Abstract
Mutations in the genes encoding for either the biosynthetic or transcriptional regulation of the anthocyanin pathway have been linked to color phenotypes. Generally, this is a loss of function resulting in a reduction or a change in the distribution of anthocyanin. Here, we describe a rearrangement in the upstream regulatory region of the gene encoding an apple (Malus × domestica) anthocyanin-regulating transcription factor, MYB10. We show that this modification is responsible for increasing the level of anthocyanin throughout the plant to produce a striking phenotype that includes red foliage and red fruit flesh. This rearrangement is a series of multiple repeats, forming a minisatellite-like structure that comprises five direct tandem repeats of a 23-bp sequence. This MYB10 rearrangement is present in all the red foliage apple varieties and species tested but in none of the white fleshed varieties. Transient assays demonstrated that the 23-bp sequence motif is a target of the MYB10 protein itself, and the number of repeat units correlates with an increase in transactivation by MYB10 protein. We show that the repeat motif is capable of binding MYB10 protein in electrophoretic mobility shift assays. Taken together, these results indicate that an allelic rearrangement in the promoter of MYB10 has generated an autoregulatory locus, and this autoregulation is sufficient to account for the increase in MYB10 transcript levels and subsequent ectopic accumulation of anthocyanins throughout the plant.
INTRODUCTION
Antioxidants are a key component of the health properties of fruit, and there is mounting epidemiological evidence that compounds such as flavonoids and anthocyanins can significantly reduce the incidence of chronic illnesses when part of a regular healthy diet (Knekt et al., 2002). It has been proposed that the combination of phytochemicals in fresh apple (Malus × domestica), including flavonoids and anthocyanins, produces the strong antioxidant activity essential to maintain good health (Eberhardt et al., 2000), and there has been a surge of interest in varieties with higher levels of antioxidants, such as phenolics. Recently, genetically engineered tomatoes (Solanum lycopersicum) showing high levels of anthocyanins have been shown to increase the longevity of cancer-susceptible mice (Butelli et al., 2008), while a study using anthocyanin-rich maize (Zea mays) showed an increase in protection against ischemia-reperfusion injury in rats (Toufektsian et al., 2008).
The fruit flesh (cortex) of most apple varieties is white or off-white in color. The skin is usually green or red, with the red anthocyanins accumulating in response to developmental, hormonal, and light signals (Ubi et al., 2006). However, there are a number of high anthocyanin, red-fleshed apple varieties originating from the wild-apple forests of Central Asia, including Malus pumila var Niedzwetzkyana (Harris et al., 2002) and Malus × domestica ‘Red Field’ Open Pollinated (OP) (‘Red Field’; Espley et al., 2007). These high anthocyanin varieties possess a dramatic phenotype with highly pigmented vegetative, floral, and fruit tissues.
Many steps in the plant anthocyanin pathway have been described through analysis of natural mutants with several examples of biosynthetic gene-related mutations leading to phenotypic changes. In particular, studies on flower color polymorphism have shown that mutations can determine flower color. However, these are mostly caused by the insertion of transposons into genes or deletion of transposons from genes, such as the anthocyanin structural genes chalcone synthase (Habu et al., 1998), dihydroflavonol 4-reductase (Inagaki et al., 1994), and anthocyanin synthase (also know as leucoanthocyanidin dioxygenase; Hisatomi et al., 1997).
There are many examples of regulation of anthocyanin biosynthesis by MYB transcription factors in diverse plant species (Allan et al., 2008). Small changes to these MYB proteins can have a marked effect on phenotype (Schwinn et al., 2006). Disruption of MYB gene expression can result in more severe phenotypes. For example, in grape (Vitis vinifera), a retrotransposon-induced mutation in the promoter region of mybA1 leads to a loss of anthocyanin accumulation in berry skin (Kobayashi et al., 2004), although further investigation has revealed that multiple mutations in an adjacent MYB gene also account for the difference in grape berry color (Walker et al., 2007). Mutations in genes encoding another major family of anthocyanin regulators, the basic helix-loop-helix (bHLH) transcription factors, also produce anthocyanin-related phenotypic changes. These include the Rc mutation in rice (Oryza sativa), which accounts for the white pericarp of most rice varieties (Sweeney et al., 2007). These characterized anthocyanin regulator mutations result in a loss or a restricted distribution of anthocyanin. In some instances within the phenylpropanoid pathway, mutations can result in gain of function. TT2 (a MYB transcription factor), TT8 (a bHLH transcription factor), and TTG1 (contains a WD40 domain) directly regulate both BANYULS, encoding a core enzyme in proanthocyanidin biosynthesis, and TT8 expression at the transcriptional level in a self-activated feedback loop (Debeaujon et al., 2003; Baudry et al., 2004, 2006).
Recently, there have been a number of studies of the transcriptional regulation of the anthocyanin pathway in apple (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007). Two genes, MYB1 (Takos et al., 2006) and MYBA (Ban et al., 2007), have been described as being responsible for apple skin color, while we previously demonstrated that the MYB10 gene regulated flesh color (Espley et al., 2007). MYB1, MYB10, and MYBA share at least 98% identity at the nucleotide residue level. At the deduced amino acid level, MYB1 and MYBA are identical and differ from MYB10 in three amino acids (Ban et al., 2007), but it is not clear whether these genes are alleles or tightly linked genes.
In the anthocyanin-accumulating red-fleshed variety ‘Red Field,’ anthocyanin levels are regulated by MYB10, with elevation of transcript levels of MYB10 correlated with higher levels of the anthocyanin biosynthetic gene transcripts (Espley et al., 2007). MYB10 has also been shown to cosegregate with the red flesh and foliage phenotype (Chagné et al., 2007), suggesting that the Rni locus that described the red flesh phenotype may be an allelic variant at the MYB10 locus. Overexpression of MYB10 in a white-fleshed, green-leaved apple variety results in transgenic plants with a red phenotype (Espley et al., 2007). We found that the white-fleshed ‘Pacific Rose’ and red-fleshed ‘Red Field’ varieties carry MYB10 alleles encoding identical proteins (Espley et al. 2007), suggesting that differences in transcriptional regulation of MYB10 are responsible for the ectopic accumulation of anthocyanin in ‘Red Field.’
Here, we report a 23-bp repeat motif in the upstream regulatory region of alleles of MYB10 found only in red-fleshed apples. This allele is autoregulatory; the MYB10 protein is able to bind and transactivate its own promoter, leading to an increase in transcript levels of MYB10 and the level of anthocyanin throughout the plant.
RESULTS
Isolation of the Upstream Regulatory Region of MYB10
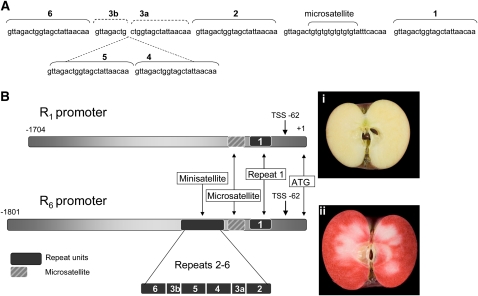
To investigate further the molecular basis of the red-fleshed phenotype, we used genome walking to isolate the upstream regulatory sequence of MYB10. This sequence was cloned from both white-fleshed apple varieties, Malus × domestica ‘Sciros’ (‘Pacific Rose’), Malus × domestica ‘Granny Smith,’ and Malus × domestica ‘Royal Gala’ and red-fleshed Malus × domestica ‘Red Field’ open pollinated (a cross between ‘Wolf River’ and Malus pumila var Niedzwetzkyana). Apples are outcrossing, and as a result they are highly heterozygous. Our isolated DNA fragments revealed two sequences of different sizes for the same region. One sequence was present in all four varieties and showed limited variation between varieties. As an example, a comparison of the sequence from ‘Granny Smith’ with ‘Pacific Rose’ revealed six single nucleotide polymorphisms (SNPs), five single base deletions, and four single base insertions over the 1700 bp of promoter analyzed. The other fragment contained an insertion of ∼100 bp. This latter version was only found in the promoter region of the red-fleshed, red-foliaged variety Malus × domestica ‘Red Field’ OP. The insertion consisted of a 23-bp sequence, duplicated in a complex pattern of five near-perfect tandem repeats (Figure 1A). We define this duplication as a minisatellite based on the size of the duplicated units. A single version of the 23-bp sequence is found in all varieties tested (both red and white-fleshed) located ∼30 bases downstream of the minisatellite insertion site. Between this potential donor sequence and the minisatellite lies a dinucleotide microsatellite. Figure 1B shows a schematic of these elements in both the promoters isolated from white-fleshed varieties with one repeat unit (R1) and those from red-fleshed varieties with six repeat units (R6). Sequence alignment of the longest cDNA clones identified through 5′ rapid amplification of cDNA ends (Espley et al., 2007) with genomic sequence locates a transcription start site at the G nucleotide at position −62 from the translational start ATG (indicated by an arrow in Figure 1B).
Figure 1.
Sequence and Schematic of the Minisatellite Region in the MYB10 Promoter in Red-Fleshed Apple Varieties.
(A) Relative positions of the repeat units in the R6 promoter allele of the MYB10 gene. Unit 1 lies downstream of the microsatellite and is predicted to be the origin of the repeat units in the minisatellite. Units 2 to 5 form the minisatellite and are found only in the MYB10 promoter from red-fleshed varieties. They are near-perfect copies of unit 1 except that unit 3 is dissected by units 4 and 5.
(B) Schematic of the differences between the R1 (found in both white- and red-fleshed apple varieties) and the R6 promoter showing positions of the ATG translation start site, repeat unit 1, the microsatellite, and the minisatellite in R6. To the right of the schematics are representative phenotypes: (i) the white-fleshed Malus × domestica ‘Sciros’ (Pacific Rose) and (ii) the red-fleshed Malus × domestica ‘Red Field’ OP. The predicted transcription start site (TSS) is indicated by an arrow at position −62 relative to the ATG start site.
A Minisatellite Is Associated with the Ectopic Anthocyanin Phenotype
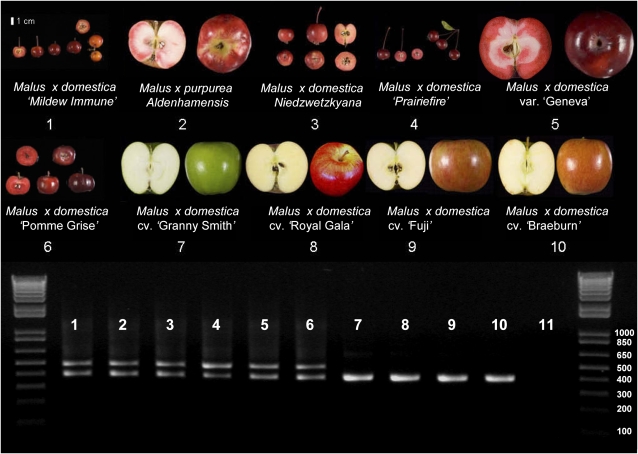
Previously, we showed that MYB10 cosegregates with Rni, a locus associated with red flesh and red foliage phenotypes in apple (Chagné et al., 2007). PCR amplification of the promoter region from red and white-fleshed varieties consistently showed that the R6 minisatellite repeat motif was amplified in all the plants with red-fleshed fruit (Figure 2). The genealogy of these apple varieties is detailed in the Supplemental Methods online. We determined the association of the minisatellite with the red-fleshed phenotype by sequencing the region encompassing the minisatellite over 19 apple varieties with diverse fruit phenotypes (11 red and eight white fleshed; Table 1). A number of sequence variations were found in the upstream region (e.g., SNPs at positions −612 and −245 from the ATG start site; Table 1), but only the minisatellite polymorphism is associated with the elevated accumulation of anthocyanins that causes red flesh and red foliage. The same region was PCR amplified from a further set of 77 apple varieties taken from two collections of Malus species, and in each case the product corresponding to the minisatellite motif was absent in the green foliage, white-fleshed varieties (see Supplemental Table 1 and Supplemental Figure 1 online). All the white-fleshed varieties tested contained only the R1 version, while the red-fleshed apple varieties contained both R1 and R6, or R6 only (Table 1).
Figure 2.
PCR Amplification of the MYB10 Promoter Region from 10 Apple Varieties.
PCR Amplification of the MYB10 promoter region gave two fragments: a 496-bp fragment corresponding to R6 that is present only in red-fleshed varieties (lanes 1 to 6) and is absent in white-fleshed varieties (lanes 7 to 10) and a 392-bp fragment corresponding to R1, present in both types. Images of fruit and corresponding gel lanes are as follows. 1, open-pollinated (OP) Malus × domestica ‘Mildew Immune Seedling’; 2, M. × purpurea ‘Aldenhamensis’; 3, M. × domestica var Niedzwetzkyana; 4, M. × domestica ‘Prairiefire’; 5, OP M. × domestica var ‘Geneva’; 6, OP M. × domestica ‘Pomme Grise’; 7, M. × domestica ‘Granny Smith’; 8, M. × domestica ‘Royal Gala’; 9, M. × domestica ‘Fuji’; 10, M. × domestica ‘Braeburn; 11, no template control.’
Table 1.
Association of the Minisatellite Motif with the Red-Fleshed Phenotype
| Variety | Flesh Color | G/T SNP Pos −612 | Minisatellite Motif | A/T SNP Pos −245 |
|---|---|---|---|---|
| Malus × ‘Babine’ | Red | G:G | R1:R6 | A:T |
| Malus × ‘Okanagan’a | Red | G:G | R1:R6 | A:T |
| Malus × ‘Simcoe’a | Red | G:G | R6:R6 | T:T |
| Malus × ‘Slocan’a | Red | G:T | R1:R6 | A:T |
| Malus × ‘Formosa’a | Red | G:T | R1:R6 | A:T |
| M. sieversii 629319a | Red | G:G | R6:R6 | T:T |
| M. sieversii FORM 35(33-01)a | Red | G:T | R1:R6 | A:T |
| M. sieversii 01P22a | Red | G:G | R6:R6 | T:T |
| M. sieversii 3563.qa | Red | G:G | R6:R6 | T:T |
| M. aldenhamii | Red | T:T | R1:R6 | A:T |
| Malus × domestica ‘Red Field’ OP | Red | G:T | R1:R6 | A:T |
| Malus × domestica ‘Close’ | White | G:T | R1:R1 | A:T |
| Malus × domestica ‘Mr Fitch’ | White | T:T | R1:R1 | A:A |
| Malus × domestica ‘Guldborg’ | White | G:T | R1:R1 | A:T |
| Malus × domestica ‘Alkmene’ | White | T:T | R1:R1 | A:A |
| Malus × domestica ‘Red Melba’ | White | T:T | R1:R1 | A:A |
| Malus × domestica ‘Rae Ime’ | White | G:G | R1:R1 | T:T |
| Malus × domestica ‘Lady Williams’ | White | T:T | R1:R1 | A:A |
| Malus × domestica ‘Granny Smith’ | White | G:T | R1:R1 | A:A |
| Association test (r2) | 0.185 | 1 | 0.491 |
Plant material and DNA samples supplied by Charles J. Simon and Philip L. Forsline (Agricultural Research Service, USDA).
A number of sequence variations were found in the promoter region, but only the minisatellite polymorphism was associated with the elevated accumulation of anthocyanins. All 11 red-fleshed varieties tested have the duplicated repeat motifs found in the MYB10 R6 promoter, while these were absent from the white-fleshed varieties. The positions shown are relative to the ATG translation start site.
Induction of Anthocyanin Pigmentation by R6:MYB10 in Tobacco and Apple
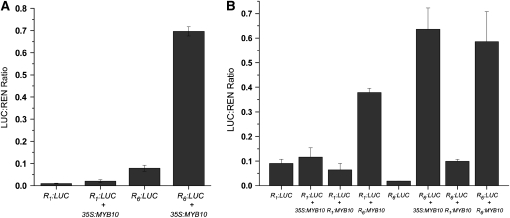
Previous studies have shown that when MYB10 was fused to the cauliflower mosaic virus 35S promoter (35S) and coinfiltrated into tobacco (Nicotiana tabacum) with a 35S:bHLH3 construct encoding a potential apple bHLH cofactor, a strong increase in anthocyanin pigment could be detected at the infiltration site (Espley et al., 2007). We therefore compared the level of anthocyanin that accumulated in tobacco leaves infiltrated with Agrobacterium tumefaciens suspensions with the MYB10 gene driven by either the R1 or R6 promoter sequences. Similar levels of anthocyanin were observed when either R6:MYB10 or 35S:MYB10 was coinfiltrated with 35S:bHLH3 (Figure 3A). However, we were unable to detect anthocyanin accumulation in leaves infiltrated with the R1:MYB10, both with and without 35S:bHLH3.
Figure 3.
The Native Apple Promoter Containing the Minisatellite Induces Ectopic Anthocyanin Accumulation.
(A) Red coloration around the infiltration site in the leaves of N. tabacum 8 d after transient transformation with (i) R6:MYB10 and (ii) 35S:MYB10 but not with (iii) R1:MYB10. All three patches were coinfiltrated with 35S:bHLH3.
(B) Photographs of regenerating ‘Royal Gala’ callus transformed with (i) R6:MYB10 and (ii) R1:MYB10. Red pigmentation was observed only on callus transformed with R6:MYB10. Emerging shoots showed a similar pigmented phenotype as shown for 35S:MYB10 in Espley et al. (2007).
(C) Representative R6:MYB10 plantlet (left), micrografted onto ‘M9’ rootstock shown next to ‘Royal Gala’ control (right). Pigmentation levels of the R6:MYB10 lines remained high.
(D) Leaves at same developmental stage taken from representative lines of (i) 35S:MYB10 (Espley et al., 2007), (ii) ‘Royal Gala’ control, and (iii) R6:MYB10.
To investigate the properties of the R6 promoter in apple, we transformed ‘Royal Gala’ (green leaves and white flesh) with MYB10 driven by the R6 promoter. While the R1 promoter is found in ‘Royal Gala,’ R6 is not (Figure 2). We have already shown that when ‘Royal Gala’ was transformed with 35S:MYB10, red callus is produced and regenerates to produce red plants (Espley et al., 2007). We observed a similar callus phenotype when ‘Royal Gala’ is transformed with R6:MYB10, with bright red areas on regenerating callus (Figure 3B), while no pigmentation was seen on regenerating apple callus transformed with R1:MYB10. Similarly, callus transformed with an empty vector cassette showed no pigmentation. To further study the development of the ‘Royal Gala’ R6:MYB10 transgenic lines, transformed plantlets were micrografted onto ‘M9’ rootstock, adapted from a previously described protocol (Lane et al., 2003). The grafted plants remained highly pigmented (Figure 3C), with a phenotype similar to the red foliaged, red-fleshed variety Malus × domestica ‘Red Field’ OP and to the previously described 35S:MYB10 transgenic lines (Espley et al., 2007) (Figure 3D). These results strongly suggest that the R6 promoter allele is responsible for the increased accumulation of anthocyanins in red-fleshed apple types.
Autoregulation of the MYB10 Promoter in the Dual Luciferase Transient Tobacco Assay
One possible result of the minisatellite insertion within the promoter of MYB10 is an increase in the basal activity of the promoter. A dual luciferase assay was used to quantify the activity of the two versions of the MYB10 promoter. R1 and R6 promoter sequences were fused to LUCIFERASE (LUC) and transactivation of the LUC gene measured relative to 35S:RENILLA (REN) by measurement of luminescence after transient expression in Nicotiana benthamiana. The R1 and R6 promoters showed little difference in activity, as determined by the ratio of luminescence produced by MYB10-promoter-LUC to 35S-REN (Figure 4A). However, when 35S:MYB10 was coinfiltrated with MYB10-promoter-LUC constructs, it transactivated the promoters. When 35S:MYB10 was coinfiltrated with R1:LUC, there was a slight elevation in transactivation (Figure 4A), while 35S:MYB10 transactivated R6:LUC >30-fold compared with background promoter activity. There was a sevenfold increase in the effect of 35S:MYB10 on R6:LUC compared with R1:LUC (0.696 ± se 0.02 compared with 0.098 ± se 0.001) (Figure 4A). This level of transactivation suggests that the presence of the repeat motifs in R6:LUC act as an enhancer of MYB10-induced transcription, resulting in the elevated LUC levels.
Figure 4.
Interaction of Native Apple Promoters and MYB10 in a Dual Luciferase Transient Tobacco Assay.
(A) Leaves of N. benthamiana were infiltrated with R1 and R6 promoter-LUC fusions on their own or coinfiltrated with 35S:MYB10, and then luminescence of LUC and REN was measured 3 d later and expressed as a ratio of LUC to REN.
(B) Comparison of the transactivation activity of R1 and R6 promoter-driven MYB10 to 35S:MYB10, when coinfiltrated with the R1 and R6 promoter- LUC fusions. The results provide a measure for the potential activity of the apple promoters, which shows a significant increase in the case of the R6-driven MYB10. Data in both panels are presented as means (± se) of six replicate reactions.
To further investigate the effect of the promoter on MYB10 transcript accumulation and predicted protein levels, we repeated this assay, replacing the 35S:MYB10 with either the R1 or R6 promoter fused to MYB10. Results indicated that the high transcript abundance of MYB10 fused to the R6 promoter enables transactivation of the reporter, particularly when the reporter is fused to R6 (Figure 4B). The results show a similar level of activity to the 35S promoter. With the R1:LUC fusion, R6:MYB10 appears to be more effective at transactivating the promoter than 35S:MYB10. The R1:MYB10 fusion did not influence transactivation to the same extent.
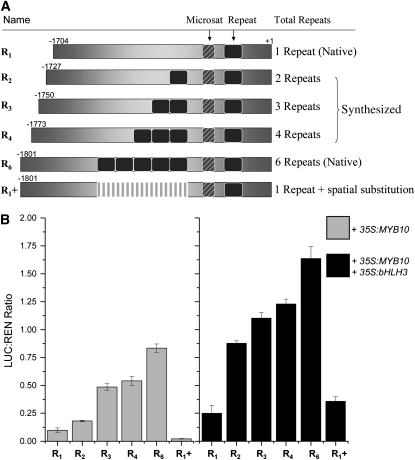
Repeat Number Influences Transactivation in the Dual Luciferase Transient Tobacco Assay
A series of constructs were built to test the influence of the number of 23-bp repeat units in the upstream region of MYB10 on MYB10-induced transcriptional activity. These constructs were based on the native promoter sequences, but with repeat units ranging from one (R1) to six (R6), and were then fused to the LUC reporter (Figure 5A) and assayed as above. To test the spatial effect that the presence of the repeat-containing minisatellite sequence might exert on other unidentified regulatory regions, a further construct (R1+) was built where the minisatellite sequence from R6 was replaced with nonspecific DNA of the same length from a cloning vector (Promega). The results show a strong positive correlation between the number of repeat units and the activation of the promoter by 35S:MYB10 (Figure 5B). When the promoter constructs were coinfiltrated with 35S:MYB10, there is basal activity from both R1 and R1+ and an increasing activation from R2 to R6. We have previously shown the dependency of MYB10 in activating promoters of the anthocyanin biosynthetic pathway on a cofactor bHLH (Espley et al., 2007). In this assay, activation by MYB10 of all the promoters (R1 through to R6) is enhanced with the addition of 35S:bHLH3 (Figure 5B).
Figure 5.
The Number of Repeat Units Affects the Transactivation Rate in a Dual Luciferase Transient Tobacco Assay.
(A) Schematic (not drawn to scale) of the different promoters with repeat units ranging from one (R1) to six (R6). The two native promoters from apple are marked, R1 as from Malus × domestica ‘Royal Gala’ and R6 from Malus × domestica ‘Red Field’ OP. The position of the repeat units (in black) relative to the microsatellite (gray diagonal box) is shown. R1+ is differentiated with gray vertical shading to represent the substituted sequence replacing the R6 minisatellite.
(B) Results of R1 to R6 promoters coinfiltrated with 35S:MYB10 alone (light gray bars) and with 35S:MYB10 and 35S:bHLH3 (black bars). Results are calculated as described in the legend to Figure 4. Data are presented as means (± se) of six replicate reactions.
Deletion Analysis of the MYB10 Promoter Using the Dual Luciferase Transient Tobacco Assay
Results from the dual luciferase transient tobacco assays described above suggest that the MYB10 transcription factor positively autoregulates its own transcription by interacting with the DNA repeat present in a single copy in the R1 promoter and in six copies in the R6 promoter. In addition, a putative cofactor bHLH3 enhances the ability of MYB10 to transactivate these promoters.
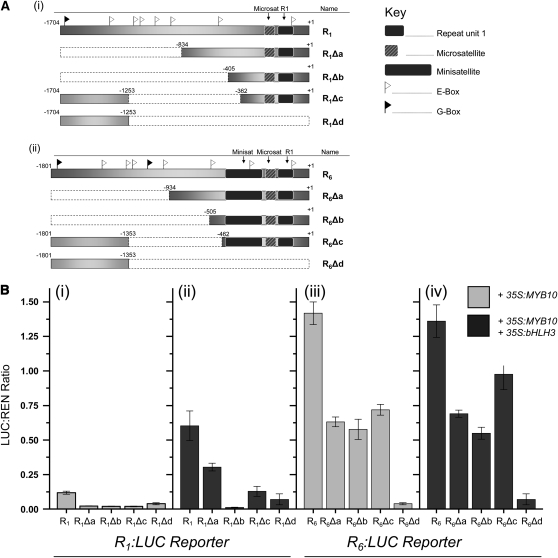
To further define regions in the promoter of MYB10 responsible for transcriptional regulation, deletion variants of the promoters (R1 and R6) were analyzed. Deletions were designed to define the distal end of the R1 and R6 promoters and the possible role of a 5′ G-box in bHLH-related transactivation (Figure 6). A second set of deletions (Figure 7) was designed to delete the native R1 repeat from the R6 promoter and to probe the role of the microsatellite, and data are presented in the next section.
Figure 6.
Deletion Studies to Identify Areas of the Promoter Critical to Transactivation by MYB10 and bHLH3 in a Dual Luciferase Transient Tobacco Assay.
(A) Schematic (not drawn to scale) of the different promoter deletions of R1 (i) and R6 (ii), denoted as Δa to Δd. Deleted areas are shown in white with dotted lines, and the relative positions of the repeat unit to the microsatellite and minisatellite are displayed. The approximate position of G-boxes (black flags) and E-boxes (white flags) are shown.
(B) Corresponding data from promoter deletion studies with luciferase fusions of R1 (i and ii) and R6 (iii and iv) coinfiltrated with 35S:MYB10 (light-gray bars) and with 35S:MYB10 and 35S:bHLH3 (dark-gray bars). Data are presented as means (± se) of six replicate reactions.
Figure 7.
Promoter Deletions to Test the Role of the First Repeat Unit and the Adjacent Microsatellite on Transactivation Levels in a Dual Luciferase Transient Tobacco Assay.
(A) Schematic (not drawn to scale) of the different promoter deletions of R1(i) and R6(ii). Deletions of the first repeat unit (R1) and microsatellite are shown in white with dotted lines. The approximate position of G-boxes (black flags) and E-boxes (white flags) are shown as Figure 6.
(B) Corresponding data from promoter deletion studies with luciferase fusions of R1 (i and ii) and R6 (iii and iv) coinfiltrated with 35S:MYB10 (light-gray bars) and with 35S:MYB10 and 35S:bHLH3 (dark-gray bars). Data are presented as means (± se) of six replicate reactions.
Figure 6A shows the proximal promoter fragments of 834 and 405 bp (R1) and 934 and 505 bp (R6) (Δa and Δb, respectively) selected for testing. The G-box identified at the extreme 5′ end of the isolated promoter fragments was absent from these variants. A third variant (R1Δc and R6 Δc) with 362- and 463-bp proximal fragments, respectively, and with a restored distal region, including the G-box, was tested. A fourth variant, Δd, contained just the distal fragment, including the G-box, and was included as a control for transactivation levels.
The combinatorial interaction of cis-acting elements that are recognized by transcription factors, such as bHLH and MYB, produce a cooperative regulation of the spatial distribution of flavonoids (Hartmann et al., 2005). Analysis of the promoter regions using the database PLACE (Higo et al., 1999) predicted many cis-acting elements. Flavonoid-related MYB binding elements were identified, including MYB26PS (Uimari and Strommer, 1997), MYB core (Planchais et al., 2002), MYBPLANT (Tamagnone et al., 1998), and MYBPZM (Grotewold et al., 1994). It was also noted that the 23-bp repeat motif starts with GTTAG, a partial sequence from reported MYB binding domains (Grotewold et al., 1994; Uimari and Strommer, 1997; Romero et al., 1998) and that this sequence is also adjacent to the microsatellite. Further bHLH-related cis-acting elements, E-boxes (CACATG) (Atchley and Fitch, 1997; Heim et al., 2003), and G-boxes (CACGTG) (Giuliano et al., 1988; Williams et al., 1992) were identified, and their location is shown in Figure 6A.
All of the R1 variants showed a reduction in the relative level of transactivation. In the presence of MYB10, transactivation was barely detectable for any of the deletion constructs. The coinfiltration of 35S:MYB10 and 35S:bHLH3 partly restored this activity for R1Δa, although it remained at half the level of the full-length R1 promoter. This suggests that elements in the distal region are critical for transactivation. This was more evident with R1Δb, which showed no transactivation, indicating that there are key elements in both the distal (−834 to −1704 from the ATG translation start site) and middle regions (−405 and −834) of the promoter. The loss of these elements from both regions in R1Δb prevented transactivation. The restoration of the distal region, including the G-box, in R1Δc does partly restore transactivation but at a lower level than R1Δa. A low level of transactivation was observed with R1Δd.
The results from the infiltration assay for R6Δa and R6Δb with 35S:MYB10 showed a similar level of transactivation, indicating that the R6 promoter can function in the most extensively deleted version. However, both variants were capable of only half the transactivation level of the intact R6 promoter, suggesting that upstream regulatory elements, beyond the minisatellite, are important for the very high level of transactivation previously seen with the R6 promoter and MYB10. Various MYB-related cis-acting elements were deleted in these variants, including MYBCORE, MYBPZM, and MYBPLANT elements. Restoration of the distal promoter region in R6Δc did not elevate transactivation. Low levels of transactivation were detected with R6Δd.
The coinfiltration of 35S:MYB10 and 35S:bHLH3 with the R6 variants resulted in little difference to transactivation levels, although restoration of the distal region R6Δc partly restored activity. It is clear from the data that transactivation by R1 is dependent on distal upstream elements that encompass the predicted bHLH binding sites. It is also clear that the R6 promoter is partly dependent on distal upstream elements but that good levels of transactivation can be initiated by just the proximal 505-bp region. This version differs from the equivalent R1 version primarily by the presence of the minisatellite.
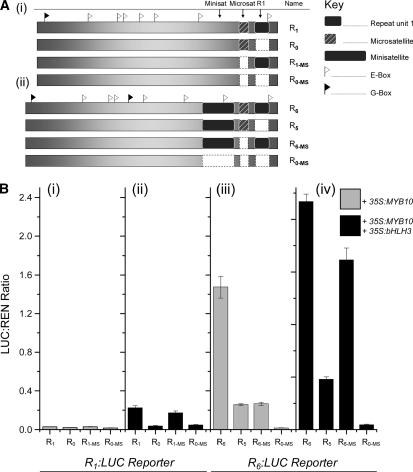
Deletion of the Primary Repeat Unit and Microsatellite in the MYB10 Promoter
Another set of constructs was built to test the importance of the primary repeat unit and the adjacent microsatellite on autoactivation of the MYB10 promoter by MYB10 (Figure 7A). The primary repeat unit, designated repeat 1 (Figure 1B), was deleted from both the R1:LUC and R6:LUC constructs, producing a version of the MYB10 promoter lacking the 23-bp unit (R0) and a version with the five units from the minisatellite-like insertion from R6 (R5). In addition, the microsatellite was independently deleted from R1 and R6, producing R1-MS and R6-MS, respectively. The partial MYB binding site (GTTAG) adjacent to the microsatellite was retained in these constructs. A construct was also built that contained neither a repeat unit nor a microsatellite, R0-MS.
For the R1 deletions (R0, R1-MS, and R0-MS), there was little detectable transactivation activity with the coinfiltration of 35S:MYB10 (Figure 7B). A similar result was seen when R0 and R0-MS were coinfiltrated with both 35S:MYB10 and 35S:bHLH3. However, transactivation was evident when R1 and R1-MS were coinfiltrated with both MYB10 and bHLH3. This result suggests that the repeat unit is necessary for autoregulation of the promoter with MYB10/bHLH3 but that the presence of the microsatellite is not critical.
Similar deletions of the R6 version of the promoter showed a large reduction in transactivation for both R5 and R6-MS when infiltrated with MYB10. Coinfiltration with both 35S:MYB10 and 35S:bHLH3 appeared to restore transactivation with R6-MS but to a much lesser extent with R5. For the R6 promoter, it appears that the first repeat unit is critical to enable high levels of transactivation and that the microsatellite is required for the highest level of transactivation.
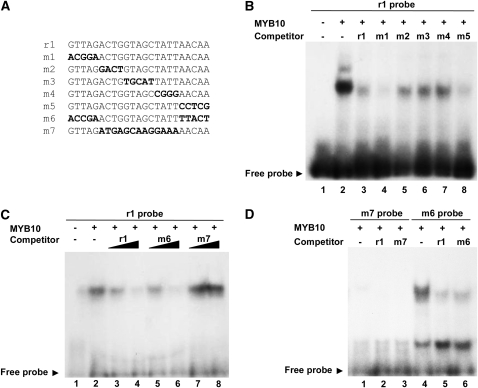
Analysis of MYB10 Protein:DNA Interaction Using Electrophoretic Mobility Shift Assays
To test directly MYB10 binding to DNA, we analyzed in vitro binding using electrophoretic mobility shift assays (EMSAs). Recombinant His-tagged MYB10 protein was purified from Escherichia coli and incubated with DNA probes representing the 23-bp repeated unit or mutations of the unit (Figure 8A). EMSA showed a band shift when the oligonucleotide probe, corresponding to the repeat motif found in the native promoter, and recombinant MYB10 were bound (Figure 8B). In control EMSA experiments, a nonspecific His-tagged protein did not complex with the repeat motif (see Supplemental Figure 2 online). When cold competitor DNA of the same sequence (r1) was added in 200-fold excess, this binding was reduced, indicating that the reaction was specific. To further determine active sites important for the oligonucleotide-protein binding, we used mutated versions of the motif as cold competitors (Figure 8B). In these versions, nucleotides were substituted across five different areas of the R1 repeat unit (Figure 8A). The results indicated that the extreme 3′ and 5′ of the repeat unit are less important for binding as both m1, which no longer contains the partial sequence from predicted MYB binding domains (GTTAG), and m5 were able to compete off the r1 probe to a similar extent as the native r1 competitor. Three other competitors carrying mutations in the inner part of the sequence (m2, m3, and m4) were less able to compete, suggesting that this region, comprising the sequence ACTGGTAGCTATT, is critical for binding.
Figure 8.
EMSAs Show That MYB10 Binds to DNA with the Repeat Motif.
(A) Sequences of the probe and competitor DNA with mutated bases shown in bold type.
(B) EMSAs of 32P-labeled probe with MYB10. Lane 1, r1 DNA probe only, no protein; lane 2, r1 probe incubated with MYB10 protein; lane 3, r1 probe with MYB10 protein and 200-fold excess of r1 cold competitor; lanes 4 to 8, as in lane 2 but with 200-fold excess m1, m2, m3, m4, and m5 cold competitor, respectively.
(C) Lane 1, r1 probe with MYB10 protein; lanes 2 and 3, r1 probe with MYB10 protein and 20- and 200-fold excess cold competitor, respectively; lanes 4 and 5, r1 probe with MYB10 protein and 20, and 200-fold excess m6 cold competitor; lanes 6 and 7, r1 probe with MYB10 protein and 20, and 200-fold excess m7 cold competitor.
(D) Lane 1, m7 probe with MYB10 protein; lane 2, m7 probe with MYB10 protein and 200-fold excess r1 cold competitor; lane 3, m7 probe with MYB10 protein and 200-fold excess m7 cold competitor; lane 4, m6 probe with MYB10 protein; lane 5, m6 probe with MYB10 protein and 200-fold excess r1 cold competitor; lane 6, m6 probe with MYB10 protein and 200-fold excess m6 cold competitor.
To confirm this finding, we substituted areas of the repeat unit sequence with randomly selected sequence from the apple actin gene (accession number CN938023) and performed binding assays. For these, substitutions were made at both the outer 3′ and 5′ sequences of the repeat unit (m6) and the entire inner sequence (m7) (Figure 8A). The signal was reduced when m6 was used as a competitor, but m7 was not able to compete off the r1 probe (Figure 8C). When the m6 and m7 oligos were themselves radiolabeled and used as probes for binding to MYB10 protein, the m7 probe failed to bind (Figure 8D), while the m6 probe bound to the protein and was partially competed off by the inclusion of r1 or m6 competitor DNA. In the m6 assay, an additional band was observed that may be due to nonspecific binding of the m6 probe with partially degraded MYB10 protein.
DISCUSSION
Our data suggest that a minisatellite in the upstream regulatory DNA region of a gene encoding an apple transcription factor, MYB10, creates a novel autoregulatory motif. This results in a massive increase in the level of anthocyanins throughout the plant. This gain-of-function mutation in the anthocyanin regulatory pathway has significant implications for the development, through both conventional and advanced breeding methods, of novel varieties of plants and fruit with enhanced anthocyanin and increased consumer appeal.
A Minisatellite Alters the Promoter of MYB10 in Red-Fleshed Apples
In the promoter from red-fleshed apples, we found a 100-bp insertion, 275 bp upstream of the ATG translation start codon. This insertion comprised a 23-bp sequence, duplicated in five tandem repeats to form a minisatellite-like repeat unit. In plants, it has been shown that minisatellites may be associated with other elements, such as miniature inverted-repeat transposable elements (Lu et al., 2008). The MYB10 minisatellite precedes a dinucleotide microsatellite. The 23-bp motif is found once in both red- and white-fleshed varieties, just downstream of the microsatellite, and is likely to be the origin of the repeat motif.
We sequenced the region encompassing the minisatellite motif in diverse apple varieties and observed an association of the repeat-containing R6 promoter of MYB10 with red flesh/red foliage in all the varieties tested. A number of sequence variations were found in the upstream region, but only the minisatellite polymorphism is associated with the elevated accumulation of anthocyanins. The same region was PCR amplified from a further set of 77 apple varieties, and in each case the product corresponding to the minisatellite motif was absent from the white-fleshed variety but present in red-fleshed varieties, suggesting that the minisatellite-containing allele of MYB10 has probably been inherited from a common ancestor in all the red-fleshed varieties tested.
Repetitive DNA can be grouped into two classes: interspersed repeats, such as retrotransposons, and local tandem repeats, such as simple sequence repeats. Simple sequence repeats are generally defined by the length of repeat unit (n = 1 to 13 bp), while minisatellites repeat units vary from 14 to 500 bp (Vergnaud and Denoeud, 2000). There are thought to be numerous mechanisms for microsatellite expansion, including replication slippage, recombination, and repair, while minisatellites appear to expand and contract as a result of recombination (Thomas, 2005). The regulatory sequence analyzed here contains examples of both a microsatellite (n = 2 bp) and, in the case of red-fleshed apple phenotypes, a series of tandem repeats forming a minisatellite-like structure (n = 23 bp). Since the first description of minisatellites in humans (Wyman and White, 1980) and in plants (Dallas, 1988), they have become a useful feature for DNA fingerprinting (Jeffreys et al., 1985), linkage studies (Nakamura et al., 1987), and genome mapping (NIH/CEPH Collaborative Mapping Group, 1992). The high rate of minisatellite polymorphism has been associated with various human pathologies and the heritability of diseases (Krontiris, 1995). Hypermutable minisatellites in promoters have been shown to have an effect on transcriptional regulation in humans. For example, the polymorphic minisatellite in the promoter region of the human insulin gene alters transcription according to minisatellite size (Kennedy et al., 1995). In plants, although minisatellites have been used for various evolutionary studies (Sykorova et al., 2006), fingerprinting (Nybom et al., 1990; Tzuri et al., 1991), and mapping (Barreneche et al., 1998), there is little evidence to date of minisatellite-induced changes to transcriptional regulation.
The MYB10 minisatellite is conserved among the red-foliaged, red-fleshed varieties tested. Although we have defined the duplication in the R6 promoter allele as a minisatellite, we have found only one version with six repeats and none of the repeat copy number variation found in human studies. This apparently stable minisatellite may be the result of an ancient rearrangement because such diverse apple varieties as M. pumila var Niedzwetzkyana, Malus marjorensis var Formosa, various Malus sieversii varieties, and Malus × purpurea 'Aldenhamensis' all carry the same mutation, and there were no exceptions in all the varieties we tested. The lack of intermediate numbers of repeats of this minisatellite might be the result of selection and domestication or might indicate that the mutation has occurred only once. The microsatellite may create a region of instability that might partially explain the presence of the repeated motifs at this particular site. In the MYB10 promoter sequences analyzed, repeat number polymorphisms (varying from 6 to 9) were detected in the microsatellite.
Analysis of the Minisatellite Effects on Transcription
We used a transient transactivation assay to identify candidate transcription factors that were able to interact specifically with the repeat-containing promoter by fusing a LUC reporter to both versions of the native apple promoter: the promoter isolated from white-fleshed apple varieties with one repeat motif (R1) and the corresponding promoter from red-fleshed varieties with six copies of the repeat motif (R6). We detected a significant increase in transactivation of the LUC-fused promoter from Malus × domestica ‘Red Field’ OP when coinfiltrated with the MYB10 transcription factor construct itself.
The constructs built to study the effect of differing numbers of repeat motifs (Figure 5A), which ranged from one motif (R1) to six (R6), showed that motif number directly affects promoter activation in the presence of MYB10. One explanation is that the MYB10 protein is able to bind its own promoter with greater efficiency when multiple motifs are present. This effect was further enhanced when an apple gene encoding a bHLH transcription factor was coinfiltrated, although the pattern of increased activation with motif number remained the same. In this assay, even R6 activity was significantly enhanced with the combination of MYB and bHLH proteins. The increase in transcript levels due to the presence of the minisatellite sequence is not merely a spatial phenomenon, as replacing the minisatellite with different sequence of the same length leads to no enhancement of transcriptional activity.
To further analyze the regions necessary for transcriptional activation, we subjected the native promoters to deletion analysis. The resulting transactivation levels for the most extreme deletions of R1 and R6 (R1Δb and R6Δb) differed; unlike the R6 variant, R1Δb was not able transactivate. The major difference between these two deletion promoters is the presence of the minisatellite in R6Δb. All the deleted promoters showed a reduction in transactivation levels, but it appears that the minisatellite in the R6 variant reduces the dependence on upstream elements for transactivation.
In the data set presented in Figure 6, there was little or no enhancement of the R6 promoter when we coinfiltrated with 35S:bHLH3. This is in contrast with the enhancement observed when 35S:bHLH3 was coinfiltrated in the experiments detailed in Figures 5 and 7. While the bHLH enhancement is usually observed with the R6 promoter, it is always observed in transient assays performed with promoters that contain fewer repeats. We believe that this incongruence may be due to the high level of MYB-mediated transcriptional activation of the R6 promoter and that endogenous levels of bHLH, which may vary in tobacco leaves under certain experimental conditions, are able to satisfy the bHLH levels required to saturate the transcriptional potential of the R6 promoter.
Targeted deletions for both versions of the MYB10 promoter included the removal of repeat unit number one and/or removal of the microsatellite (Figure 7). The removal of the repeat unit from the R1 promoter, effectively producing a promoter with none of the identified units of 23 bp (R0), resulted in barely detectable transactivation when coinfiltrated with MYB10 and bHLH3 (Figure 7). However, removal of the microsatellite (R1-MS) had little effect, with transactivation at a similar level to that of R1. Removal of both the repeat unit and the microsatellite again resulted in loss of transactivation.
Similar deletions were performed on the R6 promoter, with the removal of repeat unit number one (R5) and removal of the microsatellite (R6-MS) (Figure 7). When coinfiltrated with MYB10, both R5 and R6-MS showed a decrease in transactivation when compared with R6. The addition of bHLH3 increased the transactivation level, particularly for that of R6-MS. The data suggest that the single repeat unit is important for transactivation levels and that maximum activity is achieved only when both the microsatellite and the first repeat unit R1 are present. Future work could focus on the importance of the location of the R1 site.
Analysis of the Effect of the Minisatellite on MYB Function
In tobacco leaves, the R6-driven MYB10 cDNA was able to induce anthocyanin pigmentation when coinfiltrated with an apple gene encoding a bHLH transcription factor, similar to results previously achieved with 35S:MYB10 (Espley et al., 2007). By contrast, the R1-driven MYB10 cDNA was unable to induce detectable anthocyanin pigmentation, even when coinfiltrated with an apple gene encoding a bHLH transcription factor. Anthocyanin accumulation in this heterologous system may be dependent on a sufficient protein level of the MYB transcription factor, and when expressed under the control of the 35S or R6 promoters, this accumulation is partially dependent on a high level of bHLH transcript. However, endogenous tobacco cofactors may also interact.
For the transformation of apple with the R6 promoter driving MYB10, we used the green-leaved variety ‘Royal Gala.’ We noted intense pigmentation in the transformed callus, regenerating plants, and in grafted plants (Figure 3). This demonstrated the functionality of the R6 promoter to drive anthocyanin in a constitutive manner and is similar to the phenotype caused by overexpression of MYB10 by the 35S promoter (Espley et al., 2007).
Effect of a bHLH Transcription Factor on Transcription from the MYB10 Promoter
We have demonstrated that the presence of the minisatellite and the number of repeat units changes the level of transcription, and we predict that a MYB binding domain is located within each repeat motif. We also see differences in the level of transcriptional regulation in the presence of a bHLH transcription factor (bHLH3). Since the first descriptions of the anthocyanin-related MYB-bHLH interaction (Goff et al., 1992), considerable progress has been made into the understanding of the relationship between these two classes of transcription factors (Ramsay and Glover, 2005). The bHLH transcription factors associated with anthocyanin production may regulate the transcription initiated by their cofactor MYBs (Grotewold et al., 2000), and it has been shown that they play a key role in recruiting to the DNA a complex of proteins that regulates gene expression by histone modification (Hernandez et al., 2007).
The predicted protein sequence of MYB10 contains the conserved regions that confer dependence on a bHLH coactivator (Grotewold et al., 2000; Zimmermann et al., 2004). The apple bHLH transcription factor selected for these experiments belongs to the group of known anthocyanin regulators classed as the IIIf bHLH gene family (Heim et al., 2003), which includes TT8, reported to coregulate anthocyanin biosynthetic genes in Arabidopsis thaliana (Nesi et al., 2000). In apple, this relationship between the MYB and the bHLH transcription factors appears to conform to the proposed model, although experimental evidence from the reporter assays suggests that the reliance on a bHLH transcription factor may be less critical for the minisatellite-containing promoter (R6). The bHLH transcription factor appeared to have a decreasing influence on transcription level as the number of motifs rose (Figure 5B). However, this may be due to the limited supply of infiltrated bHLH transcription factor: while the MYB transcription factor is able to autoregulate itself to produce a constant supply of protein to perpetuate the transcript production/protein binding cycle, the bHLH transcription factor may become the limiting factor in the formation of this MYB/bHLH protein complex. In the heterologous transient assay for anthocyanin production, the R6-driven MYB10 infiltration still requires the coinfiltration of a bHLH transgene (Figure 3). There is evidence for a self-activating feedback of regulation of the Arabidopsis TT8 transcription factor (Baudry et al., 2006), where in yeast assays the TT2 MYB can bind to the promoter of a target bHLH gene only in the presence of an appropriate bHLH protein. However, the authors also suggest that in planta it may be possible for the MYB transcription factor to bind the bHLH promoter in the absence of a bHLH partner. Similarly, we cannot exclude the possibility that our candidate bHLH transcription factor may be in sufficient supply to regulate or interact with MYB10.
Deletion of the predicted G-box in the distal promoter region may account for the reduced transactivation levels seen in Figure 6, particularly in view of the partial restoration of this activity when this region was reinstated with the R1ΔC and R6ΔC variants. Overall, the data suggest some dependency between the MYB and bHLH transcription factors in apple, although overexpressing MYB10 in transgenic lines does not elevate the transcript level of candidate apple bHLH transcription factors (Espley et al., 2007).
Analysis of in Vitro Binding of DNA by the MYB10 Protein
The results from EMSA demonstrated binding between the MYB10 protein and the repeat motif, with the specificity of this reaction confirmed by competition assays. Five mutated versions of the repeat motif, representing mutated regions across the repeat unit, were also tested for their competitive capacity to identify critical nucleotide regions for binding. Mutation of the outlying nucleotides (m1 and m5) showed a similar capacity for competition when compared with the native r1 competitor. This result was confirmed by the use of the m6 competitor. The three mutated versions (m2, m3, and m4) representing the inner 13 nucleotides (ACTGGTAGCTATT) showed a reduced capacity to compete, suggesting that this region is required for binding of the MYB10 protein. These results were also verified by the use of the m7 probe, which failed to bind MYB10 protein. This indicates that there are nucleotides within the inner region of the repeat unit that specifically bind to the MYB10 protein. As this 13-nucleotide sequence does not resemble a known MYB binding motif, further research will be required to determine the core motif in this binding domain.
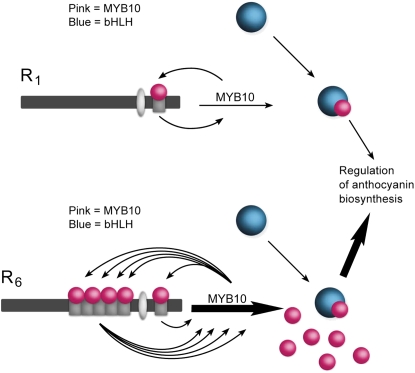
The Minisatellite Causes Autoregulation of the MYB10 Gene
We have demonstrated that the presence of the R6 minisatellite alters transcription levels and that this additional capacity for transcription of MYB10 is responsible for the MYB10-induced elevation of anthocyanin. We propose a model whereby MYB10 can influence its own expression at a specific motif (Figure 9, R1), but in varieties where this motif has been replicated (as with R6), ectopic MYB10 accumulation arises and leads to ectopic accumulation of red anthocyanin pigments (Figure 9, R6). Binding assays have shown that MYB10 does interact with the repeat motif, and the competition assays further define the region responsible for this binding.
Figure 9.
Model Showing the Autoregulation of R1 and R6 Promoters by MYB10.
The additional repeat units (shown in gray bars) in R6 create a greater capacity for MYB10 to activate its own transcription using a feed-forward mechanism that upregulates the anthocyanin pathway, leading to plants with red foliage and fruit flesh.
Previous studies, particularly in humans, suggest that minisatellites have provided an important mechanism for evolutionary change and phenotypic variation. Here, we show that such a mutation in plants has produced a remarkable phenotype, with implications for the development of novel varieties of plants and fruit with enhanced anthocyanin and increased consumer appeal.
METHODS
Isolation of the MYB10 Upstream Promoter Region
For isolation of the upstream promoter region, genomic DNA was isolated from Malus × domestica ‘Sciros’ (Pacific Rose, derived from a cross between ‘Royal Gala’ and ‘Splendour’) using the DNeasy plant mini kit (Qiagen). Nested primers to the coding region of MYB10 were designed: primary, 5′-CACTTTCCCTCTCCATGAATCTCAAC-3′, and secondary, 5′-CAGGTTTTCGTTATATCCCTCCATCTC-3′. A 1.7-kb region of upstream DNA immediately adjacent to the transcription start site was isolated from the genomic DNA by PCR genome walking using a GenomeWalker kit (Clontech), following the manufacturer's instructions. Genomic DNA was subsequently isolated from Malus × domestica ‘Granny Smith,’ Malus × domestica ‘Royal Gala,’ and Malus × domestica ‘Red Field’ OP using forward and reverse primers 5′-ACCCTGAACACGTGGGAACCG-3′ and 5′-GCTAAGCTTAGCTGCTAGCAGATAAGAG-3′, respectively. The PCR products were cloned using the TOPO TA cloning kit (Invitrogen), and the sequences aligned using Vector NTI (Invitrogen). Analysis of the promoter regions was performed using the database PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) (Higo et al., 1999)
Minisatellite Region PCR Amplification and Sequencing
Apple genomic DNA from 19 varieties was amplified using a pair of PCR primers located in the MYB10 promoter (forward: 5′-GGAGGGGAATGAAGAAGAGG-3′; reverse: 5′-TCCACAGAAGCAAACACTGAC-3′). PCR reactions were performed in 16.5 μL volume containing 1× PCR buffer mix (Invitrogen), 1.3 mM MgCl2, 100 μM of each dNTP, 0.72% formamide, 10 μM of each primer, 0.5 units of Platinum Taq DNA polymerase (Invitrogen), and 2 ng of genomic DNA. PCR amplifications were performed in a Hybaid PCR Express Thermal Cycler (Thermo Electron) with conditions as follows: 94°C for 2 min and 45 s followed by 40 cycles at 94°C for 55 s, 55°C for 55 s, and 72°C for 1 min 39 s, and a final elongation at 72°C for 10 min. The PCR products obtained were cloned using the TOPO TA cloning kit (Invitrogen). Four clones were sequenced for each PCR product. The sequences were aligned using Vector NTI (Invitrogen).
Plasmid Construction
Luciferase reporter constructs were derivatives of pGreen 0800-LUC (Hellens et al., 2005) in which the promoter sequence for MYB10 or the deletion fragments were inserted. Promoter sequences were PCR amplified using the primers 5′-ACCCTGAACACGTGGGAACCG-3′ and 5′-GCTAAGCTTAGCTGCTAGCAGATAAGAG-3′ and cloned into the multicloning region of pGreen 0800-LUC. R1 and R6 promoter fragments were cloned in as native promoter sequences, while changes to the repeat frequency for the R2, R3, and R4 promoter fragments were synthezised (Geneart AG) and cloned into R1 using the restriction enzymes SpeI and DraI. Cloning strategies for the deletion constructs are outlined in the Supplemental Methods online. The pSAK construct for 35S:MYB10 and 35S:bHLH3 was as previously described (Espley et al., 2007). The promoter sequences for R1 (EU518249) and R6 (EU518250) were PCR amplified from genomic DNA using the primers 5′-ATAGAGCTCACCCTGAACACGTGGGA-3′ and 5′-ATTGTCTTCCTCTCGAGTCCAGGCA-3′ containing SacI and XhoI restriction sites, respectively. The pSAK vector, containing the 35S:MYB10 cassette, was digested with SacI and XhoI to release the 35S promoter sequence and then religated with the PCR amplified R1 or R6 promoter fragments to produce R1:MYB10 and R6:MYB10.
Transactivation Analysis Using Transformed Tobacco Leaves
The promoter sequences for MYB10 were inserted into the cloning site of pGreen 0800-LUC (Hellens et al., 2005). In the same construct, a luciferase gene from REN, under the control of a 35S promoter, provided an estimate of the extent of transient expression. Activity is expressed as a ratio of LUC to REN activity. The promoter-LUC fusions were used in transient transformation of Nicotiana benthamiana by mixing 100 μL of Agrobacterium tumefaciens strain GV3101 (MP90) transformed with the reporter cassette with or without another Agrobacterium culture(s) (900 μL) transformed with a cassette containing MYB10 fused to the 35S, R1, or R6 promoters and bHLH3 fused to the 35S promoter. The method is further described in the Supplemental Methods online.
Induction of Anthocyanin Pigmentation in Tobacco
Nicotiana tabacum plants were grown as previously described (Espley et al., 2007) and maintained in a glasshouse for the duration of the experiment. Agrobacterium cultures were incubated as for the dual luciferase assay, and separate strains containing the MYB10 gene fused to either the 35S, R1, or R6 promoter sequences and the bHLH3 gene fused to the 35S promoter were mixed (500 μL each) and infiltrated into the abaxial leaf surface. Six separate infiltrations were performed into tobacco leaves (two plants per treatment), and changes in color were observed and digitally recorded over an 8-d period. To control for leaf-to-leaf variability, at least two leaves were infiltrated, and each leaf included positive (Agrobacterium cultures containing 35S:MYB10 + 35S:bHLH3) and negative (Agrobacterium with empty vector) controls.
Transformation of Apple
The binary vector pSAK277 containing MYB10 driven by the R6 or R1 promoters was transferred into Agrobacterium strain GV3101 by the freeze-thaw method (Holsters et al., 1978). Transgenic Malus × domestica ‘Royal Gala’ plants were generated by Agrobacterium-mediated transformation of leaf pieces using a method previously reported (Yao et al., 1995).
Production and Purification of MYB10 Protein
A synthesized version of MYB10 cDNA (Geneart) was used for recombinant protein expression (GenBank EU518248). The sequence was codon optimized for bacterial expression and cloned into the pET30a expression vector (Invitrogen) with an N-terminal His Tag. Two versions, full length (amino acids 1 to 243) and truncated (amino acids 1 to 167), were tested, and the truncated version was found to be more highly expressed. Both versions were used in EMSAs and showed similar results (see Supplemental Figure 2 online). The truncated version was used for all subsequent EMSAs (below and Supplemental Methods online). The construct was transformed into Escherichia coli BL21-CodonPlus-RIL (Stratagene), and cells were grown in 500 mL ZYM-5052 autoinducible media (Studier, 2005) at 37°C for 2 h at 300 rpm and 16°C for a further 60 h. Cells were harvested by centrifugation (3500g) and resuspended in His Trap binding buffer (30 mM imidazole, 0.5 M NaCl, 5 mM DTT, and 20 mM sodium phosphate, pH 7.4) and EDTA-free inhibitor cocktail tablets (Roche). Cells were disrupted using an EmulsiFlex-C15 high-pressure homogenizer (15,000 to 20,000 p.s.i.) (Avestin) and were then pelleted at 15,000 rpm and the supernatant filtered through a 0.25-μm filter (Millipore) before loading onto a precharged and equilibrated 5-mL His-Trap HP column (GE Healthcare) charged with Ni2+. Bound proteins were washed following the manufacturer's specifications and eluted using a continuous 0 to 500 mM imidazole gradient at 2 mL min−1. Fractions containing recombinant proteins were confirmed by SDS-PAGE and protein gel blot analysis prior to further protein purification by size-exclusion chromatography using a Superdex gel filtration 200 column (GE Healthcare) connected to an ATKA FPLC (GE Healthcare) and eluted in 20 mM Tris HCl, pH 7.0.
EMSAs
Complementary oligonucleotides for EMSA were annealed and labeled with [32P]-γ-ATP prior to binding with MYB10 protein. The forward strand sequences for the labeled probes were as follows: r1, 5′-GTTAGACTGGTAGCTATTAACAA- 3′; m6, 5′-ACCGAACTGGTAGCTATTTTACT-3′; and m7, 5′-GTTAGATGAGCAAGGAAAAACAA-3′. Forward strand sequence for the competitor oligos were as follows: r1 as above; m1, 5′-acggaACTGGTAGCTATTAACAA-3′; m2, 5′-GTTAGgactGTAGCTATTAACAA-3′; m3, 5′-GTTAGACTGtgcatTATTAACAA-3′; m4, 5′-GTTAGACTGGTAGCcgggAACAA-3′; m5, 5′-GTTAGACTGGTAGCTATTcctcg-3′; m6, 5′-accgaACTGGTAGCTATTttact-3′; and m7, 5′-GTTAGAtgagcaaggaaaAACAA-3′. Mutated bases are shown in lower case and underlined. Labeling reactions were performed using T4 Polynucleotide kinase (New England Biolabs) and [32P]-γ-ATP. Unincorporated labeled nucleotides were removed using ProbeQuant G50 micro columns (GE Healthcare). For the binding assay, 0.2 to 0.7 μg of recombinant MYB10 protein was mixed with 0.05 to 0.1 pmol of double-stranded, labeled DNA probe in binding buffer [10 mM Tris, 50 mM KCl, 2.5 mM DTT, 1 μg poly(dI-dC), 10 ug BSA, and 4% glycerol] and incubated for 30 min at 25°C. Cold competitor DNA was added at 20- or 200-fold excess versus the radiolabeled probe. The bound complexes were resolved by electrophoresis on native 5% polyacrylamide gels in 0.5% Tris-borate EDTA (TBE) buffer, pH 8.3, at 100 V for 60 min at 25°C. The gel was dried in a gel drier (Atto) at 80°C under vacuum before autoradiography with an intensifier screen at −80°C.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: Md MYB10 (‘Red Field’) OP, DQ267896; MYB10 (Pacific Rose), DQ267897; MYB10 (‘Granny Smith’), DQ267989; bHLH3, CN934367; MYB10 Codon Optimized, EU518248; MYB10 R1, EU518249; and MYB10 R6, EU518250.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gel Picture Showing Products from the Apple Germplasm PCR Assay.
Supplemental Figure 2. Electrophoretic Mobility Shift Assays Comparing Full-Length and Truncated MYB10 Protein and a Nonspecific His-Tagged Control Protein.
Supplemental Table 1. Data for the 77 Apple Varieties Tested for the Presence of the R1 and R6 Alleles.
Supplemental Methods. Plasmid Construction, Transactivation Analysis Using Transformed Tobacco Leaves, Production and Purification of MYB10 Protein, and Notes on Apple Genealogy.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Charles J. Simon and Philip L. Forsline (Agricultural Research Service, USDA) for plant material and DNA samples. We also thank colleagues at Plant and Food Research, New Zealand for their valuable contribution; William Laing for assistance with protein expression; Heather Bassett for assistance with DNA; Sakuntala Karunairetnam for vector construction of bHLH3; Julie Nichols for maintaining the glasshouse plants; and Tim Holmes, Minna Pesonen, Martin Heffer for photography and graphics, and Kimberley Snowden for helpful advice on the manuscript. We thank Inova Fruit (Geldermalsen, The Netherlands) for project support. This work was also supported by grants from the New Zealand Foundation for Research, Science, and Technology (COX0207) and PREVAR Limited, New Zealand (PREV0401).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andrew C. Allan (aallan@hortresearch.co.nz).
Online version contains Web-only data.
References
- Allan, A.C., Hellens, R.P., and Laing, W.A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13 99–102. [DOI] [PubMed] [Google Scholar]
- Atchley, W.R., and Fitch, W.M. (1997). A natural classification of the basic helix loop helix class of transcription factors. Proc. Natl. Acad. Sci. USA 94 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., and Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48 958–970. [DOI] [PubMed] [Google Scholar]
- Barreneche, T., et al. (1998). A genetic linkage map of Quercus robur L. (pedunculate oak) based on RAPD, SCAR, microsatellite, minisatellite, isozyme and 5S rDNA markers. Theor. Appl. Genet. 97 1090–1103. [Google Scholar]
- Baudry, A., Caboche, M., and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46 768–779. [DOI] [PubMed] [Google Scholar]
- Baudry, A., Heim, M.A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39 366–380. [DOI] [PubMed] [Google Scholar]
- Butelli, E., Titta, L., Giorgio, M., Mock, H.-P., Matros, A., Peterek, S., Schijlen, E.G.W.M., Hall, R.D., Bovy, A.G., Luo, J., and Martin, C. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26 1301–1308. [DOI] [PubMed] [Google Scholar]
- Chagné, D., Carlisle, C., Blond, C., Volz, R., Whitworth, C., Oraguzie, N., Crowhurst, R., Allan, A., Espley, R., Hellens, R., and Gardiner, S. (2007). Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics 8 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas, J.F. (1988). Detection of DNA 'fingerprints' of cultivated rice by hybridization with a human minisatellite DNA probe. Proc. Natl. Acad. Sci. USA 85 6831–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Nesi, N., Perez, P., Devic, M., Grandjean, O., Caboche, M., and Lepiniec, L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt, M.V., Lee, C.Y., and Liu, R.H. (2000). Nutrition: Antioxidant activity of fresh apples. Nature 405 903–904. [DOI] [PubMed] [Google Scholar]
- Espley, R.V., Hellens, R.P., Putterill, J., Stevenson, D.E., Kutty-Amma, S., and Allan, A.C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano, G., Pichersky, E., Malik, V.S., Timko, M.P., Scolnik, P.A., and Cashmore, A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., Cone, K.C., and Chandler, V.L. (1992). Functional analysis of the transcriptional activator encoded by the maize B gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6 864–875. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 543–553. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Sainz, M.B., Tagliani, L., Hernandez, J.M., Bowen, B., and Chandler, V.L. (2000). Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 97 13579–13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu, Y., Hisatomi, Y., and Iida, S. (1998). Molecular characterization of the mutable flaked allele for flower variegation in the common morning glory. Plant J. 16 371–376. [DOI] [PubMed] [Google Scholar]
- Harris, S.A., Robinson, J.P., and Juniper, B.E. (2002). Genetic clues to the origin of the apple. Trends Genet. 18 426–430. [DOI] [PubMed] [Google Scholar]
- Hartmann, U., Sagasser, M., Mehrtens, F., Stracke, R., and Weisshaar, B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57 155–171. [DOI] [PubMed] [Google Scholar]
- Heim, M.A., Jakoby, M., Werber, M., Martin, C., Bailey, P.C., and Weisshaar, B. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20 735–747. [DOI] [PubMed] [Google Scholar]
- Hellens, R., Allan, A., Friel, E., Bolitho, K., Grafton, K., Templeton, M., Karunairetnam, S., and Laing, W. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, J.M., Feller, A., Morohashi, K., Frame, K., and Grotewold, E. (2007). The basic helix loop helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc. Natl. Acad. Sci. USA 104 17222–17227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatomi, Y., Hanada, K., and Iida, S. (1997). The retrotransposon RTip1 is integrated into a novel type of minisatellite, MiniSip1, in the genome of the common morning glory and carries another new type of minisatellite, MiniSip2. Theor. Appl. Genet. 95 1049–1056. [Google Scholar]
- Holsters, M., Waele, D., Depicker, A., Messens, E., Montagu, M., and Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163 181–187. [DOI] [PubMed] [Google Scholar]
- Inagaki, Y., Hisatomi, Y., Suzuki, T., Kasahara, K., and Iida, S. (1994). Isolation of a suppressor-mutator/enhancer-like transposable element, Tpn1, from Japanese morning glory bearing variegated flowers. Plant Cell 6 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A.J., Wilson, V., and Thein, S.L. (1985). Hypervariable minisatellite regions in human DNA. Nature 314 67–73. [DOI] [PubMed] [Google Scholar]
- Kennedy, G.C., German, M.S., and Rutter, W.J. (1995). The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat. Genet. 9 293–298. [DOI] [PubMed] [Google Scholar]
- Knekt, P., Kumpulainen, J., Jarvinen, R., Rissanen, H., Heliovaara, M., Reunanen, A., Hakulinen, T., and Aromaa, A. (2002). Flavonoid intake and risk of chronic diseases. Am. J. Clin. Nutr. 76 560–568. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S., Goto-Yamamoto, N., and Hirochika, H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304 982. [DOI] [PubMed] [Google Scholar]
- Krontiris, T. (1995). Minisatellites and human disease. Science 269 1682–1683. [DOI] [PubMed] [Google Scholar]
- Lane, W.D., Bhagwat, B., Wahlgren, S., and Armstrong, J.D. (2003). Apple micrografting protocol to establish transgenic clones on field ready rootstock. Horttechnology 13 641–646. [Google Scholar]
- Lu, K., Chai, Y.R., Zhang, K., Wang, R., Chen, L., Lei, B., Lu, J., Xu, X.F., and Li, J.N. (2008). Cloning and characterization of phosphorus starvation inducible Brassica napus PURPLE ACID PHOSPHATASE 12 gene family, and imprinting of a recently evolved MITE-minisatellite twin structure. Theor. Appl. Genet. 117 963–975. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Leppert, M., O'Connell, P., Wolff, R., Holm, T., Culver, M., Martin, C., Fujimoto, E., Hoff, M., and Kumlin, E. (1987). Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 235 1616–1622. [DOI] [PubMed] [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH/CEPH Collaborative Mapping Group (1992). A comprehensive genetic linkage map of the human genome. NIH/CEPH Collaborative Mapping Group. Science 258 67–86. [PubMed] [Google Scholar]
- Nybom, H., Rogstad, S.H., and Schaal, B.A. (1990). Genetic variation detected by use of the M13 “DNA fingerprint” probe in Malus, Prunus, and Rubus (Rosaceae). Theor. Appl. Genet. 79 153–156. [DOI] [PubMed] [Google Scholar]
- Planchais, S., Perennes, C., Glab, N., Mironov, V., Inzé, D., and Bergounioux, C. (2002). Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath;CycB1;1 transcription and identification of putative regulatory proteins. Plant Mol. Biol. 50 109–125. [DOI] [PubMed] [Google Scholar]
- Ramsay, N.A., and Glover, B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. [DOI] [PubMed] [Google Scholar]
- Romero, I., Fuertes, A., Benito, M.J., Malpica, J.M., Leyva, A., and Paz Ares, J. (1998). More than 80 R2R3 MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14 273–284. [DOI] [PubMed] [Google Scholar]
- Schwinn, K., Venail, J., Shang, Y., Mackay, S., Alm, V., Butelli, E., Oyama, R., Bailey, P., Davies, K., and Martin, C. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F.W. (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41 207–234. [DOI] [PubMed] [Google Scholar]
- Sweeney, M., Thomson, M., Cho, Y., Park, Y., Williamson, S., Bustamante, C., and McCouch, S. (2007). Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 3 e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykorova, E., Fajkus, J., Meznikova, M., Lim, K.Y., Neplechova, K., Blattner, F.R., Chase, M.W., and Leitch, A.R. (2006). Minisatellite telomeres occur in the family Alliaceae but are lost in Allium. Am. J. Bot. 93 814–823. [DOI] [PubMed] [Google Scholar]
- Takos, A.M., Jaffe, F.W., Jacob, S.R., Bogs, J., Robinson, S.P., and Walker, A.R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F.A., Roberts, K., and Martin, C. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, E.E. (2005). Short, local duplications in eukaryotic genomes. Curr. Opin. Genet. Dev. 15 640–644. [DOI] [PubMed] [Google Scholar]
- Toufektsian, M.-C., et al. (2008). Chronic dietary intake of plant-derived anthocyanins protects the rat heart against ischemia-reperfusion injury. J. Nutr. 138 747–752. [DOI] [PubMed] [Google Scholar]
- Tzuri, G., Hillel, J., Lavi, U., Haberfeld, A., and Vainstein, A. (1991). DNA fingerprint analysis of ornamental plants. Plant Sci. 76 91–97. [Google Scholar]
- Ubi, B.E., Honda, C., Bessho, H., Kondo, S., Wada, M., Kobayashi, S., and Moriguchi, T. (2006). Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 170 571–578. [Google Scholar]
- Uimari, A., and Strommer, J. (1997). Myb26: A MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 12 1273–1284. [DOI] [PubMed] [Google Scholar]
- Vergnaud, G., and Denoeud, F. (2000). Minisatellites: Mutability and genome architecture. Genome Res. 10 899–907. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Lee, E., Bogs, J., McDavid, D.A.J., Thomas, M.R., and Robinson, S.P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49 772–785. [DOI] [PubMed] [Google Scholar]
- Williams, M.E., Foster, R., and Chua, N.H. (1992). Sequences flanking the hexameric G-Box core CACGTG affect the specificity of protein binding. Plant Cell 4 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman, A.R., and White, R. (1980). A highly polymorphic locus in human DNA. Proc. Natl. Acad. Sci. USA 77 6754–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J.-L., Cohen, D., Atkinson, R., Richardson, K., and Morris, B. (1995). Regeneration of transgenic plants from the commercial apple cultivar 'Royal Gala'. Plant Cell Rep. 14 407–412. [DOI] [PubMed] [Google Scholar]
- Zimmermann, I.M., Heim, M.A., Weisshaar, B., and Uhrig, J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.