Abstract
It has previously been shown that SECONDARY WALL–ASSOCIATED NAC DOMAIN PROTEIN1 (SND1) is a key transcription factor regulating secondary cell wall formation, including the biosynthesis of cellulose, xylan, and lignin. In this study, we show that two closely related SND1-regulated MYB transcription factors, MYB58 and MYB63, are transcriptional regulators specifically activating lignin biosynthetic genes during secondary wall formation in Arabidopsis thaliana. MYB58 and MYB63 are phylogenetically distinct from previously characterized MYBs shown to be associated with secondary wall formation or phenylpropanoid metabolism. Expression studies showed that MYB58 and MYB63 are specifically expressed in fibers and vessels undergoing secondary wall thickening. Dominant repression of their functions led to a reduction in secondary wall thickening and lignin content. Overexpression of MYB58 and MYB63 resulted in specific activation of lignin biosynthetic genes and concomitant ectopic deposition of lignin in cells that are normally unlignified. MYB58 was able to activate directly the expression of lignin biosynthetic genes and a secondary wall–associated laccase (LAC4) gene. Furthermore, the expression of MYB58 and MYB63 was shown to be regulated by the SND1 close homologs NST1, NST2, VND6, and VND7 and their downstream target MYB46. Together, our results indicate that MYB58 and MYB63 are specific transcriptional activators of lignin biosynthesis in the SND1-mediated transcriptional network regulating secondary wall formation.
INTRODUCTION
Secondary cell walls are deposited in specialized cells, such as tracheary elements and fibers. They are laid down next to the primary walls after the cells cease elongation. Recent molecular and genetic studies have revealed that a transcriptional regulatory network is involved in the activation of the secondary wall biosynthetic program in tracheary elements and fibers (Zhong and Ye, 2007). In this network, a group of closely related NAC domain transcription factors, including SECONDARY WALL–ASSOCIATED NAC DOMAIN PROTEIN1 (SND1), NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1), NST2, VASCULAR-RELATED NAC DOMAIN6 (VND6), and VND7, are master switches activating secondary wall biosynthesis in various Arabidopsis thaliana cell types. SND1 and NST1 function redundantly in turning on the secondary wall biosynthetic program in fibers (Zhong et al., 2006, 2007b; Mitsuda et al., 2007), whereas NST1 and NST2 are switches for secondary wall biosynthesis in the endothecium of anthers (Mitsuda et al., 2005). VND6 and VND7 are involved in regulating secondary wall thickening in vessels (Kubo et al., 2005; Zhong et al., 2008). These secondary wall NACs were proposed to act through a cascade of downstream transcription factors, which in turn lead to the activation of secondary wall biosynthetic genes (Zhong and Ye, 2007; Zhong et al., 2008). Among the downstream transcription factors, MYB46, SND3, MYB103, and KNAT7 have been proven to be direct targets of SND1 (Zhong et al., 2008), and MYB46 itself is sufficient to induce the entire secondary wall biosynthetic program (Zhong et al., 2007a). It is currently unknown which SND1-regulated downstream transcription factors are directly involved in activating secondary wall biosynthetic genes. Identification of transcription factors directly regulating the individual biosynthetic pathways of cellulose, xylan, and lignin will not only help us understand the developmental mechanisms underlying secondary wall formation but also potentially have significant impact on tree biotechnology.
The biosynthesis of the three major secondary wall components, including cellulose, xylan, and lignin, is mediated by many enzymes whose genes need to be coordinately turned on during secondary wall formation. Although it is not known whether direct activation of the individual biosynthetic pathways of secondary wall components is mediated by the same transcriptional regulators or involves different transcriptional regulators specific for each pathway, available evidence suggests that some MYB and LIM transcription factors are implicated in regulation of lignin biosynthesis (Rogers and Campbell, 2004). Promoter deletion studies have revealed that the promoter of the bean (Phaseolus vulgaris) phenylalanine ammonia-lyase 2 (PAL2) gene contains three AC elements, namely, AC-I (ACCTACC), AC-II (ACCAACC), and AC-III (ACCTAAC), which are necessary for the vascular expression of the PAL2 gene (Hatton et al., 1995). Putative AC elements are present in the majority of the promoters of phenylpropanoid biosynthetic genes, and it has been proposed that the coordinated expression of these genes is regulated by transcription factors that bind to the AC elements (Raes et al., 2003). MYB proteins involved in regulation of flavonoid biosynthesis have been shown to be able to bind to the AC elements, and this binding may directly activate the expression of the general phenylpropanoid biosynthetic genes and the flavonoid branch pathway genes (Sablowski et al., 1994; Hartmann et al., 2005). Overexpression of flavonoid-associated MYB genes results in an alteration in the expression of the general phenylpropanoid biosynthetic genes, which indirectly affects lignin biosynthesis (Borevitz et al., 2000; Deluc et al., 2006).
So far, only a few studies have been focused on MYB genes that are potentially involved in specific activation of lignin biosynthetic genes during secondary wall formation. Several MYB genes have been demonstrated to affect the expression of lignin biosynthetic genes when overexpressed (Tamagnone et al., 1998; Jin et al., 2000). Two pine (Pinus taeda) MYB genes (Pt MYB1 and Pt MYB4) and a eucalyptus (Eucalyptus grandis) MYB gene (Eg MYB2) have been shown to be expressed during secondary xylem development in wood, and their encoded proteins are able to bind to the AC elements (Patzlaff et al., 2003a, 2003b; Goicoechea et al., 2005). Although their roles in lignin biosynthesis have not yet been proven by downregulation, heterologous overexpression of Pt MYB4 or Eg MYB2 in tobacco (Nicotiana tabacum) results in ectopic deposition of lignin or increased thickening of secondary walls in xylem, respectively (Patzlaff et al., 2003a; Goicoechea et al., 2005). It has been proposed that these pine and eucalyptus MYB genes are involved in regulation of lignin biosynthesis during wood formation. However, Pt MYB4 and Eg MYB2 are close homologs of Arabidopsis MYB46 (Goicoechea et al., 2005), which has been shown to be a key regulator of the biosynthesis of all three major secondary wall components, including cellulose, xylan, and lignin (Zhong et al., 2007a). Therefore, it remains to be investigated whether Pt MYB4 and Eg MYB2 regulate the biosynthesis of all secondary wall components or specifically lignin biosynthesis.
The identification of SND1 and MYB46 as master switches for secondary wall biosynthesis provides unprecedented tools for uncovering transcription factors specifically regulating lignin biosynthetic genes during secondary wall formation. During our study of secondary wall–associated transcription factors, we have found that two SND1-regulated MYB transcription factors, MYB58 and MYB63, are specifically expressed in cells undergoing secondary wall thickening and are important regulators of lignin biosynthesis. We demonstrate that dominant repression or simultaneous RNA interference (RNAi) inhibition of MYB58 and MYB63 causes a reduction in lignin content and that their overexpression leads to ectopic deposition of lignin. We reveal that MYB58 is able to activate directly lignin biosynthetic genes and a secondary wall–associated laccase gene. In addition, we show that the expression of MYB58 and MYB63 is regulated by the SND1 close homologs NST1, NST2, VND6, and VND7 and their downstream target MYB46. Our results demonstrate that MYB58 and MYB63 are specific transcriptional activators of lignin biosynthesis in the SND1-mediated transcriptional network.
RESULTS
The MYB58 and MYB63 Genes Are Specifically Expressed in Fibers and Vessels
Fibers in Arabidopsis inflorescence stems can be used as a model to dissect the transcriptional network regulating the biosynthesis of secondary walls. To identify transcription factors involved in regulation of secondary wall biosynthesis, we searched for transcription factors that were associated with secondary wall thickening in fiber cells (Zhong et al., 2006). In this report, we studied two MYB transcription factors, MYB58 (At1g16490) and MYB63 (At1g79180), which were highly expressed in interfascicular fibers and xylem cells but not in parenchymatous pith cells (Figures 1A and 1B). At the organ level, they exhibited a predominant expression in inflorescence stems in which the secondary wall–containing fibers and xylem cells are abundant (Figure 1C).
Figure 1.
Expression and Phylogenetic Analyses of the MYB58 and MYB63 Genes.
The expression of MYB58 and MYB63 was studied using real-time quantitative RT-PCR. The quantitative differences of their expression between the wild type and transgenic lines as shown in (D) and (E) are statistically significant (P < 0.001). Error bars represent se of three biological replicates.
(A) and (B) MYB58 (A) and MYB63 (B) were expressed highly in interfascicular fibers and xylem cells but not in pith cells. RNAs used for gene expression analysis were isolated from laser-microdissected interfascicular fiber, xylem, and pith cells. The expression levels of MYB58 and MYB63 in pith cells are set to 1.
(C) MYB58 and MYB63 exhibited a predominant expression in inflorescence stems. Their expression levels in seedlings are set to 1.
(D) The expression of MYB58 and MYB63 in stems was drastically reduced by RNAi inhibition of SND1 and NST1 (SND1/NST1-RNAi). Their expression levels in the wild type are set to 100.
(E) MYB58 and MYB63 in leaves were induced by overexpression of SND1 (SND1-OE) and NST1 (NST1-OE). Their expression levels in the wild type are set to 1.
(F) Phylogenetic analysis of MYB58 and MYB63 together with other MYBs involved in regulation of secondary wall biosynthesis or phenylpropanoid metabolism. Bootstrap values are shown in percentages at nodes. A text file of the alignment used to generate this phylogeny is available as Supplemental Data Set 1 online.
(G) Alignment of the deduced amino acid sequences of MYB58 and MYB63. The R2R3 MYB domain is underlined. Identical and similar amino acid residues are shaded with black and gray, respectively. Gaps (marked with dashes) are introduced to maximize the sequence alignment.
We next investigated the developmental expression pattern of MYB58 and MYB63 using the β-glucuronidase (GUS) reporter gene. To ensure that the reporter gene expression represents the endogenous gene expression pattern, we inserted the GUS gene right before the stop codon of the MYB58 and MYB63 genes in constructs that included a 3-kb 5′ upstream sequence, the entire exon and intron sequence, and a 2-kb 3′ downstream sequence. Examination of the stems of 30 independent transgenic plants showed that in the elongating internodes, in which protoxylem vessels are the only cells with visible secondary wall thickening (Ye et al., 2002), MYB58 was predominantly expressed in protoxylem vessels (Figure 2A) and MYB63 was preferentially expressed in interfascicular fibers (Figure 2D). The expression of MYB58 and MYB63 was evident in both interfascicular fibers and xylem cells in the nonelongating internodes (Figures 2B and 2E) in which both xylem and interfascicular fibers are undergoing secondary wall thickening (Ye et al., 2002). Examination of roots revealed that MYB63 was expressed in the developing secondary xylem of roots (Figure 2F), whereas MYB58 expression was not detected in roots. These results demonstrate that MYB58 and MYB63 are both expressed in fibers and vessels albeit with different developmental patterns.
Figure 2.
Developmental Expression Study and Transactivation Analysis of MYB58 and MYB63.
The developmental expression patterns of MYB58 and MYB63 were studied using the GUS reporter gene. For transcriptional activation analysis, MYB58 and MYB63 fused with the GAL4 DNA binding domain (GAL4BD) were expressed in yeast and examined for activation of the LacZ reporter gene. co, cortex; if, interfascicular fiber; mx, metaxylem; pi, pith; px, protoxylem; sp, secondary phloem; sx, secondary xylem. Bar = 150 μm.
(A) Cross section of an elongating internode of a MYB58:GUS plant showing predominant GUS staining (blue) in protoxylem.
(B) Cross section of a nonelongating internode of a MYB58:GUS plant showing GUS staining in xylem and interfascicular fibers.
(C) Transactivation analysis showing that MYB58 and MYB63 were able to activate the expression of the LacZ reporter gene. The β-galactosidase (β-Gal) activity in yeast cells expressing GAL4BD is set to 1. Error bars represent se of three replicates.
(D) Cross section of an elongating internode of a MYB63:GUS plant showing prominent GUS staining in interfascicular fibers.
(E) Cross section of a nonelongating internode of a MYB63:GUS plant showing GUS staining in interfascicular fibers and xylem.
(F) Cross section of a root of a MYB63:GUS plant showing GUS staining in developing secondary xylem.
MYB58 and MYB63 Are SND1-Regulated Transcriptional Activators
To find out whether the expression of MYB58 and MYB63 is regulated by SND1, we analyzed their expression in transgenic plants with RNAi inhibition of SND1 and NST1 and in plants with overexpression of SND1 and NST1. Real-time quantitative RT-PCR analysis showed that the expression of MYB58 and MYB63 was drastically reduced in the plants with RNAi inhibition of SND1 and NST1 (Figure 1D). By contrast, MYB58 expression was significantly upregulated by overexpression of SND1 and NST1 (Figure 1E). MYB63 was induced by NST1 overexpression, but not by SND1 overexpression, indicating that the MYB63 expression is mainly regulated by NST1. These results suggest that MYB58 and MYB63 are likely part of the SND1/NST1-mediated transcription network involved in regulation of secondary wall biosynthesis in fibers and vessels.
A number of MYB transcription factors have previously been reported to be involved in regulation of phenylpropanoid biosynthesis, secondary xylem development, or secondary wall formation (Tamagnone et al., 1998; Borevitz et al., 2000; Patzlaff et al., 2003a, 2003b; Karpinska et al., 2004; Goicoechea et al., 2005; Deluc et al., 2006; Fornalé et al., 2006; Zhong et al., 2007a). Phylogenetic analysis demonstrated that MYB58 and MYB63 are distinct from those previously characterized MYBs that are potentially involved in phenylpropanoid or secondary wall biosynthesis (Figure 1F). MYB58 and MYB63, which contain a typical R2R3 MYB domain, are closely related to each other; they share 71% similarity and 65% identity at the amino acid level (Figure 1G).
To examine whether MYB58 and MYB63 are transcriptional activators, we fused them with the GAL4-DNA binding domain for transactivation analysis in yeast. It was shown that both MYB58 and MYB63 were able to induce the expression of the LacZ reporter gene (Figure 2C), indicating that they are transcriptional activators. Yellow fluorescent protein (YFP)–tagged MYB58 and MYB63 were expressed in Arabidopsis leaf protoplasts and shown to be localized in the nucleus, consistent with their predicted function as transcription factors (see Supplemental Figure 1 online).
Dominant Repression of MYB58 and MYB63 Results in a Reduction in Secondary Wall Thickening
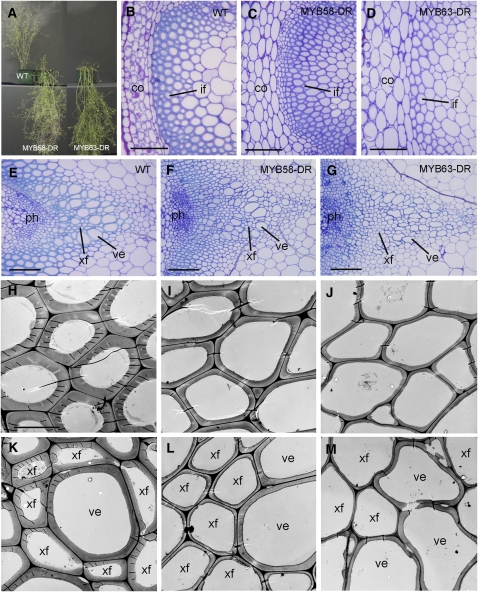
The findings that the expression of MYB58 and MYB63 is associated with secondary wall thickening and regulated by SND1 prompted us to investigate their roles in secondary wall biosynthesis. Because MYB58 and MYB63 are close homologs and both of them are expressed in vessels and fibers, we reasoned that they might function redundantly in the regulation of secondary wall biosynthesis. Therefore, we employed the EAR dominant repression approach (Hiratsu et al., 2004) for functional study of MYB58 and MYB63 as this approach has been successfully applied to facilitate the analysis of functionally redundant transcription factors involved in the regulation of secondary wall biosynthesis (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2007a). The EAR domain–induced dominant repression likely blocks the functions of both the transcription factors targeted for repression and their homologs. MYB58 and MYB63 were fused with the EAR repression domain at their C termini, and the chimeric proteins were expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter in transgenic Arabidopsis plants. It was found that out of 96 transgenic plants with MYB58 or MYB63 repression, 15 and 23 of them, respectively, showed a pendent stem phenotype (Figure 3A). Cross sections of the basal parts of stems revealed that dominant repression of MYB58 or MYB63 caused a drastic reduction in secondary wall thickness in both interfascicular fibers and xylary fibers compared with the wild type (Figures 3B to 3M; Table 1). The wall thickness of vessels was also reduced by dominant repression of MYB63 (Figures 3K to 3M; Table 1). Some of the vessels in the MYB63 repressors were apparently deformed (Figure 3G), which is likely caused by the weakening of vessel walls that renders them unable to resist the negative pressure caused by transpiration.
Figure 3.
Reduction of Secondary Wall Thickening by Dominant Repression of MYB58 and MYB63.
The bottom internodes of 8-week-old transgenic plants expressing MYB58 or MYB63 fused with the dominant EAR repression sequence (MYB58-DR and MYB63-DR) were examined for secondary wall thickening in fibers and vessels. co, cortex; if, interfascicular fiber; ph, phloem; ve, vessel; xf, xylary fiber. Bars = 76 μm in (B) to (G) and 8 μm in (H) for (H) to (M). [See online article for color version of this figure.]
(A) Wild-type plant (left) and transgenic plants expressing MYB58-DR (middle) and MYB63-DR (right). The inflorescence stems of MYB58-DR and MYB63-DR were pendent.
(B) to (D) Cross sections of interfascicular regions showing a reduction in the wall thickness of fibers in MYB58-DR (C) and MYB63-DR (D) plants compared with the wild type (B).
(E) to (G) Cross sections of vascular bundles showing a reduction in the wall thickness of xylary fibers in MYB58-DR (F) and MYB63-DR (G) plants and collapsed vessels in MYB63-DR (G) plants compared with the wild type (E).
(H) to (J) Transmission electron micrographs of interfascicular fiber walls of the wild type (H) and the MYB58-DR (I) and MYB63-DR (J) plants.
(K) to (M) Transmission electron micrographs of the walls of vessels and xylary fibers in the wild type (K) and the MYB58-DR (L) and MYB63-DR (M) plants.
Table 1.
Wall Thickness of Vessels and Fibers in the Stems of the Wild Type and Plants with Dominant Repression of MYB58 or MYB63
| Plant | Interfascicular Fibers | Vessels | Xylary Fibers |
|---|---|---|---|
| Wild type | 1.95 ± 0.31 | 0.97 ± 0.18 | 0.71 ± 0.09 |
| MYB58-DR | 0.96 ± 0.10 | 0.80 ± 0.09 | 0.45 ± 0.07 |
| MYB63-DR | 0.54 ± 0.11 | 0.62 ± 0.05 | 0.30 ± 0.06 |
The wall thickness was measured from transmission electron micrographs of fibers and vessels. Data are means (μm) ± se from 25 cells.
Overexpression of MYB58 and MYB63 Causes Ectopic Deposition of Lignin but Not Cellulose and Xylan
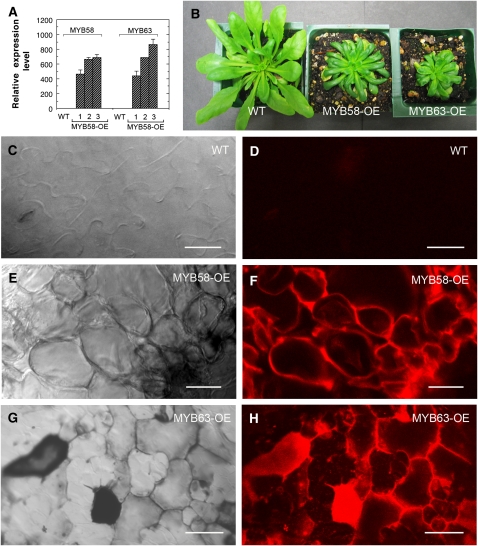
To further investigate their functions in the regulation of secondary wall biosynthesis, we overexpressed MYB58 and MYB63 under the control of the CaMV 35S promoter in wild-type Arabidopsis (Figure 4A) and examined the possible ectopic deposition of cellulose, xylan, and lignin. Among 128 transgenic plants with MYB58 or MYB63 overexpression, 58 and 44 of them, respectively, displayed a small rosette phenotype compared with the wild type (Figure 4B). Examination of leaves revealed that whereas wild-type epidermis did not show any autofluorescence signals for lignin (Figures 4C and 4D), many epidermal cells of MYB58 and MYB63 overexpressors exhibited strong signals for lignin (Figures 4E to 4H). Although MYB58 and MYB63 overexpression caused ectopic deposition of lignin, no prominent secondary wall thickening was seen in the epidermal cells, which is in sharp contrast with the ectopic secondary wall thickening induced by SND1 or MYB46 overexpression (Zhong et al., 2006, 2007a). Ectopic deposition of lignin was also observed in the mesophyll cells of leaves and the epidermis and cortical cells of stems (Figures 5A to 5C) in the MYB58 and MYB63 overexpressors. The ectopic deposition of lignin in leaves was confirmed by chemical analysis of lignin composition showing that both guaiacyl lignin and syringyl lignin were elevated in the MYB58 and MYB63 overexpressors compared with the wild type (Table 2). Consistent with these results, the amount of guaiacyl lignin and syringyl lignin was significantly reduced in the plants with dominant repression and simultaneous RNAi inhibition of MYB58 and MYB63 (Table 3).
Figure 4.
Overexpression of MYB58 and MYB63 Results in Ectopic Deposition of Lignin in the Epidermis of Leaves.
The leaves of 3-week-old transgenic plants overexpressing MYB58 or MYB63 were examined for ectopic lignin deposition. Bars = 12 μm in (C) to (H).
(A) Real-time quantitative RT-PCR analysis showing the overexpression of MYB58 (MYB58-OE) and MYB63 (MYB63-OE) in the seedlings of representative transgenic lines. The expression levels of MYB58 and MYB63 in the wild type are set to 1. Error bars represent se of three replicates.
(B) Three-week-old seedlings of the wild type (left), MYB58-OE (middle), and MYB63-OE (right). Note the reduced leaf and rosette sizes in MYB58-OE and MYB63-OE.
(C) and (D) Differential interference contrast (DIC) (C) and lignin autofluorescence (D) images of the wild-type leaf epidermis showing the absence of lignin autofluorescence signals.
(E) to (H) DIC ([E] and [G]) and lignin autofluorescence ([F] and [H]) images of the MYB58-OE ([E] and [F]) and MYB63-OE ([G] and [H]) leaf epidermis showing ectopic lignin autofluorescence signals.
Figure 5.
Overexpression of MYB58 and MYB63 Leads to Ectopic Deposition of Lignin but Not Cellulose and Xylan in Stems.
Stem sections were stained with phloroglucinol-HCl or Calcofluor White for detection of lignin or secondary wall cellulose, respectively. Xylan was immunodetected with the LM10 xylan antibody. co, cortex; ep, epidermis; if, interfascicular fiber; xy, xylem. Bars = 73 μm.
(A) Phloroglucinol-HCl staining (red color) of a wild-type stem section showing the normal lignin deposition in the walls of interfascicular fibers and xylem cells.
(B) Phloroglucinol-HCl staining of a MYB58-OE stem section showing ectopic lignin deposition in the walls of epidermal cells in addition to its normal deposition in interfascicular fibers and xylem cells.
(C) Phloroglucinol-HCl staining of a MYB63-OE stem section showing ectopic lignin deposition in the walls of cortical and epidermal cells in addition to its normal deposition in interfascicular fibers and xylem cells.
(D) to (F) Calcofluor White staining (blue color) of stem sections showing cellulose staining in the walls of interfascicular fibers and xylem cells in the wild type (D), MYB58-OE (E), and MYB63-OE (F). Note that no ectopic deposition of cellulose was detected in the MYB58-OE or MYB63-OE stem sections.
(G) to (I) Stem sections probed with the LM10 xylan antibody showing xylan staining (green color) in the walls of interfascicular fibers and xylem cells in the wild type (G), MYB58-OE (H), and MYB63-OE (I). Note that no ectopic deposition of xylan was detected in MYB58-OE or MYB63-OE stem sections.
Table 2.
Lignin Composition in the Leaves of the Wild Type and MYB58 and MYB63 Overexpressors
| Plant | G Lignin | S Lignin | Total G and S Lignin |
|---|---|---|---|
| Wild type | 0.17 ± 0.02 | 0.09 ± 0.02 | 0.26 (100%) |
| MYB58 overexpressor | 0.30 ± 0.01 | 0.25 ± 0.01 | 0.55 (212%) |
| MYB63 overexpressor | 0.22 ± 0.01 | 0.15 ± 0.03 | 0.37 (142%) |
The monolignol composition was analyzed according to Akin et al. (1993). Each data point is the mean (mg/g dry cell walls) ± se of two separate assays. Shown in the parentheses are the percentages of total G and S lignin in MYB58 and MYB63 overexpressors relative to that in the wild type.
Table 3.
Lignin Composition in the Stems of the Wild Type, Plants with Dominant Repression of MYB58 or MYB63, and MYB58/MYB63 RNAi Lines
| Plant | G Lignin | S Lignin | Total G and S Lignin |
|---|---|---|---|
| Wild type | 4.64 ± 0.13 | 2.57 ± 0.30 | 7.21 (100%) |
| MYB58 repressor | 3.00 ± 0.02 | 1.50 ± 0.03 | 4.50 (62%) |
| MYB63 repressor | 3.28 ± 0.08 | 2.06 ± 0.14 | 5.34 (74%) |
| MYB58/MYB63 RNAi | 2.99 ± 0.17 | 2.02 ± 0.08 | 5.01 (69%) |
Each data point is the mean (mg/g dry cell walls) ± se of two separate assays. Shown in the parentheses are the percentages of total G and S lignin in MYB58 and MYB63 repressors or MYB58/MYB63 RNAi lines relative to that in the wild type.
We next investigated whether MYB58 and MYB63 overexpression also resulted in ectopic deposition of cellulose and xylan. Histological staining of cellulose and immunostaining of xylan in the stems of MYB58 and MYB63 overexpressors showed no ectopic deposition of cellulose and xylan in the epidermis and cortical cells besides their normal deposition in fibers and vessels as seen in the wild type (Figures 5D to 5I). In addition, examination of leaf sections of MYB58 and MYB63 overexpressors did not reveal any ectopic deposition of cellulose and xylan.
Consistent with the histological data showing that MYB58 and MYB63 overexpression caused a specific ectopic deposition of lignin but not cellulose or xylan, it was found that overexpression of MYB58 and MYB63 resulted in a substantial induction in the expression of genes involved in the general phenylpropanoid metabolism as well as in the specific branch toward monolignol biosynthesis (Figure 6A). By contrast, the expression of the three secondary wall–associated cellulose synthase genes, including CesA4, CesA7, and CesA8 (Taylor et al., 2004), and that of two xylan biosynthetic genes, including IRX8 and IRX9 (Pena et al., 2007), were not altered by MYB58 and MYB63 overexpression (Figure 6B). In addition, MYB58 and MYB63 overexpression did not induce the expression of the chalcone synthase (CHS; At5g13930) and chalcone flavanone isomerase (CHI; At3g55120) genes in the flavonoid biosynthetic pathway or that of the aldehyde dehydrogenase (ALDH; At3g24503) and sinapoylglucose malate sinapoyltransferase (SMT; At2g22990) genes in the sinapate ester biosynthetic pathway (Besseau et al., 2007) (Figure 6B). Together, these results demonstrate that MYB58 and MYB63 are specific transcriptional activators of lignin biosynthesis during secondary wall formation.
Figure 6.
Lignin Biosynthetic Genes Were Induced by Overexpression of MYB58 and MYB63.
Leaves from 3-week-old seedlings were used for expression analysis of secondary wall biosynthetic genes with real-time quantitative RT-PCR. The quantitative differences of the expression of the tested genes between the wild type and the overexpression lines are statistically significant (P < 0.001). The expression level of each gene in the wild type is set to 1. Error bars represent se of three biological replicates.
(A) Overexpression of MYB58 and MYB63 induces the expression of lignin biosynthetic genes. A schematic diagram of the lignin biosynthetic pathway is depicted below the panel to indicate the catalytic step in which each gene is involved. The genes selected for expression analysis were previously shown to be highly expressed in stems where lignified fibers and vessels are abundant (Raes et al., 2003). PAL1, phenylalanine ammonia lyase 1; C4H, cinnamate 4-hydroxylase; 4CL1, 4-coumarate CoA ligase 1; HCT, hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyltransferase; C3H1, p-coumarate 3-hydroxylase 1; CCoAOMT1, caffeoyl CoA 3-O-methyltransferase 1; CCR1, cinnamoyl CoA reductase 1; F5H1, ferulate 5-hydroxylase 1; COMT, caffeic acid O-methyltransferase; CAD6, cinnamyl alcohol dehydrogenase 6.
(B) Overexpression of MYB58 and MYB63 does not induce the expression of secondary wall–associated cellulose synthase genes (CesA4, CesA7, and CesA8), xylan biosynthetic genes (IRX8 and IRX9), flavonoid biosynthetic genes (CHS and CHI), and sinapate ester biosynthetic genes (ALDH and SMT).
MYB58 and MYB63 Bind to the AC cis-Elements
The findings that MYB58 and MYB63 are able to induce the expression of the entire monolignol biosynthetic pathway genes and lead to ectopic deposition of lignin prompted us to investigate whether MYB58 and MYB63 directly bind to the promoters of monolignol biosynthetic genes and thereby activate their expression. To do this, we first used the 4-coumarate:CoA ligase 1 (4CL1) promoter linked with the GUS reporter gene to test its transcriptional activation by MYB58 and MYB63 (Figure 7A). Cotransfection of Arabidopsis leaf protoplasts with a CaMV 35S promoter-driven MYB58 or MYB63 expression construct and a GUS construct including 1 kb of the 4CL1 promoter showed that MYB58 and MYB63 were able to activate the expression of the 4CL1 promoter-driven GUS reporter gene (Figure 7B). Promoter deletion analysis revealed that a 260-bp 4CL1 promoter sequence was equally activated by MYB58 and MYB63 in driving the GUS reporter gene expression (Figure 7B). It has been shown previously that the 4CL1 promoter harbors one AC element at nucleotide position 159 upstream of the start codon (Raes et al., 2003). To test whether this AC element is essential for the transactivation of the 4CL1 promoter by MYB58 and MYB63, we mutated the AC element sequence in the 4CL1 promoter (Figure 7A) and examined its effect on the transactivation by MYB58 and MYB63. Mutation of the AC element completely abolished the activation of 4CL1 promoter-driven GUS expression by MYB58 and MYB63 (Figure 7B), demonstrating that MYB58 and MYB63 activate the 4CL1 promoter-driven expression through the AC element. This conclusion was further supported by the finding showing that a DNA oligonucleotide containing three copies of the AC element (Figure 7A) was sufficient to mediate the activation of the GUS reporter gene expression by MYB58 and MYB63 (Figure 7C).
Figure 7.
MYB58 and MYB63 Induce the Expression of the 4CL1 Gene through the AC Element.
The activation of the 4CL1 promoter by MYB58 and MYB63 was analyzed by cotransfecting Arabidopsis leaf protoplasts with the effector and reporter constructs. The GUS activity in the protoplasts transfected with the GUS reporter construct only (control) is set to 1. Error bars represent se of three biological replicates.
(A) Diagrams of the effector and reporter constructs used for transactivation analysis. The effector constructs contain MYB58 or MYB63 driven by the CaMV 35S promoter (35S:MYB58 and 35S:MYB63). The reporter constructs consist of the GUS reporter gene driven by the 4CL1 promoter sequences with 1 kb in length (located between −1 and −1000 relative to the start codon; 4CL1P-1kb:GUS) or 260 bp in length (located between −1 and −260 relative to the start codon; 4CL1-260bp:GUS). The location of the AC element in the 4CL1 promoter is indicated. The 4CL1P-mAC:GUS reporter construct has the 260-bp 4CL1 promoter with the AC element mutated as indicated. The 3xAC:GUS reporter construct contains the GUS reporter gene driven by a 38-bp DNA oligonucleotide containing three copies of the AC element linked with the CaMV 35S minimal promoter sequence. The sequence of the DNA oligonucleotide is shown.
(B) Transactivation analysis showing that MYB58 and MYB63 activate the expression of the GUS reporter gene driven by the native 4CL1 promoter but not by the 4CL1 promoter with mutated AC element.
(C) MYB58 and MYB63 are able to activate the expression of GUS reporter gene driven by three copies of the AC element (3xAC).
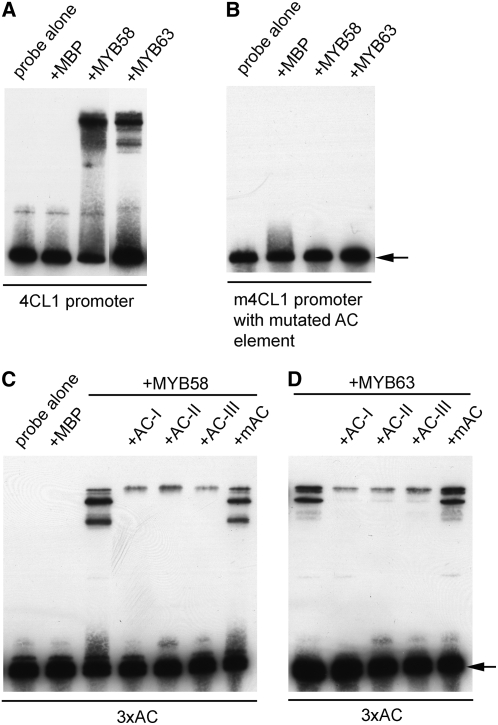
We next tested whether MYB58 and MYB63 can bind directly to the AC element using the electrophoretic mobility shift assay (EMSA). When the 260-bp 4CL1 promoter fragment was incubated with recombinant maltose binding protein (MBP) fused to MYB58 or MYB63, the mobility of the DNA fragment was apparently retarded (Figure 8A). When it was incubated with MBP alone, no mobility shift was seen, indicating that the observed mobility shift was caused specifically by MYB58 and MYB63. Mutation of the AC element in the 4CL1 promoter sequence completely abolished the mobility shift caused by MYB58 and MYB63 (Figure 8B), demonstrating that MYB58 and MYB63 bind to the 4CL1 promoter through the AC element. The direct binding of MYB58 and MYB63 to the AC element was further confirmed by the finding that MYB58 and MYB63 were able to cause a mobility shift of the DNA oligonucleotide containing three copies of the AC element (Figures 8C and 8D).
Figure 8.
MYB58 and MYB63 Bind to the AC Element in the 4CL1 Promoter.
The recombinant proteins of MYB58 and MYB63 fused with MBP were incubated with biotin-labeled DNA probes, and the samples were subjected to EMSA by PAGE and subsequent chemiluminescent detection. MBP was used as a control protein in the EMSA experiments. The free DNA probes are indicated by arrows.
(A) MYB58 and MYB63 bind to the 260-bp 4CL1 promoter fragment and result in a retardation in its mobility.
(B) Mutation of the AC element in the 260-bp 4CL1 promoter fragment abolishes its binding by MYB58 and MYB63.
(C) and (D) MYB58 (C) and MYB63 (D) bind to the 38-bp oligonucleotide containing three copies of the AC element (3xAC). The sequence of the oligonucleotide is the same as that shown in Figure 7A. For competition analysis, unlabeled oligonucleotides containing AC-I, AC-II, AC-III, or a mutated AC element (mAC) were included in the reactions.
Previous work has shown that three AC element sequences, namely, AC-I (ACCTACC), AC-II (ACCAACC), and AC-III (ACCTAAC), in the bean PAL2 gene are important for directing its vascular expression (Hatton et al., 1995). The AC element present in the 260-bp 4CL1 promoter fragment is AC-II. To examine whether MYB58 and MYB63 are able to bind to all of these three AC elements, we performed competition analysis. Addition of excess unlabeled DNA oligonucleotides containing AC-I, AC-II, or AC-III sequence effectively competed with the binding of the labeled 3xAC oligonucleotide to MYB58 and MYB63, whereas addition of an oligonucleotide containing a mutated AC element did not (Figures 8C and 8D). Together, these results suggest that MYB58 and MYB63 are able to bind to the AC elements in the promoters of monolignol biosynthetic genes.
MYB58 Directly Activates the Expression of Monolignol Biosynthetic Genes
To further substantiate the hypothesis that MYB58 and MYB63 directly activate the expression of the monolignol biosynthetic genes, we employed the steroid receptor-based inducible system, an approach successfully used for identification of direct targets of transcription factors in plants (Sablowski and Meyerowitz, 1998; Wagner et al., 1999; Baudry et al., 2004). In this system, fusion of the steroid receptor with a transcription factor renders the transcription factor inactive by sequestering it in the cytoplasm through binding to a cytoplasmic complex. Addition of steroid releases the transcription factor from retention in the cytoplasm and thus allows it to enter the nucleus to activate the expression of target genes (Sablowski and Meyerowitz, 1998). We first cotransfected Arabidopsis leaf protoplasts with the CaMV 35S promoter-driven MYB58 fused with the regulatory region of the human estrogen receptor (HER; Zuo et al., 2000) and the 260-bp 4CL1 promoter-driven GUS reporter gene (Figure 9A) to test the effectiveness of the inducible system. It was found that the GUS reporter activity driven by the 4CL1 promoter was highly induced by the addition of estradiol (Figure 9B), demonstrating that the MYB58-HER chimeric protein is able to activate the expression of downstream genes. For unknown reasons, the MYB63-HER chimeric protein exhibited high transactivation activity without addition of estradiol; therefore, it was not used in this study.
Figure 9.
Direct Activation of Monolignol Biosynthetic Genes by MYB58.
Arabidopsis leaf protoplasts were transfected with MYB58 fused with the regulatory region of HER under the control of the CaMV 35S promoter. After transfection, the protoplasts were treated with estradiol, cycloheximide (CHX), or cycloheximide plus estradiol. The expression of the monolignol biosynthetic genes was analyzed by real-time quantitative RT-PCR. The quantitative differences of the expression of the tested genes between the control and treated samples are statistically significant (P < 0.001). The expression level of each gene or GUS activity in the mock-treated (control) or cycloheximide-treated protoplasts (CHX) is set to 1. Error bars represent se of three biological replicates.
(A) Diagrams of the MYB58-HER construct used for direct target analysis and the 4CL1 promoter-driven GUS reporter gene construct.
(B) GUS activity in leaf protoplasts cotransfected with 35S:MYB58-HER and 4CL1P:GUS. Addition of the protein synthesis inhibitor cycloheximide at 2 μM completely abolished the estradiol-induced GUS activity, indicating that new protein synthesis was completely inhibited by the treatment.
(C) Induction of the monolignol biosynthetic genes by estradiol treatment of protoplasts expressing MYB58-HER. Addition of estradiol releases MYB58-HER from the cytoplasm and its subsequent entry into the nucleus leads to activation of its downstream target genes.
(D) Induction of the monolignol biosynthetic genes by estradiol activation of MYB58 occurred in the presence of cycloheximide.
To test whether MYB58 directly activates the expression of monolignol biosynthetic genes, new protein synthesis was inhibited by addition of the protein synthesis inhibitor cycloheximide. Cycloheximide at the concentration of 2 μM was shown to be sufficient to completely abolish the 4CL1 promoter-driven GUS reporter activity induced by estradiol (Figure 9B), indicating that new protein synthesis is completely inhibited by the treatment. Under this condition, the expression of MYB58 direct target genes should still be induced by estradiol activation of MYB58, but the induction of further downstream genes is blocked because of a lack of new protein synthesis. Examination of the expression of monolignol biosynthetic genes in the MYB58-HER inducible system revealed that estradiol activation of MYB58 resulted in a substantial induction in the expression of all monolignol biosynthetic genes except F5H1 (Figure 9C), and this induction in gene expression still occurred in the presence of the protein synthesis inhibitor cycloheximide (Figure 9D). These findings demonstrate that the activation of the monolignol biosynthetic genes by MYB58 does not require new protein synthesis and therefore is direct.
The Secondary Wall–Associated Laccase4 Gene Is Induced by MYB58 and MYB63
It is well established that monolignols are synthesized in the cytoplasm and then exported into the cell wall for polymerization (Boerjan et al., 2003). Both laccases and peroxidases are proposed to be involved in the oxidation of monolignols for dehydrogenative polymerization, although it is still unknown which particular peroxidase and laccase genes are directly involved in this process during secondary wall formation. Because overexpression of MYB58 and MYB63 caused an ectopic deposition of lignin, we reasoned that MYB58 and MYB63 not only activate genes for monolignol biosynthesis but also should induce genes encoding enzymes involved in monolignol polymerization. We found that two laccase genes, LAC4 (At2g38080) and LAC17 (At5g60020), were highly expressed in secondary wall–containing interfascicular fibers and xylem cells but not in parenchymatous pith cells (Figure 10A). At the organ level, they showed a predominant expression in inflorescence stems in which interfascicular fibers and xylem cells are abundant (Figure 10B). These results suggest that LAC4 and LAC17 exhibit a similar expression pattern to that of MYB58 and MYB63, and they might be involved in secondary wall biosynthesis. Expression analysis revealed that the expression of LAC4 but not LAC17 was upregulated by MYB58 and MYB63 (Figure 10C), suggesting that LAC4 is a downstream target of MYB58 and MYB63. Further study using the MYB58-HER inducible system demonstrated that MYB58 was able to activate directly the expression of the LAC4 gene (Figures 10D and 10E).
Figure 10.
Expression of the Secondary Wall–Associated Laccase4 Gene Is Induced by MYB58 and MYB63.
The expression of LAC4 and LAC17 was studied using real-time quantitative RT-PCR. The quantitative differences of their expression between the control and the treated samples are statistically significant (P < 0.001). Error bars represent se of three biological replicates.
(A) LAC4 and LAC17 were expressed highly in interfascicular fibers and xylem cells. RNAs used for gene expression analysis were isolated from laser-microdissected cells. The expression levels of LAC4 and LAC17 in the pith cells are set to 1.
(B) LAC4 and LAC17 were expressed predominantly in inflorescence stems. Their expression levels in leaves are set to 1.
(C) Overexpression of MYB58 and MYB63 induced the expression of LAC4 but not LAC17. Arabidopsis leaf protoplasts were transfected with the CaMV 35S promoter–driven MYB58 (MYB58-OE) and MYB63 (MYB63-OE) expression constructs or an empty vector (control). The expression levels of LAC4 and LAC17 in the control are set to 1.
(D) and (E) Direct activation of the LAC4 gene by MYB58. Arabidopsis leaf protoplasts were transfected with the CaMV 35S promoter–driven MYB58-HER construct. Estradiol activation of MYB58 induced the expression of LAC4 (D), and this induction still occurred when new protein synthesis was completely inhibited by cycloheximide (E). The expression level of LAC4 in the mock-treated (control) or cycloheximide-treated (CHX) protoplasts is set to 1.
Expression of the MYB58 and MYB63 Genes Is Induced by SND1 Close Homologs and MYB46
Previous study showed that SND1 and its close homologs, including NST1, NST2, VND6, and VND7, regulate the same downstream target genes (Zhong et al., 2007a, 2008). The fact that the expressions of MYB58 and MYB63 are regulated by SND1 and NST1 (Figure 1) promoted us to investigate whether other SND1 close homologs and MYB46 also regulate their expression. Expression analysis of MYB58 and MYB63 in transgenic Arabidopsis plants overexpressing NST2, VND6, VND7, or MYB46 revealed that their expression was significantly upregulated in these overexpressors compared with the wild type (Figure 11). Dominant repression or overexpression of MYB58 and MYB63 did not affect the expression of SND1 and its homologs, nor did they alter the expression of MYB46. Together, these results suggest that MYB58 and MYB63 are common downstream targets of SND1 and its close homologs and they are positioned downstream of MYB46 in the SND1-mediated transcriptional network.
Figure 11.
The Expression of MYB58 and MYB63 Is Induced by Overexpression of NST2 (NST2-OE), VND6 (VND6-OE), VND7 (VND7-OE), and MYB46 (MYB46-OE).
The expression levels of MYB58 and MYB63 in the leaves of the wild type and the overexpressors were analyzed using real-time quantitative RT-PCR. The quantitative differences of their expression between the wild type and the overexpressors are statistically significant (P < 0.001). Their expression levels in the wild type are set to 1. Error bars represent se of three biological replicates.
DISCUSSION
Lignin is the second most abundant biopolymer produced by vascular plants, and it is mainly deposited in secondary wall–containing cells, such as fibers and tracheary elements. Lignin polymer, an extensive cross-linked network, together with the cellulose-xylan network provides strength and rigidity to the secondary walls, and its impregnation into the secondary walls renders them waterproof. Being very inert, lignin also imparts a stable, protective coating to the secondary walls to shield them from chemical, physical, and biological attacks. However, uses of secondary walls for pulping and papermaking, and potentially for biofuel production, often require the removal of lignin, which leads to increased costs as well as environmental pollutions. Lignin also limits the digestibility of forage crops used to feed animals. Therefore, extensive efforts have been put into genetic modification of lignin content and composition in woody species and forage crops in the hope of generating plants with lower lignin content or altered lignin composition allowing for easy removal of lignin (Vanholme et al., 2008; Weng et al., 2008). Currently, all the genes in the monolignol biosynthetic pathway have been isolated and characterized, and some of them have been targeted for genetic modification of lignin content and composition. By contrast, much less is known about the regulatory genes controlling lignin biosynthesis during secondary wall formation (Vanholme et al., 2008; Weng et al., 2008). Uncovering lignin regulatory genes could potentially provide another means for manipulating the lignin biosynthetic pathway. The finding that MYB58 and MYB63 are SND1-regulated, secondary wall–associated transcriptional activators regulating the entire lignin biosynthetic pathway during secondary wall formation lays a foundation for further unraveling the regulatory genes controlling lignin biosynthesis.
MYB58 and MYB63 Are SND1-Regulated Transcriptional Activators of Lignin Biosynthesis during Secondary Wall Formation
We have demonstrated that MYB58 and MYB63 are transcriptional activators of lignin biosynthesis in the SND1-mediated transcription network regulating secondary wall formation based on several lines of evidence. First, the expression of MYB58 and MYB63 is specifically associated with cells undergoing secondary wall thickening and lignification, an expression pattern that is the same as that of SND1. Second, the expression of MYB58 and MYB63 is up- or downregulated by SND1/NST1 overexpression or SND1/NST1 RNAi inhibition, respectively, indicating that MYB58 and MYB63 are part of the SND1/NST1-mediated transcription network regulating secondary wall biosynthesis. This finding is important because transcription factors activating lignin biosynthesis during secondary wall formation should be regulated by SND1 or its close homologs based on previous observations that SND1 is a master switch activating secondary wall thickening and lignification and that knockout of SND1 and NST1 leads to a loss of secondary wall thickening and lignification (Zhong et al., 2006, 2007b). Third, MYB58 and MYB63 are targeted to the nucleus and can activate transcription. Fourth, overexpression of MYB58 and MYB63 results in activation of lignin biosynthetic genes and leads to an ectopic deposition of lignin. Unlike MYB46, which is a secondary wall–associated MYB transcription factor activating the biosynthetic pathways for cellulose, xylan, and lignin (Zhong et al., 2007a), MYB58 and MYB63 do not induce the biosynthetic pathways for cellulose and xylan. These results suggest that MYB58 and MYB63 are SND1-regulated MYB transcription factors specifically influencing lignin biosynthesis in cells undergoing secondary wall thickening. It should be noted that the transcriptional activation strength and sequence selectivity of transcription factors depend on their expression level (Andersson et al., 1999; Jin et al., 2000). Therefore, the observed induction of lignin biosynthetic genes by MYB58 and MYB63 overexpression needs to be further investigated under in vivo physiological conditions.
MYB58 and MYB63 are phylogenetically distinct from those MYBs that have previously been proposed to be involved in regulation of the phenylpropanoid metabolism. Overexpression of several MYB genes from antirrhinum, Arabidopsis, and grape (Vitis vinifera) has been shown to cause an alteration in lignin biosynthesis (Tamagnone et al., 1998; Borevitz et al., 2000; Deluc et al., 2006). However, it is not known whether these MYB genes are expressed in cells undergoing secondary wall thickening; therefore, it is uncertain whether they are involved in the regulation of lignin biosynthesis during secondary wall formation. Overexpression of another MYB gene from Arabidopsis, MYB61, phenocopies the ectopic lignin deposition caused by the de-etiolated3 mutation (Newman et al., 2004). Because MYB61 is involved in multiple processes, including regulation of stomatal aperture and seed coat mucilage deposition, its exact function remains to be studied (Penfield et al., 2001; Liang et al., 2005). Two other MYB genes, Pt MYB4 from pine (Patzlaff et al., 2003a) and Eg MYB2 from eucalyptus (Goicoechea et al., 2005), have been proposed to play a role in the regulation of lignin biosynthesis. Both of them are preferentially expressed in developing secondary xylem. When heterologouslly overexpressed in tobacco, ectopic lignin deposition occurred in the Pt MYB4 overexpressors (Patzlaff et al., 2003a), and increased secondary wall thickening in fibers was observed in the Eg MYB2 overexpressors (Goicoechea et al., 2005). However, the possible effects of their downregulation on lignification have not yet been examined. Since they bind to the AC elements in vitro, Pt MYB4 and Eg MYB2 appear to be strong candidates for transcriptional regulators of lignin biosynthesis. However, because both Pt MYB4 and Eg MYB2 are close homologs of Arabidopsis MYB46 (Goicoechea et al., 2005; Zhong et al., 2007a), it remains to be investigated whether they function the same as MYB46 to regulate the entire secondary wall biosynthetic program or they specifically regulate lignin biosynthesis.
Dominant repression of MYB58 and MYB63 was found to lead to a drastic reduction in secondary wall thickening and a deformation of vessel morphology. The deformed vessel phenotype is likely caused by the weakening of the vessel walls due to the reduction in the amount of lignin polymer and secondary wall thickness, which renders vessels incapable of resisting the negative pressure generated from transpiration. Similar deformed vessel phenotypes have been observed previously in plants with repression of lignin biosynthesis by downregulation of lignin biosynthetic genes (Zhong et al., 1998; Goujon et al., 2003). Although we could not exclude the possibility that MYB58 and MYB63 may also regulate the expression of some genes affecting the biosynthesis of other secondary wall components in addition to lignin, we propose that the reduced secondary wall thickening phenotype is most likely due to an indirect effect caused by the decreased lignin deposition, which may impede the overall secondary wall biosynthesis and assembly. In fact, a reduction in any one of the three major secondary wall components, including cellulose, xylan, and lignin, can result in a decrease in secondary wall thickening and a deformed vessel morphology (Zhong et al., 1998, 2005; Taylor et al., 2004; Pena et al., 2007), indicating that the biosynthesis of secondary wall components is a highly coordinated process and that an alteration in the biosynthesis of one component may lead to an impediment in the biosynthesis of other wall components.
Direct Activation of Lignin Biosynthetic Pathway Genes by MYB58
Direct target analysis using the steroid receptor–inducible system reveals that MYB58 directly activates the expression of all genes except F5H1 in the lignin biosynthetic pathway. The activation of these genes by MYB58 is most likely through its binding to the AC elements based on the findings that mutation of the AC element in the 4CL1 promoter abolishes MYB58-induced activation of the 4CL1 promoter and that MYB58 is able to bind to the AC elements in vitro. AC elements have been found to be present in the promoters of PAL1, 4CL1, hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyltransferase (HCT), p-coumarate 3-hydroxylase 1 (C3H1), caffeoyl CoA 3-O-methyltransferase 1 (CCoAOMT1), cinnamoyl CoA reductase (CCR1), and cinnamyl alcohol dehydrogenase 6 (CAD6) (Raes et al., 2003), all of which are activated by MYB58. Cinnamate 4-hydroxylase (C4H), F5H1, and caffeic acid O-methyltransferase (COMT) do not have apparent AC elements in their promoters based on stringent AC element selection criteria (Raes et al., 2003), although one could not exclude the possibility that more degenerate AC elements may be present in their promoters or other parts of the gene sequence. Consistent with this possibility, the C4H and COMT genes are also shown to be directly activated by MYB58.
The findings that MYB58 directly activates the expression of lignin biosynthetic genes and that it is able to bind to the AC elements support the hypothesis that AC elements serve as a common means for the coordinated regulation of genes in the entire lignin biosynthetic pathway. Since AC elements are also present in the promoters of lignin biosynthetic genes in tree species (Lauvergeat et al., 2002), it will be interesting to investigate whether the AC element-mediated coordination of regulation of lignin biosynthetic genes is conserved in woody plants. A number of MYB transcription factor genes have been isolated from various tree species, and some of them have been shown to bind to the AC elements (Patzlaff et al., 2003a, 2003b; Goicoechea et al., 2005). It will be important to find out which of these tree MYBs is involved in direct activation of lignin biosynthetic genes. It should be noted that although both LAC4 and LAC17 promoters contain putative AC elements, only the LAC4 gene was induced by MYB58 and MYB63. This indicates that not all gene promoters containing AC elements can be induced by MYB58 and MYB63, which supports the notion that the activation of phenylpropanoid biosynthetic genes may involve the combinatorial actions of different transcriptional activators and repressors (Jin et al., 2000).
Induction of the Secondary Wall—Associated LAC4 Gene by MYB58 and MYB63
We have demonstrated that in addition to the monolignol biosynthetic genes, the expression of a laccase gene, LAC4, is also regulated by MYB58 and MYB63. Laccases together with peroxidases have been proposed to be oxidases responsible for the oxidation of monolignols, including p-coumaryl alcohol, coniferyl alcohol, and synapyl alcohol. Previous studies have shown that laccase enzymes are able to oxidize monolignols and that their genes are expressed in lignifying cells (Boerjan et al., 2003). However, no specific laccase genes have been demonstrated to be coregulated with lignin biosynthetic genes during secondary wall formation. The findings that LAC4 is specifically expressed in cells undergoing lignification and that its expression is coregulated with lignin biosynthetic genes by MYB58 and MYB63 suggest that LAC4 is most likely involved in lignin biosynthesis in fibers and vessels. These findings also indicate that MYB58 and MYB63 regulate the coordinated expression of genes involved in both monolignol biosynthesis and polymerization, which is consistent with their overexpression phenotype showing an ectopic accumulation of lignin in the cell walls of normally unlignified cells.
Transcriptional Regulatory Network Controlling Secondary Wall Biosynthesis
Previous work has suggested that a transcriptional network is involved in the regulation of secondary wall biosynthesis. In this network, SND1 and its close homologs, including NST1, NST2, VND6, and VND7, are master switches regulating a cascade of downstream transcription factors leading to activation of the entire secondary wall biosynthesis program. It has been proposed that SND1, NST1, NST2, VND6, and VND7 are functional homologs regulating the same downstream targets, albeit in different cell types (Zhong et al., 2008). Our finding that MYB58 and MYB63 are regulated by SND1 and its close homologs further supports this hypothesis. The fact that MYB46 also regulates the expression of MYB58 and MYB63 indicates that MYB58 and MYB63 are downstream of MYB46. Because MYB58 directly activates the expression of lignin biosynthetic genes, it appears that MYB58 and MYB63 are positioned at the lowest level in the SND1-mediated transcriptional network. Further studies on how the expression of MYB58 and MYB63 is regulated will help us uncover the transcription factors that link SND1 and MYB58/MYB63 in the SND1-mediated transcriptional network.
The finding that MYB58 and MYB63 specifically activate the expression of lignin biosynthetic genes suggests that direct activation of the individual biosynthetic pathways leading to the biosynthesis of cellulose, xylan, and lignin might be mediated by pathway-specific transcription factors. Because SND1, its close homologs, and MYB46 are master activators for the biosynthesis of cellulose, xylan, and lignin, some of their downstream transcription factors other than MYB58 and MYB63 are likely involved in direct activation of the biosynthetic pathways for cellulose and xylan. Identification of transcription factors specifically regulating the biosynthesis of cellulose and xylan will provide novel tools for targeted alterations of secondary wall biosynthesis in transgenic plants.
In conclusion, we have demonstrated that MYB58 and MYB63 are SND1-regulated MYB transcription factors directly activating lignin biosynthetic genes during secondary wall formation. Uncovering MYB58 and MYB63 as lignin-specific transcriptional activators provides new insights into the SND1-mediated cascade of transcription factors regulating secondary wall biosynthesis. It is likely that the regulation of lignin biosynthesis is mediated by a combinatorial coordination of many different transcription factors, including LIM transcription factors (Rogers and Campbell, 2004) and other MYBs in addition to MYB58 and MYB63. It has recently been shown that another secondary wall–associated MYB protein, MYB85, is able to activate lignin biosynthetic genes and its overexpression results in ectopic deposition of lignin (Zhong et al., 2008). We recently found that activation of the lignin biosynthetic genes by MYB85 requires the AC elements in their promoters, indicating that MYB58, MYB63, and MYB85 may function redundantly in the regulation of lignin biosynthesis during secondary wall formation. This proposed functional redundancy may explain why dominant repression or simultaneous RNAi inhibition of MYB58 and MYB63 only results in a partial loss of lignin deposition (Table 3). In addition to transcriptional activators, such as MYB58, MYB63, and MYB85, lignin biosynthesis may also be regulated by repressors as exemplified by At MYB4 that was shown to be a transcriptional repressor of the phenylpropanoid biosynthetic pathway (Jin et al., 2000). Further dissection of transcription factors involved in the regulation of lignin biosynthesis will undoubtedly enrich our knowledge on the biosynthesis of lignin, the second most abundant biopolymer produced by land plants. Like SND1 and MYB46, close homologs of MYB58 and MYB63 are present in tree species, such as poplar, indicating that a similar transcriptional network may be involved in the regulation of secondary wall biosynthesis during wood formation. It is expected that further investigation of the transcriptional regulation of secondary wall biosynthesis in Arabidopsis will help us unravel the molecular mechanisms controlling wood formation and lignification in tree species.
METHODS
Plant Materials
Wild-type Arabidopsis thaliana (ecotype Columbia), SND1/NST1 RNAi line (Zhong et al., 2007b), and the overexpression lines of SND1, NST1, NST2, VND6, and VND7 (Zhong et al., 2006, 2007a, 2008) were grown in a greenhouse.
Real-Time Quantitative RT-PCR and GUS Reporter Gene Analyses
Inflorescence stems of 6-week-old Arabidopsis plants were subjected to microdissection of interfascicular fiber cells, xylem cells, and pith cells using the PALM microlaser system (PALM Microlaser Technologies). Total RNA was extracted from the isolated cells and used for cell type–specific gene expression analysis (Zhong et al., 2006).
Total RNA was isolated from different Arabidopsis organs or different transgenic lines using a Qiagen RNA isolation kit following the manufacturer's protocol. The seedlings used were 2 weeks old. Mature leaves and roots were from 6-week-old plants. Top (rapidly elongating internodes), middle (elongating internodes near cessation), and bottom (nonelongating internodes) parts of inflorescence stems were from 8-week-old plants.
Real-time quantitative RT-PCR analysis was done using the first-strand cDNA as a template with the QuantiTect SYBR Green PCR kit (Clontech). Primers used for PCR analysis of CesA and IRX genes were from Zhong et al. (2006), and those for lignin biosynthetic genes were from Raes et al. (2003). Additional primers are listed in Supplemental Table 1 online. The PCR threshold cycle number of each gene was normalized with that of the EF1α reference gene (primers: 5′-TGAGCACGCTCTTCTTGCTTTCA-3′ and 5′-GGTGGTGGCATCCATCTTGTTACA-3′) to calculate the relative mRNA levels.
The GUS reporter gene was employed to study the developmental expression pattern of the MYB58 and MYB63 genes. The genomic DNA fragment containing a 3-kb 5′ upstream sequence, the entire MYB58 or MYB63 exon and intron region, and a 2-kb 3′downstream sequence was used. The MYB58:GUS and MYB63:GUS constructs in the binary vector pBI101 (Clontech) were made by ligating the PCR-amplified genomic DNA fragments (primers are listed in Supplemental Table 1 online) into the SalI and SacI sites in pBI101. The constructs were transformed into wild-type Arabidopsis plants by the Agrobacterium tumefaciens–mediated transformation procedure (Bechtold and Bouchez, 1994). The first generation of 6-week-old transgenic plants was examined for the expression of the GUS reporter gene (Zhong et al., 2005).
Phylogenetic Analysis
The sequences of MYB proteins were aligned using the ClustalW program (Thompson et al., 1994), and their phylogenetic relationship was analyzed with the SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE programs using the default values provided in the PHYLIP software (Felsenstein, 1989). The statistical significance of the tree was tested by bootstrap analysis of 1000 trials. The phylogenetic tree was displayed using the TREEVIEW program (Page, 1996). The sequence alignment between MYB58 and MYB63 in Figure 1G was done using the ClustalW program.
Subcellular Localization and Transcriptional Activation Analysis
The MYB58-YFP and MYB63-YFP constructs were created by ligating the PCR-amplified (primers are listed in Supplemental Table 1 online) full-length cDNA of MYB58 or MYB63 at the BamHI site in frame with the YFP cDNA between the CaMV 35S promoter and the nopaline synthase terminator in pBI221 (Clontech). They were cotransfected with an SND1-CFP expression construct (Zhong et al., 2006) into Arabidopsis leaf protoplasts by the polyethylene glycol–mediated transfection according to Sheen (2001). The DIC images were obtained with the DIC setting using a compound microscope. Fluorescence signals (excitation = 500 nm and emission = 535 nm for YFP; excitation = 436 nm and emission = 480 nm for CFP) in the transfected protoplasts were examined using a Leica TCs SP2 spectral confocal microscope (Leica Microsystems).
The constructs for transactivation in yeast were generated by ligating the PCR-amplified cDNA (primers are listed in Supplemental Table 1 online) of MYB58 or MYB63 into the BamHI site of the pAS2-1 vector (Clontech). They were transformed into the yeast strain CG-1945 containing the His3 and LacZ reporter genes. The transformed yeast cells were grown on synthetic defined medium without His and subsequently assayed for β-galactosidase activity (Miller, 1972).
Generation of Transgenic Plants with Dominant Repression and Overexpression of MYB58 and MYB63
The full-length cDNA of MYB58 or MYB63 was fused at its C terminus in frame with the dominant EAR repression sequence (Hiratsu et al., 2004) by PCR amplification (primers are listed in Supplemental Table 1 online) to generate the MYB58 and MYB63 dominant repression constructs (MYB58-DR and MYB63-DR). The dominant EAR repression DNA sequence (Hiratsu et al., 2004) was added to the primer used for PCR amplification of the full-length MYB58 or MYB63 cDNA. The fused fragment was ligated downstream of the CaMV 35S promoter at the BamHI site in the binary vector pBI121 (Clontech). The full-length cDNA of MYB58 or MYB63 was ligated downstream of the CaMV 35S promoter at the BamHI site in pBI121 to create the MYB58 and MYB63 overexpression constructs. The dominant repression and overexpression constructs were transformed into wild-type Arabidopsis plants (Bechtold and Bouchez, 1994), and the first generation of transgenic plants was used for examination of phenotypes.
Simultaneous RNAi Inhibition of MYB58 and MYB63
The RNAi construct for inhibition of MYB58 and MYB63 expressions was created as follows. The fused MYB58 and MYB63 cDNAs (primers are listed in Supplemental Table 1 online) were cloned into pBI121 at the SalI/SacI sites in opposite orientations on both sides of the GUS spacer, which is located between the CaMV 35S promoter and the nopaline synthase terminator. The RNAi construct was transformed into Arabidopsis plants, and 96 independent transgenic lines were generated. Real-time quantitative RT-PCR analysis showed that the expression levels of MYB58 and MYB63 in 23 transgenic lines were reduced to <10% of that in the wild type. The inflorescence stems of these lines were used for analysis of lignin composition.
Histology
Tissues embedded in Low Viscosity (Spurr's) resin (Electron Microscopy Sciences) were sectioned (1-μm-thick) and stained with toluidine blue for light microscopy (Burk et al., 2006). For observation of subcellular structures, 85-nm-thick sections were poststained with uranyl acetate and lead citrate and observed using a Zeiss EM 902A transmission electron microscope (Carl Zeiss). Metaxylem vessels and the first layer of interfascicular fibers next to the endodermis were measured for their wall thickness. For each construct used for transformation, at least 10 transgenic plants with the most severe phenotypes in their first generation were examined for the wall thickness of fibers and vessels.
For visualization of lignified cell walls in leaves, methanol-cleared leaves were used for examination of lignin autofluorescence and collection of DIC images under a microscope (Zhong et al., 2006). For examination of lignified cell walls in stems, 50-μm-thick sections were stained for 5 min with 1% phloroglucinol in 6 n HCl for lignin, which was shown as a bright red color. One-micrometer-thick sections were stained for cellulose with 0.01% Calcofluor White and observed with a UV fluorescent microscope as described (Hughes and McCully, 1975). Under the conditions used, only secondary walls exhibited brilliant fluorescence.
Immunolocalization of Xylan
For examination of xylan in cell walls, 1-μm-thick sections were probed with the LM10 monoclonal antibody (1:10 dilution), which binds to 4-O-methylglucuronoxylan (McCartney et al., 2005), and detected with fluorescein isothiocyanate–conjugated goat-anti-rat secondary antibodies (Sigma-Aldrich) according to McCartney et al. (2005). The fluorescence-labeled xylan signals were visualized with a UV fluorescence microscope.
Lignin Composition Analysis
Lignin composition was determined as described by Akin et al. (1993) and Zhong et al. (1998). Leaves and stems were ground into fine powder in liquid nitrogen and extracted in 70% ethanol at 70°C. After vacuum drying at 60°C, the samples were hydrolyzed in 4 n NaOH at 170°C for 2 h. The hydrolysate was acidified with 2 n HCl to pH 2.0. The released lignin monomers were extracted into diethylether and vacuum dried. The residue was dissolved in pyridine and N,O-bis(trimethylsilyl)trifluoroacetamide and analyzed for phenolics by gas–liquid chromatography. Phenolic compounds were identified by comparison of their mass spectra with those of the authentic compounds. All samples were run in duplicate. G (guaiacyl) lignin is the sum of vanillin, acetovanillin, and vanillic acid. S (syringyl) lignin is the sum of syringaldehyde, acetosyringaldehyde, and syringic acid.
Transfection of Leaf Protoplasts for Transactivation Study
For analysis of the transactivation of the 4CL1 promoter by MYB58 and MYB63, the effector constructs were created by ligating the PCR-amplified (primers are listed in Supplemental Table 1 online), full-length cDNA of MYB58 or MYB63 into the BamHI site between the CaMV 35S promoter and the nopaline synthase terminator in pBI221 without GUS. The 4CL1 promoter fragments or a 38-bp DNA oligonucleotide containing three copies of the AC element (3xAC; 5′-agtccaccaaccgccaccaaccgccaccaaccgctgtt-3′) linked with the CaMV 35S minimal promoter were ligated at the BamHI site upstream of the GUS reporter gene in pBI221 without the CaMV 35S enhancer sequence to create the reporter constructs. The mutation of the AC element in the 4CL1 promoter was done by PCR amplification with primers containing the mutated AC element (primers are listed in Supplemental Table 1 online). The reporter and effector constructs were cotransfected into Arabidopsis leaf protoplasts (Sheen, 2001). A reference construct containing the firefly luciferase gene driven by the CaMV 35S promoter was also included in the transfection to determine the transfection efficiency. GUS and luciferase activities were assayed as described (Gampala et al., 2001) in the soluble extracts lysed from the transfected protoplasts after 20-h incubation. The GUS activity was calculated by normalizing against the luciferase activity in each transfection. The data were the averages of three biological replicates.
Estrogen-Inducible System for Testing Direct Activation of Genes by MYB58
The MYB58-HER and MYB63-HER expression constructs were created by ligating the PCR-amplified (primers are listed in Supplemental Table 1 online) full-length cDNA of MYB58 or MYB63 at the BamHI site fused with the regulatory region of HER at the C terminus (Zuo et al., 2000) between the CaMV 35S promoter and the nopaline synthase terminator in pBI221 without GUS. These constructs alone or together with the 4CL1P:GUS construct were transfected into Arabidopsis leaf protoplasts (Sheen, 2001). The protoplasts were incubated with 2 μM estradiol (Sigma-Aldrich) for 6 h to activate the MYB58 and MYB63 activities. The same concentration of ethanol used to dissolve estradiol was applied to the control protoplasts as the mock treatment. Where indicated, protoplasts were treated with cycloheximide (2 μM) 30 min before addition of estradiol to inhibit new protein synthesis. Under this condition, no new protein synthesis was detected by the GUS reporter gene analysis. The treated protoplasts were subsequently used for GUS activity analysis or subjected to total RNA isolation and real-time quantitative RT-PCR analysis. The expression level of each gene in the control protoplasts without addition of estradiol was set to 1, and the data were the averages of three biological replicates.
EMSA
The PCR-amplified MYB58 or MYB63 cDNA (primers are listed in Supplemental Table 1 online) was ligated into the BamHI site in the pMAL vector (New England Biolabs) and expressed in Escherichia coli. The recombinant protein was purified using amylose resin (New England Biolabs) according to the manufacturer's protocol. Purified recombinant MYB58 and MYB63 fused with the MBP were analyzed for their ability to bind to the 260-bp 4CL1 promoter fragment and the 38-bp oligonucleotide containing three copies of the AC element (3xAC) in EMSA. The 4CL1 promoter fragment and the 3xAC oligonucleotide were biotin-labeled at the 3′ end using the Biotin 3′ End DNA Labeling Kit (Pierce). The biotin-labeled DNA fragments were incubated for 30 min with 100 ng of MBP, MYB58-MBP, or MYB63-MBP in the binding buffer [10 mM Tris, pH 7.5, 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, 0.05% Nonidet P-40, 100 ng/μL poly(dI-dC)]. For competition analysis, unlabeled AC-I (5′-ctatttcctatattacctacctcttatac-3′), AC-II (5′-ctatttcctatattaccaacctcttatac-3′), AC-III (5′-ctatttcctatattacctaactcttatac-3′), or mAC (5′-ctatttcctatattaaatatttcttatac-3′) oligonucleotide was included in the reactions as competitors in a 20-fold molar excess relative to the labeled probes. The MYB58- or MYB63-bound DNA fragments separated by 10% PAGE were electroblotted onto nitrocellulose membrane and subsequently detected using the Chemiluminescent Detection Kit (Pierce).
Statistical Analysis
The Student's t test program (http://www.graphpad.com/quickcalcs/ttest1.cfm) was used for statistical analysis of the data in the experiments of quantitative RT-PCR, transcriptional activation of the GUS reporter gene, and measurement of cell wall thickness. In all these experiments, it was found that the quantitative differences between the two groups of data for comparison were statistically significant (P < 0.001).
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers or the GenBank database accession numbers for the genes investigated in this study are MYB58 (At1g16490), MYB63 (At1g79180), SND1 (At1g32770), NST1 (At2g46770), NST2 (At3g61910), VND6 (At5g62380), VND7 (At1g71930), MYB46 (At5g12870), MYB20 (At1g66230), MYB85 (At4g22680), MYB103 (At1g63910), MYB42 (At4g12350), MYB43 (At5g16600), MYB20 (At1g66230), MYB52 (At1g17950), MYB54 (At1g73410), MYB69 (At4g33450), MYB61 (At1g09540), MYB26 (At3g13890), MYB75 (At1g56650), LAC4 (At2g38080), LAC17 (At5g60020), PAL1 (At2g37040), C4H (At2g30490), 4CL1 (At1g51680), HCT (At5g48930), C3H1 (At2g40890), CCoAOMT1 (At4g34050), CCR1 (At1g15950), F5H1 (At4g36220), COMT (At5g54160), CAD6 (At4g34230), CesA4 (At5g44030), CesA7 (At5g17420), CesA8 (At4g18780), IRX8 (At5g54690), IRX9 (At2g37090), CHS (At5g13930), CHI (At3g55120), ALDH (At3g24503), SMT (At2g22990), Vv MYB5a (AY555190), Pt MYB1 (AY356372), Pt MYB4 (AY356371), Am MYB308 (P81393), Am MYB330 (P81395), Ptt MYB21a (AJ567345), Eg MYB1 (CAE09058), Eg MYB2 (AJ576023), Zm MYB31 (CAJ42202), and Zm MYB42 (CAJ42204).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Subcellular Localization of MYB58 and MYB63.
Supplemental Table 1. Primers Used in This Work.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Analysis in Figure 1F.
Supplementary Material
Acknowledgments
We thank the editor and the reviewers for their constructive comments and suggestions. We gratefully acknowledge the National Science Foundation (Grant ISO-0744170) for funding the molecular characterization of secondary wall–associated transcription factors and the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Contract DE-FG02-03ER15415) for funding the initial identification and expression analysis of secondary wall–associated transcription factors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zheng-Hua Ye (zhye@plantbio.uga.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Akin, D.E., Morrison, W.H., and Himmelsbach, D.S. (1993). Characterization of digestion residues of alfalfa and orchardgrass leaves by microscopic, spectroscopic and chemical analysis. J. Sci. Food Agric. 63 339–347. [Google Scholar]
- Andersson, K.B., Berge, T., Matre, V., and Gabrielsen, O.S. (1999). Sequence selectivity of c-Myb in vivo. Resolution of a DNA target specificity paradox. J. Biol. Chem. 274 21986–21994. [DOI] [PubMed] [Google Scholar]
- Baudry, A., Heim, M.A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39 366–380. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Besseau, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B., and Legrand, M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54 519–546. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2006). Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J. Integr. Plant Biol. 48 85–98. [Google Scholar]
- Deluc, L., Barrieu, F., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., Carde, J.-P., Mérillon, J.-M., and Hamdi, S. (2006). Characterization of a grapevine R3R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP – Phylogeny Inference Package (Version 3.2). Cladistics 5 164–166. [Google Scholar]
- Fornalé, S., Sonbol, F.M., Maes, T., Capellades, M., Puigdomenech, P., Rigau, J., and Caparros-Ruiz, D. (2006). Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol. Biol. 62 809–823. [DOI] [PubMed] [Google Scholar]
- Gampala, S.S., Hagenbeek, D., and Rock, C.D. (2001). Functional interactions of lanthanum and phospholipase D with the abscisic acid signaling effectors VP1 and ABI1-1 in rice protoplasts. J. Biol. Chem. 276 9855–9860. [DOI] [PubMed] [Google Scholar]
- Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., Lapierre, C., Pollet, B., Verhaegen, D., Chaubet-Gigot, N., and Grima-Pettenati, J. (2005). EgMYB2, a new transcriptional activator from eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43 553–567. [DOI] [PubMed] [Google Scholar]
- Goujon, T., Ferret, V., Mila, I., Pollet, B., Ruel, K., Burlat, V., Joseleau, J.P., Barriere, Y., Lapierre, C., and Jouanin, L. (2003). Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: Effects on phenotype, lignins and cell wall degradability. Planta 217 218–228. [DOI] [PubMed] [Google Scholar]
- Hartmann, U., Sagasser, M., Mehrtens, F., Stracke, R., and Weisshaar, B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57 155–171. [DOI] [PubMed] [Google Scholar]
- Hatton, D., Sablowski, R., Yung, M.-H., Smith, C., Schuch, W., and Bevan, M. (1995). Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 7 859–876. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Mitsuda, N., Matsui, K., and Ohme-Takagi, M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321 172–178. [DOI] [PubMed] [Google Scholar]
- Hughes, J., and McCully, M.E. (1975). The use of an optical brightener in the study of plant structure. Stain Technol. 50 319–329. [DOI] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinska, B., Karlsson, M., Srivastava, M., Stenberg, A., Schrader, J., Sterky, F., Bhalerao, R., and Wingsle, G. (2004). MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen. Plant Mol. Biol. 56 255–270. [DOI] [PubMed] [Google Scholar]
- Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., Mimura, T., Fukuda, H., and Demura, T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat, V., Rech, P., Jauneau, A., Guez, C., Coutos-Thevenot, P., and Grima-Pettenati, J. (2002). The vascular expression pattern directed by the Eucalyptus gunnii cinnamyl alcohol dehydrogenase EgCAD2 promoter is conserved among woody and herbaceous plant species. Plant Mol. Biol. 50 497–509. [DOI] [PubMed] [Google Scholar]
- Liang, Y.-K., Dubos, C., Dodd, I.C., Holroyd, G.H., Hetherington, A.M., and Campbell, M.M. (2005). AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 15 1201–1206. [DOI] [PubMed] [Google Scholar]
- McCartney, L., Marcus, S.E., and Knox, J.P. (2005). Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 53 543–546. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics, (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulates secondary wall thickening and are required for anther dehiscence. Plant Cell 17 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, L.J., Perazza, D.E., Juda, L., and Campbell, M.M. (2004). Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 37 239–250. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L.J., Smith, C., Bevan, M.W., Mansfield, S., Whetten, R.W., Sederoff, R.R., and Campbell, M.M. (2003. a). Characterization of a pine MYB that regulates lignification. Plant J. 36 743–754. [DOI] [PubMed] [Google Scholar]
- Patzlaff, A., Newman, L.J., Dubos, C., Whetten, R.W., Smith, C., McInnis, S., Bevan, M.W., Sederoff, R.R., and Campbell, M.M. (2003. b). Characterization of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 53 597–608. [DOI] [PubMed] [Google Scholar]
- Pena, M.J., Zhong, R., Zhou, G.-K., Richardson, E.A., O'Neill, M.A., Darvill, A.G., York, W.S., and Ye, Z.-H. (2007). Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Meissner, R.C., Shoue, D.A., Carpita, N.C., and Bevan, M.W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes, J., Rohde, A., Christensen, J.H., Peer, Y.V., and Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, L.A., and Campbell, M.M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164 17–30. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W.M., and Meyerowitz, E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92 93–103. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W.M., Moyano, E., Culianez-Macia, F.A., Schuch, W., Martin, C., and Bevan, M. (1994). A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F.A., Roberts, K., and Martin, C. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Gardiner, J.C., Whiteman, R., and Turner, S.R. (2004). Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11 329–338. [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme, R., Morreel, K., Ralph, J., and Boerjan, W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11 278–285. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Sablowski, R.W.M., and Meyerowitz, E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584. [DOI] [PubMed] [Google Scholar]
- Weng, J.-K., Li, X., Bonawitz, N.D., and Chapple, C. (2008). Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19 166–172. [DOI] [PubMed] [Google Scholar]
- Ye, Z.-H., Freshour, G., Hahn, M.G., Burk, D.H., and Zhong, R. (2002). Vascular development in Arabidopsis. Int. Rev. Cytol. 220 225–256. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Demura, T., and Ye, Z.-H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Lee, C., Zhou, J., McCarthy, R.L., and Ye, Z.H. (2008). A battery of transcription factors Involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Morrison, W.H., Negrel, J., and Ye, Z.-H. (1998). Dual methylation pathways in lignin biosynthesis. Plant Cell 10 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Pena, M.J., Zhou, G.-K., Nairn, C.J., Wood-Jones, A., Richardson, E.A., Morrison, W.H., Darvill, A.G., York, W.S., and Ye, Z.-H. (2005). Arabidopsis Fragile Fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Richardson, E.A., and Ye, Z.-H. (2007. a). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Richardson, E.A., and Ye, Z.-H. (2007. b). Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225 1603–1611. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (2007). Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 10 564–572. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.-W., and Chua, N.-H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.