Abstract
Postendosymbiotic evolution has given rise to proteins that are multiply targeted within the cell. Various mechanisms have been identified to permit the expression of proteins encoding distinct N termini from a single gene. One mechanism involves alternative translation initiation (aTI). We previously showed evidence of aTI activity within the Arabidopsis thaliana organellar DNA polymerase gene POLγ2. Translation initiates at four distinct sites within this gene, two non-AUG, to produce distinct plastid and mitochondrially targeted forms of the protein. To understand the regulation of aTI in higher plants, we used Polγ2 as a model to investigate both cis- and trans-acting features of the process. Here, we show that aTI in Polγ2 and other plant genes involves ribosome scanning dependent on sequence context at the multiple initiation sites to condition specific binding of at least one trans-acting factor essential for site recognition. Multiple active translation initiation sites appear to operate in several plant genes, often to expand protein targeting. In plants, where the mitochondrion and plastid must share a considerable portion of their proteomes and coordinate their functions, leaky ribosome scanning behavior provides adaptive advantage in the evolution of protein dual targeting and translational regulation.
INTRODUCTION
Complexity of the eukaryotic proteome is greatly enhanced by multiple gene products encoded by a single gene. One means of deriving this complexity is alternative translation initiation (aTI) activity. Several examples of aTI have been reported in mammalian genes (Touriol et al., 2003), operating via ribosome scanning, internal ribosome entry, and ribosome shunting. Whereas ribosome scanning allows the ribosome to pause at multiple sites of translation initiation as it scans the 5′ untranslated region (UTR) sequence, internal ribosome entry and ribosome shunting are processes allowing the ribosome to physically bypass much of the 5′ UTR sequence to position at a precise site of translation initiation. These processes can effect translational regulation in response to cellular Met levels (Hann et al., 1992), cell stress (Vagner et al., 1996; Powell et al., 2008), and developmental cues (Zhou and Cidlowski, 2005).
The coevolution of mitochondria and plastids has produced several levels of organellar crosstalk (Woodson and Chory, 2008). Several examples exist of plant genes encoding proteins targeted to both mitochondria and plastids (Mackenzie, 2005). However, one might hypothesize that the process of protein dual targeting is subject to some degree of cellular regulation to control relative protein stoichiometries in different tissues. One means to effect dual targeting control is aTI, in some cases using non-AUG initiation codons, to provide alternative N termini to the product of a single gene (Kobayashi et al., 2001; Christensen et al., 2005; Sunderland et al., 2006). Examples of aTI in plants are largely limited to individual gene studies, with little information available regarding the mechanisms controlling the process or prevalence of the phenomenon as a gene regulatory mechanism. Here, we report on the process of aTI in plants, using a dual-targeted organellar DNA polymerase as a model. We show evidence of leaky ribosome scanning with both cis- and trans-acting components of the process, together with evidence suggesting that this multi-initiation activity influences translational regulation in these genes. Our study also suggests that aTI operates in a number of plant genes.
RESULTS
In Vivo Evidence of aTI Activity Is Reproduced in Vitro
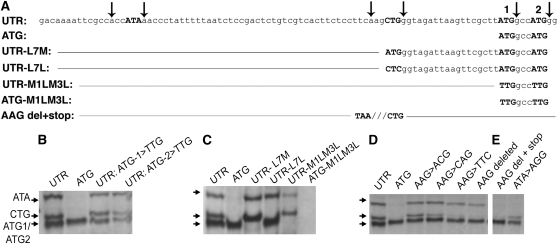
Arabidopsis thaliana Polγ1 (At3g20540) and Polγ2 (At1g50840) are highly similar, duplicate genes encoding organellar DNA polymerases (Christensen et al., 2005). In an earlier study, we demonstrated the influence of aTI on the dual targeting properties of the organellar DNA polymerase Polγ2 protein in vivo using green fluorescent protein (GFP) reporter gene constructs. To examine the aTI process in more detail, we investigated Polγ2 translation initiation in vitro. Figure 1 demonstrates evidence of in vitro aTI activity using wheat germ extracts. We identified four initiation sites: two at ATG sites (ATG1 and ATG2) separated by one codon, one at a CTG located at −21 nucleotides from ATG1, and one at an ATA at −69 nucleotides upstream of ATG1 (Figure 1A). These initiation sites were confirmed functionally by introducing mutations at each site (Figures 1B to 1E).
Figure 1.
In Vitro aTI Activity Depends on Sequence Context.
(A) The At Polγ2 UTR region that was included in the experiments is shown, together with several mutations. Each of the designed constructions extends an additional 414 bp downstream. Downward arrows indicate −3 or +4 sites in Kozak context, mutation sites are underlined, and dashes designate same as in UTR. All IVTT experiments were performed in wheat germ extracts.
(B) PAGE results showing IVTT products when the first or second ATG is mutated to TTG. UTR and ATG lanes serve as controls.
(C) IVTT results from experiments testing both CTG and ATG sites for translation initiation activity. Mutations are defined in (A). Substitution of ATG for CTG prevents the downstream ATG initiation, consistent with Kozak predictions (Kozak, 1987).
(D) IVTT results testing the importance of AAG purine triplet preceding CTG for efficiency of initiation. The AAG was subjected to nucleotide substitutions and deletion (DEL).
(E) Tests of initiation and in-frame translation from the upstream (−62 nucleotides) ATA. ATA was confirmed as an initiator codon by AGG substitution. Deletion of the AAG preceding CTG was accompanied by substitution of a stop codon (TAA), demonstrating that the ATA initiated in-frame translation accounts for the larger product. Panel arrows at left designate products from ATA, CTG, and ATG1/ATG2 translation initiation in vitro. (D) and (E) are from the same gel.
Eukaryotic protein translation initiation is known to occur primarily at AUG codons encompassed within the conserved Kozak consensus sequence (gcc)gccRccAUGG, where R is a purine three bases upstream of AUG, and the AUG is followed by another G (Kozak, 1987). Each of the four identified initiation sites contained a purine at position +4, and all but ATG1 contained a purine at position −3. This latter observation is consistent with observations of efficient, non-AUG initiation when AUG is in a weaker Kozak consensus context (Kozak, 2002). Also surprising was the upstream AUA initiation site for its distance from the initiator AUG and observation that, in vitro, each initiation site appeared approximately equal in efficiency.
In vitro transcription-translation (IVTT) experiments with the UTR sequence and with site-directed mutations showed that both AUG1 and AUG2 initiate translation (Figures 1B and 1C). Substitution of the CUG with CUC (UTR-L7L) eliminated initiation at the CUG site (Figure 1C). Similarly, mutation of the AUA to AGG eliminated initiation at the upstream AUA site (Figure 1E). These experiments demonstrate that, in vitro, four distinct sites are active in translation initiation of the Polγ2 gene.
aTI in Plants Is Cap Dependent and Involves Ribosome Scanning
Non-ATG initiation is observed in several mammalian genes in association with both leaky ribosome scanning and internal ribosome entry mechanisms (Kozak, 2002). While cap-independent internal ribosome entry has been described for various viral and mammalian genes, there is little evidence to implicate the process in plant genes. A model for aTI activity in Polγ2 via leaky ribosome scanning was supported by our initial observation that substitution of the initiator CTG with ATG enhanced initiation at the site and eliminated initiation at the downstream ATG1 and ATG2 sites (Figure 1C, UTR L7M). This observation is consistent with ribosome scanning, since strengthening the consensus ribosome binding sequence at the upstream site diverted ribosomes from the downstream site. To further test for cap dependence of aTI in our system, we evaluated translation initiation for sensitivity to transcript decapping by tobacco acid pyrophosphatase treatment, enhancement of translation initiation efficiency with addition of a cap analog, and failure of translational reinitiation in a dicistronic message.
In vitro transcription (IVT) assays using available commercial IVT preparations, followed by incubation in wheat germ extract, produce an unknown degree of transcript capping within a transcript population. Consequently, to assess cap dependence of aTI, we assayed the effect on translation initiation of both decapping, by tobacco acid pyrophosphatase, and capping with the cap analog m7G(5′)ppp(5′)G. Figure 2A shows sensitivity to transcript decapping of translation initiation at all four identified sites, suggesting that the aTI activity in Polγ2 is cap dependent. Consistent with this interpretation, translation initiation in wheat germ extract was markedly enhanced with addition of the cap analog. An RNA gel is presented to demonstrate that in vitro transcripts are stable under these treatment conditions. Figure 2B shows loss of translation initiation at all four sites with the introduction of a luciferase open reading frame upstream to the Polγ2 initiation sites. This observation suggests that internal ribosome entry does not occur within the 5′ UTR sequence of Polγ2 when preceded by upstream translated sequence.
Figure 2.
ATI in Polγ2 Appears to Involve Ribosome Scanning.
(A) mRNA preparations from the wild-type sequence (UTR) and two mutated sequences (M1LM3L altered at ATG1 and ATG2, and L7L altered at CTG) were treated with different amounts of tobacco acid pyrophosphatase prior to translation with wheat germ extracts to test for effects of transcript decapping on translation initiation efficiency at each site. Likewise, the UTR sequence was in vitro transcribed with and without the cap analog m7G(5′)ppp(5′)G to test for enhancement of initiation efficiency at each site following capping. The bottom panel shows the in vitro transcript preparations following incubation with tobacco acid pyrophosphatase and transcripts prepared in the presence of the cap analog. Note that a lower concentration of RNA, shown in the bottom panel, was used for the cap analog translation experiments.
(B) IVTT experiments comparing aTI activity of the UTR and ATG constructions (see Figure 1) versus aTI activity within the UTR sequence that contains the luciferase gene 64 bp upstream to the CTG site (LUC + UTR). Translation of the luciferase gene construction without the UTR is also shown (LUC). The dual panels showing these products were derived from the same experiment, with the image spliced to compress space between the high molecular weight luciferase products and low molecular weight UTR and ATG products.
(C) A 9-nucleotide deletion (DEL, within dashed line) and multipoint mutations (MUT, bold letters) of the polypyrimidine stretch upstream to CTG showed no effect on CTG translation initiation. Lanes labeled DEL and MUT were run on the same gels as the corresponding UTR and ATG control lanes but spliced to remove intervening lanes from the image. Arrows indicate corresponding products from initiation at ATA, CTG, and ATG1/ATG2.
Cap-independent ribosome entry can be associated with pyrimidine-rich sequence motifs proximal to the site of translation initiation (Mitchell et al., 2005). A polypyrimidine tract located upstream to the initiator CUG was subjected to both deletion and point mutation analysis to assess its importance for ribosome selection of the CUG for initiation. Neither the introduction of six purine substitutions nor the deletion of 11 nucleotides from the interval produced any detectable change in CUG initiation in vitro (Figure 2C). These observations, taken together, are uniformly consistent with ribosome behavior under conditions of leaky ribosome scanning and do not support a model of internal ribosome entry for alternative site translation initiation.
aTI Relies on Sequence Context of the Non-AUG aTI Site
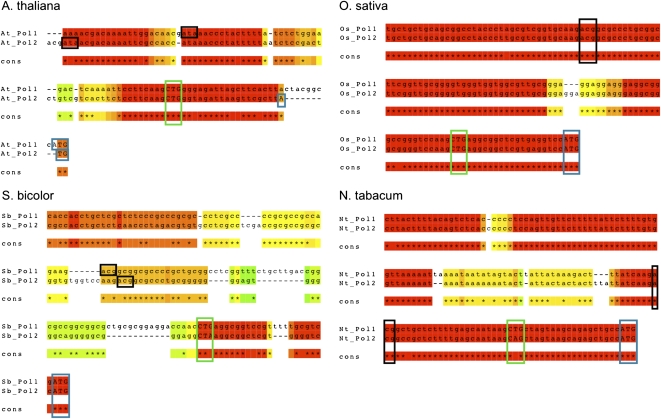
Evidence of leaky ribosome scanning implies that cis-acting sequences direct the ribosome to the non-AUG sites. Mutation of the A at the −3 position alone did not fully abolish initiation at the CUG site in Polγ2 (Figure 1D). Rather, a purine triplet preceding the alternative initiator CUG codon appeared to be essential for initiation. Purine triplets precede the CUG start site in both Polγ1 and Polγ2 genes in four different plant species investigated to date: Arabidopsis, tobacco (Nicotiana tabacum), sorghum (Sorghum bicolor), and rice (Oryza sativa) (Figure 3). The extent of 5′ UTR similarity upstream to Polγ1/Polγ2 shared by the four species is striking; each gene possesses an initiator AUG preceded six to nine codons upstream by CU(G/A), which is flanked 5′ by a purine triplet and 3′ by a purine. This observation allowed us to postulate and test the minimal sequence context for aTI activity. Introduction of a CTG, flanked 5′ by three purines and 3′ by GGT, to the 5′ UTR sequence of the cyclophilin 40 gene (At2g15790), a gene that demonstrates no evidence of aTI activity, was sufficient to create a novel aTI site (Figure 4A). Insertion of the AAGCTG sequence lacking the 3′ GGT was not sufficient for activity, however, confirming the importance of both 5′ and 3′ flanking sequence to aTI activity.
Figure 3.
Cross-Species Conservation of 5′ UTR Sequences in Polγ1 and Polγ2.
Multiple sequence alignments of the region upstream to ATG in Polγ1 and Polγ2 from Arabidopsis, O. sativa, N. tabacum, and S. bicolor were performed using the Mcoffee option of the T-Coffee program (http://www.igs.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi; Notredame et al., 2000). Blue boxes indicate annotated ATG initiator codons, green boxes indicate CTG initiator sites, and black boxes indicate upstream ATA or ACG putative initiator sites. Within the color spectrum displayed, red regions are in perfect agreement across all methods used within the analysis program, while blue regions have poor agreement, with yellow and green intermediate in alignment score.
Figure 4.
Cis-Acting Sequences Are Crucial to aTI.
(A) IVTT analysis of a novel aTI site derived by insertion of CTG with varying lengths of surrounding sequence (capitalized in bold) to the 5′ UTR of the cyclophilin-40 gene (At2g15790). Each construction also includes 444 nucleotides of cyclophilin-40 sequence 3′ to the ATG. The construction designated ATG contains no 5′ UTR sequence. The differences in size of the aTI-derived products correspond to the differences in insertion sequence length 3′ to CTG, confirming initiation at the introduced aTI site.
(B) EMSA of RNA-protein binding at the Polγ2 CTG aTI site. Bound RNA probe is shown as bands, while free RNA probe migrates to the bottom of the gel (data not shown). RNA probe sequences corresponding to the individual EMSA assays are shown, with protein binding assays conducted with wheat germ extracts. While some low level binding is detected for the antisense RNA, this may be due to similar sequence features present in the antisense comprised of three purines (GAA) followed by CTT and another purine (A). The bottom panel shows results of binding competition experiments with varying proportions of α32P-labeled/unlabeled RNA probe 2.
(C) Identical EMSA experiment to that shown in (B) with radiolabeled RNA probes 1 to 5, but substituting purified eIF4A-1 overexpression product in place of wheat germ extracts in the RNA binding assay.
Sequences flanking the aTI site influence not only ribosome scanning but also protein binding. Figure 4B shows results of RNA electromobility shift assays (EMSAs) to assess protein binding to the aTI site in wheat germ extracts. Binding to the CUG site in Polγ2 was markedly reduced by deletion of the upstream AAG purine triplet or with the deletion of sequences downstream to CUG. These experiments suggested that sequences flanking CUG were more important to binding than the CUG itself, since substitution of CUC produced little effect on binding in our experiments. This CUC substitution abolishes aTI activity at this site (Figure 1), suggesting that processes of initiation factor binding, dependent on sequence context, are distinguished from ribosome binding and translation initiation, controlled by start codon selection (Pestova et al., 2001).
Proteins associated with the observed shift in RNA mobility were extracted from the gel and identified by mass spectrometry. Cross-comparison of protein profiles derived from assays with the unmodified UTR versus site-mutated transcript sequences identified the translation initiation factor eIF4A to differentially bind the unmodified UTR sequence. In Arabidopsis, at least three genes (At3G13920, At1G54270, and At1G72730) encode eIF4A, an RNA helicase of the Asp-Glu-Ala-Asp (DEAD) box protein family. Subsequent testing showed eIF4A-1 (At3G13920) to be involved in binding at the CUG site. We cloned, overexpressed, and affinity purified this gene product, and subsequent EMSA analysis showed similar differential binding of the overexpression product to the unmodified and mutated UTR sequences (Figure 4C). This observation suggests that eIF4A-1 represents one component of the aTI process. Experiments are now needed to learn whether eIF4A-1 is essential to the process of CUG site selection.
aTI Activity Is Evident in Several Arabidopsis Genes
Information gained from analysis of Polγ2 allowed us to design a search algorithm to identify additional genes within the Arabidopsis genome that appear to implement aTI. Seventy-six of these candidates, predicted to use CTG as initiator with a −3 and +4 purine, are listed in Table 1. Three of the candidates listed were selected for further testing. Figure 5A shows results of IVTT experiments implementing parallel gene constructions with and without the entire predicted 5′ UTR. In each case tested, aTI activity was evident in vitro.
Table 1.
Predicted CTG Initiator aTI Sites within the Arabidopsis Genome
| AGI No. | Description | AGI No. | Description |
|---|---|---|---|
| AT1G21000 | Zinc binding family protein | AT4G10010 | Protein kinase family protein |
| AT1G03260 | Similar to unknown protein | AT4G02680 | EOL1, ETO1-like 1 |
| AT1G53230 | TCP3, TCP transcription factor 3 | AT4G16280 | FCA, RNA binding |
| AT1G02800 | ATCEL2, Arabidopsis cellulase 2 | AT4G23220 | Protein kinase family protein |
| AT1G50840 | POLγ2, Polymerase γ 2 | AT4G21120 | Cationic amino acid transporter |
| AT1G02180 | Ferredoxin-related | AT4G11920 | WD-40 repeat family protein |
| AT1G27840 | ATCSA-1; nucleotide binding | AT4G00730 | ANL2, Anthocyaninless 2 |
| AT1G55760 | BTB/POZ domain-containing protein | AT4G12350 | MYB42, Myb domain protein 42 |
| AT1G63820 | Similar to unknown protein | AT4G32300 | Lectin protein kinase family protein |
| AT1G78310 | VQ motif-containing protein | AT4G15850 | ATRH1, RNA helicase 1 |
| AT1G15840 | Unknown protein | AT4G26965 | NADH:ubiquinone oxidoreductase |
| AT1G34780 | ATAPRL4 (APR-LIKE 4) | AT4G37190 | Similar to Os03g0240900 |
| AT1G43160 | RAP2.6, Related to AP2 6 | AT4G17680 | Protein binding |
| AT1G11860 | Aminomethyltransferase, putative | AT5G36250 | Protein phosphatase 2C |
| AT1G71020 | Armadillo/β-catenin repeat protein | AT5G61960 | AML1, Arabidopsis MEI2-like protein 1 |
| AT2G02090 | CHR19/ETL1, Chromatin protein 19 | AT5G47190 | Ribosomal protein L19 family protein |
| AT2G20680 | Glycosyl hydrolase family 5 protein | AT5G28080 | WNK9, WNK kinase 9 |
| AT2G17990 | Similar to kinectin-related | AT5G03440 | Similar to unknown protein |
| AT2G17975 | Zinc finger (Ran binding) protein | AT5G62610 | Basic helix-loop-helix family protein |
| AT2G17760 | Aspartyl protease family protein | AT5G43960 | Nuclear transport factor 2 protein |
| AT2G01170 | Amino acid permease family protein | AT5G13360 | Auxin-responsive GH3 family protein |
| AT2G34150 | WAVE1 | AT5G14500 | Aldose 1-epimerase family protein |
| AT2G35585 | Similar to unknown protein | AT5G65470 | Similar to unknown protein |
| AT2G03980 | GDSL-motif lipase/hydrolase protein | AT5G18250 | Similar to unknown protein |
| AT2G18876 | Similar to unknown protein | AT5G60650 | Unknown protein |
| AT2G27160 | Unknown protein | AT5G44120 | CRA1 (CRUCIFERINA1) |
| AT3G22790 | Kinase interacting family protein | AT5G35330 | MBD02, Methyl-CpG-binding domain 2 |
| AT3G63000 | NPL41 (NPL4-LIKE PROTEIN1) | AT5G35630 | GS2, Gln Synthetase 2 |
| AT3G20540 | POLγ1, Polymerase γ 1 | AT5G10490 | MSL2, MSCS-like 2 |
| AT3G05580 | Ser/Thr protein phosphatase putative | AT5G14060 | Lys-sensitive aspartate kinase |
| AT3G17890 | Unknown protein | AT5G42520 | Basic Pentacysteine 6 |
| AT3G62660 | GATL7, Galacturonosyltransferase-like 7 | ||
| AT3G21180 | ACA9, Autoinhibited Ca2+-ATPase 9 | ||
| AT3G06770 | Glycoside hydrolase family 28 protein | ||
| AT3G10985 | SAG20, Wound induced protein 12 | ||
| AT3G58690 | Protein kinase family protein | ||
| AT3G25080 | Similar to unknown protein | ||
| AT3G26370 | Similar to unknown protein | ||
| AT3G51340 | Pepsin A | ||
| AT3G15030 | TCP4, TCP Transcription Factor 4 | ||
| AT3G50830 | Cold-regulated 413 plasma membrane 2 | ||
| AT3G59350 | Ser/Thr protein kinase | ||
| AT3G20070 | TTN9 (TITAN9) | ||
| AT3G54010 | PAS1, PASTICCINO1 | ||
| AT3G05380 | DNA binding |
Figure 5.
Evidence of aTI Activity in Multiple Arabidopsis Genes.
(A) Three Arabidopsis loci were tested in vitro for evidence of ATI activity based on features identified within the annotated 5′ UTR sequence. Constructions were designed to include (UTR) or omit (ATG) the 5′ UTR sequence (250 bp in each case). An additional 444 bp beyond the ATG was included in each construction. IVTT reactions with wheat germ extracts were fractionated by PAGE using similar conditions to those in Figures 1 to 3.
(B) At Polγ1 and Polγ2 gene constructions with and without the 5′ UTR sequence were subjected to IVTT to test for evidence of aTI activity at the conserved CTG initiator site.
In examining the sequence similarity that exists in various plant species between Polγ1 and Polγ2 orthologs, we found that these gene pairs shared striking 5′ UTR sequence similarity not limited to the region encompassing the CTG and ATG initiator sites, but extending a considerable distance upstream (Figure 3; Christensen et al., 2005). In Arabidopsis, the two genes diverge in sequence just upstream to the ATA initiator site. IVTT assays indicated aTI activity for the CTG initiator in Polγ1 (Figure 5B). However, in vivo protein localization experiments indicate that the POLγ1 protein is dually targeted to mitochondria and plastids when initiated at either the ATG or the CTG site (Christensen et al., 2005). Consequently, we were not able to account for the surprising extent of sequence conservation observed upstream to ATG in the two genes, across four different plant species, based on control of protein targeting alone. This observation raises the possibility that aTI activity might participate in translational regulation. Examples exist in which an N-terminal extension influences behavior or binding affinity of a protein (Fajardo et al., 1993), and small, upstream open reading frames can negatively regulate translation from a downstream initiator codon (Puyaubert et al., 2008).
aTI May Be Influenced Developmentally
Two strategies for assessing participation of aTI in translational control are to assess changes in expression during development and to examine influence on translation of aTI mutants. We developed several Arabidopsis stable transformants containing gene constructions encoding the 5′ UTR sequence of Polγ2, both in wild-type and mutant configurations, fused with a GFP reporter gene to test the influence of 5′ UTR modifications on translational control. Constructions encompassed the region from 85 nucleotides upstream to ATG1 to 240 nucleotides downstream of ATG1.
Spatial/temporal patterns of protein targeting behavior, assessed visually, showed variation in relative mitochondrial and plastid forms of the protein during plant development (Figure 6A). Evidence of developmental control was particularly striking at the stem-root junction, where plastid GFP fluorescence was especially evident in the stem, but mitochondrial fluorescence was observed in the root.
Figure 6.
aTI and Control of Translation Are Influenced by Plant Development and Point Mutations.
(A) Arabidopsis stable transformants of vector without insert (Col-0) and three Polg2-GFP constructions (construction designations indicated in Figure 1A) were evaluated for protein targeting in three tissue types by confocal laser scanning microscopy. Left panels show the green channel, right panels show the red channel, and the middle panels show merged images. Plastids autofluoresce red in photosynthetic tissue. Yellow arrows designate plastids, white arrows designate mitochondria, and red arrows designate the approximate shoot-root junction.
(B) Quantitation of GFP fluorescence in plant extracts, averaging three independent experiments, with individual plants used for each experiment and error bars indicating range in variation. Results were similar when three plants were pooled per sample. Pol2 designates the unmodified UTR construction.
(C) Arabidopsis 10-week-old stable transformants showing delayed flowering of mutants relative to wild-type (Col-0) or Polg2 5′ UTR-GFP (UTR) construction.
While dual targeting of the POLγ2 product was confirmed previously, stable transformants of the Polγ2 5′ UTR-GFP fusion produced relatively low levels of the mitochondrial form, based on visual assessments (Figure 6, UTR). Point mutations of ATG1 and ATG2 (UTR M1LM3L) produced dramatic increases in mitochondrially targeted product, suggesting that coordinate translation initiation at ATG1, ATG2, and CTG might serve to control translation activity from each individual start site. This enhancement of mitochondrially targeted product was evident in both the cotyledon and sepal tissues. An increase in plastid-targeted product was also evident upon disruption of the CTG (UTR-L7L), although this effect is less visually pronounced due to autofluorescence of the mature plastids.
GFP quantitation experiments were conducted using aboveground plant tissues from 12-d-old plants and comparing nontransformed Columbia (Col-0) to transformants with the unmodified UTR, L7L, and M1LM3L constructions. Results showed greatly enhanced (doubled) fluorescence at 488/507 nm in the transgenic lines relative to autofluorescence levels recorded in nontransformed Col-0 (Figure 6B). However, virtually no differences in GFP fluorescence levels were detected between lines containing the UTR, L7L, and M1LM3L constructions. We interpret this result to indicate that total translation levels remain unaltered in the modified constructions, so that protein levels normally divided to plastids and mitochondria in the UTR are now shuttled exclusively to mitochondria in the M1LM3L construction or to plastids in the L7L construction. Thus, the amount of protein to either organelle essentially doubles in the modified constructions, accounting for the greatly enhanced visual fluorescence observed by laser confocal microscopy. We interpret this observation as an indication that ribosome competition plays an important role in regulating translation levels of the alternate products of the gene under these conditions. How this process is modulated at developmental transitions, such as the stem-root junction shown in Figure 6, is not yet clear.
Destabilized translational control that occurred with disruption of CTG or ATG1/ATG2 sites was also evident physiologically in the derived transformants. Plants transformed with the altered constructions UTR-M1LM3L-GFP or UTR-L7L-GFP (10 plants tested from three independent transformants each) consistently displayed a delayed flowering phenotype relative to Col-0 wild-type or unmodified Polγ2 5′ UTR-GFP fusions (Figure 6C). At 12-h daylengths, flowering was delayed ∼14 d, and at 24-h daylengths, flowering was delayed 5 d. Other than the flowering delay, the transgenic lines containing the modified constructions appeared physiologically normal, and flower morphology did not appear to be altered. This apparent physiological effect on plants transformed with the mutated constructions appears to be a consequence of the higher levels of GFP protein targeting to the mitochondria or plastids in these lines. No unusual effects were observed in transformants containing the unmodified Polγ2 5′ UTR-GFP constructions.
DISCUSSION
Recent evidence has shown that 5′ UTR sequences implement important functions in the translational regulation of eukaryotic genes. Results of this study suggest that aTI occurs relatively frequently in higher plants. We found no evidence in our study of internal ribosome entry in this process; leaky ribosome scanning appears to account for all of the aTI activity that we detect in Arabidopsis Polγ2. The degree of cross-species sequence similarity discovered in Polγ1 and Polγ2 5′ UTR sequences and the number of additional genes sharing 5′ UTR features in common with Polγ2 suggest that leaky ribosome scanning participates in the translational control of additional plant genes. Genes shown in Table 1 are those found to contain a putative aTI site with CUG as initiator; we presume that a number of additional aTI sites use alternative initiation codons (Table 2). Likewise, genes shown are aTI sites with a purine at the −3 position; ∼49% of these contained a purine triplet. Of the three genes selected for in vitro testing, At3g05580 contained only the −3 purine, while AT2G20680 and AT4G15850 contained the purine triplet. Since all three displayed aTI activity, variation exists for aTI site sequence context requirements.
Table 2.
Computer-Predicted aTI Sites within the Arabidopsis Genome
| AXX—G | RRR—R | RXX—R | |
|---|---|---|---|
| CTG | 76 | 120 | 240 |
| ATC | 188 | 394 | 841 |
| ACG | 46 | 170 | 355 |
| GTG | 82 | 146 | 412 |
| ATA | 92 | 239 | 560 |
Numbers of genes identified to contain an aTI site within the 5′ UTR. Sites were characterized by start codon (rows) and sequence contexts (columns), with R designating purine.
The CTG residing six to nine amino acids upstream to the annotated ATG is active in aTI to influence POLγ2 protein targeting. However, function of the upstream ATA site has not yet been fully defined. One interesting observation suggests that the ATA site might participate in plastid targeting of the protein. Sequences immediately downstream to ATA, if translated, would be predicted to produce a plastid targeting protein. More importantly, however, we have shown in this study that the substitution of CTG with ATG prevents initiation at the downstream ATG sites, predicting only a mitochondrial product to form. In Christensen et al. (2005), this construction, designated UTR-L7M, was shown in planta to produce dual targeting of GFP. One interpretation of this result is that the plastid targeting observed is derived from the upstream ATA. However, an alternative interpretation is that the CTG-to-ATG change in UTR-L7M resulted in such increased mitochondrial targeting that we may be observing a spill-over to chloroplast targeted protein as well. We have not yet fully resolved this question.
Striking conservation of 5′ UTR sequences in plant Polγ1 and Polγ2 genes could signal coordinate translational regulation between the two genes. However, the ATA initiator appears active and in frame to Arabidopsis Polγ2 but is not in frame to At Polγ1, requiring possible initiation at an ACG 18 nucleotides further upstream, if this region is translated in Polγ1. A similar upstream ACG, flanked 5′ by the AAG purine triplet and 3′ by a +4G, is conserved within the 5′ UTR of Polγ1 and Polγ2 genes of rice, sorghum, and tobacco, suggesting at least two aTI sites in these genes as well.
One obvious means of effecting translational control of gene expression by aTI activity is the inherent translational regulation by ribosome competition at the AUG, CUG, and upstream AUA sites. The increase in mitochondrially targeted protein following disruption of the AUG sites, observed by confocal laser scanning microscopy and by measurement of GFP fluorescence, supports the assumption that ribosome competition controls alternative protein levels.
We observed evidence for sequence-dependent protein binding of at least one trans-acting factor, eIF4A-1, at the CUG aTI site in Polγ2. Translation initiation factors comprise a large number of differentially regulated genes in the Arabidopsis genome (The Arabidopsis Information Resource [TAIR]). Such factors could provide the specificity required to effect the tissue-specific aTI-associated translational regulation observed at points such as the stem-root junction.
Coevolution of mitochondria and plastids postendosymbiosis is thought to include massive nuclear transfer of genetic information, followed by selection to reduce redundancy in genes encoding organellar products (Lang et al., 1999; McFadden, 1999). Protein dual targeting mechanisms provide the nucleus with the ability to encode products essential to both mitochondria and plastids within a single gene. One caveat to this efficiency, however, is the implied necessity for a mechanism to modulate relative abundance of mitochondrial to plastid products in a particular tissue.
It is not clear what proportion of dual targeting proteins uses aTI. However, aTI-mediated multitargeting might represent an intermediate in the evolution of dual-targeting, N-terminal presequences. Two types of dual-targeting presequences, termed twin and ambiguous, are described in plants (Peeter and Small, 2001). Twin presequences comprise two distinct targeting peptide domains, one targeting plastid and one mitochondria, fused in tandem at the N terminus of the gene. Each is transcribed or translated distinctly.
The ambiguous presequence provides dual targeting from a single transcriptional and translational product but can sometimes be functionally dissected to mitochondrial and plastid targeting components (Bhushan et al., 2003). If one envisions protein dual targeting as an adaptation emerging from leaky ribosome scanning to facilitate the incorporation of novel N-terminal extensions to existing genes, then a structure such as that observed in Polγ2 might be fairly rudimentary. Inactivation of the downstream start codon, or mutation of the CTG initiator to ATG, would convert Polγ2 to an ambiguous presequence, assuming additional mutations within the N-terminal domain to permit successful targeting to both organelles (presently, mutation of CTG to ATG in Polγ2 results in exclusive targeting to the mitochondrion).
Because of the high degree of protein similarity between Polγ1 and Polγ2, it has not been feasible to use an antibody-based protein detection method to confirm mitochondrial versus plastid localization of the alternatively translated forms of these proteins in vivo. However, Ono et al. (2007) have shown presence and activity of a 116-kD DNA polymerase that appears to represent these proteins in the mitochondria and plastids of tobacco cells.
To assess the likely variation existing within the Arabidopsis genome for aTI sites, we surveyed the entire nuclear genome for 5′ UTR sequence features. Results from this computer-based analysis, summarized in Table 3, show relative frequencies of five non-ATG initiator codons as putative aTI sites and relative frequencies of purine triplets versus single purine at the −3 position. Protein targeting predictions of the 2408 putative aTI products suggest the most likely outcome of aTI to be mitochondrial targeting of an otherwise nonmitochondrial protein. Admittedly, mitochondrial targeting presequences are better defined than plastid in their amino acid composition, so protein targeting prediction programs may bias toward the calling of mitochondrial presequences. However, in an earlier study, we also pointed out that amino acids most essential to mitochondrial targeting presequences (Allison and Schatz, 1986), Ser, Arg, and Leu, are also those encoded by six codons and, therefore, most prevalent in random sequence. While it will be necessary to test many of the predicted gene candidates for aTI activity in vivo, observations to date suggest that 5′ UTR sequence features and a flexible ribosome scanning mechanism provide key links to understanding the evolution of protein dual targeting and its regulation in plants.
Table 3.
Computer-Predicted aTI Sites within the Arabidopsis Genome
| ATG-M,C | N-M | N-C | C-M | S-M | M-C | |
|---|---|---|---|---|---|---|
| AXXCTGG | 17.11 | 14.47 | 0.00 | 2.63 | 3.95 | 0.00 |
| RRRCTGR | 16.67 | 12.92 | 2.08 | 2.92 | 6.25 | 1.25 |
| RXXCTGR | 18.25 | 16.06 | 0.73 | 4.38 | 6.57 | 0.73 |
| AXXATCG | 19.15 | 9.04 | 4.79 | 0.53 | 1.06 | 1.06 |
| RRRATCR | 16.46 | 14.68 | 3.54 | 2.03 | 2.28 | 0.51 |
| RXXATCR | 17.61 | 12.06 | 4.14 | 2.36 | 2.72 | 1.18 |
| AXXACGG | 15.22 | 2.17 | 0.00 | 0.00 | 0.00 | 4.35 |
| RRRACGR | 22.94 | 6.47 | 3.53 | 0.59 | 0.59 | 1.18 |
| RXXACGR | 16.90 | 5.63 | 4.51 | 0.85 | 1.69 | 1.41 |
| AXXGTGG | 9.76 | 7.32 | 3.66 | 1.22 | 2.44 | 1.22 |
| RRRGTGR | 12.33 | 11.64 | 1.37 | 0.68 | 1.37 | 0.68 |
| RXXGTGR | 14.25 | 10.14 | 4.59 | 0.97 | 2.17 | 0.97 |
| AXXATAG | 16.13 | 5.38 | 3.23 | 1.08 | 6.45 | 1.08 |
| RRRATAR | 16.74 | 8.79 | 2.09 | 2.51 | 5.44 | 0.42 |
| RXXATAR | 16.31 | 10.11 | 2.66 | 2.30 | 4.61 | 0.89 |
Protein targeting changes predicted for aTI sites, based on aTI sequence and presented as percentages. ATG-M,C indicates the proportion of aTI-containing genes that are predicted to encode a mitochondrial (M) or chloroplast (C) targeting protein from the initiator ATG. N designates proportion of genes with no targeting predicted (cytosolic) that acquire mitochondrial (N-M) or chloroplast (N-C) targeting by initiation at the aTI site. S designates genes with predicted signal sequence (ER targeting).
METHODS
Plasmids and Strains
All constructions used for IVTT were cloned in the pET-21(+) vector (Novagen). For plant expression, binary vector pCAMBIA1302 was modified by mutating the NcoI cloning site (CCATGG) to CCAGGG to eliminate the ATG start site. Site-directed mutagenesis experiments were performed using cloned Pfu polymerase (Stratagene) for mutagenesis in pET-21(+) or the Quickchange II XL site-directed mutagenesis kit (Stratagene) for mutagenesis in pCAMBIA1302. The luciferase gene (LUC) was PCR amplified from pGL2-Basic (Promega) and cloned upstream of the Polγ2 5′ UTR (85 bp upstream to ATG and 64 bp upstream to CTG) to create the dicistronic construction in pET-21(+). For overexpression and purification of eIF4A-1 protein, At3g13290 cDNA was cloned into pGEX-5X-1 and the protein purified using the glutathione S-transferase purification kit according to manufacturer's specifications (GE Healthcare). Primer sequences for the various constructions are included in Supplemental Table 1 online.
IVTT and Transcript Modification
IVTT assays used the TNT-coupled wheat germ extract systems (Promega) with 35S-labeled Met. Translation products were separated in 16.6% Tris-Tricine gel (Schagger and von Jagow, 1987), dried, and exposed to x-ray film for 16 to 24 h. For transcript decapping experiments involving treatment with tobacco acid pyrophosphatase (Epicentre), mRNA was prepared with MAXIscript (Ambion). For transcript capping experiments, the mMESSAGE mMACHINE kit (Ambion) with the cap analog [m7G(5′)ppp(5′)G] in the reaction mix was used.
EMSA for RNA–Protein Interaction
Each RNA probe was designed in association with a T7 promoter and a spacer of a few random nucleotides upstream to the specific probe sequence. The sequences were then PCR amplified to use as template. 32P-dUTP-labeled RNA probes were prepared with the MAXIscript kit (Ambion), and RNA binding reactions were performed in a 25-μL volume containing 1 μL of labeled RNA, 4 μL of wheat germ extract (Promega), 40 units of RNasin, and 2.5 μL of 10× buffer (Massiello et al., 2006). Reactions were incubated on ice for 20 min, followed by addition of 40 μg heparin and 10 min incubation on ice. Samples were fractionated in 4% nondenaturing polyacrylamide in 0.5× TBE at constant voltage. The binding competition assay involved coincubation of cold RNA with labeled in wheat germ extracts. For specific RNA–protein interaction with eIF4A-1, 10 μg of purified protein was substituted for wheat germ extract in incubation reactions conducted as described above.
Mass Spectrometry
Tandem mass spectrometry was performed at the University of Nebraska Mass Spectrometry Core Facility using a Waters Q-TOF Ultima mass spectrometer (Waters; formally Micromass). Results were analyzed using the Mascot software package (Matrix Science).
Stable Plant Transformation
Arabidopsis thaliana stable transformations were developed with the floral dip procedure (Clough and Bent, 1998). GFP expression in plant tissues was assayed by confocal laser scanning microscopy.
Computational Analysis
The 5′ UTR, coding sequence, and protein sequences of Arabidopsis were downloaded from the TAIR website (Swarbreck et al., 2008; http://www.Arabidopsis.org). Script was written to scan the 5′ UTR of every gene and search for a CTG with −3 adenine, +4 guanine, and in frame to the annotated ATG start codon. The CTG- and ATG-translated products were used for organellar targeting prediction with Predotar (Small et al., 2004) and TargetP (Emanuelsson et al., 2007). Multiple sequence alignment of DNA Polγ1 and Polγ2 in Arabidopsis, Nicotiana tabacum, Sorghum bicolor, and Oryza sativa was constructed using the Mcoffee option of T-Coffee (Notredame et al., 2000) program at http://www.igs.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi.
GFP Quantitation
Twelve-day-old whole plants except roots were ground in liquid nitrogen and 1× Assay/Lysis buffer (GFP quantitation kit; Fluorometric, Cell Biolabs) supplemented with Complete Protease Inhibitor (Roche). Standard curve and sample preparation was performed according to the manufacturer's instructions, and fluorescence was read with a plate reader (Synergy 4 Multi-Mode Microplate Reader with Hybrid Technology; Biotek) at 488/507 nm. The graph presents the average of three independent experiments.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under accession numbers At1g50840 (POLγ2, TAIR accession locus 2036361), At3g20540 (POLγ1, TAIR accession locus 2085730), At2g15790 (CYP40, TAIR accession locus 2044596), At3g05580 (TAIR accession locus 2078087), At4g15850 (ATRH1, TAIR accession locus 2130839), At2g20680 (TAIR accession locus 2051399), At3g13930 (EIF4A1, TAIR accession locus 2088237); in the Gramene database under accession numbers Sb07g004810 (sorghum POLγ1), Sb06g030120 (sorghum POLγ2), Os08g07850 (rice POLγ1), and Os08g07840 (rice POLγ2); and in the National Center for Biotechnology Information database under accession numbers AB174898 (tobacco POLγ1) and AB174899 (tobacco POLγ2).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Table 1. Primer Information Used for This Study.
Supplementary Material
Acknowledgments
Confocal laser scanning microscopy was conducted in the Microscopy Core Facility, University of Nebraska Center for Biotechnology. This work was supported by Department of Energy Grant DE-FG02-07ER15564 to S.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sally A. Mackenzie (smackenzie2@unl.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allison, D.S., and Schatz, G. (1986). Artificial mitochondrial presequences. Proc. Natl. Acad. Sci. USA 83 9011–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan, S., Lefebvre, B., Ståhl, A., Wright, S.J., Bruce, B.D., Boutry, M., and Glaser, E. (2003). Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep. 4 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, A.C., Lyznik, A., Mohammed, S., Elowsky, C.G., Elo, A., Yule, R., and Mackenzie, S.A. (2005). Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 10 2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocols 2 953–971. [DOI] [PubMed] [Google Scholar]
- Fajardo, J.E., Birge, R.B., and Hanafusa, H. (1993). A 31-amino-acid N-terminal extension regulates c-Crk binding to tyrosine-phosphorylated proteins. Mol. Cell. Biol. 13 7295–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann, S.R., Sloan-Broun, K., and Spotts, G.D. (1992). Translational activation of the non-AUG initiated cmyc-1 protein at high cell densities due to methionine deprivation. Genes Dev. 6 1229–1240. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Dokiya, Y., and Sugita, M. (2001). Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem. Biophys. Res. Commun. 289 1106–1113. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (1987). An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, B.F., Gray, M.W., and Burger, G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33 351–397. [DOI] [PubMed] [Google Scholar]
- Mackenzie, S. (2005). Plant organellar protein targeting: A traffic plan still under construction. Trends Cell Biol. 15 548–554. [DOI] [PubMed] [Google Scholar]
- Massiello, A., Roesser, J.R., and Chalfant, C.E. (2006). SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 20 1680–1682. [DOI] [PubMed] [Google Scholar]
- McFadden, G.I. (1999). Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant Biol. 2 513–519. [DOI] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Bushell, M., Evans, J.R., Stoneley, M., Le Quesne, J.P., Spriggs, R.V., and Willis, A.E. (2005). Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 19 1556–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., and Heringa, J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302 205–217. [DOI] [PubMed] [Google Scholar]
- Ono, Y., Sakai, A., Takechi, K., Susumu, T., Takusagawa, M., and Takano, H. (2007). NtPolI-like1 and NtPolI-like2, bacterial DNA polymerase I homologs isolated from BY-2 cultured tobacco cells, encode DNA polymerases engaged in DNA replication in both plastids and mitochondria. Plant Cell Physiol. 48 1679–1692. [DOI] [PubMed] [Google Scholar]
- Peeter, N., and Small, I. (2001). Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 1541 54–63. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G., Lomakin, I.B., Pilipenko, E.V., Shatsky, I.N., Agol, V.I., and Hellen, C.U. (2001). Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, D.J., Hrstka, R., Candeias, M., Bourougaa, K., Vojtesek, B., and Fåhraeus, R. (2008). Stress-dependent changes in the properties of p53 complexes by the alternative translation product p53/47. Cell Cycle 7 950–959. [DOI] [PubMed] [Google Scholar]
- Puyaubert, J., Denis, L., and Alban, C. (2008). Dual targeting of Arabidopsis holocarboxylase synthetase1: A small upstream open reading frame regulates translation initiation and protein targeting. Plant Physiol. 146 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166 368–379. [DOI] [PubMed] [Google Scholar]
- Small, I., Peeters, N., Legeai, F., and Lurin, C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590. [DOI] [PubMed] [Google Scholar]
- Sunderland, P.A., West, C.E., Waterworth, W.M., and Bray, C.M. (2006). An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. Plant J. 47 356–367. [DOI] [PubMed] [Google Scholar]
- Swarbreck, D., et al. (2008). The Arabidopsis Information Resource (TAIR): Fene structure and function annotation. Nucleic Acids Res. 36 D1009–D1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touriol, C., Bornes, S., Bonnal, S., Audigier, S., Prats, H., Prats, A.-C., and Vagner, S. (2003). Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell 95 169–178. [DOI] [PubMed] [Google Scholar]
- Vagner, S., Touriol, C., Galy, B., Audigier, S., Gensac, M.C., Amalric, F., Bayard, F., Prats, H., and Prats, A.C. (1996). Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J. Cell Biol. 135 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson, J., and Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Natl. Rev. 9 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., and Cidlowski, J.A. (2005). The human glucocorticoid receptor: One gene, multiple proteins and diverse responses. Steroids 70 407–417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.