Abstract
The seed maturation program is repressed during germination and seedling development so that embryonic genes are not expressed in vegetative organs. Here, we describe a regulator that represses the expression of embryonic seed maturation genes in vegetative tissues. ASIL1 (for Arabidopsis 6b-interacting protein 1-like 1) was isolated by its interaction with the Arabidopsis thaliana 2S3 promoter. ASIL1 possesses domains conserved in the plant-specific trihelix family of DNA binding proteins and belongs to a subfamily of 6b-interacting protein 1-like factors. The seedlings of asil1 mutants exhibited a global shift in gene expression to a profile resembling late embryogenesis. LEAFY COTYLEDON1 and 2 were markedly derepressed during early germination, as was a large subset of seed maturation genes, such as those encoding seed storage proteins and oleosins, in seedlings of asil1 mutants. Consistent with this, asil1 seedlings accumulated 2S albumin and oil with a fatty acid composition similar to that of seed-derived lipid. Moreover, ASIL1 specifically recognized a GT element that overlaps the G-box and is in close proximity to the RY repeats of the 2S promoters. We suggest that ASIL1 targets GT-box–containing embryonic genes by competing with the binding of transcriptional activators to this promoter region.
INTRODUCTION
Precise spatial and temporal regulation of gene expression is required for proper seed maturation. Three members of the B3 family of transcription factors, LEAFY COTYLEDON2 (LEC2), ABSCISIC ACID-INSENTITIVE3 (ABI3), and FUSCA3 (FUS3), and the CBF transcription factor LEC1 have been identified as key regulators of seed maturation processes. A redundant gene regulatory network linking these major regulators has been elucidated by examining the expression of ABI3, FUS3, and LEC2 in abi3, fus3, lec1, and lec2 single, double, and triple mutants (To et al., 2006). In combination with abscisic acid (ABA), gibberellin (GA), auxin, and sugar signaling, this regulatory network governs most seed-specific traits, such as accumulation of storage compounds, acquisition of desiccation tolerance, and entry into quiescence, in a partially redundant manner (Harada, 1999; Brocard-Gifford et al., 2003; Gazzarrini et al., 2004; Kagaya et al., 2005b; Vicente-Carbajosa and Carbonero, 2005; To et al., 2006; Stone et al., 2008). LEC1 and LEC2 are expressed early in embryogenesis, and ectopic expression of these two regulators is sufficient to confer embryonic traits to vegetative organs (Lotan et al., 1998; Stone et al., 2001; Santos-Mendoza et al., 2005). ABI3 and FUS3 expression occurs late in embryogenesis, and their overexpression results in the ectopic expression of some seed maturation genes, such as 2S3 and CRC, in vegetative tissues in an ABA-dependent manner (Parcy et al., 1994; Kagaya et al., 2005a). Genetic and molecular studies have shown that ABI3, FUS3, and LEC2 regulate oleosin gene expression and lipid accumulation (Crowe et al., 2000; Santos-Mendoza et al., 2005; Baud et al., 2007). Loss of ABI3 function alters accumulation of seed storage reserves and leads to loss of desiccation tolerance, dormancy, ABA sensitivity upon germination, and chlorophyll degradation (Vicente-Carbajosa and Carbonero, 2005). In addition, the APETALA2 protein ABI4 and the bZIP domain factor ABI5 are involved in some aspects of maturation through their interactions with the major regulators LEC2, ABI3, and FUS3 (Carles et al., 2002; Brocard-Gifford et al., 2003; Lara et al., 2003; Acevedo-Hernández et al., 2005).
Seed maturation–related genes, such as those governing seed storage protein (SSP) and lipid accumulation, are controlled by the interaction of transcriptional regulators with cis-acting elements in the promoters. Functional analysis and in vitro protein–DNA interaction assays have demonstrated binding of the B3 factors LEC2, ABI3, and FUS3 to RY repeats (Reidt et al., 2000; Kroj et al., 2003; Mönke et al., 2004; Braybrook et al., 2006) and bZIP factors ABI5, bZIP10, and bZIP25 to ACGT boxes (Bensmihen et al., 2002; Lara et al., 2003). Moreover, ABI5 and its homolog EEL were shown to play antagonistic roles influencing the expression of the LEA gene Em1 through competition for the same DNA binding site (Bensmihen et al., 2002). In addition to the direct binding to DNA elements, the major regulators were also found to indirectly regulate seed maturation genes. Genetic and molecular studies have shown that LEC1 and LEC2 act upstream of ABI3 and FUS3 and control SSP gene expression through the regulation of ABI3 and FUS3 (Kagaya et al., 2005b; To et al., 2006). ABI3 functions as a seed-specific transcriptional coactivator that physically interacts with ABI5, ZIP10, and ZIP25 (Nakamura et al., 2001; Lara et al., 2003). FUS3 expression in the protoderm and its negative regulation of TRANSPARENT TESTA GLABRA1 are critical for embryogenesis (Tsuchiya et al., 2004). Moreover, FUS3 has been demonstrated to be induced by auxin and indirectly influence the seed maturation process by positive and negative regulation of ABA and GA synthesis, respectively (Gazzarrini et al., 2004).
The regulatory networks governing seed maturation in Arabidopsis thaliana are repressed prior to germination so that seed storage reserves are not accumulated during vegetative development. Chromatin modification has been implicated in the repression of the regulatory networks. Silencing of the phas gene encoding phaseolin, a bean (Phaseolus vulgaris) SSP, in vegetative tissues is associated with the presence of a nucleosome positioned over the three-phased TATA boxes of the phas promoter (Li et al., 1998). Ectopic expression of the ABI3-like factor ALF led to potentiation of the chromatin structure over the TATA region of the phas promoter (Li et al., 1999). Chromatin immunoprecipitation assays have demonstrated that histone acetylation and methylation-directed chromatin remodeling contribute to the regulation of the phas expression (Ng et al., 2006). Additionally, inhibition of histone deacetylase activity with trichostatin A during germination led to elevated expression of embryogenesis-related genes (Tanaka et al., 2008). PICKLE (PKL), a CHD3 chromatin remodeling factor that belongs to the SWI/SNF class, acts in concert with GA to ensure that embryonic traits are not expressed after germination (Ogas et al., 1997, 1999); pkl mutants express seed maturation genes in primary roots (Ogas et al., 1997, 1999; Rider et al., 2003, 2004; Henderson et al., 2004; Li et al., 2005). VP1/ABI3-LIKE (VAL) B3 proteins VAL1 and VAL2 (also referred to as HSI2 and HSL1, respectively; Tsukagoshi et al., 2007) act together with sugar signaling to repress ectopic expression of seed maturation genes in seedlings and are necessary for the transition from seed maturation to active vegetative growth (Suzuki et al., 2007; Tsukagoshi et al., 2007). VAL1 and VAL2 encode B3 domain proteins with an ERF-associated amphiphilic repression (EAR) motif. Interestingly, a CW domain of unknown function and a putative plant homeodomain–like Zn-finger domain are frequently present in chromatin remodeling factors and were identified in the VAL1 and VAL2 proteins (Suzuki et al., 2007). It was revealed that VAL1/HSI2 functions as an active repressor of a sugar-inducible reporter gene (Tsukagoshi et al., 2005). Most of the embryonic and seed maturation genes, including LEC1, ABI3, FUS3, and genes for seed storage compounds, were derepressed in seedlings of a double mutant of VAL1 and VAL2 (Suzuki et al., 2007; Tsukagoshi et al., 2007). Polycomb (PcG) group proteins were demonstrated to establish epigenetic inheritance of repressed gene expression states through methylation of Lys-27 on histone H3 (Köhler and Grossniklaus, 2002). Genetic and molecular studies have demonstrated that FUS3 is regulated by the PcG proteins; for instance, FUS3 expression is derepressed in leaves of a double mutant of CURLY LEAF and SWINGER, and chromatin immunoprecipitation corroborated the direct targeting of FUS3 by the PcG protein MEDEA (Makarevich et al., 2006). Recently, a member of BRAHMA (BRM)-containing SNF2 chromatin remodeling ATPase was found to be involved in repression of some seed maturation genes in leaves. Mutation of BRMs caused transcript accumulation of 2S, FUS3, and some other embryogenesis-related genes in leaf tissues (Tang et al., 2008).
In this study, we report the isolation and functional characterization of ASIL1 (for Arabidopsis 6b-interacting protein 1-like 1), a member of the plant-specific trihelix family of DNA binding transcription factors. ASIL1 recognizes the GT-box present in the promoter region of 2S3. We provide evidence that ASIL1 functions as a negative regulator of a large subset of Arabidopsis embryonic and seed maturation genes in seedlings. Thus, ASIL1 represents a novel component of the regulatory framework that negatively controls embryonic gene expression during vegetative growth.
RESULTS
Cloning of the ASIL1 Gene
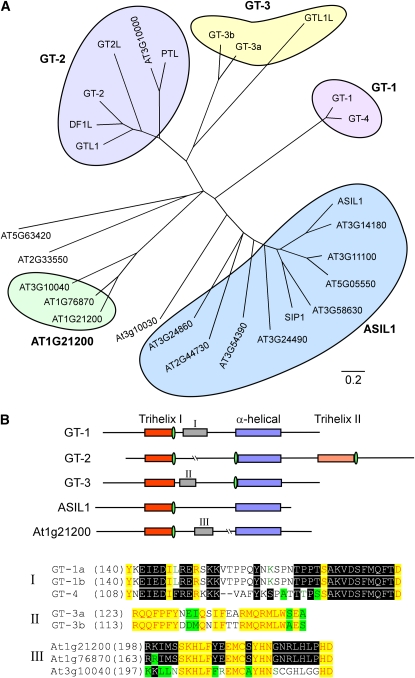
To search for transcription factors that potentially repress SSP accumulation in vegetative tissues, we identified a 49-bp element (MJ49) of the Arabidopsis 2S3 promoter between −95 and −47, which may be involved in the positive and negative regulation of SSP gene expression based on the analysis of β-conglycinin and β-phaseolin promoters (Burow et al., 1992; Chandrasekharan et al., 2003). This sequence contains two RY repeats (also referred to as the Sph element) for binding of B3 domain transcription factors (Reidt et al., 2000; Kroj et al., 2003; Mönke et al., 2004; Braybrook et al., 2006) at the 5′ and 3′ ends (RY3, CATGCATGCGTGCAT; RY4, CATGCA) and a bZIP transcription factor binding site, the ACGT-box (ACACGTG), in the center. Interestingly, an ACGT-like motif (TACACGTG) recognized by the transcriptional repressors ROM2 (Chern et al., 1996) and a trihelix GT-factor recognition site (CGTGATT) (Zhou, 1999) overlap a G-box (CACGTG) (see Supplemental Figure 1 online). Three copies of the MJ49 element fused to the HISTIDINE3 (HIS3) minimal promoter were used as bait in a yeast one-hybrid screen of an Arabidopsis seedling cDNA library linked to the yeast GAL4 activation domain. A total of 1.5 × 106 yeast transformants were screened and five confirmed positive clones were obtained (see Methods). One of these clones encoded a protein (At1g54060) predicted to be 41.7 kD with a pI of 8.6. This protein was designated as ASIL1 because of its similarity (40% amino acid identity) to Nicotiana tabacum 6b-interacting protein 1 (SIP1), an uncharacterized member of the trihelix family of DNA binding proteins that was isolated based on its interaction with the protein encoded by oncogene 6b from Agrobacterium tumefaciens (Kitakura et al., 2002). PSI-BLAST analysis (Altschul et al., 1997) of the Arabidopsis genome sequence in The Arabidopsis Information Resource database using the deduced amino acid sequence of ASIL1 revealed nine proteins with considerable similarity in their conserved trihelix motifs and α-helical regions close to the C termini (Figure 1A; see Supplemental Data Set 1 online).
Figure 1.
Comparative Analysis of Trihelix Proteins in Arabidopsis.
(A) A phylogenetic tree of ASIL proteins, GT factors, and other trihelix proteins in Arabidopsis. The tree was generated using the neighbor-joining method after sequence alignment with ClustalW. Five major subfamilies are grouped: ASIL1, GT-1, GT-2, GT-3, and At1g21200.
(B) Structure and subfamily-specific motifs of trihelix protein family. The conserved trihelix and α-helical domains are in different colors as indicated. The putative nuclear localization signals are represented in green ovals. Subfamily-specific motifs, which are shown as roman numerals I, II, and III, are aligned and indicated on the bottom of the figure.
ASIL1 Is a Nuclear Trihelix Protein
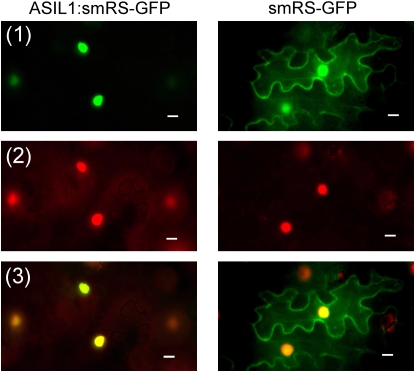
Trihelix DNA binding proteins are unique to the plant kingdom, a feature consistent with their involvement in regulating plant-specific developmental programs (Ayadi et al., 2004). In the Arabidopsis genome, 29 members encoded by 26 loci belong to the trihelix protein family that includes the transcription factors GT-1, GT-2, GT-3, and Nt SIP1-like proteins (Guo et al., 2008). As shown in Figure 1A, five distinct subfamilies can be defined in which most possess conserved N-terminal trihelix I domains and C-terminal α-helical regions. Unique motifs are identified in the GT-1, GT-3, and At1g21200 subfamilies, and the GT-2 subfamily is distinguished from other trihelix proteins by the C-terminal trihelix II domain (Figure 1B). Proteins in the ASIL1 subfamily have variable N termini and variable central regions. The ASIL1 gene encodes a polypeptide of 383 amino acids with several domains that suggest it may be a DNA binding transcription factor. The predicted protein has one DNA binding domain with the characteristics of a trihelix motif (Zhou, 1999). ASIL1 also contains putative nuclear localization signals, including SV40-like (residues 152 to 163), MAT 2-like (residues 171 to 177), and bipartite-like (residues 228 to 241) signals (Raikhel, 1992). In addition, ASIL1 contains a Gly-rich motif that is unique to the ASIL1 subfamily of trihelix proteins and a hydrophobic Pro-rich domain characteristic of a motif found in transcription repressors (Hanna-Rose and Hansen, 1996) (see Supplemental Figure 2 online). To verify that ASIL1 is localized to the nucleus in plant cells, a translational fusion was constructed between the soluble-modified red-shifted green fluorescent protein (smRS-GFP) (Davis and Vierstra, 1998) and ASIL1 expressed under the control of the cauliflower mosaic virus 35S promoter (p35S). In transient expression assay in tobacco epidermal cells, smRS-GFP alone was localized to the nucleus, plasma membrane, and cytoplasm as demonstrated previously (Figure 2; Ouelhadj et al., 2006). However, ASIL1:smRS-GFP fusion protein was localized only in the nuclei of epidermal cells (Figure 2). Collectively, these results suggest that ASIL1 is a nuclear trihelix transcription factor.
Figure 2.
Nuclear localization of ASIL1 Fused to smRS-GFP in Tobacco Epidermal Cells.
Plasmids that carried either construct p35S∷ASIL1:smRS-GFP or negative control p35S∷smRS-GFP were introduced into tobacco leaf cells by infiltration. All leaf tissues were stained with 4',6-diamidino-2-phenylindole (DAPI) and viewed using fluorescence microscopy with blue excitation to detect GFP fluorescence and UV excitation to detect DAPI. GFP is shown in green (1) and DAPI is shown in red (2). Merged images are shown at the bottom (3). Bars = 10 μm.
Phenotypic Characterization of asil1 Mutants
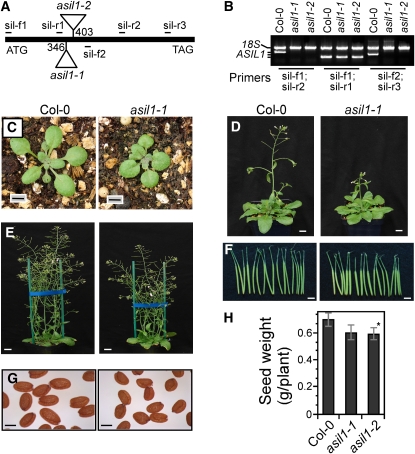
To determine the biological role of ASIL1 in plant development, we characterized two independent T-DNA–tagged alleles for the ASIL1 gene, asil1-1 (SALK_124095) and asil1-2 (SALK_074897) (Figure 3A), based on the analysis of publicly accessible collections of T-DNA insertion lines (http://signal.salk.edu/cgi-bin/tdnaexpress). Homozygous mutant plants were used to identify phenotypic differences between mutant and wild-type plants. asil1-1 and asil1-2 contain T-DNA insertions in the coding region of the intronless gene, 346 and 403 bp downstream of the translation start site, respectively, which disrupt the trihelix domain of ASIL1. The progeny of heterozygous plants exhibited a segregation of the asil1 phenotypes of ∼1:3 (x2 = 0.837, P > 0.05), indicating that the asil1-1 and asil1-2 alleles represented recessive loss-of-function mutations. RT-PCR analysis using primers that either span the T-DNA insertion or locate downstream of the insertion sites did not detect any transcript in homozygous asil1-1 and asil1-2 mutant plants (Figure 3B). However, transcripts corresponding to the region upstream of the T-DNA insertions were amplified in the asil1-1 and asil1-2 mutants (Figure 3B). These transcripts are presumably terminated inside the T-DNA. Therefore, the ASIL1 mRNA synthesis is interrupted by the T-DNA insertion in the mutant plants, and asil1-1 and asil1-2 may represent hypomorphic rather than null alleles.
Figure 3.
Phenotypes of Arabidopsis asil1 Mutant Plants.
(A) Schematic representation of the T-DNA insertion alleles of asil1-1 and asil1-2 in Arabidopsis. The ASIL1 gene has no intron. Numbers indicate positions of T-DNA insertions with respect to A of the translational start codon. The positions of PCR primers for genotyping (sil-f1 and sil-r3) and RT-PCR analysis are also indicated.
(B) RT-PCR analysis of total RNA from leaves of wild type (Col-0) and homozygous asil1-1 and asil1-2 plants with several primer pairs. 18S rRNA was used as an internal control.
(C) Phenotypes of 18-d-old wild-type and asil1-1 plantlets grown in soil. Bars = 0.5 cm.
(D) Wild-type and homozygous asil1-1 plants at 31 d of growth in soil under long days. Bars = 1 cm.
(E) Wild-type and asil1-1 adult plants at 41 d of growth in soil. Bars = 1.5 cm.
(F) Siliques of wild-type and asil1-1 mutant plants corresponding to, from right to left, 6 to 12 DPA. Bars = 0.5 cm.
(G) Mature seeds from wild-type and asil1-1 plants. Bars = 250 μm.
(H) Seed yield per plant from wild-type and homozygous asil1-1 and asil1-2 plants (n = 16). The error bars indicate ± sd of the means. Similar results were obtained from three independent experiments. Asterisk indicates a statistically significant difference between the wild type and asil1-2 mutant based on a Student's t test (P < 0.05).
The phenotypes of homozygous asil1-1 and asil1-2 plants were similar to each other but differed from the wild type at several stages of plant development. The mutants had smaller seedlings with slightly darker green and orbicular leaf blades and short leaf petioles (Figure 3C). The adult plants were ∼17% shorter (27.1 ± 0.7 cm) than the wild type (32.8± 0.8 cm) (P < 0.01) (Figure 3E). Siliques of asil1 plants were shorter (Figure 3F), seeds were slightly smaller (Figure 3G), and seed weights per plant were ∼14% less (0.5925 ± 0.0450 g) than the wild type (0.6979 ± 0.0477 g) (P < 0.05) (Figure 3H). When grown in soil, asil1 mutant plants showed a delay in flowering with respect to the wild type (wild-type and mutant plants flowered with 12.5 ± 0.6 and 13.8 ± 0.8 rosette leaves, respectively, in long days) (Figure 3D). However, asil1 mutant plants caught up quickly in subsequent growth and development and had a time span from germination to the last silique that turned completely yellow of approximately the same duration as wild-type plants. Variations in embryo morphology or developmental stages were not observed in the asil1 mutants, compared to wild-type embryos. Seed development in the asil1 mutant plants lasted the same amount of time as in the wild type, which was defined as a period of 15 d extending from anthesis to the dry seed stage.
ASIL1 Expression Pattern
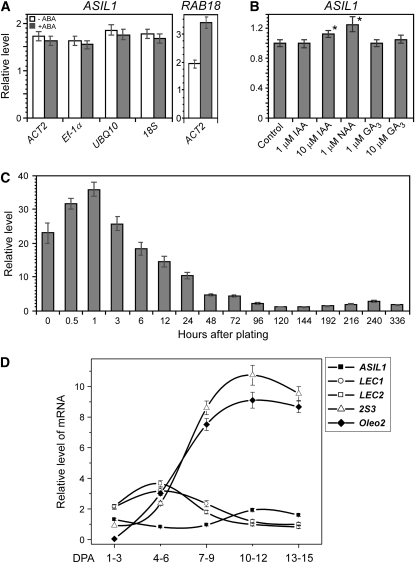
An elevated level of ABA is required during seed maturation for the accumulation of storage reserves (Finkelstein et al., 2002). In addition to ABA, other hormones, such as gibberellins (GAs) and auxins, and their coordination are also important during seed maturation and germination (Gazzarrini et al., 2004; Casson and Lindsey, 2006; Stone et al., 2008). To test the effect of ABA, GA, and auxin on ASIL1 expression, we performed quantitative real-time RT-PCR (qRT-PCR) analysis using total RNA extracted from 2-week-old seedlings grown on medium supplemented with either ABA, GA3, indole 3-acetic acid (IAA), or 1-naphthalene acetic acid (NAA). As shown in Figure 4A, the level of ASIL1 transcript was not significantly altered in response to ABA. To validate the ABA treatment, we measured mRNA for an ABA-responsive gene RAB18. The mRNA level of RAB18 was elevated significantly after ABA treatment. ASIL1 expression was also not affected by GA3 treatment (Figure 4B); however, ASIL1 expression was moderately increased by auxin treatment, suggesting that ASIL1 is an auxin-responsive transcriptional regulator, though exogenous application of a minimum of 10 μM IAA or 1 μM NAA was required to see the effect (Figure 4B).
Figure 4.
ASIL1 Expression in Response to Hormones and Temporal Regulation during Germination and Seed Development.
Real time qRT-PCR analysis was used to analyze ASIL1 transcript levels in seeds, seedlings, and siliques. The expression values of ASIL1 were normalized using the expression level of ACT2 unless otherwise indicated; housekeeping genes were considered as internal references. Results of expression represent the average of data, and sd values were calculated from the results of three independent experiments.
(A) ASIL1 expression in leaves in response to ABA. Total RNA was prepared from 2-week-old seedlings that were grown on plates of medium and incubated with 0 or 50 μM ABA for up to 24 h before harvest. ACT2, Ef-1α, UBQ10, and 18S were used as internal reference genes. Expression of RAB18, a known ABA responsive gene, was used to validate the ABA treatment.
(B) Regulation of ASIL1 expression in leaves by auxin and GA. Total RNA was isolated from 2-week-old seedlings that were grown on plates of medium then treated in liquid medium with IAA, NAA, and GA3 for 1 h at concentrations as indicated. Asterisk indicates a statistically significant difference between auxin treatment and control based on a Student's t test (P < 0.05). ASIL1 RNA levels in the untreated control were designated as onefold.
(C) Temporal expression of ASIL1 in seedlings. Total RNA was isolated from desiccated seeds or seeds that had been imbibed for up to 336 h (14 d) on plates of half-strength MS medium with 1% sucrose. Germination was completed by ∼70 h as scored by emergence of radicle from the seed coat. The lowest ASIL1 transcript level was at day 144 after imbibition and designated as onefold.
(D) Temporal expression of ASIL1 in developing siliques compared with embryonic genes LEC1, LEC2, 2S3, and Oleo2. Total RNA was isolated from siliques corresponding to five developmental stages as indicated.
PKL and VALs act to repress seed maturation genes at the very early stages of seedling development (Ogas et al., 1997, 1999; Tsukagoshi et al., 2007). Consistent with this mode of action, PKL transcript level was significantly elevated at 36 h after imbibition (Henderson et al., 2004), and accumulation of PKL protein was dramatically increased after imbibition for 48 h (Li et al., 2005). VAL gene transcripts accumulated to the highest levels in seedlings after the fifth day after imbibition (Tsukagoshi et al., 2007). We also examined the expression pattern of ASIL1 in seeds that had been imbibed for up to 14 d. Seed germination was completed by ∼60 h when radicles were fully emerged from the seed coat. ASIL1 transcript level was elevated after 0.5 h of imbibition, reached the highest level after 1 h, and then steadily decreased afterward (Figure 4C). The lowest level of ASIL1 mRNA was observed at 144 h with a slight increase in expression during seedling development (Figure 4C). Such a pattern of expression suggests that ASIL1 may establish repression of embryonic identity before the completion of seed germination.
ASIL1 expression was also examined in developing siliques during different stages of seed development: early embryogenesis (1 to 6 d postanthesis [DPA]), mid-embryogenesis (7 to 9 DPA), and late embryogenesis (10 to 15 DPA). Seeds were mature and completely dry after 15 DPA. ASIL1 transcript accumulated at a low level during embryogenesis and seed maturation, compared to LEC1, LEC2, and the seed maturation genes Oleo2 and 2S3 (Figure 4D). The lowest level of ASIL1 mRNA was at the late stage of early embryogenesis (4 to 6 DPA) when major embryonic regulators LEC1 and LEC2 were the most highly expressed (Figure 4D) and the seed maturation program has initiated (Vicente-Carbajosa and Carbonero, 2005). Based on publicly accessible microarray data (Schmid et al., 2005), ASIL1 transcript levels are lower in developing embryos than known embryonic repressors PKL, VAL1, and ESSP3, and the lowest level of ASIL1 mRNA is at stage 7 (late torpedo to early walking-stick embryos) (see Supplemental Figure 3 online) when endogenous ABA levels peak (Karssen et al., 1983; Vicente-Carbajosa and Carbonero, 2005). This expression pattern is consistent with the hypothesis that one of the primary roles of ASIL1 is to act as a transcriptional repressor of seed maturation genes.
ASIL1 Negatively Regulates Seed Maturation Genes in Seedlings
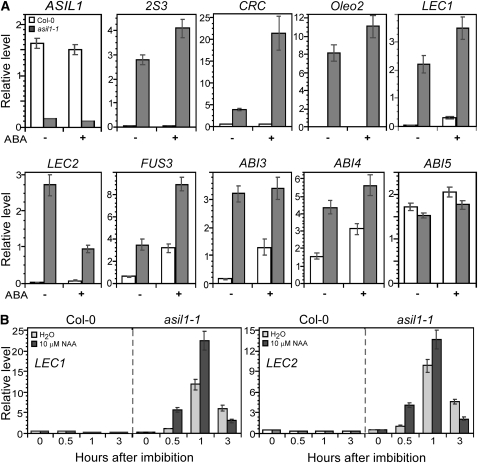
We examined various seed maturation genes in 2-week-old seedlings and seedlings treated with ABA to determine whether ASIL1 negatively regulates their expression in vegetative tissues. The accumulation of transcripts from the 2S albumin gene 2S3, 12S globulin CRC, and the major lipid-body protein oleosin Oelo2 gene (Crowe et al., 2000), which are normally expressed during the maturation phase of seed development (Parcy et al., 1994), was increased in the untreated asil1 seedlings compared to the wild type (Figure 5A). The increase in 2S3, CRC, and Oleo2 transcripts was enhanced after application of exogenous ABA, indicating that ABA is involved in the ASIL1-dependent derepression of these seed-specific genes in vegetative tissues.
Figure 5.
Effect of ASIL1 and Its Interaction with ABA and Auxin on Expression of Seed Maturation Genes in Arabidopsis Seedlings.
Expression of seed maturation genes was analyzed using RNA isolated from either 2-week-old wild-type Col-0 and asil1-1 mutant seedlings that were grown on medium and incubated with 0 (−) or 50 μM (+) ABA for 2 d (A) or wild-type and asil1-1 seeds that had been imbibed for up to 3 h in the presence or absence of 10 μM NAA (B). The mean and sd from qRT-PCR analysis were determined from three biological replicates.
(A) Expression of seed maturation genes in wild-type and asil1-1 seedlings. Total RNA was isolated from 2-week-old plants after exposure to ABA. The levels of various gene transcripts were determined by qRT-PCR using ACT2 mRNA as an internal reference.
(B) Expression of LEC1 and LEC2 in germinating wild-type and asil1-1 seeds. Total RNA was isolated from desiccated seeds or seeds that had been imbibed with the presence of NAA. Real-time RT-PCR was used to examine the levels of LEC1 and LEC2 transcripts with Ef-1α as a reference gene.
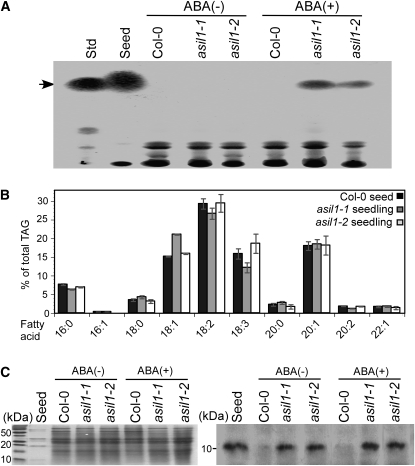
To further examine the extent to which expression of embryonic traits is modified in asil1 seedlings, we analyzed the asil1 seedlings for the presence of seed storage reserves that are normally accumulated in embryos. Total lipids were extracted and analyzed by thin layer chromatography (TLC) from 2-week-old seedlings grown in soil and treated with ABA. Upon exogenous application of ABA, the asil1 seedlings were found to accumulate triacylglycerol (TAG), which is the common storage lipid in Arabidopsis seeds; TAG was not detected in either ABA-treated or untreated wild-type seedlings or in the untreated asil1 seedlings (Figure 6A). To determine whether the profile of fatty acids in asil1 seedlings was similar to that of seeds, total lipids were separated by TLC and TAG fractions were purified and subjected to gas chromatography. The fatty acid composition of TAG from asil1 seedlings was found to be nearly identical to that of seed-derived lipids (Figure 6B). In particular, 18:2 was the most abundant fatty acid, 16:1 fatty acid was not detected in the TAG of asil1 seedlings, and a large amount of the very-long-chain fatty acid 20:1 was found in the TAG of asil1 seedlings in proportions resembling lipids derived from wild-type seeds. To investigate SSP accumulation in asil1 seedlings, proteins were isolated from 3-week-old seedlings and subjected to immunoblot analysis with a polyclonal anti-2S albumin antibody. The albumin was detected in the ABA-treated and untreated asil1-1 and asil1-2 seedlings, whereas none was found in the wild type (Figure 6C). These results suggest that metabolism in the vegetative tissues of asil1 seedlings has been modified to become similar to that of wild-type seeds.
Figure 6.
Accumulation of Seed Storage Reserves in asil1 Seedlings.
(A) TAG in wild-type (Col-0) and asil1-1 and asil1-2 mutant seedlings. Total lipids were extracted from 3-week-old seedlings grown in soil and treated with 0 (−) or 50 μM (+) ABA for 24 h before harvest. An equivalent amount of extracts from wild-type and asil1 seedlings were separated by TLC and stained with sulphuric acid. Lipid extracts from wild-type seeds were used as a control, and glyceryl trilinoleate (10 μg) was used as a standard (Std). The position of the seed-specific TAG is indicated by an arrow.
(B) Fatty acid composition of TAG fraction from wild-type seeds and asil1 seedlings treated with ABA. TAG fraction was separated by TLC, and the fatty acid composition was analyzed on a gas chromatograph and expressed as a percentage of the total TAG fraction. Results represent the average and sd from three biological replicates.
(C) Accumulation of 2S albumin in asil1 seedlings. Protein extracts (20 μg) from 3-week-old wild-type and asil1-1 and asil1-2 mutant seedlings grown in soil and treated with 0 (−) or 50 μM (+) ABA for 24 h were resolved by SDS-PAGE and analyzed by either Coomassie blue staining (left) or immunoblotting with polyclonal antibody raised against 2S albumin (right). Protein extracts (0.5 μg) from wild-type seeds were used as a control. A portion of the membrane blot is shown (right).
In Arabidopsis, the major regulators LEC1, LEC2, FUS3, and ABI3 are fundamental for governing various aspects of seed development (Vicente-Carbajosa and Carbonero, 2005; To et al., 2006). In addition, ABI4 and ABI5 interact with the major regulators in the control of seed development (Brocard-Gifford et al., 2003). To obtain insight into the role of ASIL1 in the repression of seed maturation genes in vegetative tissues, the expression of these seed regulatory genes in wild-type Col-0 and asil1 mutants was examined in 2-week-old seedlings with or without ABA treatment. In the absence of ABA, the expression of ABI5 in asil1 seedlings was slightly decreased compared with the wild type. Conversely, LEC1, LEC2, FUS3, ABI3, and ABI4 transcript levels were significantly elevated, though relative change in expression of these genes varied (Figure 5A). Upon exogenous application of ABA, the levels of LEC1, FUS3, and ABI4 transcripts were significantly enhanced in asil1 seedlings. ABI3 expression was also upregulated compared with the wild type, though at a similar level to untreated asil1, whereas ABI5 transcript level was marginally changed (Figure 5A). These results suggest that derepression of seed maturation genes such as 2S3, CRC, and Oleo2 in asil1, is either directly or indirectly mediated by these upstream regulators and that ABA signaling is involved in the upregulation of embryonic genes in asil1 seedlings.
Because the expression of ASIL1 was increased within 1 h of seed imbibition (Figure 4C) and it exhibited a modest response to auxin (Figure 4B), we asked whether ASIL1 acts before the completion of seed germination and if its mode of action is affected by auxin treatment. The expression of LEC1 and LEC2 was examined in desiccated seeds and seeds that had been imbibed for up to 3 h in the presence or absence of 10 μM NAA. During embryogenesis, LEC1 (Lotan et al., 1998) and LEC2 (Stone et al., 2001) are expressed at the earliest stage and their transcripts dramatically decrease in mature seeds (Figure 4D). In dry or imbibed wild-type seeds, LEC1 and LEC2 transcripts were not observed, whereas expression was significantly elevated in asil1 at 1 h after imbibition with the increase being enhanced by the application of auxin (Figure 5B). These results suggest that ASIL1 represses LEC1 and LEC2 at a very early stage of germination with the direct or indirect mediation of auxin signaling.
We performed microarray analysis of asil1 and wild-type seedlings to identify global gene expression profiles and to explore the expression change of embryonic and seed maturation genes in the asil1 mutant. Total RNA was isolated from 2-week-old seedlings treated with 50 μM ABA for 24 h. Labeled RNAs were hybridized to the Affymetrix ATH1 whole-genome array that represents >22,500 probe sets, representing ∼24,000 genes (Redman et al., 2004). Comparison of gene expression differences between the wild type and asil1-1 resulted in the identification of 611 and 676 genes that were more than twofold (P ≤ 0.05) up- and downregulated, respectively, in the asil1-1 mutant compared to the wild type (see Supplemental Tables 1 and 2 online). Of the derepressed genes, 63 showed more than a fivefold higher level of expression in the mutant plants. This group was comprised largely of genes involved in environmental stress responses as well as seed maturation and lipid metabolism, which include genes encoding 2S albumin and 12S globulin seed storage proteins, embryonic regulators LEC1, FUS3, ABI3, and ABI4, seed-type oleosins, and dehydrins (Table 1; see Supplemental Table 1 online). Collectively, these results indicate that ASIL1 represents a novel component of the regulatory network that downregulates a large subset of embryonic and seed maturation genes in seedlings. However, due to the application of ABA that may suppress some other gene expression, a potential role for ASIL1 in other developmental processes would not be excluded.
Table 1.
Selected Genes Repressed by ASIL1 as Identified by Microarray Analysis
| Protein | Gene | AGI Code | Fold Change |
|---|---|---|---|
| Oleosin | OLEO2 | At5g40420 | 16.16 |
| OLEO1 | At4g25140 | 7.02 | |
| 2S albumin | 2S3 | At4g27160 | 9.86 |
| 12S globulin | CRA1 | At5g44120 | 23.15 |
| CRC | At4g28520 | 19.74 | |
| Seed-specific regulator | FUS3 | At3g26790 | 7.54 |
| ABI3 | At3g24650 | 7.30 | |
| LEC1 | At1g21970 | 5.79 | |
| Embryo-abundant protein | AT3G54150 | 18.99 | |
| AT1G55450 | 6.71 | ||
| Dormancy-associated protein | DRM1-L | AT2G33830 | 13.53 |
| DRM1 | AT1G28330 | 9.78 | |
| Lipid transfer protein family | PEARLI 1-L | AT4G12470 | 8.74 |
| PEARLI 1 | AT4G12480 | 4.13 | |
| Fatty acid enzyme | SSI2-L | AT1G43800 | 68.21 |
| FAH1 | AT2G34770 | 6.33 | |
| AT2G42450 | 9.82 | ||
| AT5G24210 | 3.89 | ||
| Late embryogenesis abundant protein | AT2G03850 | 4.45 | |
| AT3G17520 | 3.48 | ||
| ABA-responsive protein | ABA3 | AT1G16540 | 12.50 |
| ABI4 | At2g40220 | 6.69 | |
| Auxin-responsive GH3 family protein | GH3.4 | AT1G59500 | 10.59 |
| GH3.3 | AT2G23170 | 8.66 | |
| ERD6 | AT1G08930 | 7.19 | |
| Dehydration-induced protein | COR47 | AT1G20440 | 7.16 |
| AT2G34300 | 4.47 | ||
| Expansin | EXPR3 | AT2G18660 | 5.45 |
| AT3G55500 | 3.76 | ||
| Chromatin remodeling factors | HDA8 | AT1G08460 | 5.15 |
| SNF7 | AT2G06530 | 4.00 | |
| Gly-rich protein | GRP7 | AT2G21660 | 5.09 |
| Expressed protein | AT5G10040 | 18.99 | |
| AT5G42900 | 10.32 | ||
| AT4G39675 | 10.16 |
ASIL1 Binds to the GT-Box
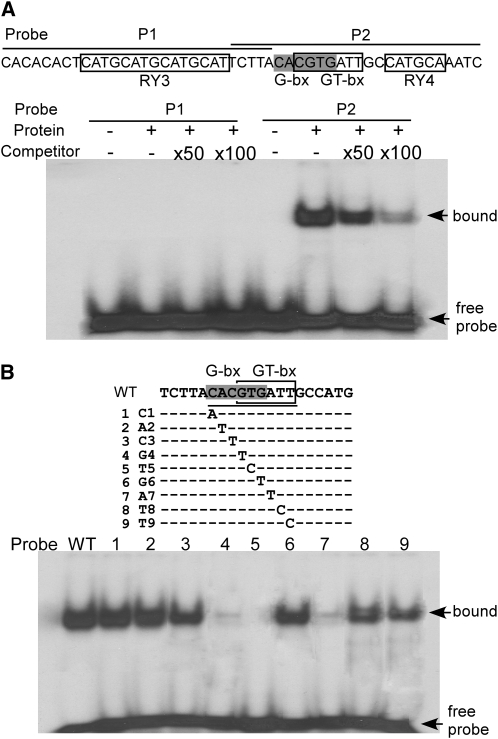
ASIL1 was isolated based on its interaction with a 49-bp element of the 2S3 gene between −95 and −47, a region that contains two RY repeats, an ACGT-box, and a GT-box–like motif (Figure 7; see Supplemental Figure 1 online). We performed in vitro DNA binding studies to confirm the DNA binding preference of ASIL1 for the 49-bp element. The entire ASIL1 coding region was inserted into the expression vector pET-28a, and ASIL1 protein extract was produced using a coupled in vitro transcription/translation reaction. The extract was incubated with probes corresponding to the RY3-motif region and G-bx/GT-bx/RY4-motif region (P1 and P2, respectively; Figure 7A) in the absence and presence of unlabeled competitor DNA. The binding of ASIL1 to the probes was analyzed using an electrophoretic mobility shift assay (EMSA). ASIL1 was unable to bind to the RY4-containing P1 probe but bound well to the P2 probe containing the ACGT- and GT-boxes; the binding to P2 was efficiently inhibited in the presence of competitor DNA in a concentration-dependant manner (Figure 7A). The G-box–containing ACGT element (ACACGTG) overlaps the GT-box–like motif (CGTGAAT). To determine the effect of specific bases within the P2 probe on the ASIL1 binding preference, a set of nine synthetic oligonucleotides containing single-nucleotide mutations within the 5′-CACGTGATT-3′ of the P2 oligonucleotide were generated and subjected to EMSA. The G residue at position 4 (G4), the T residue at position 5 (T5), and the A residue at position 7 (A7) were essential for ASIL1 binding as mutations in these positions markedly decreased binding, whereas residues C1, A2, C3, G6, T8, and T9 were not critical for ASIL1 binding (Figure 7B). These results demonstrate that the target binding sequence of ASIL1 in the 2S3 promoter is the GT-box–like domain but not the G-box as mutation of four of the six residues in the G-box had little effect on ASIL1 binding.
Figure 7.
ASIL1 Binding Specificity to the GT-Box of the 2S3 Promoter.
ASIL1 protein was produced using a coupled in vitro transcription/translation reaction and incubated with [α-32P]dATP-labeled probes (50,000 cpm/reaction) in an EMSA.
(A) Interaction of ASIL1 with sequence motifs in the 49-bp promoter region. The nucleotide sequences of probes (P1 and P2) used in the EMSA analysis are indicated on top of the figure (lines) with cis-elements indicated (boxed regions or shaded for G-box). Binding of ASIL1 to probes P1 and P2 was assayed with no (−) or 50- or 100-fold excess of unlabeled competitor DNA.
(B) DNA binding preference of ASIL1 protein to the GT-box. Binding of ASIL1 to either the G-box or GT-box was evaluated by systematically mutating the binding motifs as indicated (top panel) and assaying binding affinity by EMSA (bottom panel). The overlapping G-/GT-box motif is underlined with the G-box (shaded) and GT-box (boxed) indicated. Each of the nucleotides in the underlined motif was systematically substituted with A (1C1), T (2A2-4G4, 6G6, and 7A7) or C (5T5, 8T8, and 9T9).
Most trihelix proteins belong to the GT protein family, and most GT-binding elements are closely related though still somewhat divergent. Moreover, a single GT factor can interact with different GT elements (Zhou, 1999; see Supplemental Figure 4A online). To define the consensus sequence for GT-factor binding, we compared sequences between ASIL1 binding sites and GT elements previously identified in Arabidopsis, pea (Pisum sativum), rice (Oryza sativa), soybean (Glycine max), and bean. A consensus GT binding sequence was identified as 5′-GNNARN, where frequently N = T and R = A (see Supplemental Figure 4A online). We used this consensus sequence to search for putative ASIL1 binding sites in the promoters of seed maturation genes (2S1-5, CRC, Oleo1-4, LEC1, LEC2, FUS3, ABI3, and ABI4) that are negatively regulated by ASIL1, as determined by qRT-PCR (Figure 5) and microarray analysis (Table 1; see Supplemental Table 1 online). The putative ASIL1 binding sites were present in all the ASIL1 target genes examined. We have demonstrated that ASIL1 binding sites are closely associated with the major positive cis-elements ACGT- and G-boxes and the RY repeats in the 2S3 promoter. To investigate whether such an arrangement of ASIL1 binding sites could be a conserved feature of the 2S3 promoter, flanking sequences of the putative ASIL1 binding sites in the target gene promoters were analyzed using the PLACE program (Higo et al., 1999). Putative ASIL1 binding sites in most of the target gene promoters were consistently found either overlapping or in close proximity to the major positive cis-acting elements important for seed-specific expression, such as RY repeat, G-box, E-box, DPBF, (CA)n, and 2S elements (Higo et al., 1999) (see Supplemental Figure 4B online). This result suggests that a common mechanism exists by which ASIL1 regulates target genes by competitively inhibiting the association of transcriptional activators with these promoters.
DISCUSSION
Plant development is subject to fine regulation mediated by various developmental regulators. Embryogenesis and seed maturation are important phases of seed development. The termination of seed maturation and its transition to vegetative growth are dependant on the correct deployment of developmental programs and the maintenance of cell fates. It has been shown in Arabidopsis that many of the developmental pathways active during seed development are repressed in vegetative tissues and specific negative regulators act to shut down embryonic traits during vegetative growth (Ogas et al., 1997, 1999; Rider et al., 2003, 2004; Henderson et al., 2004; Li et al., 2005; Suzuki et al., 2007; Tsukagoshi et al., 2007; Tang et al., 2008). Here, we describe the isolation of a GT-box binding trihelix protein, ASIL1, which prevents expression of some of the foremost embryonic and seed maturation genes in seedlings.
ASIL1 Is a Nuclear Trihelix Transcription Factor
ASIL1 encodes a protein that is characteristic of the trihelix family of transcription factors. Thus far, trihelix DNA binding proteins have been found only in the plant kingdom (Ayadi et al., 2004). GT family proteins were the first group of trihelix transcription factors identified based on their binding to GT elements. GT proteins have been identified in Arabidopsis, rice, tobacco, and soybean (Zhou, 1999; O'Grady et al., 2001; Ayadi et al., 2004; Brewer et al., 2004). The rice GT-2 factor and its orthologs in Arabidopsis contain two separate trihelix (helix-loop-helix-loop-helix) motifs that function as DNA binding domains (Zhou, 1999), whereas other trihelix proteins have only one such domain (Figure 1B). The second major group of trihelix proteins in Arabidopsis is designated here as ASIL factors because of their similarity to tobacco SIP1. SIP1 is a nuclear trihelix protein that was isolated based on its interaction with the protein encoded by oncogene 6b from Agrobacterium tumefaciens (Kitakura et al., 2002). The function of SIP1 in plant growth and development remains to be determined. The ASIL1 gene encodes a DNA binding protein possessing a trihelix domain that likely facilitates its binding to DNA. ASIL1 contains three putative nuclear localization sequences, a Gly-rich motif, and a motif at the N terminus that shares some features with a repressor domain found in other transcriptional repressors (Hanna-Rose and Hansen, 1996). Experiments with smRS-GFP fused to ASIL1 demonstrated that ASIL1 localizes to the nuclei of plant cells, supporting its role as a nuclear trihelix transcriptional regulator.
ASIL1 Participates in the Repression of Seed-Specific Regulatory Network in Seedlings
Analysis of transcript levels of major seed storage product genes in asil1 mutant plants using qRT-PCR and microarray approaches revealed that ASIL1 was required for the repression of many seed maturation genes in seedlings. PKL encodes a CHD3 chromatin remodeling factor that belongs to the SWI/SNF class (Ogas et al., 1999) and acts in concert with GA to ensure that embryonic traits are not expressed after germination (Ogas et al., 1997). Many traits normally expressed during embryogenesis and seed formation have been observed in the primary roots of pkl seedlings (Ogas et al., 1997; Henderson et al., 2004; Rider et al., 2004; Li et al., 2005). LEC1, LEC2, and FUS3, which are key positive regulators for seed development and normally expressed in developing seeds (Harada, 2001), are significantly derepressed in pkl mutants during early germination (∼48 h after imbibition) as well as in the embryonic roots (Ogas et al., 1999; Rider et al., 2003). Thus, the expression of seed traits in vegetative tissues of pkl plants may be due to the elevated expression of LEC1 and LEC2 because these two major regulators of embryonic identity have the ability to induce somatic embryogenesis in vegetative tissues when expressed ectopically (Lotan et al., 1998; Stone et al., 2001; Li et al., 2005). HSI2 encodes a B3 domain protein with an EAR motif. Its expression is ubiquitous but slightly affected by sucrose. HSI2 is believed to function as a repressor of sugar-inducible reporter genes (Tsukagoshi et al., 2005). HSI2 and its homolog HSL1 (also referred to VP1/ABI3-LIKE VAL1 and VAL2) act together with sugar signaling to repress ectopic expression of seed maturation genes in seedlings and are necessary for the transition from seed maturation to active vegetative growth. Consistent with the embryonic seedling phenotypes in double mutant hsi2 hsl1, most of the embryonic and seed maturation genes, including LEC1, ABI3, FUS3, and genes for seed storage compounds, are upregulated in hsi2 hsl1 seedlings (Suzuki et al., 2007; Tsukagoshi et al., 2007). ESSP3, a member of the BRM family of chromatin remodeling ATPases, is also involved in the repression of seed maturation genes in vegetative tissues, and brm mutations cause ectopic expression of a subset of SSP and some other embryonic genes in leaves (Tang et al., 2008). However, ABI3, a key regulator for controlling seed development (Finkelstein et al., 2002) is not expressed in pkl (Rider et al., 2003), the expression of LEC2 is not detected in hsi2 hsl1 seedlings (Suzuki et al., 2007), and master regulators LEC1, LEC2, and ABI3 are not expressed in brm mutants (Tang et al., 2008), suggesting that PKL, VALs, and BRM act to selectively repress some genes that promote embryonic identity but not all. Moreover, it has not been demonstrated that these repressors directly target seed maturation genes. In contrast with PKL, VALs, and BRM regulators, the DNA binding factor ASIL1 directly regulates downstream embryonic genes in seedlings. Master regulators of seed maturation LEC1, LEC2, FUS3, and ABI3 were all substantially derepressed in 2-week-old asil1 seedlings in the absence of ABA. Furthermore, in the presence of ABA, transcript levels of SSP genes 2S3 and CRC and the major oleosin gene Oleo2, which are preferentially expressed in developing seeds, were dramatically elevated in asil1 seedlings, corresponding with a global shift in gene expression to a profile resembling late embryogenesis. Consistent with this embryonic gene expression profile, asil1 seedlings accumulated the 2S albumin with or without ABA treatment. Furthermore, after ABA treatment, seed-specific TAG was accumulated in asil1 seedlings with a fatty acid composition that was indistinguishable from that of seed lipid. Taken together, these results indicate that ASIL1 is important for repressing a large subset of embryonic and seed maturation genes in seedlings.
ABA and Auxin Are Involved in ASIL1-Dependent Derepression of Embryonic Genes
ABA is important for many aspects of plant development, including accumulation of seed storage reserves, acquisition of seed desiccation tolerance, and the inhibition of the phase transitions from seed maturation to germination and from vegetative to reproductive growth (Finkelstein et al., 2002). The seed maturation program is modulated by the endogenous ABA level, which is positively regulated by FUS3 (Gazzarrini et al., 2004) and intimately associated with the expression of major seed-specific regulators (Finkelstein et al., 2002). The fact that expression of a subset of seed maturation genes in asil1 seedlings was enhanced in response to ABA treatment suggests that ABA acts as a signal for activating embryonic genes. Induction of LEC2 in leaves results in the activation of 2S3 and oleosin S3 (Oleo1) and the accumulation of seed-specific lipids (Santos-Mendoza et al., 2005). Ectopic expression of ABI3 and FUS3 leads to the expression of 2S3 and CRC in vegetative tissues in an ABA-dependent manner (Parcy et al., 1994; Kagaya et al., 2005a). ASIL1 is, therefore, different from ABI3, FUS3, and LEC2 in terms of its mode of action in the regulation of seed-specific gene expression in seedlings. We propose that downregulation of 2S, CRC, and Oleo genes in seedlings is due to either direct binding of ASIL1 to the GT-boxes of 2S, CRC, and Oleo promoters or indirect suppression through major regulators LEC1, LEC2, FUS3, and ABI3. The fact that both mutation of ASIL1 and the ectopic expression of ABI3 affect the accumulation of seed-specific transcripts in vegetative tissues in an ABA-dependent manner (Parcy et al., 1994) suggests that ABI3 is part of the ASIL1-dependent pathway, and ABA cascade is actively involved in the ASIL1-dependent derepression of seed maturation genes in vegetative tissues. Further analysis of expression of seed maturation genes in asil1 mutants in an ABA-deficient background is required to understand the involvement of ABA in ASIL1-dependent regulation.
Genetic and molecular studies have shown that auxin plays an essential role in embryogenesis and postembryonic organ formation in Arabidopsis through its dynamic directional distribution (Tanaka et al., 2006). During embryogenesis, auxin accumulation is directed by PIN-mediated polar transportation from the apical cells to the hypophysis, the founder cell of the root stem–cell system (Tanaka et al., 2006). Given the fact that somatic embryogenesis is induced by the synthetic auxin 2,4-D (Mordhorst et al., 1998) and that derepression of LEC1 and LEC2 in imbibed asil1 seeds was enhanced by auxin, we suggest that auxin, similar to ABA, functions as a signal for the activation of embryonic genes. It has been suggested that auxin is required for the embryonic regulators FUS3, LEC1, and LEC2 to potentiate embryogenesis and seed maturation processes (Gazzarrini et al., 2004; Casson and Lindsey, 2006; Stone et al., 2008). Our findings of upregulation of embryonic genes in an auxin-dependent manner are also intriguing in light of the emerging role of auxin signaling during embryogenesis and seed maturation.
Auxin accumulation is also dynamically changed during germination and vegetative growth. For example, auxin activity was highly localized in the radical tips of germinating and germinated seeds (Liu et al., 2007), and the initiation sites of organ primordia in roots and shoots are correlated with the increased auxin levels (Tanaka et al., 2006). We demonstrated that ASIL1 exhibited a modest response to auxin and that the level of ASIL1 transcript is elevated 1 h after imbibition. We therefore suggest that the rise in ASIL1 levels may correlate with auxin distribution in cells of germinating seeds. Consistent with the ASIL1 expression pattern during germination, the accumulation of LEC1 and LEC2 transcripts in asil1 increased significantly at 1 h after imbibition. This result allows us to suggest that ASIL1 acts before the completion of seed germination. In addition to the present study, other reports also show the early repression of embryonic genes during seed germination. PKL seems to repress embryonic traits within 2 d after imbibition (Ogas et al., 1997; Li et al., 2005). Repression of a number of seed maturation genes by histone deacetylation was a transient event 24 h after imbibition (Tai et al., 2005). Recently, derepression of LEC1 was shown to occur within 12 h of treatment with the histone deacetylase inhibitor trichostatin A (Tanaka et al., 2008). LEC1 and LEC2 function upstream of other embryonic and seed maturation genes (To et al., 2006), and their ectopic expression is sufficient to provoke the embryonic program in vegetative tissues (Lotan et al., 1998; Stone et al., 2001; Santos-Mendoza et al., 2005). Therefore, expression of these two major regulators of the embryonic programming should be strictly prevented during germination and seedling development. Given the upregulation of ASIL1 by auxin and the derepression of embryonic genes in germinating asil1 seeds as well as in 2-week-old asil1 seedlings, we suggest that ASIL1 prevents ectopic expression of LEC1 and LEC2 in cells that encounter elevated auxin levels during germination and vegetative growth.
The GT-Box Is Involved in Repression of Seed Maturation Genes
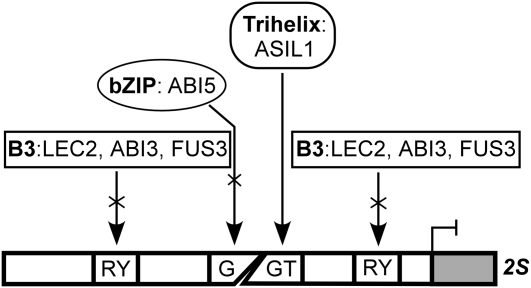
All trihelix transcription factors identified to date bind to GT-boxes. The first GT-box was identified as a GT1 binding site Box II in the pea rbcS-3A gene promoter (Green et al., 1987). Many other GT-boxes have been found in the promoter regions of genes with diverse functions, and most were shown to be highly divergent but closely related to the Box II of rbcS-3A (Zhou, 1999; Ayadi et al., 2004). GT-boxes have both positive and negative functions in the differential regulation of light responsiveness by cooperative interaction with other closely associated cis-acting elements (Zhou, 1999). Here, we demonstrate that ASIL1 exhibits binding specificity for a GT-box–like element (5′-GTGATT) in the 2S3 promoter in vitro. This GT element overlaps with the ACGT/G-box and is in close association with RY repeats. It is interesting to note that such arrangements of cis-elements are consistently found in the promoter regions of all the 2S albumin genes in Arabidopsis (see Supplemental Figure 4 online). The G-boxes are putative binding elements for the bZIP proteins bZIP10 and 25 in 2S promoters (Lara et al., 2003) and for ABI5 in the Em6 promoter (Carles et al., 2002). The RY repeats in different SSP gene promoters are specifically recognized by the B3-domain proteins LEC2, ABI3, and FUS3 (Reidt et al., 2000; Kroj et al., 2003; Mönke et al., 2004). In this work, we suggest that the relative position of the ASIL1 recognition site with respect to the ACGT/G-box and RY repeats is important for the negative regulation of 2S genes. Furthermore, we found that putative ASIL1 binding sites were consistently in close association with cis-elements that are important for the expression of seed maturation genes through the action of transcriptional activators (see Supplemental Figure 4B online). This conserved feature probably reflects a common regulatory mechanism, analogous to the sugar and ABA repression exerted by ABI4, owing to the close association of the ABI4 binding site S-box with the G-box in the minimal RBCS light-responsive unit CMA5 (Acevedo-Hernández et al., 2005). Taken together, we propose a model for the negative regulation of 2S genes mediated by the GT-box (Figure 8). In this model, ASIL1 is able to bind directly to the GT-box and repress the transcription of 2S genes by affecting the function of ABA-inducible transcriptional activators of seed development, such as LEC2, FUS3, ABI3, and ABI5. This may occur through direct competition for overlapping binding sites or closely linked sequences or be mediated by processes such as localized chromatin modifications that would prevent activator binding to the DNA.
Figure 8.
A Model for Negative Regulation of 2S Genes Mediated by the GT-Box.
ASIL1 specifically recognizes the GT-box, the bZIP proteins preferentially bind to the G-box, and the B3 regulators interact with the RY repeats. The GT-box overlaps the ACGT/G-box and is in close proximity to the RY repeats. ASIL1 represses 2S transcription by interfering with the binding of ABA-inducible bZIP and B3 proteins to the G-box and RY repeats, respectively.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seeds from ecotype Col-0 and T-DNA–tagged lines SALK_124095 (asil1-1) and SALK_074897 (asil1-2) were obtained from the ABRC. For RNA isolation from plants grown in pots of soil, Arabidopsis seeds were sown in RediEarth soil (W.R. Grace & Co.) and kept at 4°C for 3 d in the dark. Plants were grown in an environment-controlled growth chamber programmed for a 16-h long-day photoperiod with daytime temperature of 22°C and a night temperature of 16°C. Siliques were harvested in pools corresponding to early embryogenesis (1 to 6 DPA), mid-embryogenesis (7 to 9 DPA), and late embryogenesis (10 to 15 DPA) (Brocard-Gifford et al., 2003). Seeds were mature and completely dry after 15 DPA under the above growth conditions. To obtain plants in plates of medium, seeds were surface-sterilized and plated onto half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 0.55% agar and 1% sucrose. After imbibition for 3 d in the dark at 4°C, plants were grown at 22°C under continuous light. Tobacco (Nicotiana tabacum) plants were grown in pots of soil in a growth chamber at 22°C (16 h light/8h dark) for 18 d and used for leaf infiltration.
For exogenously applied ABA treatments, 2-week-old Arabidopsis seedlings aseptically grown in plates of medium under the above conditions were transferred to fresh half-strength MS medium supplemented with 0.7% agar, 1% sucrose, and 0 or 50 μM ABA (A1049; Sigma-Aldrich) and incubated for an additional 2 d before harvest. For GA and auxin treatments, 2-week-old seedlings were treated in liquid half-strength MS medium supplied with appropriate hormones for 1 h at concentrations as indicated in the figures or a mock treatment with 0 or 0.1% ethanol. NAA (N0640; Sigma-Aldrich) and GA3 (G7645; Sigma-Aldrich) were dissolved in ethanol, while IAA (I2886; Sigma-Aldrich) was dissolved in water. All tissues harvested for RNA isolation were immediately frozen in liquid nitrogen and stored at −70°C until processed.
Genotyping of T-DNA Mutants
The genotypes of asil1-1 and asil1-2 alleles were confirmed by PCR with two ASIL1-specific primers, sil-f1 and sil-r2 (Figures 3A and 3B; see Supplemental Table 3 online for all primer sequences) to detect the wild-type allele or with primer sil-r2 and the T-DNA left border–specific primer LBb1 to identify the mutant allele. The position of the T-DNA insertion in the ASIL1 gene in each mutant allele was verified by DNA sequencing of the PCR products that spanned the insertion site. The T-DNA insertion structure was verified by PCR and DNA gel blot analyses, and plants homozygous for the single insertion were used for total RNA isolation and gene expression analysis.
Yeast One-Hybrid Screening and Cloning
A yeast one-hybrid cDNA library was constructed from poly(A) mRNA from 3-week-old seedlings of Arabidopsis ecotype Col. mRNA was isolated using a mRNA extraction kit following the protocol of the supplier (GE Healthcare). cDNA was generated, and 5′ SalI and 3′ NotI sites were incorporated to enable subsequent cloning into the GAL4 activation domain vector pEXP-AD502 using the SuperScript plasmid system for cDNA synthesis and plasmid cloning with Gateway technology (Invitrogen). As bait, a DNA fragment containing three tandem copies of the MJ49 region (−95 to −47) of the At 2S3 promoter (Figure 7; see Supplemental Figure 1 online) was synthesized (Plant Biotechnology Institute) and cloned into the EcoRI/XbaI site of the pHISi vector and the EcoRI/SalI site of pLacZi vector (Clontech) to generate the plasmid constructs MJ49-HIS3 and MJ49-lacZ. Construct MJ49-HIS3 was linearized with AflII, integrated into a host chromosome of yeast strain YM4271, and used as bait to screen the cDNA library. MJ49-lacZ was integrated into the YM4271 genome in a similar fashion and used to confirm positive clones from the library screen. Approximately 1.5 × 106 transformants were screened using synthetic complete medium lacking tryptophan and histidine but including 15 mM 3-amino-1,2,4-triazole according to the instructions of the MATCHMAKER one-hybrid system (Clontech). Positive clones (HIS+) able to grow in the presence of 15 mM 3-amino-1,2,4-triazole were selected and analyzed by PCR amplification and DNA sequencing. Putative HIS+ candidates were further evaluated by transformation into YM4271 strain carrying MJ49-lacZ construct to test for the transcriptional activation of the lacZ reporter gene. β-Galactosidase activity was measured using chlorophenol red-β-d-galactopyranosidase according to the manufacturer's instructions (Invitrogen).
Amino Acid Sequence Alignment and Phylogenetic Analysis
Amino acid sequences were aligned using the AlignX program, part of the Vector NTI suite (Invitrogen), with default settings of parameters (gap opening penalty, 10; gap extension penalty, 0.05; gap separation penalty range, 8; identity for alignment delay, 40%) (Lu and Moriyama, 2004). The phylogenetic tree was constructed using the neighbor-joining algorithm with MEGA version 4 (Tamura et al., 2007) with 1000 bootstrap trials performed.
Subcellular Localization of ASIL1
The cDNA clone (accession number U70496) encoding smRS-GFP (Davis and Vierstra, 1998) was subcloned in the binary vector pCAMBIA (Hajdukiewicz et al., 1994) to generate construct p35S∷smRS-GFP. The entire coding region of ASIL1 was PCR amplified using primers sil-f4 and sil-r5 and inserted between the PstI and XbaI sites of the p35S∷smRS-GFP resulting in an in-frame translational fusion at the N terminus of smRS-GFP. This fusion was called ASIL1:smRS-GFP. The construct p35S∷ASIL1:smRS-GFP and the negative control p35S∷smRS-GFP were used for transient expression in N. tabacum epidermal cells as described by Sparkes et al. (2006). For nuclear staining, plant sections were incubated with 50 μg/mL DAPI (Invitrogen) for 15 min. DAPI-stained tissues were allowed to stand for 30 min before viewing. GFP and DAPI were excited with blue excitation and red channel, respectively, on a Zeiss Axio Imager Z1 microscope with ApoTome and AxioCam MRm (Carl Zeiss). Images were processed with AxioVision Rel. 4.7 (Carl Zeiss). The data shown were representative of three independent experiments.
RT-PCR
Total RNA was isolated from either leaves using the RNeasy plant mini kit (Qiagen) or siliques corresponding to different developmental stages as described by Suzuki et al. (2004). To remove genomic DNA, total RNA was treated with Amplification Grade DNase I (Invitrogen) following the manufacturer's instructions. Total RNA (250 ng) was used and 35 cycles of PCR were performed for analyzing ASIL1 expression in asil1-1 and asil1-2 leaves relative to wild-type Col-0. A 940-bp fragment of 18S rRNA gene that was coamplified with test gene was used as an internal standard. Reverse transcription of RNA into cDNA and the PCR amplification of both the internal standard and test gene fragment were performed in a one-step reaction using the SuperScript one-step RT-PCR system (Invitrogen). The RNA preparation and RT-PCR procedure were performed using at least two different biological replicates, and very similar results were obtained. Gene-specific primers for 18S were described by Gao et al. (2007); those for ASIL1 are listed in Supplemental Table 3 online.
Real-Time RT-PCR
Plant materials were harvested from three biological replicates, frozen immediately in liquid nitrogen, and stored at −80°C. Total RNA isolation and DNAse I treatment were as described above. RT reactions were performed with SuperScript III First-Strand Synthesis SuperMix for the qRT-PCR kit (Invitrogen) according to the manufacturer's instructions. Real-time PCR analysis was carried out using the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) on an ABI PRISM StepOnePlus real-time PCR system (Applied Biosystems), following the manufacturer's instructions. Gene-specific primers for potential reference genes ACT2, Ef-1α, UBQ10, and 18S were analyzed using total RNA isolated from developing siliques corresponding to different developmental stages and Arabidopsis seedlings treated with or without ABA. ACT2 and Ef-1α were the most stably expressed under our experimental conditions and chosen as endogenous reference genes. For each pair of primers, gel electrophoresis and melting curve analysis were performed to ensure that only a single PCR amplicon of the expected length and melting temperature was generated. The gene-specific primer sets for 2S3, ABI3, FUS3, and LEC1 were described by Santos-Mendoza et al. (2005); those for Ef-1α and UBQ10 were described by Czechowski et al. (2005); that for ACT2 was described by Tsukagoshi et al. (2005); and those for other genes used in real-time RT-PCR analysis are listed in Supplemental Table 3 online. Each sample was assayed in triplicate, and data were analyzed using the StepOne Software v2.0 (Applied Biosystems). The level of each mRNA was calculated using the mean threshold cycle (Ct) value and normalized to that of the reference gene ACT2 or Ef-1α. All results were shown as means of at least three biological replicates with corresponding standard deviations.
Microarray Analysis
Seeds from wild-type Col-0 and homozygous asil1-1 mutants were sterilized and germinated on plates with half-strength MS medium. After 14 d, seedlings were treated with 50 μM ABA for 24 h. Total RNA was extracted from the whole seedlings using the RNeasy Plant mini kit (Qiagen). For microarray analysis, we used the Affymetrix ATH1 whole-genome array that contains >22,500 probe sets representing ∼24,000 genes (Redman et al., 2004). Twenty micrograms of total RNA was used as template for double-strand cDNA synthesis, and biotin-labeled cRNA was synthesized using the GeneChip Expression One-Cycle cDNA synthesis kit (Affymetrix). Fifteen micrograms of fragmented cRNA was hybridized to the Affymetrix ATH1 GeneChip arrays according to the manufacturer's protocol (Affimetrix). Three biological replicates were used for each genotype. The GeneChip arrays were scanned using an Affymetrix GeneChip Scanner 3000. The single-array analysis and the generation of CEL image files were based on the GeneChip Expression Analysis Technical Manual (Affymetrix). The CEL files were then loaded into GeneSpring GX 9 (Agilent Technology) for extraction, transformation, quantile normalization, and background adjustment using robust multiarray analysis. For statistical analysis, gene expression data had to pass the false discovery rate multiple testing corrections (Hochberg and Benjamini, 1990). A P value cutoff of 0.05 was chosen, and differentially expressed genes were selected if they exhibited at least a twofold difference in normalized signals across all replicate experiments.
Lipid Analysis
For the investigation of TAG, total lipids were extracted from dry seeds or 3-week-old seedlings grown in soil and treated with 0 or 50 μM ABA for 24 h before harvest and analyzed by TLC as described by Rider et al. (2004) and Tsukagoshi et al. (2007) with minor modifications. Briefly, 500 mg of tissues were homogenized in 25 mL of chloroform:methanol (1:1, v/v). This mixture was extracted twice with 25 mL of 1 M NaCl/0.1 M HCl. The chloroform phase was then dried down under nitrogen, and the remaining material was redissolved in 300 μL of chloroform. Ten microliters of seed or 150 μL of leaf sample was separated on a silica Gel 60 F245 TLC plate (Merck), developed with hexane:ethyl acetate:acetic acid (6:1:0.1, v/v), and visualized by staining with sulphuric acid and heating at 120°C for 5 min. Glyceryl trilinoleate (Sigma-Aldrich) was used as a TAG standard. For the analysis of fatty acid methyl ester, TAG spots were scraped from the TLC plate, washed, and filtered into 1 mL of chloroform:methanol (1:1, v/v). This extract was dried down and redissolved in 0.5 mL of 1 n methanolic H2SO4. After heating at 80°C for 30 min, the methyl ester was extracted with 200 μL of hexane. The subsequent gas chromatography analysis of the resulting extracts was carried out as described by Young et al. (2006) on a Hewlett Packard 6890 gas chromatograph equipped with a DBWax column (10 m × 0.1 mm; Agilent Technologies Canada). Fatty acid methyl esters were detected by comparison of retention times with standard compounds.
Immunoblotting
Arabidopsis 2S albumin was purified by HPLC, and polyclonal anti-2S albumin was generated by the Animal Care Unit, Western College of Veterinary Medicine, University of Saskatchewan, Saskatoon, Canada. Protein extracts from seedlings were prepared as described by Shimada et al. (2003). Equal amounts of protein were subjected to SDS-PAGE followed by either staining with Coomassie Brilliant Blue R 250 or blotting to a polyvinylidene fluoride membrane (Fisher Scientific). The blots were incubated with 5% skim milk in PBST buffer (10 mM sodium phosphate, pH 7.2, 0.15 M NaCl, and 0.05% Tween 20). After washing three times with PBST buffer, the blots were incubated with anti-2S albumin (1:10,000 dilution) antiserum for 2 h and then washed three times in PBST. The blot was incubated with goat anti-mouse IgG (1:5000 dilution) (Bio-Rad) conjugated to horseradish peroxidase for 2 h and washed three times in PBST. The signals were detected with an ECL Plus Western Blotting Detection Kit (GE Healthcare).
In Vitro Transcription and Translation
The entire coding region of ASIL1 was amplified by PCR using primers sil-f2 and sil-r3 and cloned into the EcoRI and XhoI sites of the expression vector pET-28a (Novagen) in frame with the His-tag sequence to generate the construct pET-ASIL1. The ASIL1 protein was produced using TNT-Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions with some modifications as described previously (Gao et al., 2003). Two parallel reactions were conducted: one (25 μL) labeled with [35S]methionine to detect the incorporation by SDS-PAGE and fluorography, and the other (50 μL) in the absence of labeled methionine for the DNA binding reactions.
EMSAs
Mobility shift assays were performed as described previously (Gao et al., 2002; Sasaki et al., 2007). The sequences of all the oligonucleotides are shown in Figure 7. Both strands of the oligonucleotides were synthesized and annealed. DNA probes were generated by filling in 5′ overhangs with the Klenow fragment of DNA polymerase I (Promega) in the presence of [α-32P]dATP and purified using MicroSpin columns (GE Healthcare). Double-strand DNA filled with nonradioactive nucleotides was used as competitor DNA. DNA binding reactions were allowed to proceed in a 30 μL volume that contained 15 mM HEPES-KOH, pH 7.9, 30 mM KCl, 5mM DTT, 5 mM MgCl2, 1 mM EDTA, 5% glycerol, 2 μg of nonspecific competitor DNA poly(dI-dC), 50,000 cpm of radioactive probe, and 10 μL of in vitro translation mixture. Specific competitor DNA was added to the binding reaction as described in the legend of Figure 7. After 20 min incubation at room temperature, samples were resolved on 5% polyacrylamide gel in 0.25× Tris-borate-EDTA buffer at 4°C. The gels were subsequently dried and exposed to x-ray film.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Nicotiana tabacum SIP1, AB072391; ASIL1, At1g54060; GT-1a, At1g13450.1; GT-1b, At1g13450.2, GT-3a, At5g01380; GT-3b, At2g38250; GT-4, At3g25990; GT-2a, At1g76890.1; GT-2b, At1g76890.2; GT2L, At5g28300; GTL1, At1g33240; GTL1L, At5g47660; PTL, At5g03680; DF1L, At1g76880; 18S rRNA, X16077; 2S3, At4g27160; CRC, At4g28520; Oleo2, At5g40420; LEC1, At1g21970; LEC2, At1g28300; FUS3, At3g26790; ABI3, At3g24650; ABI4, At2g40220; ABI5, At2g36270; Em1, At3g51810; and EM6, At2g40170.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Regulatory Motifs in the 2S3 Promoter.
Supplemental Figure 2. Alignment of the Amino Acid Sequences of ASIL1 and SIP1.
Supplemental Figure 3. Expression Profiles of ASIL1 and Known Embryonic Gene Repressors PKL, VAL1, and ESSP3, in Developing Embryos Corresponding to Eight Developmental Stages.
Supplemental Figure 4. Putative ASIL1 Binding Sites and Their Close Association with cis-Acting Elements Required for Seed-Specific Expression.
Supplemental Table 1. Genes Upregulated in the asil1-1 Mutant Seedlings.
Supplemental Table 2. Genes Downregulated in the asil1-1 Mutant Seedlings.
Supplemental Table 3. Sequences of the Oligonuclotides Used in This Study.
Supplemental Data Set 1. Multiple Aignments of ASIL1 Homologs Used to Generate the Phylogenetic Tree Shown in Figure 1A.
Supplementary Material
Acknowledgments
We thank the ABRC for distribution of Arabidopsis seed lines; Brenda Oosterveen and Mark Jordan of the Affymetrix Core Laboratory (Agriculture and Agri-Food Canada, Winnipeg) for Affymetrix hybridizations; Gordon Gropp for thoughtful discussion; and Brent Mooney and Jason McEwen for technical assistance. We also thank the reviewers for providing such considerate suggestions for the article. This work was supported by the Agriculture and Agri-Food Canada Canadian Crop Genomics Initiative.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ming-Jun Gao (gaom@agr.gc.ca).
Online version contains Web-only data.
References
- Acevedo-Hernández, G.J., León, P., and Herrera-Estrella, L.R. (2005). Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 43 506–519. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi, M., Delaporte, V., Li, Y.F., and Zhou, D.X. (2004). Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. FEBS Lett. 562 147–154. [DOI] [PubMed] [Google Scholar]
- Baud, S., Mendoza, M.S., To, A., Harscoët, E., Lepiniec, L., and Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50 825–838. [DOI] [PubMed] [Google Scholar]
- Bensmihen, S., Rippa, S., Lambert, G., Jublot, D., Pautot, V., Granier, F., Giraudat, J., and Parcy, F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook, S.A., Stone, S.L., Park, S., Bui, A.Q., Le, B.H., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 103 3468–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, P.B., Howles, P.A., Dorian, K., Griffith, M.E., Ishida, T., Kaplan-Levy, R.N., Kilinc, A., and Smyth, D.R. (2004). PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131 4035–4045. [DOI] [PubMed] [Google Scholar]
- Brocard-Gifford, I.M., Lynch, T.J., and Finkelstein, R.R. (2003). Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 131 78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burow, M.D., Sen, P., Chlan, C.A., and Murai, N. (1992). Developmental control of the β-phaseolin gene requires positive, negative and temporal seed specific transcriptional regulatory elements and a negative element for stem and root expression. Plant J. 2 537–548. [Google Scholar]
- Carles, C., Bies-Etheve, N., Aspart, L., Léon-Kloosterziel, K.M., Koornneef, M., Echeverria, M., and Delseny, M. (2002). Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 30 373–383. [DOI] [PubMed] [Google Scholar]
- Casson, S.A., and Lindsey, K. (2006). The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol. 142 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan, M.B., Bishop, K.J., and Hall, T.C. (2003). Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J. 33 853–866. [DOI] [PubMed] [Google Scholar]
- Chern, M.S., Bobb, A.J., and Bustos, M.M. (1996). The regulator of MAT2 (ROM2) protein binds to early maturation promoters and represses PvALF-activated transcription. Plant Cell 8 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe, A.J., Abenes, M., Plant, A., and Moloney, M.M. (2000). The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci. 151 171–181. [DOI] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36 521–528. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S.L., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M.J., Allard, G., Byass, L., Flanagan, A.M., and Singh, J. (2002). Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol. Biol. 49 459–471. [DOI] [PubMed] [Google Scholar]
- Gao, M.J., Hegedus, D.D., Sharpe, A.G., Robinson, S.J., Lydiate, D.J., and Hannoufa, A. (2007). Isolation and characterization of a GCN5-interacting protein from Arabidopsis thaliana. Planta 225 1367–1379. [DOI] [PubMed] [Google Scholar]
- Gao, M.J., Schafer, U.A., Parkin, I.A., Hegedus, D.D., Lydiate, D.J., and Hannoufa, A. (2003). A novel protein from Brassica napus has a putative KID domain and responds to low temperature. Plant J. 33 1073–1086. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S., Tsuchiya, Y., Lumba, S., Okamoto, M., and McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7 373–385. [DOI] [PubMed] [Google Scholar]
- Green, P.J., Kay, S.A., and Chua, N.H. (1987). Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 6 2543–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, A.Y., Chen, X., Gao, G., Zhang, H., Zhu, Q.H., Liu, X.C., Zhong, Y.F., Gu, X., He, K., and Luo, J. (2008). PlantTFDB: A comprehensive plant transcription factor database. Nucleic Acids Res. 36 966–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose, W., and Hansen, U. (1996). Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 12 229–234. [DOI] [PubMed] [Google Scholar]
- Harada, J.J. (1999). Signaling in plant embryogenesis. Curr. Opin. Plant Biol. 2 23–27. [DOI] [PubMed] [Google Scholar]
- Harada, J.J. (2001). Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J. Plant Physiol. 158 405–409. [Google Scholar]
- Henderson, J.T., Li, H.C., Rider, S.D., Mordhorst, A.P., Romero-Severson, J., Cheng, J.C., Robey, J., Sung, Z.R., de Vries, S.C., and Ogas, J. (2004). PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg, Y., and Benjamini, Y. (1990). More powerful procedures for multiple significance testing. Stat. Med. 9 811–818. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y., Okuda, R., Ban, A., Toyoshima, R., Tsutsumida, K., Usui, H., Yamamoto, A., and Hattori, T. (2005. a). Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 46 300–311. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y., Toyoshima, R., Okuda, R., Usui, H., Yamamoto, A., and Hattori, T. (2005. b). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46 399–406. [DOI] [PubMed] [Google Scholar]
- Karssen, C.M., Brinkhorst-van der Swan, D.L.C., Breekland, A.E., and Koornneef, M. (1983). Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157 158–165. [DOI] [PubMed] [Google Scholar]
- Kitakura, S., Fujita, T., Ueno, Y., Terakura, S., Wabiko, H., and Machida, Y. (2002). The protein encoded by oncogene 6b from Agrobacterium tumefaciens interacts with a nuclear protein of tobacco. Plant Cell 14 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, C., and Grossniklaus, U. (2002). Epigenetic inheritance of expression states in plant development: the role of Polycomb group proteins. Curr. Opin. Cell Biol. 14 773–779. [DOI] [PubMed] [Google Scholar]
- Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073. [DOI] [PubMed] [Google Scholar]
- Lara, P., Oñate-Sánchez, L., Abraham, Z., Ferrándiz, C., Díaz, I., Carbonero, P., and Vicente-Carbajosa, J. (2003). Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 278 21003–21011. [DOI] [PubMed] [Google Scholar]
- Li, G., Bishop, K.J., Chandrasekharan, M.B., and Hall, T.C. (1999). β-Phaseolin gene activation is a two-step process: PvALF-facilitated chromatin modification followed by abscisic acid-mediated gene activation. Proc. Natl. Acad. Sci. USA 96 7104–7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Chandler, S.P., Wolffe, A.P., and Hall, T.C. (1998). Architectural specificity in chromatin structure at the TATA box in vivo: nucleosome displacement upon β-phaseolin gene activation. Proc. Natl. Acad. Sci. USA 95 4772–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.C., Chuang, K., Henderson, J.T., Rider, S.D., Jr., Bai, Y., Zhang, H., Fountain, M., Gerber, J., and Ogas, J. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P.P., Montgomery, T.A., Fahlgren, N., Kasschau, K.D., Nonogaki, H., and Carrington, J.C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52 133–146. [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. [DOI] [PubMed] [Google Scholar]
- Lu, G., and Moriyama, E.N. (2004). Vector NTI, a balanced all-in-one sequence analysis suite. Brief. Bioinform. 5 378–388. [DOI] [PubMed] [Google Scholar]
- Makarevich, G., Leroy, O., Akinci, U., Schubert, D., Clarenz, O., Goodrich, J., Grossniklaus, U., and Köhler, C. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke, G., Altschmied, L., Tewes, A., Reidt, W., Mock, H.P., Bäumlein, H., and Conrad, U. (2004). Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219 158–166. [DOI] [PubMed] [Google Scholar]
- Mordhorst, A.P., Voerman, K.J., Hartog, M.V., Meijer, E.A., van Went, J., Koornneef, M., and de Vries, S.C. (1998). Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26 627–635. [DOI] [PubMed] [Google Scholar]
- Ng, D.W., Chandrasekharan, M.B., and Hall, T.C. (2006). Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell 18 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Cheng, J.C., Sung, Z.R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Grady, K., Goekjian, V.H., Naim, C.J., Nagao, R.T., and Key, J.L. (2001). The transcript abundance of GmGT-2, a new member of the GT-2 family of transcription factors from soybean, is down-regulated by light in a phytochrome-dependent manner. Plant Mol. Biol. 47 367–378. [DOI] [PubMed] [Google Scholar]
- Ouelhadj, A., Kuschk, P., and Humbeck, K. (2006). Heavy metal stress and leaf senescence induce the barley gene HvC2d1 encoding a calcium-dependent novel C2 domain-like protein. New Phytol. 170 261–273. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giradaut, J. (1994). Regulation of gene expression program during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikhel, N. (1992). Nuclear targeting in plants. Plant Physiol. 100 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman, J.C., Haas, B.J., Tanimoto, G., and Town, C.D. (2004). Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 38 545–561. [DOI] [PubMed] [Google Scholar]
- Reidt, W., Wohlfarth, T., Ellerström, M., Czihal, A., Tewes, A., Ezcurra, I., Rask, L., and Bäumlein, H. (2000). Gene regulation during late embryogenesis: The RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 21 401–408. [DOI] [PubMed] [Google Scholar]
- Rider, S.D., Hemm, M.R., Hostetler, H.A., Li, H.C., Chapple, C., and Ogas, J. (2004). Metabolic profiling of the Arabidopsis pkl mutant reveals selective derepression of embryonic traits. Planta 219 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider, S.D., Henderson, J.T., Jerome, R.E., Edenberg, H.J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Mendoza, M., Dubreucq, B., Miquel, M., Caboche, M., and Lepiniec, L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579 4666–4670. [DOI] [PubMed] [Google Scholar]
- Sasaki, K., Mitsuhara, I., Seo, S., Ito, H., Matsui, H., and Ohashi, Y. (2007). Two novel AP2/ERF domain proteins interact with cis-element VWRE for wound-induced expression of the tobacco tpoxN1 gene. Plant J. 50 1079–1092. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Shimada, T., et al. (2003). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278 32292–32299. [DOI] [PubMed] [Google Scholar]
- Sparkes, I.A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protocols 1 2019–2025. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Braybrook, S.A., Paula, S.L., Kwong, L.W., Meuser, J., Pelletier, J., Hsieh, T.F., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 105 3151–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Wang, H.H., and McCarty, D.R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Kawazu, T., and Koyama, H. (2004). RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37 542–544. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudleym, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Dhonukshe, P., Brewer, P.B., and Friml, J. (2006). Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 63 2738–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, M., Kikuchi, A., and Kamada, H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, H.H., Tai, G.C., and Beardmore, T. (2005). Dynamic histone acetylation of late embryonic genes during seed germination. Plant Mol. Biol. 59 909–925. [DOI] [PubMed] [Google Scholar]
- Tang, X., Hou, A., Babu, M., Nguyen, V., Hurtado, L., Lu, Q., Reyes, J.C., Wang, A., Keller, W.A., Harada, J.J., Tsang, E.W., and Cui, Y. (2008). The Arabidopsis BRAHMA chromatin remodelling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 147 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, A., Valon, C., Savino, G., Guilleminot, J., Devic, M., Giraudat, J., and Parcy, F. (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, Y., Nambara, E., Naito, S., and McCourt, P. (2004). The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37 73–81. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi, H., Morikami, A., and Nakamura, K. (2007). Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 104 2543–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi, H., Saijo, T., Shibata, D., Morikami, A., and Nakamura, K. (2005). Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol. 138 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa, J., and Carbonero, P. (2005). Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49 645–651. [DOI] [PubMed] [Google Scholar]
- Young, L.W., Jalink, H., Denkert, R., and Reaney, M.T.J. (2006). Factors affecting the density of Brassica napus seeds. Seed Sci.Technol. 34 633–645. [Google Scholar]
- Zhou, D.X. (1999). Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci. 4 210–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.