Abstract
The Arabidopsis thaliana MYB5 gene is expressed in trichomes and seeds, including the seed coat. Constitutive expression of MYB5 resulted in the formation of more small trichomes and ectopic trichomes and a reduction in total leaf trichome numbers and branching. A myb5 mutant displayed minimal changes in trichome morphology, while a myb23 mutant produced increased numbers of small trichomes and two-branched trichomes. A myb5 myb23 double mutant developed more small rosette trichomes and two-branched trichomes than the single mutants. These results indicate that MYB5 and MYB23 regulate trichome extension and branching. The seed coat epidermal cells of myb5 and myb5 myb23 were irregular in shape, developed flattened columellae, and produced less mucilage than those of the wild type. Among the downregulated genes identified in the myb5 seeds using microarray analysis were ABE1 and ABE4 (α/β fold hydrolase/esterase genes), MYBL2, and GLABRA2. The same genes were also downregulated in transparent testa glabra1 (ttg1) seeds, suggesting that MYB5 collaborates with TTG1 in seed coat development. These genes were upregulated in leaves and roots by ectopically expressed MYB5. The MYBL2, ABE1, and ABE4 promoters were active in seeds, including seed coats, and the latter two also in trichomes. Models of the MYB5 regulatory networks involved in seed coat and trichome development are presented.
INTRODUCTION
The MYB gene family is one of the largest gene families encoding for transcription factors in Arabidopsis thaliana (Kranz et al., 1998; Stracke et al., 2001). MYB transcription factors are characterized by the presence of a highly conserved DNA binding domain (MYB domain) at their N termini. They are grouped into three subfamilies based on the number of imperfect repeats in the MYB domains, namely, single-repeat MYB, R2R3 MYB (two repeats), and R1R2R3 MYB (three repeats).
A number of MYB transcription factors form complexes with a basic helix-loop-helix (bHLH) transcription factor and a WD40 repeat protein to regulate various cell fate determinations (for reviews, see Schellmann and Hülskamp, 2005; Serna, 2005; Guimil and Dunand, 2006; Serna and Martin, 2006; Martin and Glover, 2007) and metabolic pathways in many plants (for reviews, see Broun, 2005; Haughn and Chaudhury, 2005; Koes et al., 2005; Ramsay and Glover, 2005; Western, 2006). In Arabidopsis, TRANSPARENT TESTA GLABRA1 (TTG1; a WD40 repeat protein) provides a scaffold to facilitate the interactions between MYB and bHLH proteins. The initiation of Arabidopsis trichomes (unicellular hairs on aerial tissues) is promoted by a multimeric complex involving GLABRA1 (GL1), GL3, ENHANCER OF GLABRA3 (EGL3), and TTG1. The multimeric complex is thought to activate the expression of the GL2 gene by binding to its promoter. GL1 codes for a R2R3 MYB protein and GL3 and EGL3 for two partially redundant bHLH proteins. GL2 encodes a homeodomain protein and is required for subsequent phases of trichome morphogenesis, such as branching, extension, and maturation of the trichome cell wall. GL1 regulates trichome initiation at leaf edges redundantly with MYB23, which also regulates trichome branching (Kirik et al., 2005). Four single-repeat MYBs, TRIPTYCHON (TRY), CAPRICE (CPC), ENHANCER OF TRY AND CPC1 (ETC1), and ETC2 act redundantly to repress trichome determination in neighboring cells. The proteins are thought to block the formation of an active GL1/GL3/EGL3/TTG1 complex by replacing GL1 in the complex and thereby inhibiting GL2 expression in neighboring cells.
Similar transcription factors specify root epidermal cell fate. In contrast with trichome initiation, the multimeric complex involving WEREWOLF (WER; a R2R3), GL3, EGL3, and TTG1 represses hair cell fate in roots by activating the GL2 gene. The four single-repeat MYBs counter the activity of WER and promote root hair formation. The CPC gene is preferentially expressed in nonhair cells, but the CPC protein moves to the nuclei of the neighboring hair-forming cells (Wada et al., 2002). In nonhair cells, CPC expression is positively regulated by WER, GL3, and EGL3. CPC is required to establish a closed chromatin conformation around the GL2 locus in the root hair–forming cells (Costa and Shaw, 2006). Similar to root hair patterning, WER, GL3, EGL3, TTG1, and GL2 inhibit stomata formation in hypocotyls (Berger et al., 1998; Martin and Glover, 2007). However, the roles of the single-repeat MYB proteins in the specification of hypocotyl stomata have not been studied in detail.
The interaction of MYB, bHLH, and TTG1 proteins is also essential for certain metabolic pathways (for reviews, see Broun, 2005; Haughn and Chaudhury, 2005; Koes et al., 2005; Ramsay and Glover, 2005; Western, 2006). Thus, PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1; MYB) and PAP2 (MYB), TRANSPARENT TESTA8 (TT8; bHLH), GL3, EGL3, and TTG1 regulate anthocyanin biosynthesis. TT2 (MYB), TT8, and TTG1 are required for proanthocyanidin (condensed tannin) biosynthesis in the seed coat. TT8, EGL3, and TTG1 are necessary for mucilage biosynthesis and epidermal cell development in the seed coat. An involvement of a single-repeat MYB protein in these pathways has not been determined.
In Arabidopsis, the outer cell layer (epidermis) of the seed coat differentiates in a process involving growth, cytoplasmic rearrangement, mucilage (primarily pectins) synthesis, and secondary cell wall production (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000). The mucilage is synthesized in the Golgi apparatus and secreted into the apoplast (the space between the plasma membrane and the primary cell wall) (Young et al., 2008). The mucilage accumulation is restricted to the outer corners of the cell where the radial and tangential walls meet. The cytoplasm moves from the edge of the cell to form a column in the center, and a spindle-shaped cellulosic secondary wall is produced between the plasma membrane and the mucilage. The resulting central elevation is known as the columella. Upon hydration, the pectinaceous mucilage expands, the radial segment of the primary cell wall ruptures, and mucilage forms a pectin hydrogel surrounding the seed. The pectinaceous slime is believed to aid germination through the retention of water surrounding the seed (Penfield et al., 2001). Arabidopsis seed coat mucilage is primarily composed of pectins, largely unbranched rhamnogalacturonan and some highly methyl esterified homogalacturonans (Penfield et al., 2001; Willats et al., 2001; Vincken et al., 2003; Somerville et al., 2004; Macquet et al., 2007b). Relatively few genes involved in pectin biosynthesis have been identified (Scheller et al., 2007).
In addition to the TT8, EGL3, and TTG1 genes, the MUM (mucilage modified) genes, APETALA2 (AP2; a homeotic gene), MYB61 (R2R3 MYB), TTG2 (WRKY), and GL2 genes also affect mucilage and/or epidermal development in the seed coat (Haughn and Chaudhury, 2005; Ramsay and Glover, 2005; Western, 2006). EGL3 interacts with TTG1 and is partially redundant with TT8 in promoting mucilage synthesis (Zhang et al., 2003). GL2 and TTG2 act downstream from EGL3/TT8/TTG1 (Johnson et al., 2002; Western et al., 2004). The effect of myb61 mutations on the seed coat is modest, with a significant amount of mucilage produced and columella formation visible (Penfield et al., 2001). MYB61 appears to function in a genetic pathway distinct from that of TTG1 (Penfield et al., 2001; Western et al., 2004). AP2 is required for early differentiation of the outer integument (Jofuku et al., 1994; Western et al., 2001). MUM4 (RHAMNOSE BIOSYNTHESIS2) encodes a putative pectin biosynthetic enzyme that is developmentally regulated by the TTG1and GL2 pathway (Western et al., 2004; Usadel et al., 2004). The mum1, mum2, and patchy mutants are defective in mucilage release. The mum3 and mum5 mutants produce a less cohesive mucilage than the wild type (Western et al., 2001). The MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with proper hydration properties (Dean et al., 2007; Macquet et al., 2007a). The MYB proteins interacting with EGL3/TT8/TTG1 are currently unknown.
The MYB5 gene is a member of the R2R3 MYB family. The MYB5 promoter is active in trichomes, stipules, leaf margins, and developing seeds (Li et al., 1996). This article further examines the MYB5 expression pattern and the effects of ectopic expression of the gene on trichome development. Two myb5 insertion mutants and a myb5 myb23 double mutant are described. The myb5 mutants display dramatically reduced mucilage production and lack columellae in the seed coat. The double mutant develops more small trichomes and two-branched trichomes on rosette leaves than the individual single mutants and the wild type. Genes that are differentially expressed between wild-type and myb5 mutant seeds have been identified using microarray and RT-PCR analysis. These genes include GL2, a single-repeat MYB, and two α/β fold hydrolase (esterase) genes. The expression patterns of the single-repeat MYB and the two hydrolase (esterase) promoters are described.
RESULTS
MYB5 Expression in Trichomes and Developing Seeds
MYB5 promoter activity was previously detected in developing leaf trichomes, stipules, epidermal cells on leaf margins, and in immature seeds using a MYB5 promoter:β-glucuronidase (GUS) construct (Li et al., 1996). Here, the MYB5 promoter:GUS activity was examined in more detail in seed sections (Figure 1). The MYB5 promoter (1459 bp) was active in developing seeds (Figures 1A and 1B). The GUS activity was detected around fertilization stage, primarily in the inner layers of the integument (Figure 1D). In the developing seed, GUS activity was expressed in all the integument layers, endosperm, suspensors, and globular and heart stage embryos (Figure 1E). However, the GUS activity appeared to be reduced in the endothelium during seed development (Figure 1F). Little GUS activity was detected in the mature embryos (Figure 1C). In situ hybridization was performed to determine the MYB5 transcript levels in trichomes and various tissues of the developing seed. The transcript was detected in developing trichomes using a MYB5 antisense probe (Figure 1G). The antisense probe hybridized to ∼90% of trichomes on the sections, whereas the sense probe exhibited no significant hybridization to the trichomes (see Supplemental Table 1 online). However, the in situ hybridization using seed sections was inconclusive as the background hybridization signal was high. Nevertheless, MYB5 transcript was detected in developing seeds using RNA gel blot hybridization (Li et al., 1996). The transcript appeared between fertilization and the 16-cell stage of embryo development and persisted beyond the heart stage (Li et al., 1996). Preliminary promoter deletion analysis was performed using MYB5 promoter:GUS constructs containing 933-, 230-, 160-, and 22-bp promoter fragments, respectively. The 933-bp MYB5 promoter:GUS construct exhibited an identical GUS expression pattern to the 1459-bp promoter (see Supplemental Figure 1 online). The other constructs failed to express GUS activity in any tissue during development (see Supplemental Figure 1 online), indicating that the regulatory sequences involved in spatial and temporal expression of GUS and presumably the endogenous MYB5 gene are positioned between −230 and −933 bp from the translational start site.
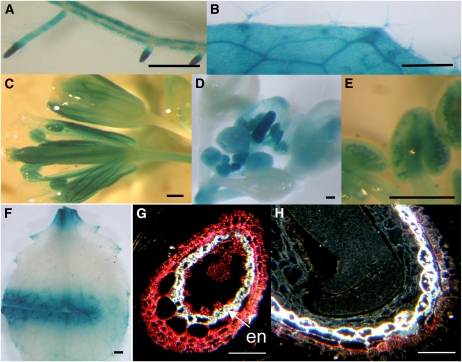
Figure 1.
MYB5 Expression in Developing Trichomes and Seeds.
MYB5 promoter:GUS expression occurs in young seeds inside siliques (A) and in more mature seeds (B) but not in mature embryos (C). GUS expression is detected in seed sections ([D] to [F]; dark-field microscopy). Pink color in seed sections represents the GUS activity. In situ hybridization of leaf sections with MYB5 antisense (G) and sense (H) probes. em, globular embryo; en, endothelium; tr, trichome. Bars = 300 μm in (A) to (C) and 50 μm in (D) to (H).
Ectopic Expression of MYB5 Affects Trichome Development
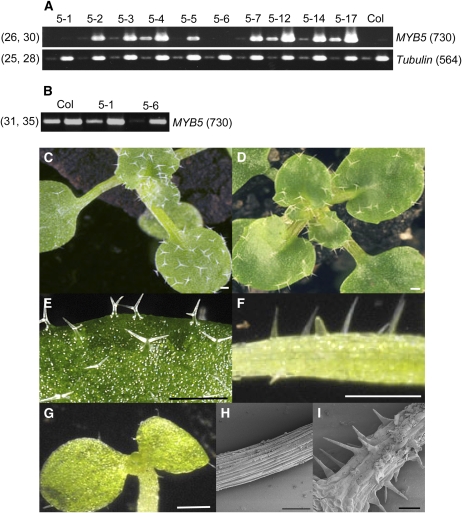
To investigate the effects of constitutive and ectopic expression of MYB5, a 35S:MYB5 construct was transformed into wild-type Arabidopsis plants and 11 lines containing the transgene were examined in detail. The total MYB5 transcript levels (transgenic and endogenous transcripts) in the seedlings of these lines were determined using RT-PCR. The MYB5 transcripts were highly expressed in lines 5-2, 5-3, 5-4, 5-5, 5-7, 5-12, 5-13, 5-14, and 5-17 compared with wild-type plants (Figure 2A, and for line 5-13, see Figure 8A). The transcript levels were reduced slightly in line 5-1 and significantly in line 5-6 compared with the wild type (Figure 2B).
Figure 2.
Phenotypic Changes Induced by Ectopically Expressed MYB5.
MYB5 transcript (A) in the 35S:MYB5 seedlings (10 lines) were amplified using RT-PCR for 26 and 30 cycles, and β-Tubulin for 25 and 28 cycles. MYB5 transcript was amplified for 31 and 35 cycles (B). The numbers in parentheses on the right sides of the gel images indicate the fragment sizes in base pairs and on the left sides, the numbers of PCR cycles. Wild-type plant (C) and the 35S:MYB5 plant (line 5-12) (D) with reduced trichome numbers. Leaf trichomes with reduced branching (line 5-13) (E) and ectopic trichomes developed on the cotyledons (line 5-12) (G) of a 35S:MYB5 seedling. Hypocotyls of the 35S:MYB5 (line 5-12) ([F] and [I]) and wild-type (H) seedlings grown in soil. Bars = 0.4 mm in (C) to (G) and 100 μm in (H) and (I).
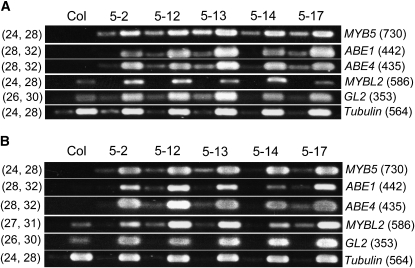
Figure 8.
RT-PCR Analysis of the Transcript Levels of Several Putative MYB5 Target Genes in 35S:MYB5 Developing Leaves and Roots.
RNA was extracted from developing leaves (A) and roots (B) of Col and 35S:MYB5 lines, OE5-2, OE5-12, OE5-13, OE5-14, and OE5-17. The samples were transcribed in RT reactions. PCR reactions (A) were performed for 24 and 28 cycles (MYB5), 28 and 32 cycles (ABE1 and ABE4), 24 and 28 cycles (MYBL2), 26 and 30 cycles (GL2, primers GL2F and GL2R), and 24 and 28 cycles (Tubulin). PCR reactions (B) were performed for 24 and 28 cycles (MYB5), 28 and 32 cycles (ABE1 and ABE4), 27 and 31 cycles (MYBL2), 26 and 30 cycles (GL2), and 24 and 28 cycles (Tubulin). The Tubulin transcript levels were included as RT-PCR controls. Primers 5p2 and 5in2 were used for MYB5 PCR, and the primers for other genes are listed in Supplemental Table 7 online. The numbers in parentheses on the right sides of the gel images indicate the fragment sizes in base pairs and on the left sides the PCR cycle numbers.
The lengths (sizes) and branching of the rosette trichomes in these lines were examined (Table 1; see Supplemental Table 2 online). The length of the longest branch plus the length of the stem represents the length (size) of a trichome. The lengths (sizes) of the trichomes on rosette leaves of the transgenic lines were compared with those of the wild type. Trichomes were grouped into three classes based on their lengths (sizes), namely, normal trichomes that are more than half the length of the average wild-type trichomes; 1/3 trichomes that are less than a third of the average lengths (sizes) of the normal wild-type (Columbia [Col]) trichomes; and 1/2 trichomes that are less than half but more than a third of the average lengths (sizes) of the normal wild-type (Col) trichomes. The lines strongly overexpressing the MYB5 transcripts produced significantly more small trichomes than the wild type did (averaging 60.7% versus 19.4% of all trichomes, respectively) (Table 1, 1/3 and 1/2). The smallest trichomes (1/3) account for 20.8% (Table 1, SL average), and the 1/2 trichomes for 39.9% of all trichomes produced by the strongly overexpressing 35S:MYB5 lines. Most of these lines also produced more trichomes with one or two branches than the wild type (averaging 29.1% of all trichomes; Table 1, 1 and 2br) (Figure 2E). The majority of the smallest trichomes possess one or two branches (13.8% for 1 and 2br, and 7.0% for 3br). Lines 5-4, 5-12, 5-13, and 5-17 exhibited a significant reduction in the number of leaf trichomes (see Supplemental Table 2 online). The reduction was most pronounced in the midvein region of leaves (Figures 2C and 2D). Lines 5-1 and 5-6 expressing reduced levels of MYB5 transcript exhibited minimal changes in trichome morphology.
Table 1.
Analysis of the Lengths and Branching of Trichomes on the 35S:MYB5 and Wild-Type Rosette Leaves
| 1/3 (%)
|
1/2 (%)
|
Normal (%)
|
1 and 2br
|
1/3 and 1/2
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1br | 2br | 3br | 4br | 1br | 2br | 3br | 4br | 1br | 2br | 3br | 4br | Total | Total | |
| Lines | ||||||||||||||
| WT | 0.0 | 1.4 | 0.4 | 0.0 | 0.0 | 4.0 | 13.6 | 0.0 | 0.3 | 2.7 | 72.7 | 5.0 | 8.4 | 19.4 |
| 5-1 | 0.0 | 0.4 | 0.4 | 0.0 | 0.0 | 1.1 | 19.3 | 0.0 | 0.0 | 2.4 | 70.7 | 5.7 | 3.9 | 21.2 |
| 5-6 | 0.0 | 1.8 | 3.3 | 0.0 | 0.0 | 3.8 | 24.8 | 0.3 | 0.0 | 2.6 | 60.2 | 3.4 | 8.2 | 34.0 |
| Strong lines | ||||||||||||||
| 5-2 | 0.0 | 4.1 | 5.6 | 0.0 | 0.0 | 4.0 | 30.1 | 1.0 | 0.0 | 3.5 | 48.3 | 3.6 | 11.6 | 44.8 |
| 5-3 | 0.3 | 4.4 | 7.4 | 0.0 | 0.1 | 2.3 | 34.8 | 0.4 | 0.0 | 2.7 | 42.3 | 5.4 | 9.8 | 49.7 |
| 5-4a | 8.5 | 11.3 | 6.9 | 0.0 | 3.5 | 11.5 | 29.1 | 0.0 | 0.9 | 3.7 | 23.8 | 0.9 | 39.4 | 70.8 |
| 5-5 | 0.0 | 12.3 | 10.6 | 0.0 | 0.0 | 7.9 | 33.4 | 0.2 | 0.0 | 0.8 | 32.8 | 2.0 | 21.0 | 64.4 |
| 5-7 | 0.0 | 7.9 | 9.5 | 0.0 | 0.0 | 9.2 | 31.5 | 0.0 | 0.3 | 2.8 | 36.1 | 2.8 | 20.2 | 58.1 |
| 5-12a | 7.1 | 5.9 | 8.1 | 0.0 | 3.8 | 6.6 | 33.3 | 0.3 | 0.3 | 3.3 | 30.3 | 0.8 | 27.0 | 65.1 |
| 5-13a | 19.1 | 19.1 | 5.8 | 0.0 | 8.8 | 12.1 | 19.1 | 0.0 | 14.5 | 1.5 | 1.0 | 0.0 | 75.1 | 84.0 |
| 5-14 | 0.0 | 9.1 | 6.4 | 0.0 | 0.0 | 5.5 | 32.8 | 0.6 | 0.1 | 2.8 | 41.0 | 1.6 | 17.5 | 54.4 |
| 5-17a | 5.8 | 9.6 | 3.1 | 0.0 | 3.9 | 9.0 | 23.8 | 0.2 | 1.6 | 10.7 | 31.4 | 1.2 | 40.6 | 55.4 |
| SL average | 4.5 | 9.3 | 7.0 | 0.0 | 2.2 | 7.6 | 29.8 | 0.3 | 2.0 | 3.5 | 31.9 | 2.0 | 29.1 | 60.7 |
Lines that produced significantly fewer trichomes (see Supplemental Table 2 online).
The trichomes of second pairs of rosette leaves (five to six pairs of leaves for each line and the wild type) were counted. The trichomes were divided into three classes (1/3, 1/2, and normal) (see Results for the classification), and each class was further divided into trichomes with one to four branches. The numbers represent the percentages of trichomes in each class (see Supplemental Table 2 online). The seedlings were grown on GM media. SL average, strong line average; br, branches.
The majority of the MYB5-overexpressing lines produced ectopic trichomes on cotyledons and hypocotyls (see Supplemental Table 3 online). No trichomes were observed on wild-type cotyledons and hypocotyls. The proportion of plants with ectopic trichomes on cotyledons and hypocotyls varied among the overexpressing lines (see Supplemental Table 3 online). Typically, only one to two trichomes were present on a single cotyledon (Figure 2G), although up to four trichomes were sometimes observed. The trichomes were unbranched and usually on the cotyledon margins. One to 20 trichomes were observed on a single hypocotyl of the MYB5-overexpressing line (Figures 2F and 2I). The hypocotyl trichomes were mostly unbranched, and the hypocotyl epidermal cells were shorter than those of the wild-type cells (Figures 2H and 2I).
The seed coat morphology of the 11 lines appeared similar to that of the wild type (Col) (see Supplemental Figure 2 online). Upon hydration, the mature seeds released large amounts of mucilage, comparable to the wild type (see Supplemental Figure 2 online).
Seed Coat Development in myb5 Insertion Mutants
To study the function of MYB5 in trichome and seed development, two myb5 T-DNA insertion mutants were characterized. One of the mutants contained a T-DNA insertion conferring kanamycin resistance in the coding region of MYB5 (myb5-1, SALK_030942). The insertion would create a truncated MYB5 protein with a 94–amino acid deletion from the C terminus and with four additional amino acids encoded by the T-DNA sequence (Figures 3A and 3B). The kanamycin-resistant plants were obtained with a 3:1 segregation ratio in heterozygous lines. A second mutant (myb5-2, SALK_105723) contained a T-DNA insertion in the MYB5 promoter region, −133 bp from the translational start (ATG) (Figure 3A).
Figure 3.
Structure of the MYB5 Gene and RT-PCR Analysis of MYB5 Transcript.
(A) Structure of the MYB5 gene and the locations of the T-DNA inserts in the myb5-1 and myb5-2 mutants. Boxes (filled and open) represent exons, the broken line an intron, and the full line the promoter region. Filled boxes indicate the region of the exon coding for the MYB domain, and triangles indicate the TDNA insertion sites. Primers LBa1 and 5p2 were used to amplify a fused TDNA-MYB5 fragment in myb5-1, and LBa1 and 5pr in myb5-2. The fused fragments were sequenced to determine the T-DNA insertion sites. Primers 5p3 and 5in2 were used to identify homozygous myb5-1 plants, and 5p1 and 5pr to identify homozygous myb5-2 plants.
(B) The amino acid sequence in frame with the MYB5 sequence at the TDNA insertion site of myb5-1 is shown and the sequence encoded by the TDNA is underlined.
(C) RT-PCR analysis of the MYB5 transcript in Col, myb5-1, myb5-2, Ler, and ttg1 seeds. Near full-length (lanes 2, 4, and 6) and 5′ partial (lane 1) transcripts were amplified using primers 5p2 and 5in2, and 5p2 and 5r2, respectively, for 30 and 34 cycles. The Tubulin transcript (lanes 3 and 5) was amplified for 26 and 30 cycles.
(D) RT-PCR analysis of the MYB5 transcript in the seeds of Col, myb5-1, and myb5-1 transformed with the MYB5 complementing construct (four lines: At5a, At5b, At5c, and At5d). The MYB5 transcript was amplified for 32 cycles (primers 5p2 and 5in2) and Tubulin for 25 and 28 cycles. The numbers in parentheses on the right sides of the gel images indicate the fragment sizes in base pairs and on the left sides the numbers of PCR cycles.
Homozygous insertion mutant plants were identified using PCR analysis with the primers shown in Figure 3A. The myb5 mutant phenotype was found to be recessive. The insertion in the coding region completely blocked the production of full-length MYB5 transcript in the myb5-1 developing seeds (Figure 3C, lane 2). However, a truncated transcript was detected at a much lower level than the full-length transcript present in the wild type (Figure 3C, lane 1). The level of the full-length transcript was also greatly reduced in myb5-2 seeds (Figure 3C, lane 4). To determine if the expression of MYB5 is regulated by TTG1, MYB5 transcript level was examined in the ttg1 developing seed using RT-PCR. The mutant ttg1-1 was used because it contains a point mutation introducing a stop codon in the coding region of TTG1 and would produce a truncated protein (Walker et al., 1999). The ttg1 mutant produces no seed coat mucilage and no leaf trichomes except at leaf margins (Koornneef, 1981). The MYB5 transcript level in the ttg1 seed was similar to that of the wild type (Figure 3C, lane 6), indicating that MYB5 is not regulated by TTG1.
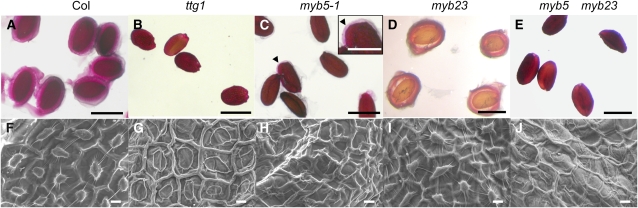
Both myb5-1 and myb5-2 mutants exhibited a similar seed coat phenotype (Figure 4; see Supplemental Figure 3 online). Wild-type seeds are surrounded by a thick layer of ruthenium red–stained mucilage (pink halo) (Figure 4A). The levels of seed coat mucilage in the mutants were dramatically reduced (Figure 4C); however, small amounts of mucilage were detected. The ttg1 mutant produces no detectable mucilage (Figure 4B). The myb5-1 seed coat epidermal cells are irregular in shape and columellae are flattened, which are present in the wild type (Figures 4F and 4H). The seed coat phenotype is similar to that of the ttg1 mutant (Figure 4G).
Figure 4.
Seed Coat Morphology and Mucilage Production.
(A) to (E) Mature seeds of Col (A), ttg1 (B), myb5-1 (C), myb23 (D), and myb5 myb23 (E) were stained with 0.05% ruthenium red for 20 min, and the pink halo represents the released mucilage. An arrowhead (see inset) indicates the remaining mucilage present in myb5-1 seed coats.
(F) to (J) Scanning electron microscopy images of the Col (F), ttg1 (G), myb5-1 (H), myb23 (I), and myb5 myb23 (J) seed coat show the normal ([F] and [I]) and flattened ([G], [H], and [J]) columellae. Bars = 0.5 mm (A) to (E), 0.25 mm for the inset in (C), and 10 μm in (F) to (J).
The seed coat development in the myb5-1 mutant was restored by transformation with a complementing MYB5 construct that includes the 1459-bp promoter region and the coding region of the MYB5 gene. The construct was transformed into the myb5-1 mutant, and transformed plants were selected on hygromycin media. Four of the transformed lines were characterized in detail, and all were found to contain the homozygous myb5-1 mutation and the complementing transgene. The MYB5 transcript was restored to levels similar to those in the wild type in developing seeds (Figure 3D). The columellae and the levels of seed coat mucilage produced by these lines were similar to those of wild-type seeds (see Supplemental Figure 3 online).
To examine seed coat development in more detail, cross sections of wild-type and mutant seed coat stained with toluidine blue were compared (Figure 5). At the start of stage 3 of seed coat development (classified according to Western et al., 2000), the initial mucilage (toluidine blue–stained pink material) production in myb5-1 seed coat epidermal cells appeared similar to that of the wild type (Col) (Figure 5A). Cytoplasmic rearrangement (cytoplasm moving toward the cell center) also occurred normally in the mutant epidermal cells. At stage 4, the secondary cell wall thickened to form columellae in wild-type epidermal cells, while in the mutant, the secondary cell wall synthesis was initiated but failed to proceed to form columellae (Figure 5B). The levels of mucilage in the mutant appeared to be lower than in the wild type (Figure 5B). By stage 5, large amounts of mucilage had accumulated in the wild-type epidermal cells (Figure 5C), and columellae were fully formed (Figure 5C). In the mutant, only small amounts of mucilage were detected, and no columellae had formed (Figure 5C). Upon hydration, the mucilage was released from the mature wild-type seed coat, and columellae became clearly visible (Figure 5D). Thus, the myb5 mutation primarily affected the later stages of mucilage synthesis. For comparison, sections of the ttg1 seed coat are also shown (Figure 5). In the ttg1 seed coat epidermal cells, mucilage was barely detectable and columellae were flattened.
Figure 5.
Sections of Developing Seed Coat.
(A) Seed coat sections (2 μm) stained with 0.05% toluidine blue at stage 3 for Col and myb5-1 and stage 2 for ttg1 (stages according to Western et al., 2000). An arrowhead indicates the cytoplasm undergoing rearrangement.
(B) Seed sections at stage 4.
(C) Seed sections at stage 5 (mature seed). An arrow points to the secondary cell wall and an arrowhead to columellae.
(D) Sections of mature seed coat after hydration and release of mucilage.
(E) Seed sections of the myb5 myb23 double mutant at stage 3 (left panel) and 5 (middle panel) and of the myb5 mutant at stage 5. The pink material in the outer layers (top layers) represents mucilage.
Bars = 20 μm.
Trichome and Seed Coat Development in a myb5 myb23 Double Mutant
The MYB23 gene is expressed in developing trichomes and regulates trichome branching (Kirik et al., 2001, 2005). While loss-of-function myb23 mutations did not affect seed coat development, including mucilage production (Kirik et al., 2005; Matsui et al., 2005), ectopic expression of MYB23 fused to a repression motif sequence reduced seed coat mucilage production (Matsui et al., 2005). However, it is not clear whether the MYB23 gene plays a role in seed coat development and mucilage production.
To determine if the MYB5 gene is partially redundant with the MYB23 gene in regulating trichome and seed coat development, a myb5 myb23 double mutant was created by crossing a myb23 mutant (Salk_ 048592) with the myb5-1 mutant. The myb23 mutant was shown to contain a T-DNA insertion in the second intron of the MYB23 gene (Kirik et al., 2005). The homozygous myb23 mutant was identified using PCR with gene-specific primers. The double homozygous mutant was identified using PCR analysis of the two loci.
The lengths (sizes) and branching patterns of the rosette trichomes in myb5 myb23, myb23, myb5, and wild-type young plants were examined in two experiments (Table 2; see Supplemental Table 4 online). While the myb5-1 mutant exhibited minimal changes in trichome lengths (sizes) and branch numbers compared with the wild type (Table 2, 2br and 1/3, and 1/2), the myb23 mutant displayed a significant increase of small trichomes (53.0 and 59.6% for experiments 1 and 2, respectively). The majority of trichomes in the myb23 mutant possess two branches (58.1 and 64.9% for experiments 1 and 2, respectively), which is consistent with a previous report (Kirik et al., 2005). The smallest trichomes (1/3) of the myb23 mutant account for 23.2 and 22.3% of all trichomes measured in experiments 1 and 2, respectively. Almost all of the trichomes (99.8 and 99.9% for experiments 1 and 2, respectively) (Figure 6) in the myb5 myb23 double mutant are small trichomes (1/3 and 1/2), with a majority being of class 1/3 (Table 2, 1/3) (81.5 and 56% for experiments 1 and 2, respectively). More trichomes in the double mutant contain two branches (86.8 and 82.6%) than in either of the two single mutants, myb5 (17.9 and 4.2%) and myb23 (58.1 and 64.9%). The small trichomes remained small throughout leaf development. While the number of trichomes produced by each rosette leaf varied between experiments, the double mutant developed significantly fewer rosette trichomes than the wild type (see Supplemental Table 4 online). Similar results were obtained from the analysis of trichomes at rosette leaf margins of myb5 myb23, myb23, myb5, and the wild type (see Supplemental Table 5 online). The double mutant produced significantly fewer margin trichomes than the wild type. Almost all of the margin trichomes of the double mutant are of class 1/3 and possessed fewer branches than the wild type (see Supplemental Table 5 online). These results indicate that the MYB5 and MYB23 genes act redundantly in regulating trichome branching and extension.
Table 2.
Analysis of the Lengths and Branching of Trichomes on the Mutant and Wild-Type Rosette Leaves
| 1/3 (%)
|
1/2 (%)
|
Normal (%)
|
2br
|
1/3 and 1/2
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1br | 2br | 3br | 4br | 1br | 2br | 3br | 4br | 1br | 2br | 3br | 4br | Total | Total | |
| myb5 myb23 | ||||||||||||||
| Exp1 | 0.0 | 73.0 | 8.5 | 0.0 | 0.0 | 13.8 | 4.3 | 0.2 | 0.2 | 0.0 | 0.0 | 0.0 | 86.8 | 99.8 |
| Exp2 | 0.1 | 51.9 | 4.0 | 0.0 | 0.1 | 30.6 | 13.2 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 82.6 | 99.9 |
| myb23 | ||||||||||||||
| Exp1 | 0.3 | 18.2 | 4.7 | 0.0 | 0.1 | 14.0 | 15.6 | 0.1 | 0.0 | 25.9 | 20.8 | 0.3 | 58.1 | 53.0 |
| Exp2 | 0.3 | 19.4 | 2.6 | 0.0 | 0.6 | 23.0 | 13.7 | 0.0 | 1.0 | 22.5 | 16.7 | 0.2 | 64.9 | 59.6 |
| myb5 | ||||||||||||||
| Exp1 | 0.2 | 2.9 | 7.2 | 0.0 | 0.0 | 6.8 | 22.7 | 0.0 | 0.3 | 8.2 | 50.5 | 1.2 | 17.9 | 39.8 |
| Exp2 | 0.0 | 0.3 | 2.3 | 0.0 | 0.0 | 1.9 | 17.8 | 0.0 | 0.1 | 2.0 | 73.6 | 2.0 | 4.2 | 22.3 |
| Col (wild type) | ||||||||||||||
| Exp1 | 0.0 | 1.8 | 1.7 | 0.0 | 0.0 | 3.1 | 11.8 | 0.0 | 0.0 | 2.4 | 76.5 | 2.6 | 7.3 | 18.4 |
| Exp2 | 0.0 | 0.6 | 1.2 | 0.0 | 0.0 | 1.0 | 10.0 | 0.0 | 0.0 | 3.3 | 78.0 | 6.0 | 4.9 | 12.4 |
The results of two experiments are presented (Exp1 and Exp2). The trichomes of second pairs of rosette leaves (seven pairs/14 leaves for each mutant and the wild type) were counted. The trichomes were divided into three classes (1/3, 1/2, and normal), and each class was further divided into trichomes with one to four branches. The numbers represent the percentages of trichomes in each class (see Supplemental Table 4 online). The seedlings were grown on GM media. br, branches.
Figure 6.
Trichome Morphology of myb23 and myb5 myb23 Rosette Leaves.
Bars = 0.5 mm.
The myb23 mutant seed coats released a large amount of mucilage upon hydration, while little mucilage was released from the double mutant seed coats (Figures 4D and 4E). The seed coat columellae failed to develop in the double mutant (Figure 4J). At stage 3 of seed coat development, the initial mucilage synthesis of the double mutant (Figure 5E) appeared similar to those of the myb5 mutant and the wild type. Hence, the seed coat development of the double mutant is similar to that of the myb5 mutant.
Identification of Genes Differentially Expressed in the myb5-1 Mutant and Wild-Type Seed
To identify genes that are differentially expressed in the wild-type and myb5-1 seeds, microarray analysis of RNA transcripts in developing seeds was performed using Affymetrix chips. The microarray analysis identified 76 genes with P values < 0.02 and fold changes > 1.5 (see Supplemental Table 6 and Supplemental Figure 4 online). Twenty-one genes were downregulated and 55 genes were upregulated in the mutant. These genes are grouped based on their putative functions (see Supplemental Table 6 and Supplemental Figure 4 online). For example, there are eight genes coding for putative transcription factors and nine genes that may be involved in signal transduction or protein interaction. Twenty-three genes displayed fold changes greater than two, and only nine genes changes greater than 3 (Table 3; see Supplemental Table 6 online). The MYB5 gene is one of the most strongly downregulated genes in the myb5 mutant (Table 3). The GL2 gene, which is known to regulate mucilage synthesis, was downregulated slightly. A single repeat MYB gene, which was previously named MYBL2 (Sawa, 2002), was downregulated 6.3-fold. Inspection of the amino acid sequence of MYBL2 identified a motif (PDLNIGLIP) similar to the EAR repressive motifs (e.g., LDLDLELRL) (Hiratsu et al., 2003) found in many transcriptional repressors (Ohta et al., 2001; Kazan 2006). Hence, it is likely that MYBL2 acts as a transcriptional repressor. A gene (GH) coding for a glycosyl hydrolase 10 protein was downregulated 5.3-fold. Glycosyl hydrolases have been suggested to play a role in the modification of mucilage structure and the seed coat secondary cell wall (Western, 2006). A gene coding for a putative small RNA recognition motif protein and another gene for a putative long-chain-fatty-acid-CoA ligase (LCFACL) were upregulated. The expression of two α/β fold hydrolase genes, designated ABE1 (At2g23550) and ABE4 (At2g23580) (see Discussion), was reduced 47-and 9-fold, respectively. The two α/β fold hydrolase genes were highly expressed in developing wild-type seed with high array signal readings. It is possible that the reduced expression of the two ABE genes is the major contributing factor for reduced mucilage synthesis in the myb5-1 seed coat.
Table 3.
The Most Differentially Expressed Genes Identified in the Microarray Analysis Using myb5 and Wild-Type (Col) Seeds
| AGI ID | Gene Title | P Value | F/Change | atmyb5 |
|---|---|---|---|---|
| At3g13540 | MYB5 transcription factor | 0.000 | 31.3 | Down |
| At1g71030 | MYBL2 transcription factor | 0.004 | 6.3 | Down |
| At1g79840 | Homeobox-leucine zipper (GL2) | 0.011 | 1.8 | Down |
| At2g23550 | α/β Fold hydrolase (ABE1) | 0.000 | 47.5 | Down |
| At2g23580 | α/β Fold hydrolase (ABE4) | 0.001 | 8.9 | Down |
| At4g33810 | Glycosyl hydrolase (family 10) (GH) | 0.002 | 5.3 | Down |
| At3g52550 | Hypothetical protein | 0.006 | 4.5 | Down |
| At5g60630 | Expressed protein | 0.010 | 3.1 | Down |
| At2g14160 | RNA recognition motif protein | 0.007 | 5.4 | Up |
| At1g64400 | Long-chain-fatty-acid CoA ligase | 0.000 | 3.8 | Up |
To further analyze the expression of the genes identified in the microarray experiments, the transcript levels of the two ABE genes, MYBL2 and LCFACL, were determined using RT-PCR analysis of wild-type (Col) and myb5-1 seed. As the TTG1 gene is essential for seed coat development, these transcripts were also examined in wild-type (Landsberg erecta [Ler]), ttg1 (Ler background), and myb5 ttg1 seeds. The myb5 ttg1 double mutant was created by crossing myb5-1 into the ttg1 mutant. The double mutant displayed a leaf trichome phenotype (a few trichomes at leaf margins) and a seed coat phenotype similar to those of ttg1 (Figures 4B and 4G).
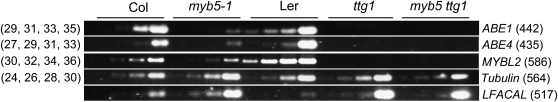
The transcript level of ABE1 was greatly reduced in the myb5-1 developing seed compared with in the wild-type (Col) seed (Figure 7). The transcript levels of the ABE4 and MYBL2 genes were also reduced, while the LCFACL gene was upregulated in the myb5-1 seed (Figure 7). These results are consistent with those obtained using microarray analysis (Table 3). Similarly, the transcript levels of the ABE1, ABE4, and MYBL2 genes were greatly reduced, while the levels of LCFACL were strongly increased in ttg1 and myb5 ttg1 seeds compared with the wild type (Ler) (Figure 7). The changes of expression levels of the four genes were more pronounced in the ttg1 mutant than in myb5. These results indicate that these four genes (i.e., ABE1, ABE4, MYBL2, and LCFACL) are regulated by both the MYB5 and TTG1 genes.
Figure 7.
RT-PCR Analysis of the Transcript Levels of Several Putative MYB5 Target Genes in Developing Seeds.
RNA was extracted from the developing seeds of Col, myb5, Ler, ttg1, and myb5 ttg1 and transcribed in RT reactions. The PCR reactions were performed for 29, 31, 33, and 35 cycles (ABE1, primers Hy550F2 and Hy550R2); 27, 29, 31, and 33 cycles (ABE4, primers Hy580F2 and Hy580R2); 30, 32, 34, and 36 cycles (MYBL2, primers My030F and My030R), and 24, 26, 28, and 30 cycles (Tubulin, primers Tub1 and Tub2). For LFACAL (primers CoAF and CoAR), the reactions were performed for 29, 31, 33, and 35 cycles with the Col and myb5 samples and 26, 28, 30, and 32 cycles for the Ler, ttg1, and myb5 ttg1 samples. The Tubulin transcript levels were included as RT-PCR controls. The primers used are listed in Supplemental Table 7 online. The numbers in parentheses on the right sides of the gel images indicate the fragment sizes in base pairs and on the left sides the PCR cycle numbers.
Expression of Putative MYB5 Target Genes in 35S:MYB5 Plants and Mutants
In wild-type plants, the expression of the ABE1 and ABE4 genes was barely detectable in leaves and roots, while the GL2 and MYBL2 genes were expressed in both tissues (https://www.genevestigator.ethz.ch). To determine if these genes are regulated by ectopically expressed MYB5, their expression levels in leaves and roots were examined in five 35S:MYB5 lines using RT-PCR. All five lines exhibited the trichome phenotype described above. In wild-type leaves and roots (Col), the MYB5 transcript was not detectable with limited (i.e., up to 28) amplification cycles (Figures 8A and 8B). In the 35S:MYB5 leaves and roots, high levels of the MYB5 transcripts were detected in all five lines (Figures 8A and 8B). No RT-PCR products of the ABE1 and ABE4 transcripts were amplified in the wild-type tissues, while high levels of these transcripts were induced in the 35S:MYB5 tissues (Figures 8A and 8B). Both the MYBL2 and GL2 genes were expressed in the wild-type tissues, and their expression was increased in the 35S:MYB5 tissues (Figures 8A and 8B). Hence, ectopically expressed MYB5 upregulates these four genes in leaves and roots.
To determine if the MYB23 regulates GL2, MYBL2, and ABE4 expression, their transcript levels in developing seeds were examined in the wild type and in the myb23, myb5, and myb5 myb23 mutants using RT-PCR. The transcript levels of the four genes in the myb23 mutant were similar to those in the wild type, and the levels in the myb5 mutant were similar to those in the myb5 myb23 double mutant (see Supplemental Figure 5A online). As expected, the MYB5 transcript was not detected in the myb5 and myb5 myb23 seeds, and the MYB23 transcript was not in the myb23 and myb5 myb23 seeds (see Supplemental Figure 5B online). These results indicate that the MYB23 gene is not essential for the expression of these genes.
Expression of ABE1 and ABE4 Promoters in Trichomes and Seed
The transcription start sites of the ABE1 and ABE4 genes were mapped using RT-PCR with overlapping forward primers. An ABE1 cDNA product was detected using the forward primer 550F1 (−18 bp from start codon) but not with the primer 550F2 (−40 bp) and other upstream primers (see Supplemental Figure 6 and Supplemental Table 7 online). This result indicates that the ABE1 transcript initiates between positions −18 and −40 bp from the translation start site. Likewise, the ABE4 transcript initiates between positions −24 and −42 bp (see Supplemental Figure 6 online).
The expression patterns of the ABE1 and ABE4 promoters were examined using the GUS gene driven by the promoters (proABE1:GUS and proABE4:GUS, respectively). The expression patterns of the two promoters were similar (Figure 9). The two promoters were active in the tips of cotyledons, in developing trichomes, and in seeds (Figures 9A to 9D and 9H to 9J). The GUS expression in developing seeds commenced after fertilization and became clearly visible around the globular stage (Figures 9E and 9F). Activity was detected in the integument layers, endosperm, suspensors, and embryos (Figures 9E and 9F). The expression was much lower in the endothelium than in other integument layers (Figure 9E). In the mature seed, the GUS activity was detected only in the outer integument layers but not in the mature embryos (Figure 9G). Hence, the expression patterns of the ABE1 and ABE4 promoters were similar to that of the MYB5 promoter (Figure 1).
Figure 9.
Expression Patterns of the ABE4 and ABE1 Promoters.
GUS expression driven by the 296-bp ABE4 promoter ([A] to [G]) and by the 568-bp ABE1 promoter ([H] to [J]). The GUS expression in the tip of a cotyledon ([A] and [J]), developing trichomes ([B], [C], and [H]), and seeds ([D] and [I]). Seed sections (dark-field microscopy) showing GUS expression (pink) in early globular (E), late globular (F), and mature (G) stages. en, endothelium. Bars = 0.4 mm in (A) to (D), (H), and (J) and 50 μm in (E) to (G) and (I).
Expression of the MYBL2 Promoter
The MYBL2 gene is expressed in leaves, roots, and developing seeds as shown using RT-PCR (Figure 7, Figures 8A and 8B). The expression pattern of the MYBL2 promoter was examined using a 947-bp promoter fused with the GUS gene. The GUS activity was expressed in roots and strongly in secondary root tips (Figure 10A). In flowers, GUS was expressed in developing siliques, anthers, and mature sepals (Figures 10C to 10E). GUS activity was only detected in the trichomes of the strongly expressing lines (Figure 10B) but not the weaker ones and was induced by wounding (Figure 10F). Activity was detected in seeds' integument layers, endosperms, suspensors, and embryos (Figure 10G). However, the levels of GUS activity in the endothelial layer were much lower than in the other integument layers as the seeds became more mature, and no expression was detected in the mature embryo (Figures 10G and 10H). Hence, the expression pattern driven by the MYBL2 promoter in developing seed is similar to that of the MYB5 promoter.
Figure 10.
GUS Expression Driven by the 947-bp MYBL2 Promoter.
GUS expression (average lines) in roots (A), trichomes and a rosette leaf ([B]; a strongly expressing line), flower buds ([C] and [D]), anthers (E), and a wounded leaf (F). Seed sections (dark-field microscopy) showing GUS expression (pink) in seeds of globular (G) and mature (H) stages. en, endothelium. Bars = 0.4 mm in (A) to (F) and 50 μm in (G) and (H).
DISCUSSION
Using a MYB5 promoter:GUS construct and RNA gel blot hybridization, the MYB5 gene was shown previously to be expressed in developing leaf trichomes, stipules, epidermal cells on leaf margins, and immature seeds (Li et al., 1996). This article examines the ProMYB5:GUS activity in more detail during seed development and confirms that MYB5 is expressed in developing trichomes using in situ hybridization. MYB5 promoter activity was detected in integument layers, endosperms, suspensors, and developing embryos.
Ectopic expression of MYB5 using the double 35S promoter resulted in the increased production of small trichomes on rosette leaves and a reduction in trichome branching. The majority of the overexpressing lines developed ectopic trichomes on cotyledons and hypocotyls, and many of these lines produced fewer leaf trichomes than the wild type. The myb5 insertion mutant displayed minimal changes in trichome morphology, while the myb23 mutant developed increased numbers of small trichomes and trichomes possessing two branches relative to the wild type. The double myb5 myb23 mutant produced more small trichomes and trichomes containing two branches than either of the single mutants. These results indicate that the MYB5 and MYB23 genes regulate trichome branching and extension in a partially redundant manner.
Seed coat mucilage production was greatly reduced in the myb5 mutant, and seed coat epidermal cells were irregular in shape and lacked columellae. The initial mucilage production, the onset of cytoplasmic rearrangement, and the initiation of secondary cell wall synthesis appeared normal in the myb5 mutant, but the mucilage and secondary cell wall synthesis were prevented from proceeding to completion. Similar seed coat development was observed for the double myb5 myb23 mutant.
Genes differentially expressed in the wild type and myb5 seeds were identified using microarray and RT-PCR analyses. Among them were the ABE1, ABE4, MYBL2, GL2, and LCFACL genes. GL2 is known to be regulated by TTG1 during seed development. The other four genes were also found to be regulated by TTG1 in developing seeds, suggesting that MYB5 collaborates with TTG1 in seed coat development. The ABE1, ABE4, MYBL2, and GL2 genes were downregulated in myb5 seeds and upregulated in leaves and roots by ectopically expressed MYB5. The MYBL2, ABE1, and ABE4 promoters were active in developing seeds, including in the integument layers, endosperms, suspensors, and developing embryos. The ABE1 and ABE4 promoters were also active in trichomes. Consequently, these genes are likely to be the direct targets of the MYB5 protein.
Comparison of MYB Sequences
Although MYB5 is not classified as part of the GL1, WER, and MYB23 subgroup and does not contain the WVxxDxFELSxL motif characteristic of this subgroup (Kranz et al., 1998; Stracke et al., 2001), its MYB DNA binding domain is very similar to those of GL1, WER, and MYB23. Limited similarity also occurs between the C termini of MYB5 and MYB23 (see Supplemental Figure 7 online).
Sequence comparison shows that the single repeat in CPC is ∼40% identical to the R3 repeat of the GL1 protein. CPC and its homologs are predicted to have no DNA binding activity, and gel mobility shift and yeast one-hybrid assays showed that CPC failed to bind DNA (Koshino-Kimura et al., 2005; Tominaga et al., 2007). Thus, these proteins act as passive repressors by competing with GL1 for GL3/EGL3 binding. However, the single repeat in MYBL2 exhibits a much higher identity (65%) to the R3 of MYB5 than to the single repeat of CPC and its homologs (∼40%). It is unclear if MYBL2 possesses any DNA binding activity. CPC and its homologs lack the long C-terminal sequences present in most of the MYB proteins, including MYBL2 (Figure 11A). The presence of a putative repression motif (EAR) and a second novel repression motif in MYBL2 (Matsui et al., 2008) suggests that it acts as a dominant transcriptional repressor. The EAR motif can convert various transcription factors into dominant repressors (Hiratsu et al., 2003; Li et al., 2007).
Figure 11.
Structures of the MYB Proteins and a Phylogenetic Analysis for ABE Proteins.
(A) The structures of the MYB5, MYBL2, and CPC proteins. The filled boxes represent MYB repeats. The gray boxes indicate the putative repression motifs in MYBL2.
(B) A phylogenetic tree for ABE1 to 20. The Arabidopsis proteins were identified using a BLAST search (http://www.Arabidopsis.org) with the ABE1 sequence and then confirmed by second and third BLAST searches with ABE12 and ABE19 sequences, respectively. The proteins displaying significant identity over their full-length sequences to the three proteins are included. The genes in the eight-gene tandem array are designated ABE1 to ABE8. ABE3 appears to encode a truncated peptide. The sequences were aligned using ClustalW, and the phylogenetic analysis was performed using PHYLIP. The phylogenetic tree was generated using 1000 bootstrapped replicates. The numbers on the branches indicate the number of times the partition of the sequences into two sequence sets at the nodes occurs among the bootstrapped trees (out of 1000 trees). The more solid nodes are represented by higher bootstrap values (the highest value being 1000 in this analysis).
MYB–bHLH Interactions
The MYB5 protein has been shown to interact with three BHLH proteins of subgroup IIIf, namely, TT8, EGL3, and BHLH012, in a yeast two-hybrid system. MYB23 interacts with EGL3 and BHLH012 but not with TT8 (Zimmermann et al., 2004). The interactions of these proteins with GL3 were not tested. Clones expressing MYBL2 were isolated using TT8, EGL3, and BHLH012 as baits in three different screenings, indicating that MYBL2 is able to interact with the three proteins (Zimmermann et al., 2004). MYBL2 also interacts with GL3 in a yeast two-hybrid system (Sawa, 2002). The R3 repeats in the MYB domains are the structural basis for MYB and BHLH interactions (Zimmermann et al., 2004). So MYB5, MYB23, and MYBL2 are likely to form multimeric regulatory complexes with the BHLH proteins and TTG1 in vivo (Figure 12).
Figure 12.
The Proposed Models of the MYB5 Regulatory Network in Developing Seeds and Trichomes.
In the developing seed (A), the MYB5/TTG1/TT8/EGL3 complex could directly regulate the ABE1, ABE4, GH, GL2, and MYBL2 genes. The MYBL2 protein could repress the synthesis of PA biosynthesis in developing seeds except in the endothelium layer. MYBL2 could also constitute a negative feedback loop possibly by downregulating TT8. In developing trichomes (B), the GL1 complex regulates trichome initiation via GL2 and activates TRY, CPC, and the ETCs, which suppress trichome initiation in neighboring cells. GL1 and MYB23 redundantly regulate trichome initiation on leaf margins. MYB5 and MYB23 regulate trichome branching and extension in a partially redundant manner.
Seed Coat Development
Mutations in MYB23 genes did not affect seed coat development, including mucilage production (Figure 4; Kirik et al., 2005; Matsui et al., 2005). Seed coat mucilage was substantially reduced in the myb5 mutants, although a small amount was produced. This may be a consequence of partial redundancy of similar ABEs expressed in the seed coat or due to partial redundancy of MYB5, MYB23, and MYB61. However, the double myb5 myb23 mutations also failed to completely block the mucilage production. Furthermore, the effects of the double mutations on the expression of the GL2, MYBL2, and ABE4 genes were similar to the effects of the single myb5 mutation. These results indicate that MYB23 is not essential for the expression of these genes and for seed coat mucilage production. While the MYB23 transcript was detected in developing seed, its expression levels were relatively low (https://www.genevestigator.ethz.ch), and its expression pattern in various cell layers of the developing seed has not been reported. Consequently, the role of the MYB23 in seed coat development has yet to be resolved.
The target gene set of MYB5 may partially overlap with the genes regulated by MYBL2. Hence, MYBL2 would participate in a negative feedback loop for seed coat development (Figure 12A). If MYB5 acts as an activator and MYBL2 a repressor, blocking the expression of both should result in minimal changes of the overlapping target genes. Thus, in the myb5 mutant, the expression of MYB5 and MYBL2 genes was reduced. One such overlapping gene may be GL2. The negative feedback regulated by MYBL2 may be accomplished by the combination of the following three mechanisms: reducing the transcription of the MYB5, TTG1, EGL3, and TT8 genes; inactivating the MYB5/TTG1/EGL3/TT8 complex by replacing MYB5; and downregulating the overlapping target genes. The negative feedback loop may ensure an optimal mucilage synthesis. Indeed, MYBL2 has recently been shown to act as a negative regulator of anthocyanin biosynthesis in Arabidopsis seedlings, and TT8 expression was enhanced in a mybl2 mutant (Dubos et al., 2008; Matsui et al., 2008). Anthocyanin biosynthesis resembles proanthocyanidin (PA) biosynthesis, as the latter branches off from the anthocyanin pathway at a very late stage (Koes et al., 2005). Hence, one of the functions of MYBL2 in the developing seed may be to restrict PA biosynthesis in the endothelium layer (Figure 12A) where the MYBL2 promoter activity is lower than in other layers in later stages of seed development.
Glycosyl hydrolases are required for the modification of polysaccharides in muro (Western, 2006). The mum1 and patchy mutants appear to produce normal amounts of mucilage but are unable to release their mucilage upon seed hydration. Recent cloning of PATCHY reveals that the gene encodes a putative glycosyl hydrolase (Western, 2006). It remains to be seen if the glycosyl hydrolase 10 (GH) gene is mutated in any of the two mutants. The GH gene is a member of a tandem array comprising six genes and exhibits significant identity in amino acid sequence to an Arabidopsis xylanase that has been implicated in the secondary cell wall metabolism of vascular bundle cells (Suzuki et al., 2002).
The ABE1 and ABE4 genes are developmentally regulated and primarily expressed in the developing seed and siliques, according to the microarray database (https://www.genevestigator.ethz.ch). The expression patterns of their promoters are similar to those of the MYB5 promoter, and the two genes are downregulated in the myb5 mutant and upregulated by the ectopic expression of MYB5. These results suggest that the two genes are the direct targets of the MYB5 protein. The two genes are very similar with 80% identity in a 210-bp promoter region (see Supplemental Figure 6 online) and 76% identity in the encoded amino acid sequences (see Supplemental Figure 8 online). Their promoter regions contain two putative MYB binding sites and a bHLH binding site (see Supplemental Figure 6 online). The two putative MYB binding sites in each promoter are part of two almost identical repeat sequences and are similar to the MYB.Ph3 binding site (AACTAACT; Solano et al., 1995). The ABE1 and ABE4 genes may be partially redundant.
The α/β Fold Hydrolases/Esterases
The ABE1 and ABE4 proteins are members of the α/β hydrolase fold-1 family. There are 167 protein entries in this family in Arabidopsis (InterPro: IPR000073; http://www.ebi.ac.uk/interpro/). The α/β hydrolase fold is the core structure of this family of hydrolytic enzymes, which have widely differing phylogenetic origins and catalytic functions (Ollis et al., 1992). The enzymes contain a conserved catalytic triad (see Supplemental Figure 8 online). ABE1 and ABE4 are part of an eight-gene array linked in tandem. The eight encoded proteins are very similar to each other and to 12 other α/β fold hydrolases (Figure 11B; see Supplemental Data Set 1 and Supplemental Figure 8 online). We designated the 20 α/β fold hydrolase genes ABE1 to ABE20 (for alpha/beta fold esterase) (Figure 11B) as they display significant identity to other α/β fold hydrolases with proven esterase activity (see Supplemental Figure 8 online; Fridman et al., 2005). The peptides of ABE15 to ABE19 are significantly longer than those of the other ABE peptides and may have acquired additional functions.
Little is known about the function of these proteins in Arabidopsis. However, several ABE-like proteins in other plant species have been characterized and their substrates identified. These are methylketone synthase 1 (MKS1) from wild tomato (Lycopersicon hirsutum), methyl jasmonate esterases (MJE) from tomato (Solanum lycopersicum), and salicylic acid binding protein 2 (SABP2) from tobacco (Nicotiana tabacum) (Stuhlfelder et al., 2004; Forouhar et al., 2005; Fridman et al., 2005). MKS1 deesterifies β-ketoacyl-acyl-carrier protein to β-ketoacid and thus leads to the formation of methyl-ketones after decarboxylation (Fridman et al., 2005). MJE and SABP2 demethylesterify their respective substrates (Stuhlfelder et al., 2004; Forouhar et al., 2005). The peptide sequences of ABE1 and ABE4 exhibit 40 to 50% identity to the esterases. Furthermore, many residues in SABP2 that contact the substrate are conserved in ABE1 and ABE4 (i.e., 7 out of 15 in ABE1, and 10 out of 15 in ABE4) (see Supplemental Figure 8 online). Hence, it is likely that ABE1 and ABE4 target the carboxyl esters of their substrates in a similar way to the esterases. They may be involved in the activation, transport, and localization of the ester substrates required for mucilage (pectin) synthesis. Many of the galacturonic acids present in pectin are esterified. These esters include methyl esters and acetyl esters (Scheller et al., 2007). It is unclear if ABE1 and ABE4 play a role in the modification of seed coat mucilage (pectin) as they exhibit little sequence identity to a family of known pectin methylesterases.
Trichome Development
Overexpressing the MYB5 gene leads to changes in trichome development similar to those observed in some 35S:GL1 and 35S:MYB23 plants (Larkin et al., 1994; Kirik, et al., 2001). Overexpression of GL1 or MYB23 driven by the 35S promoter results in ectopic formation of trichomes on cotyledons and the abaxial surface of the first leaves in some plants (Larkin et al., 1994; Kirik, et al., 2001). Reduction of leaf trichomes in the midvein region was observed in a portion of the plants, although most of the 35S:MYB23 plants exhibited normal trichome density and distribution on leaves. The epidermal cells on the hypocotyls of 35S:MYB23 seedlings were shorter than those of the wild type and possessed a roundish or conical surface.
Abundant trichomes are produced on hypocotyls of an etc1 try cpc triple mutant, indicating that one function of these genes is to repress the initiation of hypocotyl trichomes (Kirik et al., 2004). However, a try cpc double mutant develops hypocotyl trichomes in only some plants (on average less than one trichome per hypocotyl) (Kirik et al., 2004). Overexpressing MYB5 may partially overcome such repression, resulting in ectopic trichome development in only some hypocotyls.
The myb23 insertion mutants displayed reduced trichome branching and did not affect trichome initiation, indicating that the gene is required for trichome branching but has no essential function in trichome patterning (Kirik et al., 2005). We found that the myb23 mutant produced more small trichomes than the wild type. The double myb5 myb23 mutations increased the production of small trichomes and reduced the trichome branching compared with the two single mutations. These results indicate that MYB5 and MYB23 are partially redundant in regulating trichome branching and extension (Figure 12B). The gl1 null mutant developed a small number of trichomes at the edges of late rosette leaves and petioles, while the gl1 myb23 double mutant was completely smooth (Kirik et al., 2005). This indicates that the development of trichomes on leaf edges and petioles is regulated redundantly by the two genes (Figure 12B). MYB23 appears to act on leaf margin trichome morphogenesis at the same genetic level as GL2 (Kirik et al., 2005).
Many 35S:GL2 plants displayed a gl2-like phenotype (i.e, aborted leaf trichomes and a few remaining unbranched trichomes) (Ohashi et al., 2002). Transgenic 35S:MYBL2 plants also produced fewer trichomes than wild-type plants (Sawa, 2002). It is not clear how overexpression of these genes reduces trichome number. One possible mechanism is squelching, in which an overexpressed protein titrates a partner factor whose interaction with its endogenous counterpart is essential for its function (Larkin et al., 1994; Kirik, et al., 2001; Ohashi et al., 2002). Alternatively, the overexpressed proteins may affect the spatio-temporal expression pattern of GL2, which is thought to be important for trichome morphogenesis. Thus, overexpressing MYBL2 reduced GL2 expression (Sawa, 2002). Overexpressing MYB5 increased both GL2 and MYBL2 expression. Similarly, the expression of GL2 was induced in ectopic trichomes resulting from 35S:MYB23 expression (Kirik et al., 2001). However, it remains to be determined if GL2 is required to mediate trichome branching and extension regulated by MYB5 and MYB23. The ABE1 and ABE4 promoters are active in developing trichomes, while the expression of MYBL2 transcript in trichomes has yet to be determined. Further studies are required to establish the roles of these three genes in trichome morphogenesis. Many genes and mutants affecting trichome cell morphogenesis have been identified (for reviews, see Schellmann and Hülskamp, 2005; Mathur, 2006), and some of them may be target genes regulated by MYB5 and MYB23.
Models of a MYB5 Regulatory Network
We propose that the MYB5 protein could form a complex with TTG1/TT8/EGL3 in the developing seed, which could directly regulate the expression of ABE1, ABE4, GL2, MYBL2, GH, and other genes downregulated in the myb5 seed (Figure 12A). The ABE1 and ABE4 genes are candidate downstream genes playing a role in seed coat mucilage synthesis. The MYBL2 protein could be part of a negative feedback loop modulating seed coat development and could also restrict PA synthesis to the endothelial layer. A similar complex of MYB5/TTG1/GL3/EGL3 could regulate trichome branching and extension (Figure 12B).
METHODS
Plant Materials and Growth
Arabidopsis thaliana Col-0 and Ler accessions were used as wild-type controls. Plants were grown on soil under constant illumination or on germination medium containing appropriate selective antibiotics. Two myb5 insertion mutant lines (SALK_030942 and SALK_105723) and a myb23 mutant line (Salk_ 048592; Kirik et al., 2005) were obtained from the SALK population (http://signal.salk.edu; Alonso et al., 2003) at the ABRC (http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/abrchome.htm). The seeds were germinated in medium containing kanamycin (50 μg/mL) to select for the T-DNA inserts. The ttg1-1 mutant (Walker et al., 1999) was obtained from the ABRC.
Plasmid Construction and Plant Transformation
The pMYB5:GUS construct was described previously (Li et al., 1996). To construct p35S:MYB5, a MYB5 genomic fragment (−13 to +1275) was amplified using PCR and cloned into the XhoI-SacI sites of the pKYLX71 binary vector under the control of a double 35S promoter (Schardl et al., 1987). The sequence of the insert was verified by sequencing. The construct was transformed into Agrobacterium tumefaciens strain GV3101, and Arabidopsis plants (Col-0) were transformed by vacuum infiltration (Bechtold et al., 1993). The transformed plants were selected on media containing kanamycin (50 μg/mL) and timentin (200 μg/mL), and the presence of the transgene was verified using PCR with primers 35S2F and 35S2R (see Supplemental Table 7 online). For the complementation experiment, a BamHI-EcoRI MYB5 fragment, including a 1459-bp promoter region and a 1.2-kb coding region, was inserted into BamHI-EcoRI sites of pGreen0179. The resultant complementing construct was transformed into the myb5-1 mutant using Agrobacterium harboring plasmid pSOUP (Hellens et al., 2000). T1 transformed plants (15 lines) were isolated on hygromycin (15 μg/mL) plus timentin (200 μg/mL) media and T2 plants on hygromycin (15 μg/mL) plus kanamycin media. The homozygous myb5-1 insertion mutation was verified using PCR, as described in Figure 3. The presence of the complementing transgene was confirmed using PCR.
To construct pABE1:GUS and pABE4:GUS, a PCR fragment (primers 550PF and 550PR; see Supplemental Table 7 online) containing a 568-bp ABE1 promoter region (the entire intergenic region) and another PCR fragment (primers 580PF and 580PR; see Supplemental Table 7 online) containing 296 bp of the ABE4 promoter region (the entire intergenic region) were cloned into pENTR/D-TOPO (Gateway Entry Vector; Invitrogen), respectively. The two inserts were then transferred into pKGWFS7,0 (Gateway destination vector) to generate translational fusions ABE1:GFP-GUS and ABE4:GFP-GUS using the Invitrogen protocol. The resultant fused GUS proteins would contain one and six amino acids from ABE1 and ABE4, respectively. The two inserts were sequenced.
To construct the MYBL2 promoter:GUS construct, a 978-bp MYBL2 promoter fragment (primers MYBL2F and MYBL2R), including 31 bp downstream of the ATG and 947 bp upstream of the translational start site, was cloned into pENTR/D-TOPO (Invitrogen). The promoter fragment was then transferred to pKGWFS7,0 through recombination (Invitrogen Gateway Technology). The promoter fragment in the vectors was sequenced.
In Situ Hybridization, Seed Sectioning, and Trichomes
In situ hybridization on transverse sections of expanding leaves was performed as previously described by Li et al. (1999). A PCR fragment (+ 905 to +1244) coding for the C terminus of MYB5 was cloned into SphI-XhoI sites (antisense) and XhoI-BamHI sites (sense) of plasmid pGEM7 (Promega), respectively. The probes were generated using T7 RNA polymerase. Preparation of tissue sections and GUS staining of plant tissues and tissue sections were described previously (Li et al., 1999). Developing seeds were staged according to Western et al. (2000). The morphology of mature seed coats was examined using a JEOL JSM 6340F field emission scanning electron microscope. Leaf trichomes were examined using a dissecting microscope, and the statistical analysis of trichomes was performed using Microsoft Excel.
RT-PCR Analysis
Developing seeds (globular to walking-stick stages) were isolated from siliques in RNAlater solution (Ambion) for RNA extraction. Total RNA samples were extracted from plant tissues using the RNeasy plant mini kit (Qiagen), and 1 to 2 μg of the samples were converted into cDNA using Superscript III RNaseH− Reverse Transcriptase (Invitrogen). The cDNA samples were amplified by PCR using the following conditions: first cycle, 94°C for 3 min; second cycle, 94°C for 30 s, 56°C for 30 s, and 72°C for 50 s; third cycle, 72°C for 10 min. The second cycle was repeated for a number of times. Two biological replicates were used for the RT-PCR analyses. All the DNA gels were stained using CYBR Gold (Invitrogen).
Microarray Analysis
Five groups of wild-type plants (Col) and five groups of myb5-1 plants were grown at the same time. Developing seeds (globular to walking-stick stages) were collected from each group of plants, resulting in five pools (biological replicates) of wild-type seeds and five pools of myb5 seeds. The seeds were dissected out from siliques in RNAlater solution (Ambion), and total RNAs (five wild-type and five myb5-1 samples) were extracted using the RNeasy plant mini kit. RNA quality and integrity were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies). Four wild-type samples and three myb5-1 samples were used for microarray analysis. Additional wild-type and myb5-1 samples (different from the microarray samples) were used for validation RT-PCR analysis.
Microarray experiments were performed according to the Affymetrix GeneChip Expression Analysis Manual (http://www.affymetrix.com). Approximately 100 ng of RNA was used as template for the synthesis of labeled cRNA probe using the Two Cycle Amplification Kit (Affymetrix). Briefly, the RNA samples were spiked with a set of bacterial RNAs (controls) of known concentrations. Examining the hybridization intensities of these controls on GeneChip arrays helps to monitor the labeling process independently. The first cycle of cDNA synthesis was performed using an oligo(dT) primer fused to a T7 RNA polymerase promoter and was followed by the first cycle of in vitro transcription using T7 RNA polymerase to generate cRNA. The cRNA was used as a template for the second cycle of cDNA synthesis followed by the second cycle of in vitro transcription to generate the biotin-labeled antisense cRNA. The labeled cRNA was fragmented and then injected into the GeneChips containing the Arabidopsis ATH1 Genome array (four chips for four wild-type [Col] biological replicates and three for three myb5-1 biological replicates) (Affymetrix). After hybridization, the GeneChips were stained with streptavidin-phycoerythrin and biotinylated anti-streptavidin antibody. The array was scanned with an Affymetrix GeneChip Scanner 3000 (Victoria Agribiosciences Centre). The Arabidopsis gene annotation data set was imported from NetAffx (Affymetrix). GeneChip operating software was used to produce CEL files containing raw probe intensities for the arrays and to incorporate the annotation data set.
Data Analysis and Gene Annotation
The CEL files for the arrays were analyzed using the ArrayAssist software recommended by Affymetrix and developed by Strand Life Sciences (http://www.strandgenomics.com) and available in Sratagene (http://www.stratagene.com; ArrayAssist Manual). The wild-type and myb5 CEL files were grouped and processed with the PLIER algorithm (http://mbi.osu.edu/2004/ws1abstracts.html). The PLIER algorithm provides background subtraction, normalization, and probe summarization (i.e., averaging multiple probe values within a probe set into a single expression value). Variance stabilization was performed to add a fixed quantity (16) to all linear scale signal values, which suppresses noise at log signal values. The addition of the fixed quantity is required by the ArrayAssist software (see ArrayAssist Manual). The linear scale data were then transformed into log scale, where logs are taken to base 2. The log data set was then subjected to significance analysis (unpaired t test) to obtain a P value, a fold change, and a direction of change (up or down) for each gene (for detail on these algorithms, see ArrayAssist Manual). The Benjamini-Hochberg FDR method was used to obtain P values corrected for multiple testing. The genes with P values > 0.02 and fold changes > 1.5 are included in Supplemental Table 6 online. Genes were assigned to functional categories following the classification schemes used at the NetAffx Analysis Center (Affymetrix) and gene function annotation at The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org).
The statistical analysis for Supplemental Table 2, Supplemental Table 4, and Supplemental Table 5 online was performed using Microsoft Excel.
Phylogenetic Analysis
The 20 ABE protein sequences (Figure 11B) were obtained from TAIR (http://www.Arabidopsis.org/tools/bulk/sequences/index.jsp) and directly imported into ClustalW for alignment (http://www.Arabidopsis.org/cgi-bin/bulk/sequences/seqtoclustalw.pl) (http://www.ebi.ac.uk/clustalw/index.html). The resultant aligned sequences in a PHYLIP format (see Supplemental Data Set 1 online) were used to construct a phylogenetic tree (unrooted) with the phylogenetic analysis software PHYLIP (PHYLIP 3.68; http://evolution.genetics.washington.edu/phylip.html). PHYLIP contains many programs (e.g., SEQBOOT, PROTPARS, CONSENSE, and DRAWGRAM). SEQBOOT was used to process the 20 aligned sequences and produced 1000 bootstrapped sequence sets. PROTPARS was then used to run the parsimony method with the 1000 bootstrapped sequence sets. The resultant tree file was processed using CONSENSE, which computes consensus trees by the majority-rule consensus tree method and also allows one to easily find the strict consensus tree. A phylogenetic tree was plotted using DRAWGRAM.
Accession Numbers
Sequence data and microarray data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: GL1 (At3G27920), WER (At5g14750), MYB23 (At5G40330), MYB5 (At3G13540), TTG1 (At5g24520), GL2 (At1g79840), MYBL2 (At1g71030), ABE1 (At2g23550), ABE4 (At2g23580), SABP2 (AAR87711), ABE1-20 (Figure 11), other genes (Table 3; see Supplemental Table 6 online), and microarray data (GSE14229, http://www.ncbi.nlm.nih.gov/geo/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemetal Figure 1. The GUS Expression Patterns Driven by MYB5 Promoters.
Supplemental Figure 2. Seed Coat SEM Images and Released Mucilage of Wild-Type (Col) and 35S:MYB5 Lines.
Supplemental Figure 3. Seed Coat SEM Images and Released Mucilage of the Wild Type and Mutants.
Supplemental Figure 4. Pie Chart Categorizing Genes That Are Differentially Expressed in Col and myb5-1 Seeds.
Supplemental Figure 5. RT-PCR Analysis of the Transcript Levels of MYB5, MYB23, GL2, MYBL2, and ABE4 in Developing Mutant and Wild-Type Seeds.
Supplemental Figure 6. Transcription Start Sites of ABE1 and ABE4 Genes and Sequence Alignment of Their Promoter Regions.
Supplemental Figure 7. Alignment of the Deduced Amino Acid Sequences of GL1, WER, MYB23, and MYB5.
Supplemental Figure 8. Alignment of 10 Deduced ABE Amino Acid Sequences.
Supplemental Table 1. Analysis of Trichome Hybridization Using the MYB5 Antisense and Sense Probes.
Supplemental Table 2. Analysis of the Lengths and Branching of Trichomes in the 35S:MYB5 and Wild-Type Rosette Leaves.
Supplemental Table 3. The Percentages of 35S:MYB5 Seedlings Producing Ectopic Trichomes on Cotyledons and Hypocotyls.
Supplemental Table 4. Analysis of the Lengths and Branching of Trichomes in the Mutant and Wild-Type Rosette Leaves.
Supplemental Table 5. Analysis of the Lengths and Branching of Trichomes at the Mutant and Wild-Type Rosette Leaf Margins.
Supplemental Table 6. Differentially Expressed Genes Identified in the Microarray Analysis Using myb5 and Wild-Type (Col) Seeds.
Supplemental Table 7. Primer Sequences.
Supplemental Data Set 1. Sequences Used to Generate the Phylogenetic Tree Presented in Figure 11B.
Supplementary Material
Acknowledgments
We thank Edgar Pelcmanis-Sakers (Botany, La Trobe University) for help in the preparation of the seed sections and Tracie Webster (Victorian Agribiosciences Centre, La Trobe University) for assistance with the microarray experiments. The project was supported in part by the Grains Research and Development Corporation (ACT, Australia).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Roger W. Parish (r.parish@latrobe.edu.au).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. Paris. Life Sci. 316 1194–1199. [Google Scholar]
- Beeckman, T., De Rycke, R., Viane, R., and Inze, D. (2000). Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 113 139–148. [Google Scholar]
- Berger, F., Linstead, P., Dolan, L., and Haseloff, J. (1998). Stomata patterning on the hypocotyls of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol. 194 226–234. [DOI] [PubMed] [Google Scholar]
- Broun, P. (2005). Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8 272–279. [DOI] [PubMed] [Google Scholar]
- Costa, S., and Shaw, P. (2006). Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature 439 493–496. [DOI] [PubMed] [Google Scholar]
- Dean, G., Zheng, H., Tewari, J., Huang, J., Young, D.S., Hwang, Y.T., Western, T.L., Carpita, N.C., McCann, M.C., Mansfield, S.D., and Haughn, G.W. (2007). The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19 4007–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos, C., Gourrierec, J.L., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., Routaboul, J.-M., Alboresi, A., Weisshaar, B., and Lepiniec, L. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55 940–953. [DOI] [PubMed] [Google Scholar]
- Forouhar, F., Yang, Y., Kumar, D., Chen, Y., Fridman, E., Park, S.W., Chiang, Y., Acton, T.B., Montelione, G.T., Pichersky, E., Klessig, D.F., and Tong, L. (2005). Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA 102 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, E., Wang, J., Iijima, Y., Froehlich, J.E., Gang, D.R., Ohlrogge, J., and Pichersky, E. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosythesis of methylketones. Plant Cell 17 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimil, S., and Dunand, C. (2006). Patterning of Arabidopsis epidermal cells: Epigenetic factors regulate the complex epidermal cell fate pathway. Trends Plant Sci. 11 601–609. [DOI] [PubMed] [Google Scholar]
- Haughn, G., and Chaudhury, A. (2005). Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci. 10 472–477. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takahi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G.W., Montagu, M.V., and Okamuro, J.K. (1994). Control of Arabidopsis flower and seed development by the domeotic gene APETALA2. Plant Cell 6 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C.S., Kolevski, B., and Smyth, D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. (2006). Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11 109–112. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Lee, M.M., Wester, K., Herrmann, U., Zheng, Z., Oppenheimer, D., Schiefelbein, J., and Hülskamp, M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132 1477–1485. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Schnittger, A., Radchuk, V., Adler, K., Hülskamp, M., and Bäumlein, H. (2001). Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev. Biol. 235 366–377. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Simon, M., Hülskamp, M., and Schiefelbein, J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268 506–513. [DOI] [PubMed] [Google Scholar]
- Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10 236–242. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1981). The complex syndrome of ttg mutants. Arabidopsis Inf. Serv. 18 45–51. [Google Scholar]
- Koshino-Kimura, Y., Wada, T., Tachibana, T., Tsugeki, R., Ishiguro, S., and Okada, K. (2005). Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 46 817–826. [DOI] [PubMed] [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16 263–276. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Oppenheimer, D.G., Lloyd, A.M., Paparozzi, E.T., and Marks, M.D. (1994). Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell 6 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.F., Higginson, T., and Parish, R.W. (1999). A novel MYB-related gene from Arabidopsis thaliana expressed in developing anthers. Plant Cell Physiol. 40 343–347. [DOI] [PubMed] [Google Scholar]
- Li, S.F., Iacuone, S., and Parish, R.W. (2007). Suppression and restoration of male fertility using a transcription factor. Plant Biotechnol. J. 5 2970312. [DOI] [PubMed] [Google Scholar]
- Li, S.F., Santini, J.M., Nicolaou, O., and Parish, R.W. (1996). A novel myb-related gene from Arabidopsis thaliana. FEBS Lett. 379 117–121. [DOI] [PubMed] [Google Scholar]
- Macquet, A., Ralet, M.-C., Kronenberger, J., Marion-Poll, A., and North, H. (2007. b). In-situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 48 984–999. [DOI] [PubMed] [Google Scholar]
- Macquet, A., Ralet, M.-C., Loudet, O., Kronenberger, J., Mouille, G., Marion-Poll, A., and North, H. (2007. a). A naturally occurring mutation in an Arabidopsis accession affects a β-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19 3990–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., and Glover, B.J. (2007). Functional aspects of cell patterning in aerial epidermis. Curr. Opin. Plant Biol. 10 70–82. [DOI] [PubMed] [Google Scholar]
- Mathur, J. (2006). Trichome cell morphogenesis in Arabidopsis: A continuum of cellular decisions. Can. J. Bot. 84 604–612. [Google Scholar]
- Matsui, K., Hiratsu, K., Koyama, T., Tanaka, H., and Ohme-Takagi, M. (2005). A chimeric AtMYB23 repressor induces hairy roots, elongation of leaves and stems, and inhibition of the deposition of mucilage on seed coats in Arabidopsis. Plant Cell Physiol. 46 147–155. [DOI] [PubMed] [Google Scholar]
- Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55 954–967. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y., Oka, A., Ruberti, I., Morelli, G., and Aoyama, T. (2002). Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J. 29 359–369. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis, D.L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S.M., Harel, M., Remingston, S.J., Silman, I., and Schrag, J. (1992). The alpha/beta hydrolase fold. Protein Eng. 5 197–211. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Meissner, R.C., Shoue, D.A., Carpita, N.C., and Bevan, M.W. (2001). MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, N.A., and Glover, B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. [DOI] [PubMed] [Google Scholar]
- Sawa, S. (2002). Overexpression of the AtmybL2 gene represses trichome development in Arabidopsis. DNA Res. 9 31–34. [DOI] [PubMed] [Google Scholar]
- Schardl, C.L., Byrd, A.D., Benzion, G., Altschuler, M.A., Hilderbrand, D.F., and Hunt, A.G. (1987). Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61 1–11. [DOI] [PubMed] [Google Scholar]
- Scheller, H.V., Jensen, J.K., Sorensen, S.O., Harholt, J., and Geshi, N. (2007). Biosynthesis of pectin. Physiol. Plant. 129 283–295. [Google Scholar]
- Schellmann, S., and Hülskamp, M. (2005). Epidermal differentiation: Trichomes in Arabidopsis as a model system. Int. J. Dev. Biol. 49 579–584. [DOI] [PubMed] [Google Scholar]
- Serna, L. (2005). Epidermal cell patterning and differentiation throughout the apical-basal axis of the seedling. J. Exp. Bot. 56 1983–1989. [DOI] [PubMed] [Google Scholar]
- Serna, L., and Martin, C. (2006). Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 11 274–280. [DOI] [PubMed] [Google Scholar]
- Solano, R., Nieto, C., Avila, J., Canas, L., Diaz, I., and Paz-Ares, J. (1995). Dual DNA-binding specificity of petal epidermis specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 14 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., Osborne, E., Paredez, A., Persson, S., Raab, T., Vorwerk, S., and Youngs, H. (2004). Toward a systems approach to understanding plant cell walls. Science 306 2206–2211. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Stuhlfelder, C., Mueller, M.J., and Warzecha, H. (2004). Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. Eur. J. Biochem. 271 2976–2983. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Kato, A., Nagata, N., and Komeda, Y. (2002). A xylanase, AtXyn1, is predominantly expressed in vascular bundles, and four putative xylanase genes were identified in the Arabidopsis thaliana genome. Plant Cell Physiol. 43 759–767. [DOI] [PubMed] [Google Scholar]
- Tominaga, R., Iwata, M., Okada, K., and Wada, T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel, B., Kuschinsky, A.M., Rosso, M.G., Eckermann, N., and Pauly, M. (2004). RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol. 134 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken, J.-P., Schols, H.A., Oomen, R.J.F.J., McCann, M., Ulvskov, P., Voragen, A.G.J., and Visser, R.G.F. (2003). If homogalacturonan were a side chain of rhamnogalacturonan I. Implications for cell wall architecture. Plant Physiol. 132 1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., Marks, M.D., Shimura, Y., and Okada, K. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129 5409–5419. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, D.M., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L. (2006). Changing spaces: The Arabidopsis mucilage secretory cells as a novel system to dissect cell wall production in differentiating cells. Can. J. Bot. 84 622–630. [Google Scholar]
- Western, T.L., Burn, J., Tan, W.L., Skinner, D.J., Martin-McCaffrey, L., Moffatt, B.A., and Haughn, G.W. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 127 998–1011. [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Skinner, D.J., and Haughn, G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Young, D.S., Dean, G.H., Tan, W.L., Samuels, A.L., and Haughn, G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats, W.G.T., McCartney, L., and Knox, J.P. (2001). In-situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213 37–44. [DOI] [PubMed] [Google Scholar]
- Windsor, J.B., Symonds, V.V., Mendenhall, J., and Lloyd, A.L. (2000). Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J. 22 483–493. [DOI] [PubMed] [Google Scholar]
- Young, R.E., McFarlane, H.E., Hahn, M.G., Western, T.L., Haughn, G.W., and Samuels, A.L. (2008). Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20 1623–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Gonzalez, A., Zhao, M., Payne, C.T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zimmermann, I.M., Heim, M.A., Weisshaar, B., and Uhrig, J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.