Abstract
Early endosperm development involves a series of rapid nuclear divisions in the absence of cytokinesis; thus, many endosperm mutants reveal genes whose functions are essential for mitosis. This work finds that the endosperm of Arabidopsis thaliana endosperm-defective1 (ede1) mutants never cellularizes, contains a reduced number of enlarged polyploid nuclei, and features an aberrant microtubule cytoskeleton, where the specialized radial microtubule systems and cytokinetic phragmoplasts are absent. Early embryo development is substantially normal, although occasional cytokinesis defects are observed. The EDE1 gene was cloned using a map-based approach and represents the pioneer member of a conserved plant-specific family of genes of previously unknown function. EDE1 is expressed in the endosperm and embryo of developing seeds, and its expression is tightly regulated during cell cycle progression. EDE1 protein accumulates in nuclear caps in premitotic cells, colocalizes along microtubules of the spindle and phragmoplast, and binds microtubules in vitro. We conclude that EDE1 is a novel plant-specific microtubule-associated protein essential for microtubule function during the mitotic and cytokinetic stages that generate the Arabidopsis endosperm and embryo.
INTRODUCTION
The endosperm that surrounds the embryo is a triploid tissue generated from the product of fertilization of the diploid central cell of the embryo sac. The endosperm plays an important role in the development of angiosperms, acting primarily as a nurse tissue to support the growth of the embryo in the developing seed, but also playing an important role in communicating signals between the embryo and maternal integuments (Berger, 2003; Costa et al., 2004). In some species, such as Zea mays, storage reserves in the endosperm are vital for seedling growth after germination (Berger, 2003). In other species, including the model plant Arabidopsis thaliana, much of the endosperm is degraded as the embryo expands during seed development (Penfield et al., 2004). However, the early stages of endosperm development are remarkably similar in both Arabidopsis and cereals (Berger, 2003). Following fertilization, waves of multiple mitoses without cytokinesis form a syncitium that is organized into nuclear-cytoplasmic domains (NCDs) by radial microtubule systems (RMSs; Olsen, 2004). At this stage, nuclei and associated cytoplasm migrate and three zones of endosperm are observed: the micropylar endosperm that surrounds the embryo, the adjacent chalazal endosperm, and the peripheral endosperm (Brown et al., 2003). The nuclei in the micropylar and chalazal regions are embedded in common cytoplasm, but the peripheral endosperm NCDs are clearly separated (Scott et al., 1998). At the heart stage of embryo development in Arabidopsis seeds, the syncitium contains ∼400 nuclei (Scott et al., 1998). In a second phase, the syncitium becomes cellularized by a separate round of cytokinesis involving microtubule polar configurations and miniphragmoplasts (Brown et al., 1999; Olsen, 2004).
Mutants defective in endosperm development (e.g., disorganized endosperm cell division, failure of cellularization, and seed death) have provided an effective way to identify novel genes involved in cell divisions and the microtubular cytoskeleton (Liu et al., 2002; Sorensen et al., 2002; Steinborn et al., 2002; Dickinson, 2003). The titan and pilz mutants are characterized by the presence of greatly enlarged nuclei in the endosperm resulting from successive rounds of DNA replication without cytokinesis (Steinborn et al., 2002; Tzafrir et al., 2002). Many such mutations affect functions in mitosis that are essential for plant growth. The PILZ group of genes encodes proteins of the tubulin-folding complex required for microtubule formation (Steinborn et al., 2002), and some TITAN genes (TTN3, TTN7, and TTN8) encode Structural Maintenance of Chromosome proteins that are involved in chromosome dynamics, whereas others (TTN5) are responsible for regulation of intracellular vesicle transport (Liu et al., 2002; Tzafrir et al., 2002). Thus, endosperm-defective mutants provide a means of dissecting basic cellular processes in plants.
In this study, a forward genetics approach identified a novel plant-specific protein, ENDOSPERM DEFECTIVE1 (EDE1), and we show that EDE1 is essential for microtubule function and nuclear proliferation during endosperm development. Moreover, EDE1 colocalizes with mitotic microtubules in vivo and binds microtubules in vitro. We conclude that EDE1 is a novel plant-specific microtubule-associated protein essential for seed development and for microtubule function in the endosperm.
RESULTS
The ede1 Mutation Affects Seed Development
A microscopy screen of a collection of wrinkled seed mutants (Focks and Benning, 1998) identified the first allele, ede1-1, as defective in seed development. The mutation leads to 66% seed abortion (n = 244) in homozygous ede1-1 siliques, while mature plants showed no other obvious phenotype and were indistinguishable from the wild type. Aborted seeds fall into two categories: brown shrunken seeds, which indicate late abortion (40%), and small white desiccated seeds, indicating early abortion (26%). No aborted seeds were observed (n = 270) in crosses of ede1-1 × wild type and of wild type × ede1-1, confirming that ede1-1 is a recessive mutation.
Analysis of Feulgen-stained ede1-1 endosperm at 6 d after pollination (DAP) revealed a dramatically reduced number of enlarged endosperm nuclei that contained multiple oversized nucleoli compared with the wild type (Figures 1A to 1D). Staining with 4',6-diamidino-2-phenylindole (DAPI) confirmed that the increased nuclear volume was accompanied by a rise in DNA content. The amount of DNA in each nucleus at 4 DAP, estimated from the intensity of DAPI staining, was more than threefold greater in ede1-1 (373 units per nucleus ±116; n = 7) compared with the wild type (113 units per nucleus ±10; n = 7). The nucleoli from ede1-1 were also >4 times larger than those from the wild type (1357 pixels ± 246; n = 7 and 307 pixels ±33.5; n = 7 respectively). The ede1-1 mutation leads to a range of seed phenotypes, from mild to severe, within a single homozygous ede1-1 silique. In the mild phenotypes, the endosperm contained approximately one-tenth of the number of NCDs in the peripheral endosperm compared with the wild type at equivalent stages of development, and both micropylar and chalazal endosperm had formed (Figures 1A and 1B). In ede1-1 seeds displaying a moderate effect, the NCDs of the central peripheral endosperm were very large and multinucleate (Figure 1C), and the embryo had associated endosperm (see Supplemental Figure 1 online). In the most extreme examples, only one highly stretched NCD containing connected nuclei was present (Figure 1D). Despite the variable number of NCDs, cellularization of ede1-1 endosperm was not observed.
Figure 1.
EDE1 Is Essential for Seed Development.
(A) to (D) Confocal micrographs of Feulgen- stained seeds of the wild type (A) and ede1-1 mutant ([B] to [D]) at 6 DAP. Note the NCDs that invade the central vacuole (CV) of the endosperm. Only few enlarged NCDs are present in the ede1-1 mutant when compared with the wild type. The range of ede1-1 phenotypes is also shown: mild (B), moderate (C), and extreme (D). EM, embryo; CE, chalazal endosperm. Bars = 20 μm.
(E) Developmental stages of the wild type (top row) and of ede1-1 mutant endosperm (bottom row). Whole mounts of cleared seeds were observed at different developmental stages (0 to 8 DAP) using differential interference contrast optics. The enlarged nuclei in the mutant are marked by asterisks. Bar = 20 μm (0 to 2 DAP) and 50 μm (4 to 8 DAP).
To identify the developmental stage at which the endosperm defect first arises in the ede1-1 mutant, we examined mutant and wild-type seed development with Nomarski optics. Up to 2 DAP, ede1-1 seeds were indistinguishable from the wild type. Enlarged mutant nuclei started to be visible from 4 DAP onwards (Figure 1E). At 6 DAP, the mutant endosperm displayed on average between 1 and 10 enlarged nuclei, whereas the wild-type endosperm contained an excess of 100 normal nuclei. An extreme example is shown in Figure 1E, where at 6 DAP, only one nucleus is visible in the mutant endosperm. ede1-1 embryos develop normally up to 8 DAP and at a rate similar to that of the wild type (Figure 1E) and show no obvious morphological defects. Beyond the heart stage of embryo development, seed shrinkage and collapse were observed in the late aborted ede1-1 seeds.
EDE1 Is Required for Microtubule Organization in the Endosperm
The nuclear division defect in ede1-1 endosperm suggested a possible defect in cytoskeletal organization. To evaluate this, developing mutant and wild-type seeds were analyzed by fluorescence microscopy (Figure 2). Immunolabeling with antitubulin and counterstaining with DAPI demonstrated that ede1-1 endosperm lacks the organized microtubule arrays typical of wild-type endosperm. In wild-type endosperm, the nuclei of the syncitium are evenly spaced by RMSs (Brown et al., 1999) that radiate from each nucleus and define the NCDs (Figures 2A and 2B). By contrast, in early (4 DAP) ede1-1 endosperm, only a few enlarged and unevenly spaced nuclei lacking associated microtubule structures are present (Figure 2D). Later in development (6 to 8 DAP), giant ede1-1 endosperm nuclei displayed several enlarged nucleoli, lacked associated RMSs, and failed to cellularize (Figures 2E and 2F). Despite a careful analysis of the mutant and wild type (∼800 ede1-1 and 400 wild-type seeds at 4 to 6 DAP), we never observed chromatin condensation, mitotic spindles, and cytokinetic phragmoplasts in the mutant. By contrast, mitotic figures, such as spindles and phragmoplasts, were observed at a frequency of ∼1 in 20 in wild-type endosperm (Figure 2C), indicating at least a 40-fold decrease in the mitotic index of the ede1-1 mutant endosperm.
Figure 2.
EDE1 Is Required for Microtubule Function in Endosperm.
Wild-type ([A] to [C]) and mutant ([D] to [F]) endosperm stained with DAPI (red) and immunolabeled with antitubulin (green). Bar = 20 μm in (A), (B), (D), (E), and (F) and 40 μm in (C).
(A) and (B) Syncytial phase of wild-type endosperm development at 4 DAP with microtubules radiating from evenly spaced nuclei around the periphery of the embryo sac.
(C) Cytokinesis during endosperm cellularization at 6 DAP. The asterisks indicate phragmoplasts.
(D) The mutant syncytial endosperm at 4 DAP is characterized by few enlarged nuclei displaying numerous nucleoli and lacking organized microtubule structures.
(E) At 6 DAP, the mutant endosperm displays giant nuclei without radial microtubule systems and that never cellularize.
(F) At 8 DAP, when the wild-type endosperm is fully cellularized, ede1-1 endosperm contains only giant nuclei with several nucleoli.
Cloning of the EDE1 Gene and Identification of Additional ede1 Alleles
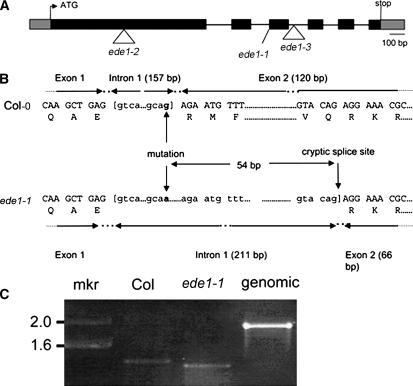
The EDE1 gene was identified by positional cloning using a population created from a cross between the homozygous ede1-1 mutant and Landsberg erecta (Ler). The mapping revealed that the mutation lies on BAC F6E13, and sequencing of the nonrecombinant region revealed a G-to-A single base transition at the intron1/exon2 boundary in gene At2g44190. The gene is 1925 bp long from the start ATG to the stop codon and consists of six exons (Figure 3A), defining a 474–amino acid protein with a predicted molecular mass of 53 kD and a calculated pI of 9.8. RT-PCR using RNA purified from young siliques of homozygous ede1-1 and the wild type showed that mRNA from the gene is present in the mutant but is smaller than the wild type (Figure 3C). Cloning and sequencing of the RT-PCR products showed that the mutant uses a cryptic splice site that is 54 bases after the 3′ of the intron1/exon2 boundary (Figure 3B). This results in a small deletion during processing of the mRNA that leaves the reading frame intact but is predicted to code for a protein that is 18–amino acid residues shorter than the wild type and missing the residues between Arg-304 and Gln-321. The ede1-1 mutation was complemented using a binary construct containing the full-length genomic sequence of the wild-type gene At2g44190. Endosperm from mature seeds (n = 284) of 10 independent T2 ede1-1 homozygous plants that were homozygous for the transgene had a wild-type appearance, confirming that At2g44190 encodes the EDE1 protein.
Figure 3.
Structure of the EDE1 Gene.
(A) Blocks denote exons, and lines denote introns. The ede1-1 allele contains a G-to-A base transition at +1066, ede1-2 contains an insertion of T-DNA into exon 1 (+414), and ede1-3 contains an insertion of T-DNA into intron 2 (+1231). The sequence of a cDNA (GenBank BX818775) was used to define the 5′ and 3′ untranslated regions, shown as gray boxes.
(B) Detail of DNA and amino acid sequences surrounding the ede1-1 mutation in Columbia (Col) and ede1-1. Comparison of ede1-1 and Col cDNA sequences shows that the G-to-A polymorphism at +1066 removes the exon1/intron1 splice site and that the ede1-1 mutant plant uses a cryptic splice site at +1120 instead.
(C) RT-PCR of At2g44190 transcript from total RNA isolated from young siliques of Col wild-type and homozygous ede1-1 plants. Product from genomic DNA is shown as control. mkr, marker (kb).
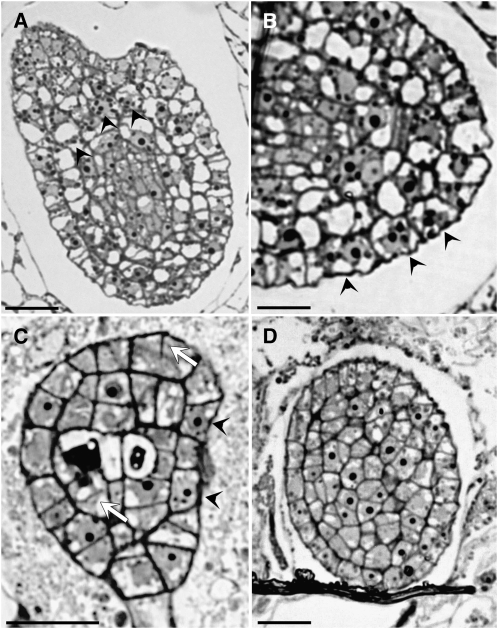
A search of the SALK T-DNA insertion mutant collection (Alonso et al., 2003) identified two further EDE1 alleles that have insertions in the first exon and second intron, respectively, and were called ede1-2 and ede1-3 (Figure 3A). Disruption of the EDE1 gene by T-DNA insertion appears to be lethal: we were unable to identify any plants that were homozygous for the insertion from progeny derived from selfing hemizygous ede1-2 and ede1-3 plants. Hemizygous ede1-2 and ede1-3 have siliques containing ∼38% (n = 288) and 36% (n = 471) abnormal seeds, respectively. Of these abnormal seeds, ∼20% aborted early and 16 to 18% aborted late at the heart-torpedo embryo stage. The late-aborting seeds failed to germinate and displayed defects similar to those described for ede1-1 (see Supplemental Figure 2 online). Similar to what was observed in ede1-1 seeds (Figure 1E), embryo development in ede1-2 and ede1-3 proceeds up to at least the heart stage (see Supplemental Figure 3 online). To determine whether ede1-2 and ede1-3 alleles display defects associated with aberrant mitosis or cytokinesis, we examined histochemically stained sections of mutant embryos from globular to heart stage by light microscopy. Enlarged nuclei containing multiple nucleoli and occasional cell wall stubs were observed in mutant embryos (Figure 4) in association with the previously described endosperm defects.
Figure 4.
ede1 Null Mutant Embryos Have Cell Division Defects: Histological Sections Reveal Multinucleate Cells and Cell Wall Stubs.
(A) and (B) ede1-2 embryos at heart stage. Arrowheads point to enlarged nuclei containing multiple nucleoli.
(C) ede1-3 globular embryo revealing multinucleate cells (arrowheads) and cell wall stubs (white arrows).
(D) Section across a wild-type embryo at heart stage.
Bars = 50 μm in (A) and (D) and 20 μm in (B) and (C).
To determine whether the maternal or paternal sporophytes are implicated in the ede1-3 mutation, we conducted reciprocal crosses between heterozygous (EDE1/ede1-3) and wild-type plants. In a cross between a heterozygous female (EDE1/ede1-3) and a wild-type male (EDE1/EDE1), 20% of the seed aborted at a heart-torpedo stage; while in a cross between a wild-type female (EDE1/EDE1) and a heterozygous male (EDE1/ede1-3), only 4% seed abortion was observed, similar to wild-type values (Table 1). In neither case were early aborted seeds observed. These results suggest that the ede1-3 mutant displays incomplete maternal effect seed lethality. Alternatively, but not exclusively, the paternal allele could suppress 60% of the lethality expected in case of maternal effect only.
Table 1.
Seed Phenotype of Reciprocal Crosses between Heterozygous ede1-3 and Col-0 Wild-Type Plants
| Female Male | Col-0 (+/+) (Selfed) | ede1-3 (+/−) (Selfed) | ede1-3 (+/−) × Col-0 | Col-0 (+/+)× ede1-3 (+/−) |
|---|---|---|---|---|
| Early aborted | 0% | 20% | 0% | 0% |
| Normal | 96% | 64% | 80% | 96% |
| Late aborted | 3% | 16% | 20% | 4% |
| n (seeds) | 186 | 471 | 101 | 99 |
Finally, F1 plants with the genotype ede1-1/ede1-2 or ede1-1/ede1-3 have a similar seed phenotype to homozygous ede1-1 plants, indicating that the different mutants belong to the same complementation group and that ede1-2 and ede1-3 are recessive to ede1-1 (Table 2).
Table 2.
Live and Total Aborted Seeds from Crosses between ede1-1, ede1-2, and ede1-3 Mutant Genotypes.
| ede1-1 | ede1-2 | ede1-3 | |
|---|---|---|---|
| ede1-1 | 66% ab 34% nor n = 251 | 64% ab 35% nor n = 230 | 56% ab 22% nor n = 234 |
| ede1-2 | 32% ab 68% nor n = 218 | 40% ab 60% nor n = 198 | |
| ede1-3 | 36% ab 64% nor n = 201 |
Ab, aborted; nor, normal.
The EDE1 Gene Defines a Novel Family of Plant-Specific Proteins
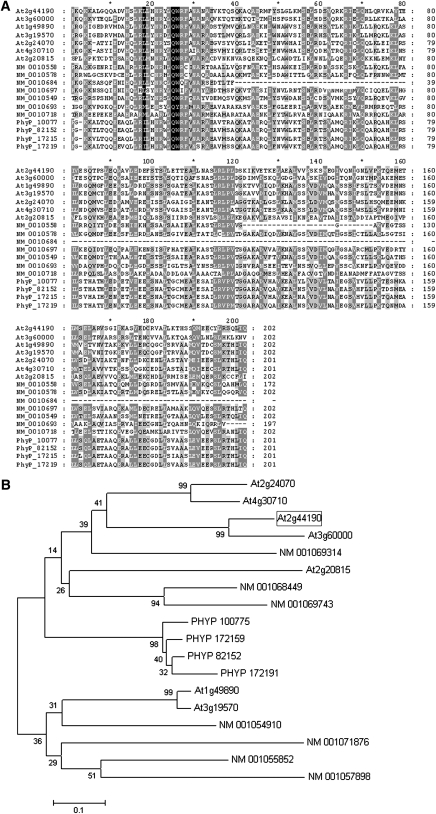
Sequence analysis identified the EDE1 protein as a member of a family of related, plant-specific proteins, all containing the InterPro domain of unknown function DUF566 (InterPro:IPR007573). A BLAST search (Altschul et al., 1990) revealed six similar proteins in Arabidopsis, one of which (At3g60000) shares 60% amino acid sequence identity. Seven similar proteins were identified in Oryza sativa and four in the moss (Physcomitrella patens) (Figure 5A). Sequence alignments of all EDE1-like proteins revealed that the highest similarity is located at the C-terminal half of the protein, suggesting that important and conserved functional domains exist in this region (Figure 5A). Phylogenetic analysis of the C-terminal sequences of all members of the EDE1 family identified four types, one of which is confined to moss. EDE1, together with At3g60000, At2g24070, At4g30710, and a rice protein (NM_001069314), fall into the same type, suggesting that NM_001069314 could be the ortholog of EDE1 (Figure 5B; see Supplemental Data Set 1 online).
Figure 5.
The EDE1 Gene Defines a Novel Family of Plant-Specific Proteins.
(A) Multiple sequence alignment of the C-terminal region of all EDE1-like proteins found in Arabidopsis (At), O. sativa (NM_), and P. patens (PhyP_). Shading indicates amino acid conservation: black (100%), dark gray (80 to 99%), light gray (50 to 80%), and white (<50%). The sequences were aligned using ClustalW.
(B) Predicted evolutionary relationship among members of the EDE1 family. The phylogenetic tree was generated using MEGA version 4. Bootstrap values from 1000 trials are indicated.
Searches of the Chlamydomonas genomic sequence (http://www.chlamy.org/) revealed no significant similarities, and no similar sequences were identified in any bacterial, fungal, or animal genomes, indicating that the EDE1 family is specific to land plants.
EDE1 Is Highly Expressed during Early Seed Development
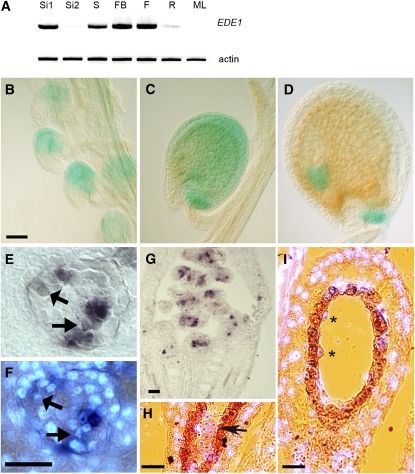
The developmentally restricted phenotype of the ede1 mutant alleles suggested that expression of the EDE1 gene might be specific to young proliferating tissues and to the reproductive phase. The expression pattern of the EDE1 gene was determined using RT-PCR, and expression was found to be strongest in young siliques (2 to 4 DAP), 4-d-old seedlings, flower buds, and open flowers (Figure 6A). The expression was weaker in roots and below the detection level in older siliques (8 to 10 DAP) and in mature leaves. This pattern of gene expression was confirmed by analyzing the large number of expression microarray experiments currently available (https://www.genevestigator.ethz.ch), which also shows that the strongest expression of EDE1 occurs in the inflorescence, although expression could also be detected in the shoot apices, cotyledons, and radicles of embryos at globular and heart stages of development. Otherwise, the EDE1 gene is expressed at very low levels in mature tissues. The pattern of gene expression during development was confirmed in transgenic plants containing EDE1 promoter–β-glucuronidase (GUS) constructs. Analysis of 10 independent lines showed that expression of the GUS reporter was detected in the embryo sac in prefertilization ovules (Figure 6B) and in seeds following fertilization. GUS expression occurred in both embryo and endosperm throughout most of the syncytial phase of endosperm development (Figure 6C) but became undetectable in the cellularized endosperm, when cell division had ceased (Figure 6D). Expression was maintained in the embryo up to heart stage (Figure 6D). Because GUS is a stable protein, it cannot provide accurate information on highly dynamic changes in gene expression. We therefore confirmed and extended these observations by mRNA in situ hybridization analysis. Hybridization of an antisense EDE1 probe to sections of floral meristems indicated that the level of expression was strongest in unfertilized ovules (Figures 6E to 6G) and weaker in embryo and endosperm nuclei in developing seeds (Figures 6H and 6I). When CYCLIN B was used as a control probe in similar sections, the signal in endosperm nuclei was comparable to that obtained with EDE1 as a probe. By contrast, the signal in the embryo was much stronger with CYCLIN B than with EDE1 (see Supplemental Figure 4 online). However, the EDE1 signal was not restricted to embryonic tissues per se but could be detected in other actively proliferating tissues, such as young unfertilized ovules (Figure 6G). This produced a spotty pattern in which some cells displayed high levels of signal and neighboring cells did not (Figure 6E), a pattern consistent with the accumulation of mRNA in a cell cycle–dependent manner (Fobert et al., 1994, 1996). When sections of unfertilized ovules were counterstained with the DNA-specific dye DAPI, to reveal the stage of the cell cycle, the cells containing the strongest EDE1 signal were all in interphase (Figures 6E and 6F). Prophase cells contained only a weak signal, and metaphase cells had no detectable signal. These results are consistent with a G2 phase–specific expression of the EDE1 gene (Ito et al., 2001).
Figure 6.
Expression of EDE1 in Tissues of Arabidopsis.
(A) RT-PCR of EDE1 transcript from total RNA isolated from 2- to 4-DAP siliques (Si1), 8- to 10-DAP siliques (Si2), seedlings (S), flower buds (FB), flowers (F), roots (R), and mature leaves (ML). Actin was used as control.
(B) to (D) GUS expression under the control of EDE1 promoter in ovules and developing seeds. Prefertilization ovules (B), ovule at 4 DAP (C), and ovule at 8 DAP (D). Bar = 40 μm.
(E) to (G) Expression of EDE1 in unfertilized ovules by in situ hybridization. Bar = 40 micron.
(E) Unfertilized ovule expressing EDE1 (purple brown signal) in a patchy pattern.
(F) DAPI counterstaining of the same section as in (E) to reveal the nuclei. Early mitotic nuclei are indicated by arrows in (E) and (F).
(G) Prefertilization silique showing patches of purple signal within unfertilized ovules.
(H) Signal (purple) on an embryo (arrow) at 4 DAP with DAPI counterstaining (light blue). Bar = 40 micron.
(I) Section through a 4-DAP ovule showing signal in the endosperm nuclei (asterisks). DAPI counterstaining is in light blue. The seed coat is shown in orange-brown. Bar = 40 μm.
To gain further insight into the pattern of EDE1 expression, we interrogated the Arabidopsis coresponse database (http://csbdb.mpimp-golm.mpg.de/csbdb/dbcor/ath.html) to find those genes that show similar patterns of expression. This resource compiles microarray expression data from numerous experiments and can provide clues to gene function by association with genes of known function. Eighteen from the top 20 genes, whose expression most closely corresponded with EDE1 over a range of experimental conditions, are either known or predicted to be involved in mitosis and cytokinesis (see Supplemental Table 1 online). These include KNOLLE (Lauber et al., 1997) and several cyclins with predicted mitotic function. Notably, two microtubule associated proteins, a homolog of end binding 1 protein (EB1) (Chan et al., 2003; Mathur et al., 2003) and a mitotic kinesin, NACK1/HINKEL (Nishihama et al., 2002), are also among the top 20, along with the chromosomal protein SMC4 (Liu et al., 2002). Many of these coexpressed genes contain one or more M-specific activator (MSA) elements that are believed to enhance gene expression during G2 and M phase (see Supplemental Table 1 online). Sequence analysis identified two MSA elements in the EDE1 promoter sequence and putative anaphase-promoting complex (APC) target motifs within the EDE1 coding sequence. Moreover, a study to identify genes that are coregulated during the cell cycle in Arabidopsis cell suspension cultures (Menges et al., 2005) revealed that EDE1 (termed a hypothetical protein in that report) is one of 82 genes that peak in their expression during the G2/M phase of the cell cycle.
Our expression studies, together with the tight coresponse of EDE1 with many genes that show transcriptional upregulation during G2/M, strongly suggest that EDE1 expression is highly regulated during the cell cycle.
The GFP-EDE1 Fusion Protein Decorates Microtubules
To gain insight into the cellular role of EDE1, we fused the EDE1 genomic sequence with a green fluorescent protein (GFP) reporter gene. The functionality of the GFP-EDE1 fusion protein was demonstrated by expressing it under the EDE1 promoter in homozygous ede1-1 mutant plants, where full complementation was observed in eight out of eight independent transgenic lines (see Supplemental Table 2 online). Although the GFP fusion confirmed that EDE1 was expressed in the endosperm of developing ovules (see Supplemental Figure 5 online), the complexity of the tissue interfered with high-quality imaging. Therefore, we studied the dynamic localization of GFP-EDE1 in cell suspensions. In Arabidopsis cells transiently expressing GFP-EDE1, the GFP signal was associated with spindles and phragmoplasts (see Supplemental Figure 6 online). To test if the association depended on microtubules, we treated Arabidopsis cells expressing GFP-EDE1 with taxol (10 μM) or oryzalin (10 μM), drugs that are specific for the stabilization or destabilization of microtubules, respectively. Oryzalin treatment caused a loss of GFP fluorescence (data not shown), whereas the GFP-EDE1 signal in taxol-treated cells remained fibrillar (see Supplemental Figure 7 online).
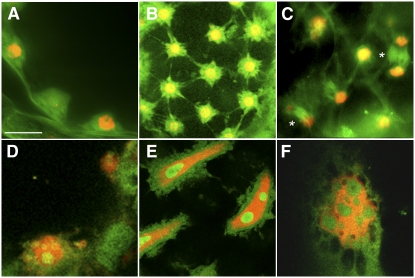
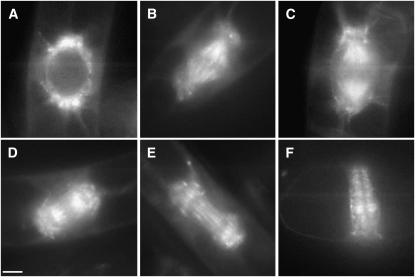
To examine the dynamics of GFP-EDE1 redistribution during the cell cycle, we stably transformed tobacco (Nicotiana tabacum) BY-2 cells with the EDE1 promoter:GFP-EDE1 construct. GFP-EDE1 fluorescence in proliferating transgenic tobacco BY-2 lines was virtually undetectable during interphase, but, at the onset of mitosis, GFP-EDE1 began to accumulate into opposing polar caps in the perinuclear region from which the microtubule arrays radiate toward the cell cortex (Figure 7A). As cells progressed into mitosis, GFP-EDE1 decorated microtubules of the spindle and spindle poles at metaphase and anaphase (Figures 7B and 7C). In anaphase, a clear gap developed in the central zone of the spindle, and fluorescence was largely restricted to the region between the chromatids and each pole, suggesting that GFP-EDE1 decorates pole-to-chromatid kinetochore microtubules at this stage (Figure 7C). At the anaphase/telophase transition and during telophase, GFP-EDE1 strongly associated with midzone microtubules out of which the phragmoplast emerges (Figures 7D and 7E) and remained associated with the phragmoplast throughout cell plate formation (Figure 7F; see Supplemental Figure 8 online). At the end of cell division, GFP fluorescence was no longer detectable. Notably, GFP-EDE1 was not detected on cortical microtubules, either in the interphase array or the preprophase band.
Figure 7.
GFP-EDE1 Localizes to Microtubules during Mitosis.
Living tobacco BY-2 suspension cells transformed with EDE1 promoter:GFP-EDE1. Bar = 10 μm.
(A) GFP-EDE1 accumulates in nuclear caps from which radial microtubules emanate in premitotic cells.
(B) During mitosis, fluorescence accumulates in metaphase spindle and spindle poles.
(C) and (D) Labeling of the kinetochore microtubules in anaphase, together with labeling around the spindle poles.
(E) GFP-EDE1 strongly associated with midzone microtubules from which the early columnar phragmoplast develops.
(F) Fluorescence remains associated with the phragmoplast throughout cell plate formation.
EDE1 Is a Microtubule Binding Protein
The above observations indicate that the EDE1 protein associates with microtubules during cell division. To further verify this association, we performed costaining experiments using antitubulin antibodies to visualize the microtubules in BY-2 tobacco cells expressing GFP-EDE1. Tubulin colocalized with GFP-EDE1 in a fibrous pattern on mitotic structures (Figure 8A). Colocalization could not be observed in interphase cells because GFP-EDE1 was not expressed at detectable levels during interphase. Colocalization of EDE1 with tubulin could result either from direct binding of EDE1 to microtubules or from indirect binding via other microtubule-associated proteins. We tested the ability of EDE1 to bind directly to microtubules by an in vitro cosedimentation experiment. In vitro–translated [35S] methionine-labeled EDE1 was incubated with or without taxol-polymerized mammalian brain microtubules. Microtubules were pelleted through a sucrose cushion, and both supernatants and pellets were analyzed by autoradiography. EDE1 was significantly enriched in the tubulin pellet compared with a nonmicrotubule binding control (Figure 8B), indicating that, in vitro, EDE1 binds directly to microtubules.
Figure 8.
EDE1 Is a Microtubule Binding Protein.
(A) GFP-EDE1 colocalizes with spindle microtubules in tobacco BY-2 cells. Left panel, GFP-EDE; middle panel, antitubulin indirect immunofluorescence; right panel, merged image. Bar = 10 μm.
(B) EDE1 cosediments with microtubules in vitro. Equal amounts of radiolabeled EDE1 were used in pull-down experiments with (+) or without (−) microtubules that had been stabilized by taxol treatment. EDE1 protein preferentially copurified with microtubules (found in the pellet [P] rather than the supernatant [S] after centrifugation). A control protein (At5g16050) showed no preferential copurification with microtubules.
DISCUSSION
EDE1 Is a Novel Microtubule-Associated Protein and Preferentially Associates with Nuclear Microtubules during Mitosis
EDE1 defines a novel microtubule-associated protein that is specific to land plants. We were able to detect clear structural homologs in lower plants, such as moss, but not in single celled plants, microbes, or animals. Many plant microtubule-associated proteins have substantial homology to animal and yeast proteins (reviewed in Hussey et al., 2002; Chan et al., 2003; Van Damme et al., 2004), indicating that the functionality of the microtubule cytoskeleton is largely conserved between plants and animals. However, other plant microtubule-associated proteins have little or no apparent similarity to animal and yeast proteins. These proteins have been identified either by biochemical isolation (Korolev et al., 2005, 2007; Buschmann et al., 2006; Wang et al., 2007) or by genetic screens (Buschmann et al., 2004; Nakajima et al., 2004; Ambrose et al., 2007; Walker et al., 2007; Perrin et al., 2007), supporting the notion that the plant cytoskeleton has distinctive features defined by novel microtubule-associated proteins (Lloyd and Hussey, 2001). This is consistent with the fact that plants have specific microtubule arrays not shared with other organisms. The cortical array, which is composed of parallel plasma membrane-associated microtubules, the preprophase band, and the cytokinetic phragmoplast are all distinct and all are involved in organizing the growing cell wall or the new cross wall. In addition, the acentric mitotic spindle of plants differs from the centrosome-containing animal spindle, highlighting further differences between plants and other eukaryotes (Lloyd and Chan, 2006).
EDE1 is unusual in that is localized preferentially on the nucleus-associated microtubule arrays, being undetectable in cortical microtubules and the preprophase band. Its accumulation in polar caps around the premitotic nucleus is similar to that reported for proteins associated with microtubule nucleation, such as γ-tubulin (Liu et al., 1994; Murata et al., 2005). This suggests a role for EDE1 in microtubule function and/or organization during division. Not only does its association with microtubules appear to be cell cycle regulated, but its overall expression is strictly dependent on cell cycle progression, being detectable only in a narrow window during G2/M. Once cytokinesis has been completed, the phragmoplast-associated EDE1 is no longer detectable. Such a protein expression profile, together with the cell cycle–dependent mRNA accumulation shown by our in situ analysis, indicates that regulatory mechanisms are in place to ensure EDE1 accumulation only during cell division. The low-level accumulation of EDE1 and CYCLIN B transcripts in the syncytial endosperm of wild-type seeds detected by in situ hybridization may be attributed to the narrow mitotic peak of synchronous nuclear divisions in endosperm. Sequence analysis identified two MSA elements in the EDE1 promoter. MSA-like sequences have been found in promoters of several other genes expressed during G2 and M phases, including B-type cyclins from tobacco, soybean (Glycine max), and Arabidopsis and At NACK1 and At NACK2 that encode plant kinesin-like proteins (Ito et al., 1998; 2001). Moreover, sequence analysis revealed the presence of several putative APC targets within the EDE1 sequence (seven RXXL-like D-box motifs and one KEN box motif). APC-dependent degradation of spindle-associated proteins during mitotic exit has been reported in animals and yeast and provides a mechanism to ensure that their activity on spindle microtubule dynamics is strictly regulated in the cell cycle (Juang et al., 1997; Seki and Fang, 2007; Woodbury and Morgan, 2007). That EDE1 functions in division is consistent with its expression in proliferating tissues (such as seedlings and young siliques), the cell cycle–specific transcript accumulation, and its coexpression with other G2/M-regulated genes. Of particular significance, the coexpressed genes include two other previously defined microtubule-associated proteins that are involved in different aspects of microtubule function during cell division, such as EB1a,b and the At NACK1/HINKEL kinesin, which is known to be involved in cytokinesis (Strompen et al., 2002). A further clue that EDE1 functions in division is provided by seeds (particularly of the T-DNA insertion mutants ede1-2 and ede1-3, and the more severely affected examples in ede1-1; Figure 1D), in which the nuclei are both enlarged and stretched. Such stretched nuclei are characteristic of defective mitosis, where mitosis has been attempted but fails to complete successfully (Sorensen et al., 2002; Pitt et al., 2004).
EDE1 Is Required for Endosperm Nuclear Divisions and Cellularization and Is Essential for Seed Development
Since organized nuclear proliferation and cellularization of the endosperm are both prerequisites for successful seed development, the EDE1 gene is essential and abrogation of its function prevents completion of the life cycle. The lethality of the ede1-2 and ede1-3 mutations indicates that EDE1 protein is indispensable for seed viability and that the ede1-1 is a weak allele that allows for seed viability (albeit at a reduced level), either by retaining partial functionality of the EDE1 protein or by a variable splicing event. Furthermore, the ede1 knockout mutants display a maternal effect, incompletely penetrant seed lethality, and a zygotic/endosperm requirement for either pollen or egg sac–transmitted EDE1. The expression of EDE1 in early stages of flower development, in unfertilized ovules, and in both endosperm and embryo in developing seeds suggests that EDE1 is expressed both maternally and zygotically. Also, because the endosperm inherits two maternal copies but only one paternal copy of the genome, ede1-3 could cause a maternal dosage-sensitive effect on endosperm development. To determine definitely that seed abortion in ede1 siliques is caused in a maternal effect dosage-sensitive manner, additional wild-type EDE1 copies should be introduced using a triploid or tetraploid wild-type pollen donor as previously described for other genes involved in maternal regulation of embryogenesis (Grossniklaus et al., 1998). Although embryo lethality in homozygous embryos cannot be ruled out at this stage, the endosperm defects, together with the maternal-effect observed, would suggest that a dosage-dependent endosperm defect is the most likely source of the seed lethality observed in ede1 siliques.
The presence of more than one enlarged nucleus in some ede1-1 endosperm indicates that mitosis, at least up to stage IV of endosperm development (one to eight endosperm nuclei, according to Boisnard-Lorig et al., 2001), is not completely abolished, but its relative frequency is decreased by at least 40-fold compared with the wild type. This would explain why, despite careful observation, we failed to observe any mitotic figures in young mutant seeds. After stage IV of endosperm development, the mutant nuclei continue to enlarge without cytokinesis, generating giant nuclei with several nucleoli. This could be explained by several rounds of failed mitosis, by fusion of multiple postmitotic nuclei, or by endoreduplication. However, the fact that chromosome condensation or aberrant mitotic structures were never observed in the ede1 endosperm suggests that the latter is most likely and that enlarged ede1 nuclei arise by endoreduplication. Agents that disrupt cytoskeleton assembly, such as oryzalin and colchicine (Grandjean et al., 2004), or mutations in proteins involved in microtubule function, such as PILZ and the kinesin KIF14 (Mayer et al., 1999; Carleton et al., 2006), have been reported to induce endoreduplication in animal and plant cells, although the mechanism behind it is still unclear. Enlarged endosperm nuclei have previously been reported in the ttn3 mutant endosperm (Liu and Meinke, 1998). However, unlike ede1, ttn3 nuclei continue to progress through the mitotic cycle, as indicated by the fact that both condensation of prophase chromosomes and giant mitotic figures are visible in ttn3 endosperm. Moreover, cellularization of ttn3 endosperm still occurs despite the presence of giant nuclei and viable seeds are produced, while ede1 mutant endosperm never cellularize and seed abortion occurs after the heart stage of embryo development.
The absence of microtubule arrays in ede1 is reminiscent of the pilz mutants, which encode orthologs of mammalian tubulin-folding cofactors specifically involved in the synthesis of tubulin polymers (Steinborn et al., 2002). However, unlike the abnormally shaped pilz embryos consisting of only one or few grossly enlarged cells, ede1 embryos of late aborted seeds develop up to heart stage in a manner similar to the wild type and without alterations in cell architecture. Despite the apparent normal development, defects associated with defective cytokinesis, such as enlarged nuclei containing multiple nucleoli in dividing cells, and cell wall stubs, were occasionally found in embryos of ede1 null alleles. Such defects are typically associated with genes required for the execution of cytokinesis, such as KNOLLE (Lukowitz et al., 1996), KEULE (Assaad et al., 1996), HINKEL (Strompen et al., 2002), RUNKEL (Nacry et al., 2000), and PLEIADE (Hauser and Bauer, 2000). Interestingly, these cytokinesis-defective mutants (with the exception of keule) are similar to ede1, in also being impaired in endosperm cellularization (Sorensen et al., 2002). Cytokinesis defects have also been described in titan and pilz mutants (Liu and Meinke, 1998; Mayer et al., 1999). Moreover, ede1 embryo cells do not display radial swelling, suggesting that the cortical microtubule array is not affected and that the most likely source of the defective cytokinesis resides in the malfunctioning of the mitotic and/or cytokinetic microtubule arrays. Unlike the TTN and PILZ group genes, members of phylogenically dispersed gene families, the EDE1 family is unique to land plants, and no functional data or associated mutant phenotypic information have yet been described for any other members of the family.
The defects observed in ede1 mutant embryos are consistent with our expression data, showing that EDE1 is expressed equally in the embryo and in the endosperm. However, the embryo-associated defects appear to be less severe than those observed in the endosperm, allowing for embryo development to proceed up to at least heart stage. It is possible that functional redundancy among the members of the EDE1 family could account for the mild embryo phenotype, or else cytokinetic defects associated with the ede1 mutation might possibly become more severe in later stages of embryo development. However, the most likely explanation, supported by our genetic analyses (Table 1) is that the developing endosperm is the primary target of EDE1 function. In this scenario, seed abortion beyond heart stage of embryo development could be explained by the inability of the aberrant endosperm to support and nurture the growing embryo. The higher sensitivity of the endosperm tissue to the ede1 mutation might be explained by the fact that the microtubule cycle is different in endosperm compared with somatic cells (Brown and Lemmon, 2001). The latter, more conventional, microtubule cycle involves the bunching of cortical microtubules into a preprophase band that predicts the division plane; the phragmoplast then develops out of the spindle remnant and lays a cell plate in the predicted plane. The alternative microtubule cycle of endosperm involves no cortical microtubules or preprophase bands. Instead, a branched phragmoplast spreads along the interzone formed by the overlap of radial microtubule systems emanating from each nuclear-cytoplasmic domain. Significantly, GFP-EDE1 does not label cortical or preprophase band microtubules in somatic-type tobacco BY-2 cells, but it does label the nuclear caps from which microtubules radiate in premitotic cells, along with the spindle and phragmoplast. These differences could account for the greater impact of EDE1 on endosperm development, although common structural similarities between the two types of microtubule arrays could account for effects on other tissues, such as those of growing embryos.
In summary, this article describes the isolation of a novel microtubule-associated protein essential for microtubule function during cell division in Arabidopsis. Although the most striking phenotype associated with the ede1 mutation remains confined to endosperm tissue, cytokinesis defects in mutant embryos reveal a wider role for EDE1 in microtubule function during somatic division. EDE1 is the founding member of a conserved group of proteins found only in land plants. The question of how plant-specific microtubule arrays are organized remains open, but this study suggests that EDE1 clearly plays a part in this.
METHODS
Plant Material and Media
The ede1-1 mutant originated from a previously described ethyl methanesulphonate–mutagenized M2 population of the Col-2 ecotype (Dörmann et al., 1995). The ede1-2 (SALK 047950) and ede1-3 (SALK 048191) alleles in the Col-0 ecotype are T-DNA insertion mutants that were obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003). Plants were grown in a climate-controlled glasshouse at 20°C day/16°C night with 16 h total lighting between October and March. For aseptic growth, seeds were surface sterilized and plated on Murashige and Skoog medium containing 0.8% (w/v) phytoagar. Primary transformants were selected on plates described but containing either kanamycin (50 mg·L−1) or hygromycin (50 mg·L−1).
Molecular Cloning of EDE1
ede1-1 plants were crossed to plants of the Ler laboratory strain, and 400 F2 progeny were obtained for phenotypic analysis. Genomic DNA was purified using the DNeasy plant mini kit (Qiagen) and used for gene mapping. Mapping of the ede1-1 locus was achieved using the cleaved-amplified polymorphic sequence (CAPS) markers ADHa and UFOa (chromosome I); Ve017a and PhyB/hy3 (II); TSA1a, AtDMC1a, and GAPCa (III) Det 1 (IV); and LFY3a, ASA1a, and R89998a (V). The results showed unambiguously that ede1-1 was linked with Ve017a on the lower arm of chromosome II since none of the plants showing the ede1 phenotype were Ler for Ve017e. Fine mapping using the CAPS markers m429, AthBIO2, ML, AthUIQUE, and 90J19T7 and the simple sequence length polymorphic marker nga 168 showed that the locus lay between AthBIO2 and ML (Konieczny and Ausubel, 1993). Single-nucleotide polymorphism (SNP) and InDel markers were identified to further resolve the locus (see Supplemental Table 3 online) and showed that the mutation lay in a 70-kb region between nucleotide 47048 on BAC F6E13 and nucleotide 9510 on BAC F4I1. DNA from the 70-kb nonrecombinant region of ede1-1 containing the mutation was amplified using PCR in 14 5000-bp overlapping fragments and cloned into pGEMT-Easy (Promega). The DNA was sequenced and all differences with the reference sequence were checked by sequencing independently amplified fragments. CAPS and simple sequence length polymorphic markers were used are described at http://www.Arabidopsis.org/, and the PCR analysis was done as previously described (Konieczny and Ausubel, 1993). The mutation was identified and was confirmed by complementation of the homozygous ede1-1 mutant using a genomic DNA fragment containing the wild-type EDE1 gene, including 5′ and 3′ untranslated regions (Ch2: 18278467 to 18281873). This sequence was excised from binary cosmid clone 53E16 (GeTCID; John Innes Centre) using BamHI and ligated into the BamHI site of the pBIN+ binary vector (van Engelen et al., 1995). The pBIN-EDE1 vector was confirmed by sequencing using M13 forward and reverse primers. This clone was introduced into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986), and ede1-1 homozygous plants were transformed using the floral dip method (Clough and Bent, 1998). The wild-type version of the gene and 5′ regulatory regions were cloned by PCR amplification of genomic DNA. The EDE1 genomic sequence and open reading frame were amplified from Arabidopsis thaliana Col-0 genomic DNA and cDNA using the following primers pairs: EDE1-ATG (5′-ATGGAGGCGAGAATCGGCCGATC-3′) and EDE1-stop (5′-TCAAACAGAAGTTGTGCACTC TTG-3′); or EDE1-5′-GUS (5′-GGCAATCAAATTTCTTCGAA-3′) and EDE1-3-GUS (5′-GTTGCGTCACGCGAAGCTTC-3′), each containing attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTAT- 3′) or attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTC- 3′) recombination sequences (Invitrogen), respectively, as adapter sites at the 5′ end, according to the manufacturer's instructions.
Characterization of the ede1-2 and ede1-3 Alleles and Segregation Analysis
The presence of the T-DNA insertions in ede1-2 and ede1-3 was confirmed by PCR using the T-DNA left border primer 5′-TGGTTCACGTAGTGGGCCATCG-3′ and the At2g44190 gene-specific primer EDE1-5, 5′-CTTCTTTGTATCGGCTTGAATCTTCG-3′, for SALK line 047950 (ede1-2) and EDE1-4, 5′-TTTGAAGCGACAACAAGTGCTTCTGC-3′, for SALK line 048191 (ede1-3). For self-crossing analysis, heterozygous plants were allowed to self-pollinate, and progeny seed was analyzed. For reciprocal cross analysis, mutant plants were crossed among themselves (Table 2) or with the wild type as indicated in Table 1. In all cases, progeny seeds were assessed phenotypically in the silique.
Expression of GFP-EDE1 Fusions in Tobacco and Arabidopsis Cell Cultures
Expression of GFP-EDE1 driven by the EDE1 promoter was obtained using the Multisite Gateway Three Fragment System (Invitrogen) according to the manufacturer's instructions. Briefly, a DNA fragment containing 800 bp of the 5′ region of EDE1 leading to the ATG translation start codon was amplified from genomic DNA with the following primers: 5′-GGGGACAACTTTGTATAGAAAAGTTGCAAGAACACACGAAAGAGACCAAAG-3′ and 5′-GGGGACTGCTTTTTTGTACAAACTTGTTCAATCAAATTTCTTCGAAATTGAC-3′and cloned into pDONR P4-P1R. GFP 2.5 was amplified from pDEST02 with the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGAGTAAAGGAGAAGAACTTTTCACTGG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTATTTGTATAGTTCATCCATGCCATGTG -3′ and cloned into pDONR221. The genomic sequence of EDE1 was amplified from genomic DNA with the primers 5′-GGGGACAGCTTTCTTGTACAAAGTGGAAATGGAGGCGAGAATCGGCCGATC-3′ and 5′-GGGGACAACTTTGTATAATAAAGTTGATCAAACAGAAGTTGTGCACTCTTGCTGATG-3′ and cloned into pDONR P2R-P3. All primers contained the respective attB recombination sequences (Invitrogen) as adapter sites at the 5′ end, according to the manufacturer's instructions. LR Clonase mix (Invitrogen; for recombination of attL sites with attR sites) was used to insert each DNA fragment into pMULTISITE GW Vector. Electrocompetent Agrobacterium cells (strain LBA4404.pBBR1MCSvirGN54D) were transformed with EDE1 promoter:GFP-EDE1, and BY-2 tobacco (Nicotiana tabacum) and Arabidopsis cells were transformed as previously described (An, 1985). Transformed cells were treated with taxol (10 μM; Sigma-Aldrich) or oryzalin (10 μM; Chem Service) for 12 h prior to microscopy observation.
Fluorescent and phase-contrast images of 3-d-old BY-2 and Arabidopsis suspension cells were recorded using a ×60 oil immersion objective on a Nikon E600 equipped with a Hamamatsu Orca CCD camera and Metamorph image software. Image stacks were processed using ImageJ (http://rsb.info.nih.gov/ij/download.html) and figures prepared in Adobe Photoshop
Expression of EDE1-GUS Fusions in Arabidopsis Plants
PCR products produced with the primers EDE1-5-GUS and EDE1-3-GUS (sequences indicated above) were cloned into the Entry vector pDONR 207 via the BP reaction that allows for recombination between attB and attP sites and to destination vector pBGWFS7 (gift from Ben Trewaskis, Max Planck Institute, Germany) according to the manufacturer's instructions (Invitrogen). Electrocompetent Agrobacterium cells (strain GV3101) were transformed with the EDE1-GUS plasmids and Arabidopsis plants transformed using the floral dip method (Clough and Bent, 1998). Ovules were incubated overnight in GUS-staining solution (80 mM NaPO4, pH 7.0, 0.4 mM K-ferrocyanide, 8 mM EDTA, 0.05% Triton X-100, and 0.8 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) at room temperature. Chlorophyll was then removed with repeated washes with 95% ethanol and samples observed with a Nikon E800 microscope and recorded using Viewfinder 3.0.1 image software (Pixera).
Microscopy Examination of Developing Seeds
For Feulgen staining, siliques at 6 DAP were fixed with ethanol/acetic acid, (3:1 [v/v]) for 16 h and rinsed three times with distilled water for 15 min each. They were treated with 5 n HCl for 1 h, rinsed three times for 5 min with distilled water, and stained with Schiffs Reagent (Sigma-Aldrich) for 3 h. The siliques were rinsed three times with cold (4°C) distilled water and washed in 70% (v/v) and then 95% (v/v) ethanol for 10 min and then three times 10 min in 100% (v/v) ethanol. They were repeatedly washed in 100% (v/v) ethanol for >1 h, until the ethanol remained colorless. The samples were incubated for 1 h in ethanol/LR White (London Resin) (1:1 [v/v]) and 16 h in pure LR White. The siliques were dissected on a slide, in a drop of fresh LR White under a dissecting microscope, and baked at 60°C overnight. The material was viewed using a Leica TCS NT/SP microscope (Leica Microsystems) with ×20 and ×63 water immersion lenses using wide Fluorescein isothiocyanate (FITC) settings (argon ion laser excitation at 488 nm, emission viewed at 520 nm). For DAPI staining, seeds were excised from siliques and placed in small glass vials with a solution containing 4% (v/v) paraformaldehyde, 25 mM PIPES (pH 6.9, H2SO4), 2.5 mM MgSO4, and 2.5 mM EDTA) and then subjected to vacuum infiltration. The fixative was replaced and the tissue left overnight at 4°C. The fixative was replaced with 0.85% (w/v) saline and the seeds left on ice for 30 min. The seeds were then dehydrated at 4°C with solutions containing increasing amounts of ethanol for 90 min each (50, 70, 85, 95, and 100% [v/v]). The ethanol was replaced with 50% ethanol:50% Histoclear (v/v) for 60 min at room temperature, 100% (v/v) Histoclear for another 60 min at room temperature, and then twice more with 100% Histoclear, at room temperature for 60 min each. Finally, the seeds were placed into 50% Histoclear/50% paraffin wax (v/v) and kept overnight at 50°C. The samples were subsequently kept at 60°C and the wax changed every morning and evening for the following 3 d. A few seeds were placed in a suitably sized warm mold, followed by fresh wax. The wax blocks were sectioned to 8 μm. The sections were dewaxed and stained in the dark with DAPI (1 μM/mL) for 20 to 30 min and then washed with water. Images were taken using a Bio-Rad confocal scanning laser microscope with a ×40 oil immersion lens and analyzed on an Apple Macintosh using the program NIH image (National Institutes of Health).
For seed clearing, wild-type and mutant siliques were collected at different stages of development (measured as DAP) and immersed in Carnoy's fixative (3 parts 95% ethanol to 1 part glacial acetic acid [v/v]) overnight at 4°C. After several changes of 100% (v/v) ethanol, siliques were hydrated in ethanol series and finally the material was transferred to water. Seeds were excised from siliques under a dissecting microscope and immersed in Hoyers solution (7.5 g gum Arabic, 100 g chloral hydrate, and 5 mL glycerol in 30 mL water). Cleared seeds were examined with a Nikon Microphot-SA equipped with Nomarski optics and supplied with Nikon Coolpix 990 digital camera. For light microscopy of developing embryos, siliques were fixed in 4% (w/v) paraformaldehyde in phosphate buffer (PBS) at room temperature under vacuum for 1 h and then overnight at 4°C. Samples were washed three times with PBS, dehydrated in ethanol series, and gradually infiltrated in LR White resin (Electron Microscopy Sciences), ending with three changes of pure resin. Samples were polymerized for 16 h at 60°C, sectioned at 50-nm thickness on a EMUC6 ultramicrotome (Leica Microsystems), and stained with toluidine blue (Sigma-Aldrich).
Immunoflorescence Localization
For endosperm immunofluorescence, siliques were fixed in 4% (w/v) paraformaldehyde in microtubule-stabilizing buffer/DMSO (MTSB; 50 mM PIPES, 5 mM EGTA, 5 mM MgCl2, and 5% DMSO, pH 6.7 to 7.0) at room temperature under vacuum for 1 h and then overnight at 4°C, according to Brown and Lemmon (1995). Fixed siliques were mounted to specimen holders and sectioned at 30 μm with a Vibratome series 1000 (Warner Instruments). Sections of ovules were processed as described in Lauber et al. (1997). Briefly, cell walls were partially digested with 2% (w/v) driselase (Sigma-Aldrich) for 30 min, and the plasma membrane was permeabilized with 1% (v/v) Triton X-100 in 10% (v/v) DMSO-MTSB for 30 min at room temperature. After blocking with 1% (w/v) BSA in MTSB for 1 h at room temperature, antitubulin YOL1/34 (Oxford Biosciences) was diluted 1:50 in 3% (w/v) BSA in MTSB and incubated overnight at 4°C. After washing in MTSB, anti-rat Fluorescein isothiocyanate (FITC)-conjugated secondary antibody (Sigma-Aldrich) was used at 1:300 dilution in 3% (w/v) BSA in MTSB. DNA was stained with 1 μg/mL DAPI (Sigma-Aldrich), samples were mounted in Citifluor (Amersham), and confocal laser scanning was performed using a Leica SP2 DM IRB inverted microscope (Leica Microsystems) with a ×60 oil immersion objective.
Tobacco BY-2 cells were fixed for 30 min in PME buffer (50 mM PIPES, 5 mM EGTA, and 5 mM MgSO4, pH 7.0) plus 0.025 M sorbitol (PMES), containing 4% (w/v) paraformaldehyde and labeled with 1:50 antitubulin YOL1/34, as previously described (Chan et al., 2003). Images were recorded using a ×60 oil immersion objective on a Nikon E600 equipped with a Hamamatsu Orca CCD camera and Metamorph image software. All image stacks were processed using ImageJ (http://rsb.info.nih.gov/ij/download.html) and figures prepared in Adobe Photoshop.
Expression Analysis
RNA was isolated from different tissues using the Plant RNeasy extraction kit (Qiagen). One microgram of RNA was treated with 10 units of RNase free DNaseI (Amersham Biosciences) for 10 min at 37°C and reverse transcribed using the Superscript III Reverse Transcriptase (Invitrogen) and oligo(dT)12-18 according to the manufacturer's instructions. RT-PCR was performed using the following EDE1-specific primers: EDE1-F1101, 5′-TTTGTTGAATGCTTCATTGCGGTTGCC-3′, and EDE1-R1425, 5′-TCAAACAGAAGTTGTGCACTCTTGCTG-3′. The actin gene was used as a control using the following primers: 5′-CGCGAAAAGATGACTCAAATC-3′ and 5′-AGATCCTTTCTGATATCCACG-3′.
In Situ Hybridization
Tissue was fixed and processed as described by Drea et al. (2005), with the following modifications: tissue was fixed in ethanol:acetic acid:formalin (50:5:10 [v/v]), the slides were processed in the slide processor InsituPro VS Intavis, including the hybridization step-up to the signal detection, and the signal was developed using Western-Blue Stabilized Substrate. Full-length EDE1 and CYCLIN B were amplified from Arabidopsis seedling cDNA using EDE1-ATG and EDE1-stop primers (indicated above) and CYC-5 (5′-ATGGCGACAGGACCAGTTGTTCATC-3′) and CYC-3 (5′-TCATGGAGCAGATGACATAAGAGAC-3′) primers. The PCR products were cloned into pGEM-T vector (Promega) according to the manufacturer's instructions. EDE1 and CYCLIN B cDNAs were then amplified from pGEM vectors with universal forward and reverse primers for subsequent transcription with T7 RNAP. PCR reactions were performed with the following cycle: 94°C for 3 min, then 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1.5 min, with a final extension of 72°C for 6 min. In vitro transcription was performed in 10-μL reactions for 2 h at 37°C in the presence of digoxigenin-UTP nucleotides (0.35 mM). Hydrolysis was performed immediately in 100 mM carbonate buffer, pH 10.2, at 60°C for 30 min, and products precipitated in 2.5 M ammonium acetate and 3 volumes of absolute ethanol for 1 h at 4°C. Pellets were resuspended in 30 μL TE (100 mM Tris and 10 mM EDTA) buffer. Dilutions (100×) were made in water, and 1 μL of each spotted on nitrocellulose for dot-blot: 30 min in blocking solution (Sigma-Aldrich), 30 min in anti-DIG-alkaline phosphatase (Roche); 5 min wash in TBS (10 mM Tris and 250 mM NaCl); 5 min in AP buffer (100 mM Tris, 100 mM NaCl, pH 9.5, and 50 mM MgCl2), and developed as described above until signal was sufficient. All probes were then diluted 100-fold in hybridization solution (300 mM NaCl, 10 mM Tris, pH 6.8, 10 mM NaH2PO4, 5 mM EDTA, 50% [v/v] formamide, 5% [w/v] dextran sulfate, 0.5 mg/mL tRNA, 1× Denhardt's, and 0.1 mg/mL salmon testis DNA) and maintained stably at −20°C until hybridization. Images were captured with a Nikon E800 microscope and recorded using Viewfinder 3.0.1 image software (Pixera).
Microtubule Cosedimentiation Assays
Microtubules were obtained by incubating 5 mg/mL bovine brain tubulin (Cytoskeleton) in GTB buffer (80 mM PIPES, pH 6.9, 2 mM Mg Cl2, and 0.5 mM EGTA) with 30% (v/v) glycerol, 1 mM GTP, and 20 μM paclitaxel (Sigma-Aldrich) for 40 min at 37°C. Full-length EDE1 cDNA was amplified using the primers EDE1-ATG and EDE1-stop (indicated above) and cloned into the Gateway Entry vector pDONR 207 (Invitrogen) via the BP reaction according to the manufacturer's instructions. Using the LR recombination reaction, EDE1 cDNA was then transferred from pDONR 207 into pDEST14 (Invitrogen) and used for in vitro translation with the T7-TNT rabbit reticulocyte lysate system (Promega). Twenty-five microliters of in vitro–translated [35S] methionine-labeled EDE1 was diluted in 200 μL of GTB buffer containing complete protease inhibitor mixture (Roche) and spun at 16,000g for 1 h at 4°C. One hundred microliters of supernatant was incubated with or without 50 μg of microtubules and 20 μM paclitaxel for 30 min at 37°C. The microtubule and EDE1 mixtures were layered over 1 mL of 15% (w/v) sucrose in GTB buffer and spun at 16,000g for 30 min at 4°C. Pellets were analyzed for the presence of radiolabeled EDE1 by SDS-PAGE followed by autoradiography. Equal loading of the gel was verified by Coomassie Brilliant Blue staining. A control protein, known not to associate with microtubules (At5g16050), was amplified from cDNA using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATATGTCTTCTGATTCGTCCCGGGAAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGTGGGTCTCACTGCGAAGGTGGTGGTTGGGC-3′ and used as a control in all experiments.
Bioinformatics
Alignments were performed with ClustalW using default settings (www.ebi.ac.uk/clustalx), and phylogenetic trees were generated using MEGA version 4 (Tamura et al., 2007) with bootstrap values from a minimum of 1000 trials. Protein domains were predicted using ELM (elm.eu.org) and ScanProsite (expasy.org/tools/scanprosite/).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At2g44190, At3g60000, At1g49890, At3g19570, At2g24070, At4g30710, At2g20815, At5g16050, At3g11520, NM_001055852, NM_001054910, NM_001057898, NM_001068449, NM_001069314, NM_001069743, NM_001071876, PhP_100775, PhP_172159, PhP_82152, and PhP_172191.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Embryo-Associated Endosperm in ede1-1 Seeds with Moderate Phenotype.
Supplemental Figure 2. Phenotype of the ede1-3 Allele.
Supplemental Figure 3. Embryo Development in ede1-2 and ede1-3 Alleles.
Supplemental Figure 4. Expression of CYCLIN B in Developing Seeds of Arabidopsis.
Supplemental Figure 5. GFP-EDE1 Expression in Ovules.
Supplemental Figure 6. Localization of GFP-EDE1 in Transiently Transformed Arabidopsis Cells.
Supplemental Figure 7. Optical Sections of Two Transiently Transformed Arabidopsis Mitotic Cells Treated with Taxol.
Supplemental Figure 8. Time-Lapse Series of GFP-EDE1 Dynamics during Phragmoplast Expansion in Stable Tobacco BY2 Cells.
Supplemental Table 1. Coresponse of Transcription of Genes with EDE1.
Supplemental Table 2. GFP-EDE1 Expressed under EDE1's Own Promoter Complements the ede1-1 Mutation.
Supplemental Table 3. SNPs Developed and Used in Mapping ede1-1 to BAC F6E13 and F4l1.
Supplemental Data Set 1. Alignment Used to Generate the Phylogenetic Tree in Figure 5B.
Supplementary Material
Acknowledgments
We thank Henrik Buschmann (John Innes Centre) for useful advice and discussions, Grant Calder, Kim Findlay, and Susan Bunnewell (John Innes Centre) for their help and support with microscopy analysis, Roy Brown (University of Louisiana) for his advice on endosperm techniques, and the horticultural John Innes Centre team. This work was supported by the Biotechnology and Biological Science Research Council through its Core Strategic Grant to John Innes Centre and Project Grant BB/D52189X/1, Syngenta UK, a Marie Curie Research Training Network (RTN) Grant (“SY-stem” MRTN-CT-2004-005336), and the European Commission Framework Programme 6 (Integrated Project “AGRON-OMICS” LSHG-CT-2006-037704).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: John H. Doonan (john.doonan@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Ambrose, J.C., Shoji, T., Kotzer, A.M., Pighin, J.A., and Wasteneys, G.O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, G. (1985). High efficiency transformation of cultured tobacco cells. Plant Physiol. 79 568–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad, F.F., Mayer, U., Wanner, G., and Jürgens, G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253 267–277. [DOI] [PubMed] [Google Scholar]
- Berger, F. (2003). Endosperm: The crossroad of seed development. Curr. Opin. Plant Biol. 6 42–50. [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that sncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.C., and Lemmon, B.E. (1995). Methods in plant immunolight microscopy. Methods Cell Biol. 49 85–107. [DOI] [PubMed] [Google Scholar]
- Brown, R.C., and Lemmon, B.E. (2001). Phragmoplasts in the absence of nuclear division. J. Plant Growth Regul. 20 151–161. [Google Scholar]
- Brown, R.C., Lemmon, B.E., and Nguyen, H. (2003). Events during the first four rounds of mitosis establish three developmental domains in the syncytial endosperm of Arabidopsis thaliana. Protoplasma 222 167–174. [DOI] [PubMed] [Google Scholar]
- Brown, R.C., Lemmon, B.E., Nguyen, H., and Olsen, O.A. (1999). Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12 32–42. [Google Scholar]
- Buschmann, H., Chan, J., Sanchez-Pulido, L., Andrade-Navarro, M.A., Doonan, J.H., and Lloyd, C.W. (2006). Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr. Biol. 16 1938–1943. [DOI] [PubMed] [Google Scholar]
- Buschmann, H., Fabri, C.O., Hauptmann, M., Hutzler, P., Laux, T., Lloyd, C.W., and Schaffner, A.R. (2004). Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr. Biol. 14 1515–1521. [DOI] [PubMed] [Google Scholar]
- Carleton, M., Mao, M., Biery, M., Warrener, P., Kim, S., Buser, C., Marshall, C.G., Fernandes, C., Annis, J., and Linsey, P.S. (2006). RNA interference-mediated silencing of mitotic kinesis KIF14 disrupts cell cycle progression and induces cytokinesis failure. Mol. Cell. Biol. 26 3853–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Calder, G.M., Doonan, J.H., and Lloyd, C.W. (2003). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5 967–971. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Costa, L.M., Gutierrez-Marcos, J.F., and Dickinson, H.G. (2004). More than a yolk: The short life and complex times of the plant endosperm. Trends Plant Sci. 9 507–514. [DOI] [PubMed] [Google Scholar]
- Dickinson, H. (2003). Plant cell cycle: Cellularization of the endosperm needs Spätzle. Curr. Biol. 13 R146–R148. [DOI] [PubMed] [Google Scholar]
- Dörmann, P., Hoffmann-Benning, S., Balbo, I., and Benning, C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea, S., Corsar, J., Crawford, B., Shaw, P., Dolan, L., and Doonan, J.H. (2005). A streamlined method for systematic, high resolution in situ analysis of mRNA distribution in plants. Plant Methods 1 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert, P.R., Coen, E.S., Murphy, G.J., and Doonan, J.H. (1994). Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 13 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert, P.R., Gaudin, V., Lunness, P., Coen, E.S., and Doonan, J.H. (1996). Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell 8 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks, N., and Benning, C. (1998). wrinkled1: A novel, low-seed-Oil mutant of arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean, O., Vernoux, T., Laufs, P., Belcram, K., Mizukami, Y., and Traas, J. (2004). In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb-group gene in Arabidopsis. Science 280 446–450. [DOI] [PubMed] [Google Scholar]
- Hauser, M.T., and Bauer, E. (2000). Histochemical analysis of root meristem activity in Arabidopsis thaliana using a cyclin:GUS (beta-glucuronidase) marker line. Plant Soil 226 1–10. [Google Scholar]
- Hussey, P.J., Hawkins, T.J., Igarashi, H., Kaloriti, D., and Smertenko, A. (2002). The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 50 915–924. [DOI] [PubMed] [Google Scholar]
- Ito, M., Araki, S., Matsunaga, S., Itoh, T., Nishihama, R., Machida, Y., Doonan, J.H., and Watanabe, A. (2001). G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, M., Iwase, M., Kodama, H., Lavisse, P., Komamine, A., Nishihama, R., Machida, Y., and Watanabe, A. (1998). A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang, Y.L., Huang, J., Peters, J.M., McLaughlin, M.E., Tai, C.Y., and Pellman, D. (1997). APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275 1311–1314. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of Tl-DNA Gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Korolev, A.V., Buschmann, H., Doonan, J.H., and Lloyd, C.W. (2007). AtMAP70-5, a divergent member of the MAP70 family of microtubule-associated proteins, is required for anisotropic cell growth in Arabidopsis. J. Cell Sci. 120 2241–2247. [DOI] [PubMed] [Google Scholar]
- Korolev, A.V., Chan, J., Naldrett, M.J., Doonan, J.H., and Lloyd, C.W. (2005). Identification of a novel family of 70 kDa microtubule-associated proteins in Arabidopsis cells. Plant J. 42 547–555. [DOI] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jurgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., Joshi, H.C., Wilson, T.J., Silflow, C.D., Palevitz, B.A., and Snustad, D.P. (1994). γ-Tubulin in Arabidopsis: Gene sequence, immunoblot, and immunofluorescence studies. Plant Cell 6 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.M., McElver, J., Tzafrir, I., Joosen, R., Wittich, P., Patton, D., Van Lammeren, A.A.M., and Meinke, D. (2002). Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 29 405–415. [DOI] [PubMed] [Google Scholar]
- Liu, C.M., and Meinke, D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16 21–31. [DOI] [PubMed] [Google Scholar]
- Lloyd, C., and Chan, J. (2006). Not so divided: The common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 7 147–152. [DOI] [PubMed] [Google Scholar]
- Lloyd, C., and Hussey, P. (2001). Microtubule-associated proteins in plants–Why we need a MAP. Nat. Rev. Mol. Cell Biol. 2 40–47. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jürgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84 61–71. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., Srinivas, B.P., and Hulskamp, M. (2003). A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr. Biol. 13 1991–1997. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Herzog, U., Berger, F., Inze, D., and Jürgens, G. (1999). Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur. J. Cell Biol. 78 100–108. [DOI] [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A.H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Murata, T., Sonobe, S., Baskin, T.I., Hyodo, S., Hasezawa, S., Nagata, T., Horio, T., and Hasebe, M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7 961–968. [DOI] [PubMed] [Google Scholar]
- Nacry, P., Mayer, U., and Jürgens, G. (2000). Genetic dissection of cytokinesis. Plant Mol. Biol. 43 719–733. [DOI] [PubMed] [Google Scholar]
- Nakajima, K., Furutani, I., Tachimoto, H., Matsubara, H., and Hashimoto, T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama, R., Soyano, T., Ishikawa, M., Araki, S., Tanaka, H., Asada, T., Irie, K., Ito, M., Terada, M., Banno, H., Yamazaki, Y., and Machida, Y. (2002). Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109 87–99. [DOI] [PubMed] [Google Scholar]
- Olsen, O.A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S., Rylott, E.L., Gilday, A.D., Graham, S., Larson, T.R., and Graham, I.A. (2004). Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16 2705–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin, R.M., Wang, Y., Yuen, C.Y., Will, J., and Masson, P.H. (2007). WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 49 961–971. [DOI] [PubMed] [Google Scholar]
- Pitt, C.W., Moreau, E., Lunness, P.A., and Doonan, J.H. (2004). The pot1(+) homologue in Aspergillus nidulans is required for ordering mitotic events. J. Cell Sci. 117 199–209. [DOI] [PubMed] [Google Scholar]
- Scott, R., Spielman, M., Bailey, J., and Dickinson, H. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341. [DOI] [PubMed] [Google Scholar]
- Seki, A., and Fang, G. (2007). CKAP2 is a spindle-associated protein degraded by APC/C-Cdh1 during mitotic exit. J. Biol. Chem. 282 15103–15113. [DOI] [PubMed] [Google Scholar]
- Sorensen, M.B., Mayer, U., Lukowitz, W., Robert, H., Chambrier, P., Jurgens, G., Somerville, C., Lepiniec, L., and Berger, F. (2002). Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129 5567–5576. [DOI] [PubMed] [Google Scholar]
- Steinborn, K., Maulbetsch, C., Priester, B., Trautmann, S., Pacher, T., Geiges, B., Kuttner, F., Lepiniec, L., Stierhof, Y.-D., Schwarz, H., Jurgens, G., and Mayer, U. (2002). The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 16 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen, G., El Kasmi, F., Richter, S., Lukowitz, W., Assaad, F.F., Juergens, G., and Mayer, U. (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell-cycle dependent manner. Curr. Biol. 12 153–158. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Ne, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tzafrir, I., McElver, J.A., Liu, C.-m., Yang, L.J., Wu, J.Q., Martinez, A., Patton, D.A., and Meinke, D.W. (2002). Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128 38–51. [PMC free article] [PubMed] [Google Scholar]
- Van Damme, D., Van Poucke, K., Boutant, E., Ritzenthaler, C., Inze, D., and Geelen, D. (2004). In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136 3956–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.P., Pereira, A., and Stiekema, W.J. (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 4 288–290. [DOI] [PubMed] [Google Scholar]
- Walker, K.L., Müller, S., Moss, D., Ehrhardt, D.W., and Smith, L.G. (2007). Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 6 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Zhy, L., Liu, B., Wang, C., Jin, L., Zhao, Q., and Yuan, M. (2007). Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in destabilising cortical microtubules. Plant Cell 19 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury, E.L., and Morgan, D.O. (2007). Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9 106–112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.