsorbed against human or mouse IgG). In competitive inhibition assays, LT-IIb-B5 was used in biotinylated form and bound protein was probed with peroxidase-conjugated streptavidin.
Cellular activation assays
Cytokine induction in stimulated macrophage or BMDC culture supernatants was measured by ELISA, using kits from eBioscience (6, 7). THP1-Blue™ cells, stably transfected with NF-κB-inducible reporter system, were used for colorimetric determination of NF-κB activation. This involved measuring the activity of NF-κB-inducible alkaline phosphatase secreted in stimulated culture supernatants, using a Synergy HT multi-mode microplate reader (Bio-Tek) (26). In certain experiments, the cells were pretreated for 30 min with blocking Abs to TLR2 (clone TL2.1), TLR1 (polyclonal), TLR6 (polyclonal) or TLR4 (clone HTA125) (InvivoGen). The mAbs were used at 0.25 μg/ml and the polyclonal Abs at 1 μg/ml.
TLR1 mutants, TLR1/6 chimeras, and reporter gene assays
A common variant of human TLR1 represented by the NCBI accession no. AAI09095 is referred to as “wild-type” TLR1 in this paper and was used as the basis for generating various mutants and variants. TLR1 point mutants (F314D, Q316K, Y320N, E321V, I328N, R337G, M338W, V339S, H340G) were generated by random mutagenesis using error-prone PCR (27) followed by sequencing. A polymorphic variant (P315L) of TLR1 was generated as previously described (28) based on the technique of overlap extension PCR (29). The mutants/variants were generated as N-terminal FLAG-tagged constructs within pFLAG-CMV (Sigma-Aldrich) and were verified by sequencing. Wild-type or genetically altered TLR1 or TLR6, as well as previously constructed and characterized TLR1/6 chimeric receptors that display normal levels of expression (10), were used as TLR2 signaling partners in reporter assays of inducible luciferase activity. Briefly, SW620 cells (which are deficient in TLR-1, -2, and -6 expression (10)) were cotransfected with various combinations of wild-type, point mutants, or chimeric TLRs, along with a firefly luciferase reporter gene and a Renilla luciferase transfection control, as previously described (10). Two days post-transfection, the cells were stimulated for 6h and the Renilla and firefly luciferase activities were measured in cell lysates using the Promega luciferase reporter assay system. Luciferase activity was calculated as a ratio of firefly luciferase activity to Renilla luciferase activity, to correct for transfection efficiency.
Protein-protein docking analysis
The crystal structures of human TLR2/TLR1-lipopeptide complex [PDB 2z7x] and of LT-IIb [PDB 1QB5] were previously determined by Jin et al., at 2.1 Å resolution (13), and by Van Den Akker et al., at 1.9 Å resolution (1), respectively. The TLR2/TLR1 and LT-IIb-B5 structures were submitted as receptor and ligand, respectively, to GRAMM-X (http://vakser.bioinformatics.ku.edu/resources/gramm/grammx) (30), ZDOCK (http://zdock.bu.edu/db_insert.php) (31) and ClusPro (http://nrc.bu.edu/cluster) (32) protein-protein docking servers for complex prediction. The analysis was carried out using default parameter, except that the DOT docking program was selected in the ClusPro server. The Firedock Server (33) (http://bioinfo3d.cs.tau.ac.il/FireDock/index.html) was used for further refinement and scoring for global energy values and the model with the lowest energy was selected for further analysis. The LigPlot software (34) (http://www.csb.yale.edu/userguides/graphics/ligplot/ligplot_descrip.html) was used for plotting predicted intermolecular interactions, and the Crystallography & NMR System (CNS) program (35) (http://cns.csb.yale.edu/v1.1) was used for buried surface calculation. Chains D-H of the original LT-IIb-B5 structure file in PDB were renamed B1-5, respectively, for convenience and clarity.
Statistical analysis
Data were evaluated by ANOVA and the Dunnett multiple-comparison test using the InStat program (GraphPad Software). Where appropriate (comparison of two groups only), unpaired two-tailed t tests were performed. The level of significance was set at 0.05. All experiments were performed at least twice for verification.
Results
Critical residues in the “upper region” of LT-IIb-B5 involved in TLR binding
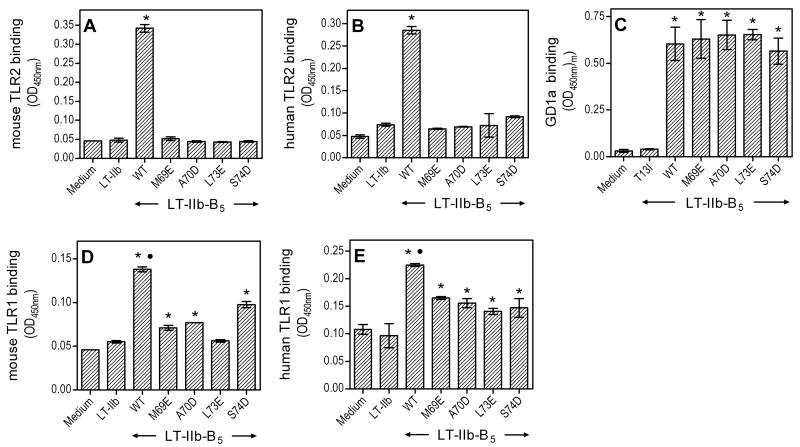
We hypothesized that the solvent-accessible hydrophobic surface in the “upper region” of LT-IIb-B5 is critical for TLR2 binding, since this interaction is abrogated when this surface is buried by the A subunit in the LT-IIb holotoxin (7). We generated point-substitution mutations rendering the involved hydrophobic residues hydrophilic (M69E, A70D, L73E, and S74D) and reasoned that the hydrophilic/charged nature of the mutated residues would abrogate the interaction of the modified LT-IIb-B5 molecules with TLR2. The hypothesis was supported by the findings from TLR2 binding assays comparing the mutants with wild-type LT-IIb-B5. Indeed, all mutants lost the ability to bind mouse or human TLR2, as seen with the LT-IIb holotoxin which served as negative control (Fig. 1 A,B). In stark contrast, the mutants retained the capacity to bind GD1a (Fig. 1C), an interaction that is mediated by the “lower region” of the doughnut-shaped B pentamer (1). For control purposes, we also tested a lower-region point mutant (T13I) of LT-IIb-B5 which, as expected, did not bind GD1a (Fig. 1C); however, T13I could bind TLR2 comparably to the wild-type (OD450nm binding values were 0.468 ± 0.031 and 0.413 ± 0.016, for mutant and wild-type, respectively, in side-by-side comparison). The figure 1C findings suggest that the upper-region point mutations (M69E, A70D, L73E, and S74D) do not cause global alterations in the B pentamer, since successful assembly of the B pentamer is a requirement for GD1a binding (1). Assembly of wild-type and mutant B pentamers, yielding molecules of about 60 kDa, was also confirmed by their similar fractionation patterns in size-exclusion chromatography (not shown).
Figure 1. Hydrophilic point mutations in the upper region of LT-IIb-B5 inhibit binding to TLRs but not to GD1a.
Binding of wild-type (WT) LT-IIb-B5 or upper-region point mutants (M69E, A70D, L73E and S74D) was determined on microtiter wells coated with mouse or human TLRs as indicated (A,B and D,E) or with GD1a ganglioside (C). The LT-IIb holotoxin and a lower-region point mutant of LT-IIb-B5 (T13I) were used as negative controls in the TLR (A,B and D,E) and GD1a (C) binding assays, respectively. All ligands were used at 10 μg/ml, and bound protein was detected colorimetrically after probing with anti-LT-IIb Ab, followed by addition of peroxidase-conjugated secondary Ab. Data are shown as means ± standard deviations (SD) (n = 3) from one of three independent sets of experiments that yielded similar results. Asterisks denote statistically significant (p < 0.05) binding compared to negative controls, and black circles in D and E indicate significantly higher (p < 0.05) binding by WT compared to all upper-region point mutants.
By analogy to Pam3CSK4, agonists that activate the TLR2/1 heterodimer may be expected to contact both TLR components (13). Our previous studies identified TLR1 as the signaling partner of TLR2 in response to LT-IIb-B5, although the possibility for a direct LT-IIb-B5/TLR1 interaction was not addressed (6, 7). We thus sought to determine whether LT-IIb-B5 binds TLR1 and, if so, to investigate the role of the upper region hydrophobic surface in this interaction. The LT-IIb holotoxin, which does not activate TLR2/1 (7), was used as a negative control. Although LT-IIb-B5 displayed statistically significant binding to mouse or human TLR1 (p < 0.01 vs. LT-IIb negative control; Fig. 1 D,E), this was not as pronounced as binding to TLR2 (contrast with Fig. 1 A,B). Interestingly, the mutants showed moderate-level, intermediate TLR1 binding between that of the LT-IIb holotoxin and the wild-type B pentamer (Fig. 1 D,E). As expected, none of the ligands, wild-type or mutant, could bind TLR6 (not shown). The figure 1 data collectively indicate that LT-IIb-B5 interacts with both TLR2 and TLR1 and, moreover, a defined hydrophobic site in its upper region plays a critical role in TLR2 binding but a relatively modest role in TLR1 binding.
Hydrophilic point mutations in the LT-IIb-B5 upper region abrogate TLR2/1-dependent immunostimulation
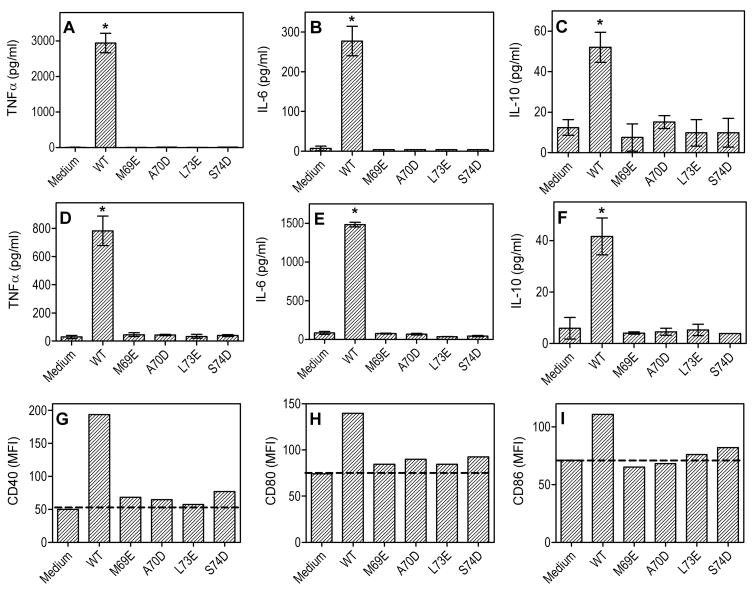
We next determined whether the upper-region point mutants also lost the ability to induce TLR2/1-dependent immunostimulation. Moreover, since these mutants retained GD1a-binding activity, they could be appropriately used as tools for investigating whether LT-IIb-B5 exhibits GD1a-dependent immunostimulatory properties that are independent of TLR2/1 activation. We found that all four hydrophilic mutants (M69E, A70D, L73E, and S74D) failed to induce NF-kappa;B activation in reporter THP1-Blue cells, in contrast to wild-type LT-IIb-B5 which readily activated NF-κB in a TLR2-dependent way (Fig. 2A). Consistent with the notion that LT-IIb-B5 binds TLR1 but not TLR6, LT-IIb-B5-induced NF-κB activation was inhibited by anti-TLR1 but not by anti-TLR6 (Fig. 2B). The specificity of anti-TLR blocking Abs was confirmed by including appropriate assay controls, i.e., Pam3CSK4 (TLR2/1 agonist), FSL-1 (TLR2/6 agonist), and LPS (TLR4 agonist) (Fig. 2 A,B). Moreover, all four hydrophilic point mutations inhibited the ability of LT-IIb-B5 to induce cytokine production in mouse peritoneal macrophages (Fig. 3 A–C) or bone marrow-derived dendritic cells (BMDC) (Fig. 3 D–F), suggesting that GD1a binding is not sufficient per se for these activities. Similarly, wild-type LT-IIb-B5 upregulated the expression of costimulatory molecules (CD40, CD80, and CD86) in BMDC, although the mutants showed little or no activity compared to no-agonist controls (Fig. 3 G–I). In summary, the hydrophobic upper region of LT-IIb-B5, defined by M69, A70, L73, and the Cβ of S74, is critical for TLR2/1-dependent immunostimulation, in contrast to the GD1a-binding site in the lower region of the molecule.
Figure 2. Hydrophilic point mutations abrogate the ability of LT-IIb-B5 to induce TLR2/1-dependent NF-κB activation.
Wild-type (WT) LT-IIb-B5 and indicated point mutants (all at 10 μg/ml) were tested for their capacity to activate NF-κB in reporter THP-1-Blue cells in a TLR2-or TLR1-dependent way (A and B, respectively). Prior to stimulation, the cells were pretreated for 30 min with anti-TLR2 (A) or anti-TLR1 (B) (or other anti-TLR Abs, as indicated, for control purposes). Activation was determined colorimetrically by measuring the activity of NF-κB-inducible alkaline phosphatase secreted in the culture supernatants upon 24-h incubation. Pam3CSK4, LPS, and FSL-1 (which activate TLR2/1, TLR4, and TLR2/6, respectively) were used for monitoring the specificity of the blocking Abs. Results are presented as means ± SD (n = 3) from one of two independent sets of experiments yielding similar findings. Asterisks indicate statistically significant (p < 0.05) activation of NF-κB compared to no-agonist control and black circles show significant (p < 0.05) inhibition of activation.
Figure 3. Wild-type LT-IIb-B5, but not the upper-region hydrophilic mutants, activates antigen-presenting cells.
Mouse peritoneal macrophages (A–C) or BMDC (D–I) were stimulated for 18–20 h with wild-type (WT) LT-IIb-B5 or the indicated point mutants (10 μg/ml). Induction of release of the indicated cytokines (A–F) in culture supernatants was measured by ELISA. Data are means ± SD (n = 3) from one of three independent sets of experiments yielding similar findings, and asterisks indicate significantly (p < 0.05) higher cytokine production compared to medium-only control. Upregulation of the indicated costimulatory molecules was assayed by FACS and is reported as mean fluorescent intensity (MFI) (G–I); the data are representative of five independent experiments yielding similar results.
Effects of TLR1 alterations on cooperative TLR2-dependent cell activation by LT-IIb-B5
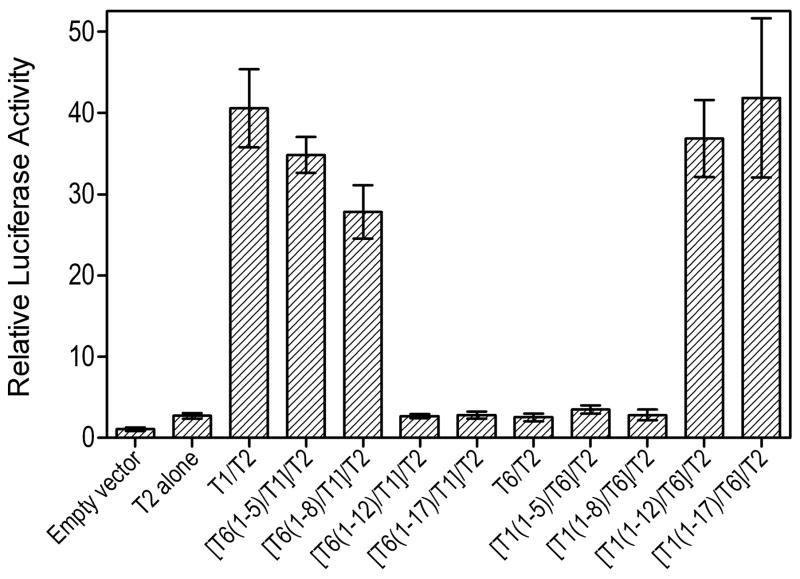
The ectodomain of both TLR2 and TLR1 comprises 19 leucine-rich repeat (LRR) modules (13). Using TLR1/6 chimeric receptors, constructed by reciprocally exchanging increasing segments of N-terminal LRR modules between these two highly homologous receptors (10), we determined which segment(s) of TLR1 are required for cooperative TLR2-induced cell activation in response to LT-IIb-B5. We found that as an increasing number of N-terminal LRRs of TLR1 were replaced by corresponding TLR6 LRRs, the cells progressively lost their ability for TLR2-dependent activation in response to LT-IIb-B5 (Fig. 4). Specifically, although replacement of up to the first eight N-terminal LRRs (construct [T6(1–8)/T1]/T2) had a relatively minor effect, the additional replacement of the subsequent four LRRs (construct [T6(1–12)/T1]/T2) completely abrogated the response (Fig. 4). The testing of the reverse chimeric constructs showed that replacement of up to the first eight N-terminal LRRs of TLR6 with those of TLR1 (construct [T1(1–8)/T6]/T2) was incapable to rescue LT-IIb-B5-induced cell activation (Fig. 4). This required the replacement of LRR1–12 of TLR6 by the corresponding TLR1 LRRs (construct [T1(1–12)/T6]/T2) and the response was completely restored by replacement of LRR1–17 of TLR6 with those of TLR1 (Fig. 4). Taken together, these data indicate that the LRR9-12 region of TLR1 is critical for cooperative TLR2-induced cell activation by LT-IIb-B5, whereas regions above or below LRR9-12 play a relatively minor role.
Figure 4. Effect of chimeric TLR1/6 receptors, generated by reciprocal domain exchange, on TLR2-dependent cell activation by LT-IIb-B5.
SW620 cells were cotransfected with human TLR2 and chimeric TLR1/6 receptors or full-length TLR1 or TLR6 (in the X-axis labels, TLR is shown as T for simplicity and clarity). The cells additionally received a firefly luciferase reporter gene and a Renilla transfection control. After 48h, the cells were stimulated for 6h with LT-IIb-B5 (2 μg/ml) or Pam3CSK4 (20 ng/ml). Cellular activation is reported as relative luciferase activity, normalized to that of cells transfected with reporter and empty vectors. Data are presented as means ± SD (n = 3).
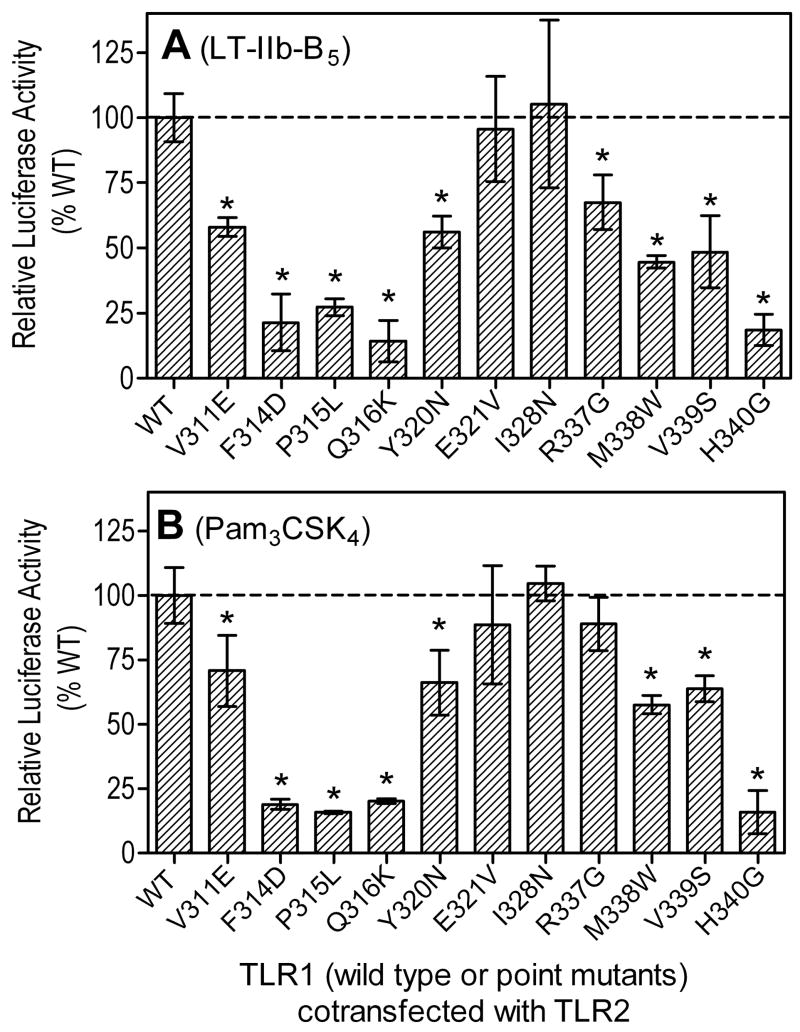
The crystallographic structure of the Pam3CSK4-induced active conformation of the TLR2/1 heterodimer has revealed a number of residues involved in the dimerization interface and/or ligand binding (13). These include P315 of TLR1 in the hydrophobic core of the dimer interface, which when mutated to L (to mimic the P315L natural polymorphism) leads to inhibition of Pam3CSK4-induced signaling (28). We found that the P315L variant, as well as a number of other TLR1 point mutations in the dimer interface, inhibited TLR2-dependent activation of transfected SW620 cells by LT-IIb-B5 (Fig. 5A). In contrast, mutation of a residue (I328N) that is not involved in dimer interface (or ligand binding) had no effect on the ability of LT-IIb-B5 to activate the transfectants (Fig. 5A). The effects of the same TLR1 point mutations on Pam3CSK4-induced cell activation followed a similar pattern (Fig. 5B), except for R337G which had no significant influence on cell activation by Pam3CSK4, although it modestly inhibited activation by LT-IIb-B5 (Fig. 5). Interestingly, a mutated residue (H340G) that was not predicted to be involved in dimer interface or ligand binding (13) inhibited both LT-IIb-B5- and Pam3CSK4-induced cell activation (Fig. 5). Since most of the TLR1 interface residues influenced the ability of both Pam3CSK4 and LT-IIb-B5 to induce cell activation, it appears that the reported Pam3CSK4-induced “m”-shaped structure of TLR2/1 may constitute a predominant activation conformation, although subtle variations are possible depending on the structural nature of the agonist. This, in turn, provides justification for using the Pam3CSK4-induced active conformation of TLR2/1 (13) to model the interaction of this structure with LT-IIb-B5.
Figure 5. Effects of TLR1 point mutations on cooperative TLR2-dependent cell activation by LT-IIb-B5.
SW620 cells were cotransfected with human TLR2 and TLR1 (the latter in wild-type or point mutant versions, as indicated), as well as with a firefly luciferase reporter gene and a Renilla transfection control. After 48 h, the cells were stimulated for 6h with LT-IIb-B5 (2 μg/ml) (A) or Pam3CSK4 (20 ng/ml) (B). Cellular activation is reported as relative luciferase activity, normalized to that of cells transfected with reporter and empty vectors. Results are presented as means ± SD (n = 3). Asterisks indicate significantly (p < 0.05) reduced activities compared to transfectants that received wild-type (WT) TLR1.
Model for LT-IIb-B5 binding to TLR2/1
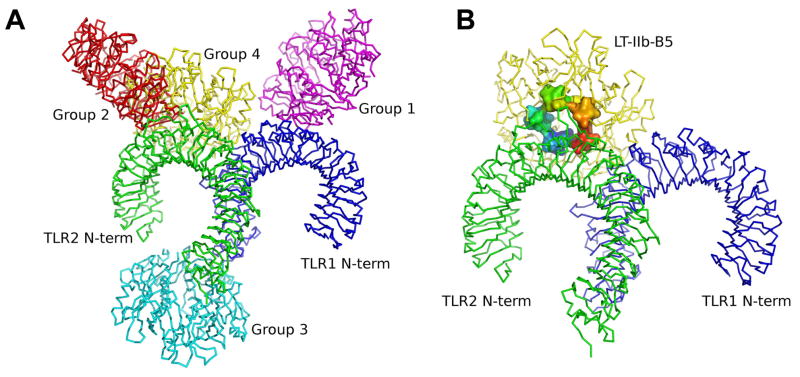
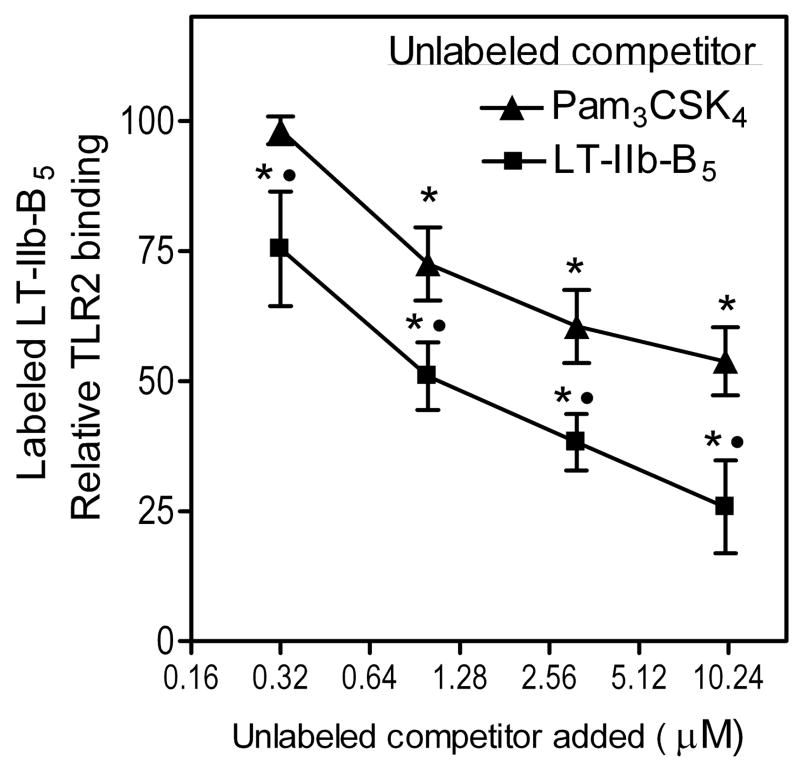
Although the crystallographic structure of the TLR2/1-Pam3CSK4 complex has provided a groundbreaking understanding of ligand recognition by these TLRs (13), it is uncertain how relatively large protein ligands may interact with the TLR2/1 heterodimer. Indeed, TLR2/1-activating protein ligands that do not bear acyl chains are unlikely to be accommodated within the heterodimer’s hydrophobic pockets, which are essential for binding lipopeptides (13). To glean an insight into this issue, we modeled the interaction of LT-IIb-B5 with TLR2/1. The top 10 scored TLR2/1-LT-IIb-B5 models were retrieved from GRAMM-X (30), ZDOCK (31), and ClusPro (32) protein-protein docking servers, and were assigned into four groups on the basis of TLR2 and/or TLR1 sites involved (Table I and Fig. 6A). These models are discussed in detail in the following section, where we support that the most possible model involves interaction between the hydrophobic surface in the upper region of LT-IIb-B5 and the convex surface of the central domains of the TLR2/1 heterodimer (Fig. 6B). According to this model, LT-IIb-B5 interacts primarily with the TLR2 component and the putative contact points (supplemental Fig. 1) are located in regions that extend from LRR5 to LRR10, thus partially overlapping with the Pam3CSK4-binding site (13). If this theoretical prediction is true, then Pam3CSK4 should be able to compete with LT-IIb-B5 for TLR2 binding. Indeed, we found that the binding of biotinylated LT-IIb-B5 is inhibited by Pam3CSK4 in a dose-dependent manner, although unlabeled LT-IIb-B5 exhibited stronger inhibitory activity (Fig. 7).
Table I.
Classification of different models of LT-IIb-B5 binding to TLR2 and/or TLR1 based on the interactive sites involved.
| Groups | Interactive sites | Models | ||
|---|---|---|---|---|
| GRAMMX | ZDOCK | ClusPro | ||
| 1 | TLR1 only | 1, 2, 4, 6, 7, 9 | 1, 2, 4, 6, 7 | 3, 6, 8, 9 |
| 2 | TLR2 only | 10 | 3, 8 | 7, 10 |
| 3 | C-terminus of TLR1 and TLR2 | 3, 8 | 1, 2, 4, 5 | |
| 4 | Convex surface of TLR2 and TLR1 central domains | 5 | 5, 9, 10 | |
Figure 6. Model of LT-IIb-B5 interactions with TLR2/1.
(A) The TLR2/1 and LT-IIb-B5 structures were submitted as receptor and ligand, respectively, to GRAMMX, ZDOCK and ClusPro protein-protein docking servers for complex prediction and the top ten scored models were categorized into four groups (group 1, purple; group 2, red; group 3, cyan; and group 4, yellow). (B) The ZDOCK model 9 from group 4 (lowest global energy) after refinement. The TLRs are shown in surface mode with TLR2 colored green and TLR1 colored blue. Residues 69 to 74 in LT-IIb-B5 are shown in surface mode and colored by chain from blue to red.
Figure 7. Competitive inhibition of LT-IIb-B5 binding to TLR2 by Pam3CSK4.
Binding of biotinylated LT-IIb-B5 (0.16 μM) to plate-bound TLR2 in the presence of increasing concentrations of unlabeled LT-IIb-B5 or Pam3CSK4. Bound protein was probed with peroxidase-conjugated streptavidin and binding was determined colorimetrically. The data (means ± SD; n = 3) were normalized to the binding activity of labeled LT-IIb-B5 in the presence of 10 mg/ml BSA only (uninhibited control), the mean activity of which was taken as 100. Asterisks indicate significant (p < 0.05) inhibition of binding, and black circles denote significant differences (p < 0.05) between the unlabeled competitors.
Discussion
This study has investigated the mode whereby a non-lipidated protein ligand may interact with the TLR2/1 heterodimer. First, we showed that LT-IIb-B5 employs its hydrophobic upper pore region to directly bind TLR2. Specifically, a ring of four residues (M69, A70, L73, and S74) defines a TLR2-binding site for induction of TLR2/1-dependent NF-κB activation, cytokine production, and costimulatory molecule upregulation in antigen-presenting cells. Intriguingly, M69, A70, L73, and S74 are exactly shared by LT-IIa-B5, the only other known enterotoxin B pentamer that activates TLR2 (5), but not by the Type I B pentamers, LT-I-B5 and CT-B5 (1). Although figure 6B depicts the most probable model of the LT-IIb-B5–TLR2/1 interaction (see also below), protein-protein docking analysis suggested additional models which were assigned into four groups (Table I and Fig. 6A).
In groups 1 and 2, LT-IIb-B5 is predicted to bind to TLR1 only or TLR2 only, respectively. These models are not favored because our experimental data show that LT-IIb-B5 can bind to both TLR2 and TLR1 (though not to TLR6). In group 3, LT-IIb-B5 is predicted to bind to the C-terminus of both TLR2 and TLR1. This appears unlikely given the proximity of these sites to the cell membrane and the size of LT-IIb-B5 that could cause serious space constraints. Similar space constraints would probably be encountered if LT-IIb-B5 was to interact with the concave surfaces of the horseshoe-like structures of the TLR2/1 heterodimer, i.e., in a mode similar to MD-2 binding to TLR4 (36). In this regard, the doughnut-shaped LT-IIb-B5 is about 35 Å thick and the diameter of its upper region is 66 Å, whereas the inner diameter of the TLR2 and TLR1 “horseshoes” is about 38 Å with openings of about 18 Å. It is thus unlikely that the TLR2 or TLR1 concave faces could accommodate LT-IIb-B5 or other ligands of similar size and shape. Not surprisingly, such models were not anticipated by our analysis.
According to the group 4 models, LT-IIb-B5 binds to the convex surface of TLR2 and TLR1. The Z dock models 9, 10 and GrammX model 5 predict the involvement of the upper hydrophobic region of LT-IIb-B5 in TLR2/1 interactions, as also supported by our experimental data; the three models were further refined in the Firedock server (33) and gave global energies of 8.26, 17.92 and 24.27, respectively. As the lowest global energy represents the most energetically favorable model (i.e., closest to the native structure of the complex), the refined Z dock model 9 (Fig. 6B) was selected for further analysis. LigPlot analysis (supplemental Fig. 1) of possible interactions in this model predicts that LT-IIb-B5 binds to TLR2 mainly through hydrophobic interactions involving several residues, including L73 and S74 which are supported by our experimental data. Although not predicted to directly participate in these interactions, M69 and A70 are located close to the LT-IIb-B5/TLR2 interface and, moreover, are key residues of the hydrophobic surface of LT-IIb-B5 (1); the M69E and A70D mutations are thus likely to abrogate the LT-IIb-B5-TLR2/1 interaction by disrupting the hydrophobic nature of the interface.
The TLR2/1 heterodimer presents an “m”-shaped activation conformation upon Pam3CSK4 binding (13). This model bears similarities with that of TLR3 binding to double-stranded RNA, although it is currently uncertain whether the TLR2/1 signaling complex forms by cooperative receptor dimerization as does the TLR3 signaling complex (37). A similar “m”-shaped activation conformation may be retained in TLR2/1 interactions with LT-IIb-B5, since point mutations of TLR1 residues involved in the Pam3CSK4-induced TLR2/1 dimerization interface also inhibited LT-IIb-B5-induced TLR2/1 signaling. The TLR2 residues predicted to interact with LT-IIb-B5 (Supplemental Fig. 1) are located in regions encompassing LRR5 to LRR10 and, therefore, overlap partially with the Pam3CSK4 binding site, which comprises residues from LRR9 to LRR12 (13). This prediction is consistent with our findings that Pam3CSK4 competitively inhibits the binding of LT-IIb-B5 to TLR2, albeit not as potently as seen in the self-competitive assay.
The observation that the upper-region point mutants retained partial TLR1 binding activities suggests that the corresponding hydrophobic surface of LT-IIb-B5 may indirectly contribute to TLR1 interactions but is not critically involved. It is possible that the substitution of hydrophobic residues with hydrophilic ones may cause short-range conformational changes that may partially affect TLR1 binding. Any such conformational alterations on LT-IIb-B5, however, would not have a global impact on the protein since the GD1a-binding activity of the distally located lower region was preserved. Consistent with these notions, no obvious hydrophobic LT-IIb-B5/TLR1 interface was predicted by running the LigPlot program (34). Moreover, the figure 6B model predicts that LT-IIb-B5 interacts primarily with the TLR2 component of the TLR2/1 heterodimer. Indeed, using the Crystallography & NMR System software (35), the surface area buried at the LT-IIb-B5/TLR2 interface was calculated to be 1663 Å2 compared to 546 Å2 for the surface area buried at the LT-IIb-B5/TLR1 interface. However, it is conceivable that this contact with TLR1 may enable LT-IIb-B5 to bridge the two receptors in a signaling-competent conformation. Alternatively, or in addition, LT-IIb-B5 may induce a conformational change in TLR2 which contributes to the formation of the active heterodimer. In this regard, it is known that the formation of TLR9 homodimers is not sufficient per se for receptor activation, which is actually regulated by conformational changes induced by agonistic DNA ligands (38).
Previous studies by one of the coauthors have shown that LRR9–12 of TLR1 or TLR6 are minimally required for discrete recognition of TLR1- or TLR6-specific lipopeptides, respectively (10). These findings anticipated the crystallographic demonstration that the LRR9-12 segments of the TLR2/1 heterodimer contain the Pam3CSK4 binding site (13). We found that the LRR9-12 region of TLR1 is also essential for cooperative TLR2-induced cell activation by LT-IIb-B5. Importantly, this TLR1 region forms part of the TLR1/LT-IIb-B5 interface (Fig. 6B) and, moreover, contains critical residues involved in the TLR2/1 dimer interface, such as V311, P315, Y320, R337, V339 (13), which were confirmed experimentally in this study.
By contrast to the TLR2/1-interactive upper region, the lower region of the doughnut-shaped LT-IIb-B5 contains a GD1a-binding site where T13 and T14 play critical roles (1, 39). We previously showed that GD1a binding by LT-IIb-B5 enhances its ability to induce TLR2/1-mediated NF-κB activation (6). However, the role of GD1a is to facilitate the LT-IIb-B5 interaction with TLR2/1, probably by concentrating the agonist in lipid rafts and rendering it more available to the TLR2/1 signaling complex, and does not seem to contribute by itself to immunostimulatory signal transduction. Indeed, all upper-region point mutants were inert in all activation assays tested, despite maintaining full GD1a-binding capacity.
The same LT-IIb-B5 residues involved in TLR2 binding (M69, A70, L73, and S74) are also critical for hydrophobic interactions with the A2 segment of the A subunit in the fully-assembled LT-IIb holotoxin (1). As a result, the TLR2-binding site is blocked in the intact holotoxin, which consequently does not bind or activate TLR2 (7). Since evolution does not work with preconceived plans to generate a fully assembled AB5 toxin by design, the expression of the B pentamer in free form is likely to predate the fully assembled LT-IIb molecule. It is thus intriguing to speculate that, at least in part, the evolutionary pressure for the development of fully assembled LT-IIb holotoxin was to suppress TLR2-mediated innate immunity in response to the free B pentamer. By contrast, the four identified critical residues mediating the TLR2/1-dependent immunostimulatory activity of LT-IIb-B5 may be exploited for improved adjuvant development. In this respect, appropriate genetic engineering of the TLR2-interactive site of LT-IIb-B5 may further promote its interactions with TLR2/1 and enhance its potential as a vaccine adjuvant.
Although the term “pattern-recognition receptor” has been extremely useful in introducing the concept of multiple ligand recognition by TLRs and other innate immune receptors (40, 41), there is no precise definition for “pattern” in biological interactions. Although such a term may involve or imply accurate spatial arrangement of repeated interacting groups, it has been argued that it is specific microbial molecules with exact known structures, and not “patterns”, that interact with TLRs and innate host receptors in general (42). This notion is clearly supported by our current work describing involvement of specific residues of a microbial ligand in TLR2 interactions. Moreover, we and others have previously shown that TLRs and other pattern-recognition receptors from mammals or plants can be activated by specific peptide sequences (14, 43, 44).
In summary, our experimental data and model predictions suggest that LT-IIb-B5 interacts with the convex surface of the central domains of the TLR2/1 heterodimer. This mode of TLR interaction is distinct from that of Pam3CSK4, which also binds TLR2/1 albeit involving insertion into hydrophobic pockets, and quite different compared to MD-2 binding to the concave surfaces of the TLR4/4 homodimer (13, 20, 36). The notion that different ligands may bind TLRs in different ways, suggests that the TLR binding sites may be determined primarily by the properties of the ligands. Furthermore, the possible existence of multiple (though possibly overlapping) TLR binding sites may explain, at least in part, the relatively promiscuous binding capacity of these receptors.
Supplementary Material
Footnotes
This work was supported by U.S. Public Health Service R01 Grants AI052344 and AI056148 (to R.I.T.), DE013833 (to T.D.C), and DE015254 and DE017138 (to G.H.), from the NIH.
Abbreviations used in this paper: LT-IIb-B5, pentameric B subunit of type IIb E. coli enterotoxin; LRR, leucine-rich repeat.
References
- 1.Van Den Akker F, Sarfaty S, Twiddy EM, Connell T, Holmes RK, Hol WGJ. Crystal structure of a new heat-labile enterotoxin, LTIIb. Structure. 1996;4:665–678. doi: 10.1016/s0969-2126(96)00073-1. [DOI] [PubMed] [Google Scholar]
- 2.Holmes R, Jobling MG, Connell T. Cholera toxin and related enterotoxins of gram-negative bacteria. Bacterial toxins and virulence factors in disease. In: Moss J, Iglewski B, Vaughn M, Tu AT, editors. Handbook of natural toxins. Marcel Dekker, Inc; New York: 1995. pp. 225–255. [Google Scholar]
- 3.Connell TD. Cholera toxin, LT-I, LT-IIa and LT-IIb: the critical role of ganglioside binding in immunomodulation by type I and type II heat-labile enterotoxins. Expert Rev Vaccines. 2007;6:821–834. doi: 10.1586/14760584.6.5.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Arce S, Gockel CM, Connell TD, Russell MW. Immunomodulation with enterotoxins for the generation of secretory immunity or tolerance: applications for oral infections. J Dent Res. 2005;84:1104–1116. doi: 10.1177/154405910508401205. [DOI] [PubMed] [Google Scholar]
- 5.Hajishengallis G, Tapping RI, Martin MH, Nawar H, Lyle EA, Russell MW, Connell TD. Toll-like receptor 2 mediates cellular activation by the B subunits of type II heat-labile enterotoxins. Infect Immun. 2005;73:1343–1349. doi: 10.1128/IAI.73.3.1343-1349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang S, Wang M, Tapping RI, Stepensky V, Nawar HF, Triantafilou M, Triantafilou K, Connell TD, Hajishengallis G. Ganglioside GD1a is an essential coreceptor for toll-like receptor 2 signaling in response to the B subunit of Type IIB enterotoxin. J Biol Chem. 2007;282:7532–7542. doi: 10.1074/jbc.M611722200. [DOI] [PubMed] [Google Scholar]
- 7.Liang S, Wang M, Triantafilou K, Triantafilou M, Nawar HF, Russell MW, Connell TD, Hajishengallis G. The A subunit of Type IIb enterotoxin (LT-IIb) suppresses the proinflammatory potential of the B subunit and its ability to recruit and interact with TLR2. J Immunol. 2007;178:4811–4819. doi: 10.4049/jimmunol.178.8.4811. [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill LA. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity. 2008;29:12–20. doi: 10.1016/j.immuni.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Omueti KO, Beyer JM, Johnson CM, Lyle EA, Tapping RI. Domain exchange between human Toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J Biol Chem. 2005;280:36616–36625. doi: 10.1074/jbc.M504320200. [DOI] [PubMed] [Google Scholar]
- 11.Uematsu S, Akira S. Toll-Like receptors (TLRs) and their ligands. Handb Exp Pharmacol. 2008:1–20. doi: 10.1007/978-3-540-72167-3_1. [DOI] [PubMed] [Google Scholar]
- 12.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 13.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Hajishengallis G, Ratti P, Harokopakis E. Peptide mapping of bacterial fimbrial epitopes interacting with pattern recognition receptors. J Biol Chem. 2005;280:38902–38913. doi: 10.1074/jbc.M507326200. [DOI] [PubMed] [Google Scholar]
- 15.Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, Adams LG, Baumler AJ. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol. 2005;58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 16.Epelman S, Stack D, Bell C, Wong E, Neely GG, Krutzik S, Miyake K, Kubes P, Zbytnuik LD, Ma LL, Xie X, Woods DE, Mody CH. Different domains of Pseudomonas aeruginosa exoenzyme S activate distinct TLRs. J Immunol. 2004;173:2031–2040. doi: 10.4049/jimmunol.173.3.2031. [DOI] [PubMed] [Google Scholar]
- 17.Massari P, Visintin A, Gunawardana J, Halmen KA, King CA, Golenbock DT, Wetzler LM. Meningococcal porin PorB binds to TLR2 and requires TLR1 for signaling. J Immunol. 2006;176:2373–2380. doi: 10.4049/jimmunol.176.4.2373. [DOI] [PubMed] [Google Scholar]
- 18.Sing A, Reithmeier-Rost D, Granfors K, Hill J, Roggenkamp A, Heesemann J. A hypervariable N-terminal region of Yersinia LcrV determines Toll-like receptor 2-mediated IL-10 induction and mouse virulence. Proc Natl Acad Sci U S A. 2005;102:16049–16054. doi: 10.1073/pnas.0504728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, Sharma A. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodsky I, Medzhitov R. Two modes of ligand recognition by TLRs. Cell. 2007;130:979–981. doi: 10.1016/j.cell.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 21.Okusawa T, Fujita M, Nakamura J, Into T, Yasuda M, Yoshimura A, Hara Y, Hasebe A, Golenbock DT, Morita M, Kuroki Y, Ogawa T, Shibata K. Relationship between structures and biological activities of mycoplasmal diacylated lipopeptides and their recognition by toll-like receptors 2 and 6. Infect Immun. 2004;72:1657–1665. doi: 10.1128/IAI.72.3.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishiguchi M, Matsumoto M, Takao T, Hoshino M, Shimonishi Y, Tsuji S, Begum NA, Takeuchi O, Akira S, Toyoshima K, Seya T. Mycoplasma fermentans lipoprotein M161Ag-induced cell activation is mediated by Toll-like receptor 2: role of N-terminal hydrophobic portion in its multiple functions. J Immunol. 2001;166:2610–2616. doi: 10.4049/jimmunol.166.4.2610. [DOI] [PubMed] [Google Scholar]
- 23.Hajishengallis G, Nawar H, Tapping RI, Russell MW, Connell TD. The Type II heat-labile enterotoxins LT-IIa and LT-IIb and their respective B pentamers differentially induce and regulate cytokine production in human monocytic cells. Infect Immun. 2004;72:6351–6358. doi: 10.1128/IAI.72.11.6351-6358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 25.Nawar HF, Arce S, Russell MW, Connell TD. Mucosal adjuvant properties of mutant LT-IIa and LT-IIb enterotoxins that exhibit altered ganglioside-binding activities. Infect Immun. 2005;73:1330–1342. doi: 10.1128/IAI.73.3.1330-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Shakhatreh M-AK, James D, Liang S, Nishiyama S-i, Yoshimura F, Demuth DR, Hajishengallis G. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 2007;179:2349–2358. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- 27.Cirino PC, Mayer KM, Umeno D. Generating mutant libraries using error-prone PCR. Methods Mol Biol. 2003;231:3–9. doi: 10.1385/1-59259-395-X:3. [DOI] [PubMed] [Google Scholar]
- 28.Omueti KO, Mazur DJ, Thompson KS, Lyle EA, Tapping RI. The polymorphism P315L of human Toll-like receptor 1 impairs innate immune sensing of microbial cell wall components. J Immunol. 2007;178:6387–6394. doi: 10.4049/jimmunol.178.10.6387. [DOI] [PubMed] [Google Scholar]
- 29.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 30.Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34:W310–314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 32.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 33.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DC, Monks BG, McKnight CJ, Lamphier MS, Duprex WP, Espevik T, Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 39.Connell TD, Holmes RK. Mutational analysis of the ganglioside-binding activity of the type II Escherichia coli heat-labile enterotoxin LT-IIb. Mol Microbiol. 1995;16:21–31. doi: 10.1111/j.1365-2958.1995.tb02388.x. [DOI] [PubMed] [Google Scholar]
- 40.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 41.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Beutler B. Not “molecular patterns” but molecules. Immunity. 2003;19:155–156. doi: 10.1016/s1074-7613(03)00212-7. [DOI] [PubMed] [Google Scholar]
- 43.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 44.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.