Abstract
Expression of miR398 is induced in response to copper deficiency and is involved in the degradation of mRNAs encoding copper/zinc superoxide dismutase in Arabidopsis thaliana. We found that SPL7 (for SQUAMOSA promoter binding protein–like7) is essential for this response of miR398. SPL7 is homologous to Copper response regulator1, the transcription factor that is required for switching between plastocyanin and cytochrome c6 in response to copper deficiency in Chlamydomonas reinhardtii. SPL7 bound directly to GTAC motifs in the miR398 promoter in vitro, and these motifs were essential and sufficient for the response to copper deficiency in vivo. SPL7 is also required for the expression of multiple microRNAs, miR397, miR408, and miR857, involved in copper homeostasis and of genes encoding several copper transporters and a copper chaperone, indicating its central role in response to copper deficiency. Consistent with this idea, the growth of spl7 plants was severely impaired under low-copper conditions.
INTRODUCTION
Copper is an essential micronutrient for most living organisms as a cofactor for proteins involved in various functions, including photosynthesis, scavenging reactive oxygen species, and ethylene sensing (Pilon et al., 2006). The most abundant copper protein in higher plants is plastocyanin (PC), which is involved in photosynthetic electron transport in the thylakoid lumen. Another major copper protein, Cu/Zn superoxide dismutase (CSD), scavenges superoxide (Bowler et al., 1994). The Arabidopsis thaliana genome encodes three isozymes of CSD, which are localized to the cytoplasm (CSD1), chloroplast stroma (CSD2), and peroxisome (CSD3) (Kliebenstein et al., 1998).
In response to copper deficiency, plants modify the allocation of copper for the efficient utilization of the limited resource. The molecular mechanisms underlying the response to copper deficiency have been best characterized in a unicellular green alga, Chlamydomonas reinhardtii (Merchant et al., 2006). In Chlamydomonas, the function of PC is taken over by that of cytochrome c6, which contains iron rather than copper as a cofactor (Merchant et al., 1991). The transcription factor Copper response regulator1 (Crr1), containing a SBP (for SQUAMOSA promoter binding protein) domain, mediates this switching of photosynthesis machinery in response to copper deficiency and is also hypothesized to be somehow involved in copper sensing (Kropat et al., 2005). However, the Arabidopsis genome does not encode an ortholog to the algal cytochrome c6 (Weigel et al., 2003), and it is likely that during evolution flowering plants abandoned this strategy of response to copper deficiency.
Instead, Arabidopsis, Physcomitrella patens, and Barbula unguiculata switch their type of superoxide dismutase (SOD) to adapt to copper deficiency (Shikanai et al., 2003; Abdel-Ghany et al., 2005; Yamasaki et al., 2007, 2008; Nagae et al., 2008). In Arabidopsis, the production of CSD1 and CSD2 is downregulated and their function is replaced by Fe SOD in low-copper conditions (Abdel-Ghany et al., 2005). Previously, we demonstrated that the miR398 family was involved in this regulation (Yamasaki et al., 2007). miR398 is expressed only under copper deficiency and mediates the degradation of CSD1 and CSD2 mRNAs directly. Consequently, the limited copper is preferentially transferred to PC, which is essential for photosynthesis in higher plants (Weigel et al., 2003).
Simultaneously, plants respond to copper deficiency by elevating copper uptake. In Arabidopsis, the expression of some copper transporter genes is upregulated in response to copper deficiency (Sancenón et al., 2003; Wintz et al., 2003; Mukherjee et al., 2006). In addition, plants mobilize the limited copper between tissues. A copper chaperone, CCH (for copper chaperone), may be involved in this process during senescence (Mira et al., 2002). Production of CCH is also upregulated under copper deficiency. Although the physiology of adaptation to copper deficiency has been studied in higher plants, the molecular mechanism regulating gene expression in response to copper deficiency is unclear.
In this study, we present evidence that Arabidopsis SPL7, a member of the SPL (for SQUAMOSA promoter binding protein–like) family, which has a SBP domain (Cardon et al., 1999), activates the transcription of multiple genes involved in copper homeostasis, including miR398, some copper transporters, and CCH. We propose that SPL7 is a central regulator for copper homeostasis in Arabidopsis.
RESULTS
The cis Elements of the miR398 Promoter Are Essential for Response to Copper Deficiency
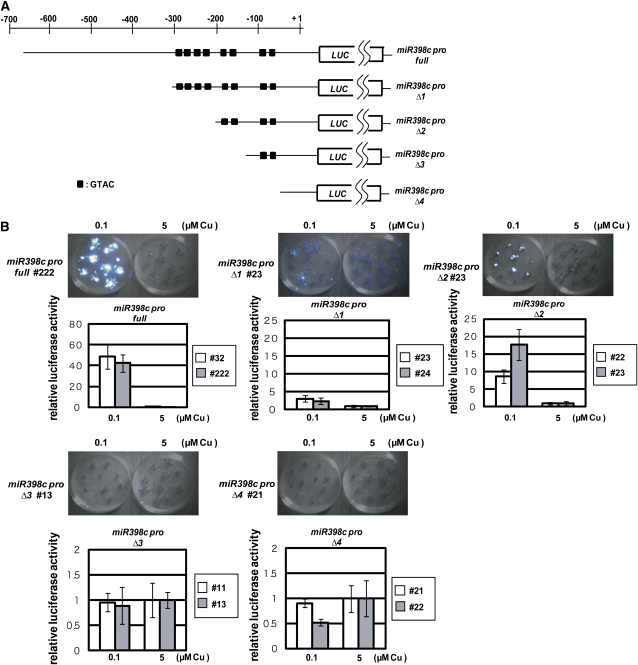
In Arabidopsis, the miR398 family consists of three genes, miR398a, -b, and -c. Copper deficiency upregulates the expression of miR398 and leads to the degradation of CSD1 and CSD2 mRNAs, which is essential for efficient utilization of limited copper (Yamasaki et al., 2007). To study the molecular mechanism underlying the copper-dependent transcriptional regulation of miR398, we focused on its promoter. For the promoter analysis, we selected miR398c, as its transcription initiation site was determined by 5′ rapid amplification of cDNA ends (Xie et al., 2005). The PLACE prediction software (http://www.dna.affrc.go.jp/PLACE/signalscan.html) identified eight GTAC motifs in a region of <300 bp upstream of the miR398c transcription initiation site (Figure 1A). The miR398b promoter also contains eight GTAC motifs in the proximal region (−300 to −1), whereas there are no GTAC motifs in the corresponding region of miR398a. miR398b and miR398c respond similarly to copper deficiency (see Figure 4 below).
Figure 1.
Identification of cis Elements in the Promoter Region of miR398c.
(A) Construction of miR398c pro full:LUC and various truncated derivatives, miR398c pro Δ1–4:LUC. The black boxes indicate the positions of GTAC motifs.
(B) Luminescence images and relative luciferase activities in 2-week-old seedlings of transgenic lines. Each graph shows relative levels of luminescence in two independent transgenic lines (white and gray bars). Data are averages of four independent seedlings with sd. Note that the scales of the y axes differ among panels.
Figure 4.
SPL7 Activates Transcription of miR398.
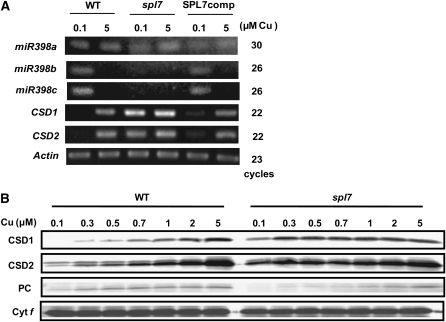
(A) RT-PCR analysis of genes responding to copper deficiency. The plants were grown for 3 weeks on MS agar medium containing 0.1 or 5 μM CuSO4. The number of PCR cycles was 30 for miR398a, 26 for miR398b, 26 for miR398c, 20 for CSD1 and CSD2, and 23 for Actin (control). SPL7comp identifies spl7 seedlings transformed with 35S pro:SPL7 cDNA.
(B) Immunoblot analysis of CSDs and PC in plants cultured on MS medium containing various concentrations of copper. Cytochrome f was detected as a loading control.
The GTAC motif is essential for transcriptional activation in response to copper deficiency in Chlamydomonas (Quinn et al., 2000). This suggests that copper deficiency activates the transcription of miR398b and miR398c via these GTAC motifs also in Arabidopsis. To test this possibility, we constructed transgenic Arabidopsis plants that express the luciferase gene (LUC) driven by full and truncated versions of the miR398c promoter (Figure 1A). LUC driven by the putative full promoter, covering the −650 to +31 region (miR398c pro full:LUC), contained all eight GTAC motifs. LUC driven by the truncated promoter, covering the −650 to −296 region (miR398c pro Δ1:LUC), also contained the eight motifs. miR398c pro Δ2:LUC contained four motifs, miR398c pro Δ3:LUC contained two motifs, and miR398c pro Δ4:LUC contained none.
LUC activity in seedlings grown on MS medium (Murashige and Skoog, 1962) containing low (0.1 μM) and high (5 μM) copper (Figure 1B) was monitored with a charge-coupled device camera. In miR398c pro full:LUC lines, high LUC activity was detected only under low copper, consistent with the previous result of RNA gel blot analysis (Yamasaki et al., 2007). LUC activity also responded strictly to copper status in miR398c pro Δ1:LUC and miR398c pro Δ2:LUC lines, although at a lower level than in the miR398c pro full:LUC lines. These results indicate that the −193 to +31 region containing four GTAC motifs is sufficient for the response to copper deficiency, although the further upstream region, especially −650 to −296, enhances the transcription in a copper-dependent manner. The miR398c pro Δ1:LUC lines responded to copper deficiency with smaller amplitude than the miR398c pro Δ2:LUC lines (Figure 1B). This result suggests that the −296 to −193 region on its own, containing four GTAC motifs, reduces the amplitude of the transcriptional response to copper deficiency, although it does not affect the response. In contrast, LUC expression did not respond to copper deficiency in the miR398c pro Δ3:LUC or miR398c pro Δ4:LUC lines.
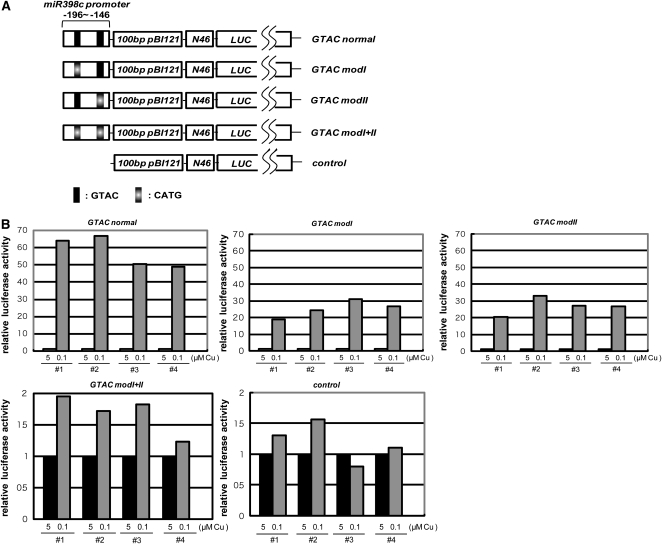
Subsequently, we constructed transgenic Arabidopsis plants that express LUC driven by the −196 to −146 region of the miR398c promoter followed by the N46 minimum promoter (Odell et al., 1985) (Figure 2A). This region contains two GTAC motifs that were shown to be essential for the response to low copper (Figure 1). LUC activity responded to copper status in the GTAC normal line (Figure 2B). In GTAC modI and GTAC modII lines, in which the GTAC motif located at −185 to −182 or −157 to −154 was modified to CATG, LUC activity also responded to copper status. However, LUC activity did not respond more to copper status in the GTAC modI+II line, in which both GTAC motifs were modified to CATG, as in the line lacking the miR398 promoter region (control). We conclude that the GTAC motif located at −185 to −182 or −157 to −154 is sufficient for the response to copper deficiency and also necessary at least in the absence of upstream GTAC motifs (see Discussion).
Figure 2.
The GTAC Motifs Located at −185 to −182 and −157 to −154 Are Sufficient for the Copper Response.
(A) The −196 to −146 region of the miR398c promoter was fused with the minimal promoter of 46N. To adjust the distance from GTAC motifs to the transcriptional start site, the 100-bp unrelated sequence derived from pBI121 was inserted between them. The chimeric promoter was placed in front of the LUC gene. The GTAC motif was replaced by the mutant CATG as indicated.
(B) Relative LUC activity in four independent T1 plants. The measurements are normalized so that luciferase activity at 5 μM copper (black) is set to 1. Note that the scales of the y axes differ among panels.
SPL7 Activates miR398 Transcription under Low-Copper Conditions
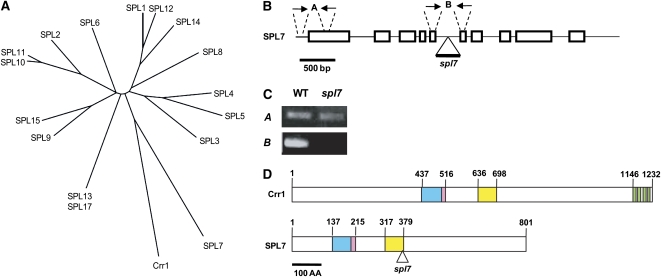
Members of the SPL family were predicted to recognize and bind to GTAC motifs in Arabidopsis (Birkenbihl et al., 2005; Liang et al., 2008). The SPL family is encoded by 16 genes in Arabidopsis (Figure 3A). Because Crr1, which is involved in copper homeostasis in Chlamydomonas (Kropat et al., 2005), recognized and bound to a GTAC motif, it is also possible that a member of the SPL family is involved in the copper response of miR398 expression via binding of its GTAC motifs in Arabidopsis. To assess this possibility, we examined a T-DNA insertion line (SALK_093849, At5g18830) of SPL7, which showed the highest similarity to Chlamydomonas Crr1 (Figure 3A). The SPL7 gene consists of 10 exons and 9 introns and encodes a larger protein than the other members (Figure 3B). In spl7, T-DNA was inserted into the fifth intron, and RT-PCR indicated that the intron was not precisely spliced (Figures 3B and 3C, primer set B). If the mutant version of SPL7 is expressed, as suggested by a result of RT-PCR using primer set A (Figures 3B and 3C), the C-terminal 422 amino acids are truncated in the mutant protein in spl7 (Figure 3D). The severe phenotypes reported here (see Figures 4, 7, and 10 below) indicate that the C-terminal part of SPL7 is essential for the function or stabilization of SPL7. Except for the SBP domain that is predicted to bind to a GTAC motif (Figure 3D, blue box), a nuclear localization signal (Figure 3D, pink boxes), and a well-conserved unknown motif (Figure 3D, yellow boxes), there was no similarity between Crr1 and SPL7. Although Crr1 has four putative metal binding motifs, CXXC, in the C-terminal region (Figure 3D, green boxes), the motifs were not conserved in SPL7.
Figure 3.
Structure of SPL7, a Member of the SPL Family.
(A) A phylogenetic tree of the Arabidopsis SPL family members, including Chlamydomonas Crr1, based on their SBP domain sequences.
(B) Exon–intron structure of SPL7. The triangle labeled spl7 indicates the site of T-DNA insertion, and arrows (A and B) indicate primer sets for amplifying cDNAs in Figure 3C. The primer sequences are described in Supplemental Table 5 online.
(C) RT-PCR analysis of SPL7 mRNA with primer pairs A and B in 4-week old seedlings cultured on standard MS medium.
(D) Diagram of the motif structures of Crr1 and SPL7. A triangle indicates the position from which the C-terminal part is truncated by the T-DNA insertion in spl7. The blue boxes indicate the SBP domain, and the pink boxes describe a putative nuclear localization signal. The yellow boxes show an unknown motif that is conserved between Crr1 and SPL7, and the green boxes indicate putative metal binding motifs.
Figure 7.
SPL7 Activates a Set of Genes Involved in Copper Homeostasis.
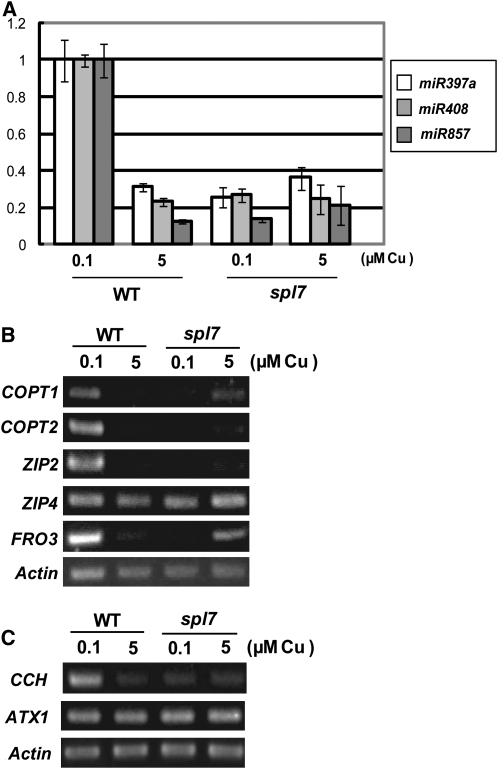
(A) Quantitative RT-PCR analysis for microRNAs. Aerial parts of seedlings grown for 3 weeks on MS agar medium containing 0.1 or 5 μM CuSO4 were used for RNA extraction. Error bars indicate sd (n = 3). The primer sequences are described in Supplemental Table 6 online.
(B) RT-PCR analysis of copper transporter genes. Roots were harvested from the seedlings used in (A) and RNA was extracted. The number of PCR cycles was 21 for COPT1, 22 for COPT2, 25 for ZIP2, 27 for ZIP4, 23 for FRO3, and 20 for Actin (control).
(C) RT-PCR analysis of copper chaperone genes. RNA samples extracted in (A) were used. The number of PCR cycles was 19 for CCH, 22 for ATX1, and 23 for Actin (control).
Figure 10.
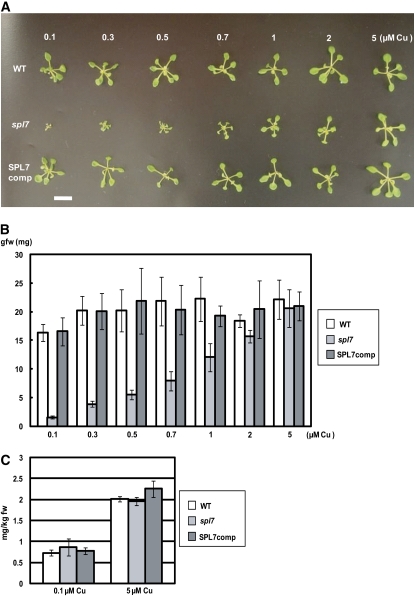
The spl7 Phenotype in Various Concentrations of Copper.
(A) Growth phenotypes of the wild type, spl7, and SPL7comp lines grown for 3 weeks on MS medium containing various concentrations of copper. Bar = 10 mm.
(B) Fresh weights of seedlings. Data are averages of four independent seedlings. Error bars indicate sd (n = 4).
(C) Copper contents in the wild type, spl7, and SPL7comp lines grown for 3 weeks on MS medium containing 0.1 or 5 μM CuSO4. Data are averages of three independent experiments. Error bars indicate sd (n = 3).
We grew the spl7 mutant in copper-deficient and -sufficient conditions and determined the levels of miR398 mRNA in the seedlings. Although the sequences of miR398b and miR398c were almost the same except for four nucleotides even in the precursors, we successfully detected microRNA precursors separately by the specific primer sets recognizing the minor differences in their sequences (see Supplemental Figure 1 online). RT-PCR analysis showed that the levels of miR398b and miR398c mRNA were below the detection limit in the spl7 mutant even under low copper (Figure 4A). For more quantitative analysis, we performed real-time PCR (see Supplemental Figure 2A online); because of the high degree of sequence similarity, in this experiment we detected miR398b and miR398c as a mixture. In contrast to miR398b and miR398c, the level of miR398a was constant in all copper conditions tested in both the wild type and spl7; this is consistent with the observation that the miR398a promoter region does not contain any GTAC motifs. The impact of the constant expression of miR398a on the copper response is likely to be subtle, since its expression level is quite low compared with that of miR398b and miR398c. Although we previously reported that miR398a also responded to copper (Yamasaki et al., 2007), it is likely that we detected miR398b and miR398c due to their high similarity to miR398a.
Consistent with the absence of miR398b and miR398c expression, CSD1 and CSD2 mRNAs accumulated even under low copper in spl7 (Figure 4A; see Supplemental Figure 2A online). Protein gel blot analysis also showed high levels of CSD1 and CSD2 even under low copper (Figure 4B; see Supplemental Figure 3 online). The response to copper status was almost restored in spl7 transformed with SPL7 cDNA driven by the cauliflower mosaic virus 35S promoter (Figure 4A, SPL7comp). These results indicate that SPL7 activates the transcription of miR398b and miR398c in response to copper deficiency.
The level of PC is restricted by copper supply via conversion from its apoprotein rather than via transcriptional control in Arabidopsis (Abdel-Ghany et al., 2005). In spl7, the PC level is lower especially at low copper concentrations (Figure 4B), suggesting the competition of copper acquisition between PC and other copper proteins, including CSD1 and CSD2.
The SBP Domain of SPL7 Binds to the GTAC Motifs of the miR398c Promoter
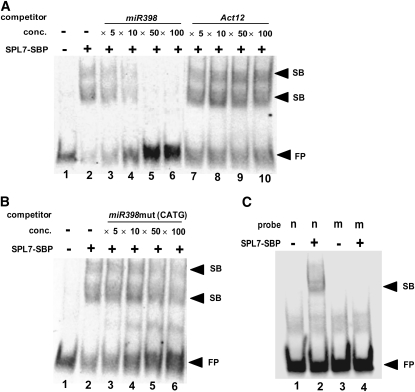
To study whether SPL7 interacts directly with the miR398 promoter via its GTAC motifs, we produced the SBP domain of SPL7 as a fusion protein with a His tag in Escherichia coli and used the purified recombinant protein in an electrophoretic mobility shift assay. The 40-nucleotide promoter sequence, including one of the GTAC motifs (−185 to −182) that are sufficient for the copper response (Figure 1), was labeled by digoxigenin and used as a probe. Addition of the 300 nM SBP domain protein efficiently retarded the mobility of the probe, revealing two shift bands (Figure 5A). The binding was impaired by the addition of a cold competitor probe containing the same sequence, and the shifted band was completely negated by the addition of 50× competitor. However, the addition of 100× cold Act12 DNA, which is not related to the miR398c promoter sequence, did not alter the shift pattern.
Figure 5.
The SBP Domain of SPL7 Binds to a GTAC Motif.
(A) Recombinant SBP domain of SPL7 was incubated with digoxigenin-labeled miR398c promoter (−203 to −163; lane 2). The mixture was also incubated with various concentrations of nonlabeled miR398c promoter (lanes 3 to 6) and Act12 promoter (lanes 7 to 10). Lane 1 contains only the probe. The molar ratio of protein to DNA is 125:1.
(B) The mixture was incubated with an unlabeled mutant version of the miR398c promoter (lanes 3 to 6). The GTAC motifs were substituted with CATG. Lane 1 contains only the probe.
(C) Recombinant SBP domain of SPL7 was incubated with the digoxigenin-labeled wild-type miR398c promoter (−203 to −163; lane 2) and with the mutated probe that was used in (B) as a competitor (lane 4). Lanes 1 and 3 contain controls without SPL7-SBP.
Arrowheads indicate the positions of two shift bands (SB) and the free probe (FP).
To test whether the SBP domain interacts with the miR398c promoter via its GTAC motifs, we used a mutant version of the sequence, in which the GTAC motif was altered to CATG, as a competitor (Figure 5B). Although addition of 50× to 100× mutated competitor slightly reduced the extent of the shift, the reduction was much lower than that of the wild-type competitor. Similarly, no shift band was detected when the digoxigenin-labeled mutant version of miR398c promoter was directly used as a probe (Figure 5C). This result and the genetic evidence (Figures 1 to 4) show that SPL7 interacts directly with the miR398c promoter via its GTAC motifs and activates the transcription of miR398c under copper deficiency.
SPL7 Is Constitutively Expressed Mainly in Roots
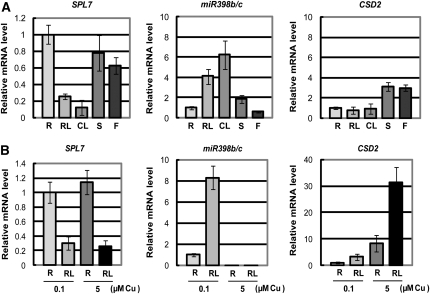
Quantitative RT-PCR analysis indicates that SPL7 mRNA accumulates mainly in roots but also in stems and flowers (Figure 6A). A trace amount of RNA was detected in rosette leaves, but the level was less than the detection limit in cauline leaves. In contrast, miR398b and miR398c were expressed mainly in leaves (Figure 6A) (Sunkar et al., 2006). Consistent with the previous report (Yamasaki et al., 2007), CSD2 mRNA accumulates in tissues where miR398b/c RNA does not accumulate (Figure 6A). Although aerial tissues were harvested from plants grown on soil, roots came from seedlings grown on MS medium containing 5 μM CuSO4 (Figure 6A). It is apparent that at least some aerial tissues experience mild copper deficiency in plants grown on soil, even when copper is replete in roots. To exclude the effect of different culture conditions and to study the impact of the different copper levels on gene expression, we harvested roots and rosette leaves from seedlings grown on MS medium containing 0.1 and 5 μM CuSO4 (Figure 6B). Although the SPL7 mRNA accumulates mainly in roots, a low level of the transcript was detected in leaves from seedlings cultured on the medium. The difference in copper level did not affect the SPL7 mRNA levels in roots or leaves. In contrast, the level of miR398b/c and consequently the level of CSD2 mRNA responded strictly to the difference in copper level, as described previously (Yamasaki et al., 2007). We conclude that the level of SPL7 mRNA is not as strongly affected by the copper level as Chlamydomonas CRR1 mRNA (Kropat et al., 2005), suggesting that SPL7 is involved in the primary process of copper sensing.
Figure 6.
SPL7 Expression Patterns.
(A) Quantitative RT-PCR analysis of tissue-specific expression. Plants were grown for 3 weeks on MS agar medium containing 5 μM CuSO4 for the preparation of roots (R) or for 5 weeks on soil for the preparation of rosette leaves (RL), cauline leaves (CL), stems (S), and flowers (F). Error bars indicate sd (n = 3).
(B) Quantitative RT-PCR analysis of the response to copper deficiency. Roots (R) and rosette leaves (RL) were harvested from seedlings grown for 3 weeks on MS medium containing 0.1 or 5 μM CuSO4. Error bars indicate sd (n = 3). The primer sequences are described in Supplemental Table 6 online.
SPL7 Regulates the Accumulation of a Set of Copper Proteins via Multiple MicroRNAs
SPL7 activated the transcription of miR398b and miR398c in response to copper deficiency (Figures 1 to 5). It is possible that SPL7 regulates the expression of other genes involved in copper homeostasis under copper deficiency. As well as miR398b and miR398c, the expression of miR397 and miR408 was upregulated specifically under low copper (Abdel-Ghany and Pilon, 2008). Whereas miR397 targets laccases lac2, lac4, and lac17 mRNAs, miR408 is involved in degrading lac3 and plantacyanin mRNAs (Abdel-Ghany and Pilon, 2008). Laccases encoded by lac genes and plantacyanin are apoplastic copper proteins that are believed to mediate lignin polymerization (Sterjiades et al., 1992; Bao et al., 1993; Nersissian et al., 1998). Additionally, miR857 was predicted to target lac7 mRNA from a genome-wide bioinformatic analysis (http://asrp.cgrb.oregonstate.edu/db/analyzeList.html). We analyzed the induction of these microRNAs in response to copper deficiency in the wild type and spl7 (Figure 7A). Consistent with an earlier report (Abdel-Ghany and Pilon, 2008), expression of miR397a and miR408 was upregulated under low copper in the wild type, as was expression of miR857. In the spl7 background, however, the levels remained low even under copper deficiency (Figure 7A). This result indicates that SPL7 also activates the transcription of miR397, miR408, and miR857. We conclude that SPL7 is a central gene regulating the levels of many copper proteins, including Cu/Zn SOD, laccases, and plantacyanin, via microRNAs such as miR397, miR398b, miR398c, miR408, and miR857.
SPL7 Regulates the Level of Copper Uptake and the Delivery System
One of the strategies by which plants respond to nutrient deficiency is to upregulate genes encoding nutrient uptake proteins (Puig et al., 2007). The expression of some copper transporter genes responded to copper deficiency (Sancenón et al., 2003). COPT1 (for Copper transporter1) and COPT2 are high-affinity copper transporters (Sancenón et al., 2003), and ZIP2 (for ZRT, IRT-like protein) and ZIP4 are putative divalent cation uptake transporters for copper and zinc (Wintz et al., 2003). Expression of the ferric-chelate reductase gene FRO3, involved in the reduction of divalent copper to monovalent copper, was also upregulated upon copper limitation (Mukherjee et al., 2006). We analyzed the response of these genes to copper deficiency (Figure 7B). We did not detect clear upregulation by copper deficiency in ZIP4 even in roots of the wild type (Figure 7B). In contrast, the expression of COPT1, COPT2, ZIP2, and FRO3 was increased under copper deficiency in wild-type roots. In contrast, their expression levels remained low under copper deficiency in spl7 roots (Figure 7B). Interestingly, FRO3 mRNA accumulated in spl7 under high copper (5 μM), although it was not detected in the wild type. This result suggests that SPL7 mediates the transcriptional upregulation of COPT1, COPT2, ZIP2, and FRO3 in response to copper deficiency and probably also the transcriptional suppression of FRO3 under copper sufficiency. All of the results of RT-PCR were consistent with the information independently obtained by microarray analysis (see Supplemental Table 1 online).
The copper chaperone CCH donates copper to a P-type ATPase, RAN1, and its production also is upregulated under copper deficiency (Mira et al., 2002). It has been proposed that CCH is also involved in the mobilization of copper between tissues during senescence. We observed the accumulation of CCH mRNA under low copper in the wild type (Figure 7C). In spl7, however, the mRNA level remained low under low copper. Another copper chaperone, ATX1 (for Antioxidant 1), was expressed constitutively, and its expression was not disturbed in spl7. This result indicates that SPL7 regulates the expression of CCH in response to copper deficiency. These results are consistent with the information obtained by microarray analysis (see Supplemental Table 1 online). Taking all results together (Figures 6 and 7), we conclude that SPL7 is a central regulator of copper homeostasis in Arabidopsis, modulating the levels of copper proteins, copper transporters, and a chaperone directly or indirectly via microRNAs.
SPL7 Regulates Many Genes in Response to Copper Deficiency
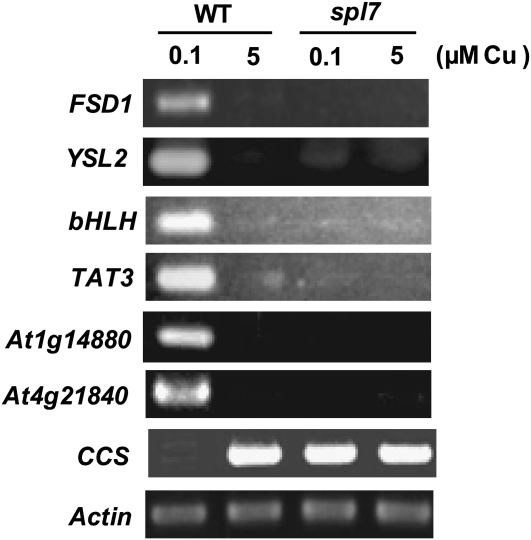
To characterize the copper-responsive gene network regulated by SPL7 more extensively, we performed microarray analysis in the wild type and spl7 grown in the copper-deficient and -sufficient conditions. In addition to the genes detected by RT-PCR (Figures 4 and 7), we identified 19 candidate genes that were upregulated >20 times in the wild type grown in low-copper conditions compared with spl7 cultured in the same conditions (see Supplemental Table 2 online). The genes discovered in RT-PCR (Figure 7) are not included in this table, since the induction by copper deficiency was less than 20 times that in the wild type and is separately listed in Supplemental Table 1. We also performed the RT-PCR analysis for some representative genes (Figure 8). The Fe SOD gene, FSD1, is upregulated in response to copper deficiency in the wild type (Abdel-Ghany et al., 2005) (Figure 8), but FSD1 did not respond to copper deficiency in spl7. YSL2 encodes a metal-nicotianamine transporter, involving the lateral movement of metals, especially iron and also copper, in the vasculature (DiDonato et al., 2004). Its copper response was also shown to be SPL7-dependent. In addition, the transcript levels of a novel basic helix-loop-helix (bHLH)–type transcription factor (At1g71200), TAT3 encoding tyrosine amino transferase, and two unknown genes (At4g05616 and At4g21840) were low in spl7 even in low-copper conditions compared with those in the wild type (Figure 8).
Figure 8.
Genome-Wide Analysis of the Copper-Responsive Network Regulated by SPL7.
The representative genes identified by transcriptome analysis (see Supplemental Tables 1 to 3 online) were analyzed by RT-PCR. Aerial parts of seedlings grown for 3 weeks on MS agar medium containing 0.1 or 5 μM CuSO4 were used for RNA extraction. The number of PCR cycles was 20 for FSD1, 25 for YSL2, 25 for bHLH, 20 for TAT3, 25 for At1g14880, 26 for At4g21840, and 23 for Actin (control).
On the contrary, we detected some candidate genes that were upregulated in spl7 grown in low-copper conditions compared with wild-type plants cultured in the same conditions (see Supplemental Table 3 online). As a representative, the RT-PCR data are shown for CCS, encoding copper chaperone for CSD1 and CSD2 (Figure 8). This type of response may be due to the secondary effects of drastic phenotypes of spl7 cultured in low-copper conditions (see Figure 10A below) in some candidate genes. These results indicate that SPL7 regulates the expression of many genes directly or indirectly in response to copper deficiency.
Nickel Mimics Copper in Response to Copper Deficiency
In Chlamydomonas, nickel induces a copper-deficient response even under copper sufficiency (Kropat et al., 2005). This result suggests that nickel has the potential to interfere with copper sensing, possibly by directly binding to Crr1, causing misregulation of the response to copper deficiency. To test whether nickel interferes with copper sensing also in Arabidopsis, we analyzed the accumulation of CSD1 and CSD2 in the presence of nickel plus high-level copper (5 μM). In contrast to the observation in Chlamydomonas (Kropat et al., 2005), both CSDs were not downregulated in the presence of nickel under copper sufficiency, implying that miR398b/c were not upregulated in these conditions (see Supplemental Figure 3 online).
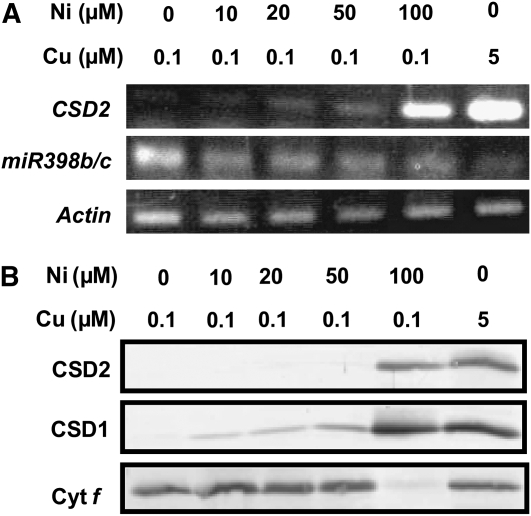
Subsequently, we analyzed the expression of miR398b/c in seedlings grown on medium containing low copper and various concentrations of nickel. RT-PCR analysis showed that expression of miR398b/c was suppressed by nickel even under low copper (Figure 9A). Consequently, the level of CSD2 mRNA was increased under high nickel because of the reduction in miR398b/c level. The levels of CSD1 and CSD2 protein were also increased by nickel feeding, reflecting the increase in RNA levels (Figure 9B). Cytochrome f was not detected in the presence of 100 μM NiCl2, possibly because of secondary effects. Our results are in contrast with the observation in Chlamydomonas, where nickel inhibited copper sensing (Kropat et al., 2005): in Arabidopsis, nickel mimics copper in the plant response to copper deficiency.
Figure 9.
Excess Nickel Mimics Copper in Regulating the Expression of miR398.
(A) RT-PCR analysis of CSD mRNAs in seedlings grown for 3 weeks on MS medium containing the indicated concentrations of copper and nickel. The number of PCR cycles was 22 for CSD2, 25 for miR398b/c, and 23 for Actin (control).
(B) Immunoblot analysis of CSDs and cytochrome f in the seedlings used in (A).
spl7 Is Hypersensitive to Copper Deficiency
SPL7 is essential to activate a series of gene expression in response to the copper deficiency (Figures 4, 7, and 8; see Supplemental Tables 1 to 3 online). To study the physiological significance of this regulatory process, we grew spl7 on media containing various concentrations of copper (Figure 10). As reported previously (Abdel-Ghany et al., 2005), the 0.1 μM copper in the original MS medium was not sufficient for Arabidopsis seedlings, and wild-type seedlings were slightly smaller than those grown with higher levels of copper (Figures 10A and 10B). In the presence of 0.3 μM copper, however, seedling growth was almost saturated, suggesting that this copper level was optimal. Consistently, the level of miR398b was drastically decreased upon an increase in copper from 0.1 to 0.5 μM in the wild type (Yamasaki et al., 2007). In contrast, seedling growth was drastically impaired in spl7, especially at copper levels of <0.5 μM (Figures 10A and 10B), indicating that the response to copper deficiency via SPL7 is physiologically significant.
Since SPL7 is essential to the expression of genes involved in copper uptake (COPT1, COPT2, ZIP2, and FRO3) (Figure 7B), it is likely that the copper content is reduced in spl7 seedlings grown under low copper. To study this possibility, we determined the copper contents in wild-type and spl7 seedlings grown with various concentrations of copper (Figure 10C). Despite the severe phenotype of spl7 under low copper, the copper level was comparable between spl7 and the wild type at all concentrations of copper (Figure 10C). This result suggests that copper uptake was compensated by other copper transporters (COPT3, COPT4, COPT5, and possibly ZIP4) in spl7 and that SPL7 is required for allocating limited copper in plants under low copper, rather than activating copper uptake in roots.
Seedling growth was still slightly impaired even in the presence of 2 μM copper in spl7, although it was almost completely suppressed by 5 μM copper (Figure 10B). This result is consistent with the fact that a trace amount of miR398b RNA was detected in the presence of 2 μM copper, although the amount was less than the detection limit at 5 μM copper (Yamasaki et al., 2007). This result suggests that the function of SPL7 is required even under copper sufficiency, although its function is more important in copper deficiency.
DISCUSSION
A prototype of transcription factors containing a SBP domain is SQUAMOSA promoter binding protein, which regulates the expression of MADS box genes during early flower development in Antirrhinum majus (Klein et al., 1996). Family members have been reported in many plants, including Chlamydomonas (Kropat et al., 2005) and P. patens (Riese et al., 2007), as well as in higher plants such as Zea mays (Moreno et al., 1997) but not in animals or fungi. In Arabidopsis, the SPL family is encoded by 16 genes (Figure 2A). SPL3 is involved in flowering (Cardon et al., 1997), and SPL8 is involved in pollen sac development via the mediation of gibberellin signaling (Unte et al., 2003; Zhang et al., 2007). In this study, we demonstrate that SPL7 is a central regulator for copper homeostasis, operating especially under copper deficiency. In contrast to other members containing a C3H-type N-terminal zinc finger, SPL7 has a C4-type zinc finger and is classified into subgroup A1.4 (Yang et al., 2008). A1.4 members are most closely related to Chlamydomonas Crr1 (Figure 3A).
The SBP domain of the SPL family members binds to the GTAC motif (Birkenbihl et al., 2005). Using the in vitro assay (Figure 5), we also demonstrated that the GTAC motif in the miR398c promoter is essential for binding with SPL7. Including the GTAC motif used in this assay (−185 to −182), the miR398c promoter contains eight GTAC motifs within 300 nucleotides (Figure 1A). The presence of multiple GTAC motifs also characterizes the promoters of genes that are shown to be under the control of SPL7. Multiple GTAC motifs are included in the promoter region (within 600 nucleotides) of miR397 (7), miR408 (10), miR857 (6), COPT1 (4), COPT2 (4), ZIP2 (5), and CCH (4). Although the expression of FRO3 responds to copper, the FRO3 promoter contains no GTAC motifs, implying that an unknown transcription factor secondarily mediates this signaling from SPL7. Actually, we showed that the transcript level of some transcription factors is elevated in low-copper conditions via the function of SPL7 (Figure 8; see Supplemental Table 2 online). Recently, the GTAC motifs present in the Fe SOD gene of a moss, B. unguiculata, were shown to be responsible for repressing the gene expression in the presence of copper (Nagae et al., 2008). The promoter of an Arabidopsis Fe SOD gene, FSD1, is also characterized by the presence of multiple GTAC motifs, but they are likely to be involved in the activation of gene expression in the absence of copper (Figure 8).
Although we demonstrated that two GTAC motifs localized at −185 to −182 and −157 to −154 are sufficient for the response to copper deficiency (Figure 2) and also necessary at least in the absence of upstream GTAC motifs (Figure 1), it is possible that the distal six GTAC motifs also contribute to binding to SPL7 in vivo. In fact, the miR398c pro full∷LUC lines containing mutations in the proximal third and fourth GTAC motifs (GTAC was substituted by CATG) still responded to copper deficiency (see Supplemental Figure 4 online). These mutations suppress the binding of the SBP domain to the promoter region in vitro (Figure 5B), suggesting that the mutant motifs are also inactive in vivo. It is probable that SPL7 can alternatively bind to some GTAC motifs to activate transcription.
Arabidopsis SPL7 is closely related to Chlamydomonas Crr1 in structure and function (Figure 3A). Both activate the transcription of a set of genes in response to copper deficiency via binding to the GTAC motifs and are possibly also involved in copper sensing (Kropat et al., 2005). However, the strategies of response to copper deficiency show diversity between green algae and flowering plants. Crr1 activates a set of genes including CYC6, encoding cytochrome c6, a heme-containing substitute for copper-containing PC (Eriksson et al., 2004). During their evolution, land plants are likely to have abandoned this strategy, and PC is essential for photosynthesis in Arabidopsis (Weigel et al., 2003). Instead, Arabidopsis switches SODs in response to copper deficiency (Abdel-Ghany et al., 2005). However, the Chlamydomonas genome does not encode Cu/Zn SOD (Merchant et al., 2007), and Fe SOD is likely to function constitutively. Thus, although the central gene for copper homeostasis is conserved between Arabidopsis and Chlamydomonas, SPL7 and Crr1, respectively, activate different sets of genes in response to copper deficiency, leading to different strategies. To downregulate the expression of several genes under copper deficiency, Arabidopsis uses microRNAs: miR397, miR398, miR408, and miR857 (Figures 4 and 7).
Although SPL7 mRNA is constitutively detected in some tissues, SPL7 activates transcription only under low copper (Figure 6). This result implies that SPL7 is involved in the primary sensing of copper concentration. What is the mechanism of copper sensing for regulating transcription factors? In Schizosaccharomyces pombe, the cellular localization of a central transcription factor, Cuf1, is regulated by copper (Beaudoin and Labbé, 2007), and its C-terminal Cys-rich region is involved in copper sensing (Beaudoin and Labbé, 2006). In Chlamydomonas, however, Kropat et al. (2005) proposed a model of copper sensing via Crr1, a member of the plant-specific family of transcription factors. Under copper sufficiency, copper may occupy the zinc binding site of the SBP domain of Crr1 or bind to the Cys-rich C-terminal region, resulting in inactivation of Crr1. In SPL7, however, the putative Cys-rich region is not conserved (Figure 3D). Therefore, inactivation of SPL7 may be mediated by some unidentified copper-sensing protein in Arabidopsis. It is also possible that the stability of SPL7 is regulated by the monitoring of copper status. Although SPL7 mRNA accumulated under any copper conditions (Figure 6B), we could not detect the fusion of the SPL7 protein with a T7 tag even under the control of the 35S promoter. SPL7 protein may be degraded via an unknown mechanism such as ubiquitination under high copper. It may be directly modified by copper to be recognized for degradation. Alternatively, SPL7 binding protein may be modulated by copper to activate the degradation of SPL7.
In Chlamydomonas, nickel activates Crr1 function even under copper sufficiency (Quinn et al., 2003). This result is explained by the binding of nickel to the putative copper binding site of Crr1, disturbing the copper sensing. However, nickel mimics copper under low copper in Arabidopsis (Figure 9). This discrepancy is most simply explained by the structural difference between SPL7 and Crr1. Nickel binding instead of copper may not inhibit Crr1 function in transcriptional activation, thus causing the disruption of copper sensing under copper sufficiency in Chlamydomonas. In contrast, binding of nickel as well as copper may inhibit SPL7 function in transcriptional activation, resulting in the disruption of copper sensing under copper deficiency in Arabidopsis. It is also possible that nickel binds to the other protein involved in primary copper sensing and the information is transferred to SPL7 in Arabidopsis. This difference in nickel sensitivity may have a physiological reason: although nickel is not essential for Chlamydomonas, it is essential for Arabidopsis, especially for nitrogen metabolism as a cofactor of urease (Witte et al., 2005). Despite its significance, excess nickel is toxic because of the production of reactive oxygen species, leading to lipid peroxidation (Freeman et al., 2004). The strategy for managing excess nickel may partly overlap that for managing excess copper, and SPL7 may have dual functions.
spl7 plants showed a severe phenotype under low copper (Figure 10). Since the seedlings were grown on medium containing sucrose, the phenotype cannot be explained by the lack of holo-PC, which is essential for photosynthesis. Thus, this phenotype clearly indicates the significance of copper homeostasis maintained by SPL7 in Arabidopsis. Since the expression of other metal transporters was reduced in spl7 (Figure 7B), it is also possible that zinc or ion levels were also affected. Actually, our microarray analysis clarified the gene network that responds to copper deficiency (Figure 8; see Supplemental Tables 1 to 3 online). YSL2 is upregulated in response to copper deficiency, which is probably required for appropriate copper redistribution and also for the iron trafficking in low-copper environments. Since the YSL2 promoter region contains five GTAC motifs within 1000 bp, YSL2 may be directly regulated by SPL7. In spl7, the transcript levels of some transcription factors were reduced. The candidates include the putative bHLH-type transcription factor (At4g25100) containing six GTAC motifs within its 1000-bp promoter region. Therefore, SPL7 may indirectly regulate several genes that do not contain GTAC motifs by directly regulating these transcription factors.
Unexpectedly, spl7 seedlings grown on low-copper media contained the same level of copper as the wild-type seedlings (Figure 10C), although they could not upregulate the expression of copper transporter genes (Figure 7B). The levels of COPT1 and COPT2 mRNA were much higher in leaves than in roots (Sancenón et al., 2003). Upregulation of these copper transporter genes under low copper may be relevant to appropriate copper distribution in plants rather than to incorporation into root cells. Even in the presence of 2 μM CuSO4, spl7 seedling growth was somewhat suppressed, suggesting that some tissues experience copper deficiency even when roots are replete. Although SPL7 is expressed mainly in roots, where plants probably sense copper availability, the mRNA for its direct target gene, miR398, is found mainly in leaves (Figure 6). We still cannot explain this discrepancy. Recently, miR398 RNA was found in the phloem of Brassica napus (Buhtz et al., 2008), suggesting the movement of the microRNA. However, it is puzzling that many candidates for the direct target of SPL7 are expressed also in shoots except for miR398b and miR398c (Figures 7 and 8; Supplemental Tables 1 and 2 online). To understand the mechanism of copper homeostasis in multicellular plants, we cannot model the function of SPL7 based on simple analogy with Chlamydomonas Crr1.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia gl1 was used as the wild type. A mutant defective in the SPL7 gene was obtained from the Salk T-DNA collection (http://signal.salk.edu). Seedlings were grown on agar-solidified MS medium including 1% sucrose and the indicated concentrations of CuSO4 and NiCl2 under controlled conditions (light intensity of 40 μmol·m−2·s−1, 16/8-h light/dark cycle at 23°C). For LUC assay using the T1 generation, 2-week-old T1 plants grown on MS medium containing 5 μM CuSO4 and 40 mg/L kanamycin were transferred to MS medium containing 5 μM CuSO4 and cultured for 3 d. After monitoring LUC activity, plants were transferred again to standard MS medium containing 0.1 μM CuSO4 and were cultured for an additional 3 d, then Luc activity was monitored again.
Plasmid Construction
Primer sequences can be found in Supplemental Table 4 online. To construct miR398c pro full, Δ1, Δ2, Δ3, and Δ4:LUC, we PCR-amplified the miR398c promoter region using primers miR398c pro-F and miR398c pro-R for miR398cp full:LUC, miR398c pro-F1 and miR398c pro-R for miR398c pro Δ1:LUC, miR398c pro-F2 and miR398c pro-R for miR398c pro Δ2:LUC, miR398c pro-F3 and miR398c pro-R for miR398c pro Δ3:LUC, and miR398c pro-F4 and miR398c pro-R for miR398c pro Δ4:LUC. The PCR products were digested with BglII and NcoI and cloned into the pBI221LUC+ vector (Ono et al., 2004). The resulting plasmids were digested with SalI and EcoRI, and the inserts were cloned into pBIN19. For SPL7comp, the SPL7 cDNA sequence was amplified by RT-PCR using primers SPL7ox-F and SPL7ox-R. The PCR products were digested with XbaI and SacI and then cloned into pBI121, which contained the hygromycin resistance gene. For construction of miR398c pro modI:LUC, we used primers miR398c pro-F and miR398c modI-R for template 1 and miR398c modI-F and miR398c pro-R for template 2. The resulting products were fused by recombinant PCR (Rashtchian, 1995) using primers miR398c pro-F and miR398c pro-R and then cloned into pBIN19 in the same way as miR398c pro full:LUC. miR398c pro modII:LUC was constructed in the same way as miR398c pro modI:LUC. We used primers miR398c pro-F and miR398c modII-R for template 1 and miR398c modII-F and miR398c pro-R for template 2. To construct miR398c pro modI+II:LUC, the miR398c promoter region was amplified from the modII:LUC plasmid using primers miR398c pro-F and miR398c modII-R for template 1 and miR398c modII-F and miR398c pro-R for template 2 and then cloned into pBIN19. For the control, we amplified the sequence of pBI121 using primers pBI121-F and pBI121-R, and the resulting PCR product was digested with SalI and BglII and cloned into the pBI221LUC+ vector. Finally, the plasmid was digested with HindIII and EcoRI and cloned into pBIN19. For the GTAC normal, the promoter region of miR398c was amplified by primers miR398c proHindIII-F and miR398c proSalI-R. The PCR products were digested with HindIII and SalI and cloned into the control vector. For GTAC modI, modII, and modI+II construction, the vector sequences of miR398c pro modI, modII, and modI+II:LUC were amplified by primers miR398c proHindIII-F and miR398c proSalI-R and cloned into pBIN19 in the same way as for the GTAC normal.
In Vivo Luciferase Assay
One millimolar luciferin (Invitrogen) was sprayed on 2-week-old seedlings, and the luminescence was captured by a charge-coupled device camera. The luciferase activity was calculated by Aquacosmos software (Hamamatsu Photonics).
Production of the Recombinant SBP Domain of SPL7
The sequence encoding the SBP domain of SPL7 was amplified by RT-PCR using primers SBP-NcoI-F and SBP-XhoI-R and was cloned into pET-30a(+) (Novagen). The resulting plasmid was introduced into Escherichia coli BL21-Gold (Stratagene). Production of the SBP domain fused with the His tag was induced by 1 mM isopropyl-β-d-thiogalactopyranoside at 37°C, and the recombinant protein was purified with the HisTrap system (GE Healthcare).
Electrophoretic Mobility Shift Assay
The miR398c promoter region containing 40 nucleotides (5′-TTGTAAATCAGTTTCGCAGTACACAATTCTGTAGGTTTTT-3′) was labeled by digoxigenin in the DIG Gel Shift kit, second generation (Roche). SBP domain protein (300 ng) was incubated with 1 ng of probe DNA and various concentrations of competitor DNA at room temperature for 15 min. The sequences of competitors are 5′-GAAAATGGCCTTTTAACCTTTTTCTTAGGCCCAAATGGGC-3′ for Act12 and 5′-TTGTAAATCAGTTTCGCACATGACAATTCTGTAGGTTTTT-3′ for miR398mut (the underlined area indicates sequence modification). The samples were loaded onto 7.5% (w/v) polyacrylamide gels and run at 4°C for 1.5 h at 0.8 V/cm2 in 0.5× TBE electrophoresis buffer (44.5 mM Tris base, 44.5 mM boric acid, and 1 mM EDTA). DNA was blotted onto a nylon membrane and detected with digoxigenin-specific antibodies.
RNA Extraction and RT-PCR Analysis
Total RNA was isolated from many 2-week-old seedlings grown on MS agar medium supplemented with CuSO4 by Sepazol reagent (Nakarai). Five micrograms of total RNA was used for cDNA synthesis in the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). To detect PCR products, ethidium bromide staining was used. Primer sequences used for RT-PCR are described in Supplemental Table 5 online. The results were confirmed using two or three independent biological replicates. The number of PCR cycles was optimized for each primer set to be in the linear range of amplification.
Immunoblot Analysis
Rosette leaves (0.1 g) in 3-week-old seedlings were harvested and ground in protein extraction buffer as described (Abdel-Ghany et al., 2005). The total protein concentrations were determined by protein assay (Bio-Rad). Twenty micrograms of proteins was loaded onto 15% SDS-PAGE gels. Proteins were blotted onto nitrocellulose membranes and incubated with specific antibodies. Then the blots were incubated with alkaline phosphatase–conjugated secondary antibody (Sigma-Aldrich), and signals were detected by 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium color reaction and developed to be in the linear range of detection (Sigma-Aldrich). Detection of dilution series of the protein samples indicates that the detection of CSDs and PC is 50 and 25% sensitive, respectively.
Copper Ion Measurement
Three-week-old seedlings were dried overnight at 60°C and then acid-digested in 1 mL of HNO3 for 5 min in a 500-W microwave oven at full power. Samples diluted with water were analyzed by atomic absorption spectrophotometry (Unicam).
Microarray Analysis
Total RNA was isolated from 2-week-old wild-type and spl7 seedlings grown on MS agar medium and MS agar medium supplemented with 5 μM CuSO4 by the RNeasy Plant Mini kit (Qiagen). Microarray analysis was performed as recommended by the manufacturer's instructions (Agilent Technologies). Total RNA was used to produce Cy3-labeled complementary RNA probes. The labeled probes were hybridized to an Agilent Arabidopsis 3 Oligo Microarray (Agilent Technologies). Feature extraction software and GeneSpring (Agilent Technologies) were used to locate and delineate every spot in the array and to integrate the intensity, filtering, and normalization of each spot. Three RNA samples isolated from independent plant materials were analyzed.
Quantitative RT-PCR Analysis
Transcript levels were assayed using the qPCR system (Mx30000p, Stratagene) and the Brilliant SYBR Green QPCR Master Mix (Stratagene). Three RNA samples isolated from independent plant materials were analyzed. Transcript abundance for wild-type plants grown on MS medium (a and d), roots (b), and roots of wild-type plants grown on MS medium (c) was normalized to 1 for each gene. Primer sequences are described in Supplemental Table 6 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative and GenBank/EMBL databases under the following accession numbers: SPL7, AT5G18830 and Q8S9G8; CSD1, AT1G08830 and Q9FRQ6; CSD2, AT2G28190 and O78310; ACT12, AT3G46520 and P53497.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Specific Amplification of miR398b and miR398c.
Supplemental Figure 2. Quantification of Gene Expression by Real-Time PCR.
Supplemental Figure 3. Excess Nickel Does Not Disturb the Copper Regulation under Copper Sufficiency.
Supplemental Figure 4. Identification of cis Elements in the Promoter Region of miR398c.
Supplemental Table 1. Microarray Data for Genes Detected by RT-PCR.
Supplemental Table 2. A List of Genes Upregulated >20 Times under Copper Deficiency in the Wild Type.
Supplemental Table 3. A List of Genes Downregulated under Copper Deficiency to <5% of Normal Transcript Levels in the Wild Type Compared with spl7.
Supplemental Table 4. Primer Sequences Used for Plasmid Construction.
Supplemental Table 5. Primer Sequences Used for RT-PCR Analysis.
Supplemental Table 6. Primer Sequences Used for Quantitative RT-PCR Analysis.
Supplementary Material
Acknowledgments
We thank Takahiro Nakamura for his critical advice on motif analysis by computer and Yuki Mori and Kazutoshi Saeki for their technical advice on measuring copper content. Marinus Pilon gave us his microRNA results before publication. T.S. was supported by Grants-in-Aid for Creative Scientific Research (Grant 17GS0316) from the Japan Society for the Promotion of Science and for Scientific Research on Priority Areas (Grant 16085206) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. T.S. was also supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation; Grant GPN0008). H.Y. was supported by a Grant-in-Aid for Research Fellowship for Young Scientists (Grant 20-2979) from the Japan Society for the Promotion of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Toshiharu Shikanai (shikanai@pmg.bot.kyoto-u.ac.jp).
Online version contains Web-only data.
References
- Abdel-Ghany, S.E., Müller-Moulé, P., Niyogi, K.K., Pilon, M., and Shikanai, T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17 1233–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany, S.E., and Pilon, M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283 15932–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, W., O'Malley, D.M., Whetten, R., and Sederoff, R.R. (1993). A laccase associated with lignification in loblolly pine xylem. Science 260 672–674. [DOI] [PubMed] [Google Scholar]
- Beaudoin, J., and Labbé, S. (2006). Copper induces cytoplasmic retention of fission yeast transcription factor cuf1. Eukaryot. Cell 5 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, J., and Labbé, S. (2007). Crm1-mediated nuclear export of the Schizosaccharomyces pombe transcription factor Cuf1 during a shift from low to high copper concentrations. Eukaryot. Cell 6 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl, R.P., Jach, G., Saedler, H., and Huijser, P. (2005). Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352 585–596. [DOI] [PubMed] [Google Scholar]
- Bowler, C., Van Camp, W., Van Montagu, M., and Inze, D. (1994). Superoxide dismutase in plants. Crit. Rev. Plant Sci. 13 199–218. [Google Scholar]
- Buhtz, A., Springer, F., Chappell, L., Baulcombe, D.C., and Kehr, J. (2008). Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53 739–749. [DOI] [PubMed] [Google Scholar]
- Cardon, G., Höhmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237 91–104. [DOI] [PubMed] [Google Scholar]
- Cardon, G.H., Höhmann, S., Nettesheim, K., Saedler, H., and Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 12 367–377. [DOI] [PubMed] [Google Scholar]
- DiDonato, R.J., Jr., Roberts, L.A., Sanderson, T., Eisley, R.B., and Walker, E.L. (2004). Arabidopsis Yellow Stripe-Like2 (YSL2): A metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. 39 403–414. [DOI] [PubMed] [Google Scholar]
- Eriksson, M., Moseley, J.L., Tottey, S., Del Campo, J.A., Quinn, J., Kim, Y., and Merchant, S. (2004). Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics 168 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, J.L., Persans, M.W., Nieman, K., Albrecht, C., Peer, W., Pickering, I.J., and Salt, D.E. (2004). Increased glutathione biosynthesis plays a role in nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Cell 16 2176–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, J., Saedler, H., and Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250 7–16. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Monde, R.A., and Last, R.L. (1998). Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat, J., Tottey, S., Birkenbihl, R.P., Depege, N., Huijser, P., and Merchant, S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102 18730–18735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X., Nazarenus, T.J., and Stone, J.M. (2008). Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry 47 3645–3653. [DOI] [PubMed] [Google Scholar]
- Merchant, S., Allen, M.D., Kropat, J., Moseley, J.L., Long, J.C., Tottey, S., and Terauchi, A.M. (2006). Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763 578–594. [DOI] [PubMed] [Google Scholar]
- Merchant, S., Hill, K., and Howe, G. (1991). Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. EMBO J. 10 1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, H., Martínez, N., and Peñarrubia, L. (2002). Expression of a vegetative-storage-protein gene from Arabidopsis is regulated by copper, senescence and ozone. Planta 214 939–946. [DOI] [PubMed] [Google Scholar]
- Moreno, M.A., Harper, L.C., Krueger, R.W., Dellaporta, S.L., and Freeling, M. (1997). liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 11 616–628. [DOI] [PubMed] [Google Scholar]
- Mukherjee, I., Campbell, N.H., Ash, J.S., and Connolly, E.L. (2006). Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223 1178–1190. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nagae, M., Nakata, M., and Takahashi, Y. (2008). Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol. 146 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersissian, A.M., Immoos, C., Hill, M.G., Hart, P.J., Williams, G., Herrmann, R.G., and Valentine, J.S. (1998). Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: Plant-specific mononuclear blue copper proteins. Protein Sci. 7 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell, J.T., Nagy, F., and Chua, N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313 810–812. [DOI] [PubMed] [Google Scholar]
- Ono, S., Tanaka, T., Watakabe, Y., and Hiratsuka, K. (2004). Transient assay system for the analysis of PR-1a gene promoter in tobacco BY-2 cells. Biosci. Biotechnol. Biochem. 68 803–807. [DOI] [PubMed] [Google Scholar]
- Pilon, M., Abdel-Ghany, S.E., Cohu, C.M., Gogolin, K.A., and Ye, H. (2006). Copper cofactor delivery in plant cells. Curr. Opin. Plant Biol. 9 256–263. [DOI] [PubMed] [Google Scholar]
- Puig, S., Andrés-Colás, N., García-Molina, A., and Peñarrubia, L. (2007). Copper and iron homeostasis in Arabidopsis: Responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ. 30 271–290. [DOI] [PubMed] [Google Scholar]
- Quinn, J.M., Barraco, P., Eriksson, M., and Merchant, S. (2000). Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275 6080–6089. [DOI] [PubMed] [Google Scholar]
- Quinn, J.M., Kropat, J., and Merchant, S. (2003). Copper response element and Crr1-dependent Ni(2+)-responsive promoter for induced, reversible gene expression in Chlamydomonas reinhardtii. Eukaryot. Cell 2 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashtchian, A. (1995). Novel methods for cloning and engineering genes using the polymerase chain reaction. Curr. Opin. Biotechnol. 6 30–36. [DOI] [PubMed] [Google Scholar]
- Riese, M., Höhmann, S., Saedler, H., Münster, T., and Huijser, P. (2007). Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 401 28–37. [DOI] [PubMed] [Google Scholar]
- Sancenón, V., Puig, S., Mira, H., Thiele, D.J., and Penarrubia, L. (2003). Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 51 577–587. [DOI] [PubMed] [Google Scholar]
- Shikanai, T., Müller-Moulé, P., Munekage, Y., Niyogi, K.K., and Pilon, M. (2003). PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterjiades, R., Dean, J.F., and Eriksson, K.E. (1992). Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 99 1162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., Kapoor, A., and Zhu, J.K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unte, U.S., Sorensen, A.M., Pesaresi, P., Gandikota, M., Leister, D., Saedler, H., and Huijser, P. (2003). SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, M., Varotto, C., Finazzi, G., Rappaport, F., Salamini, F., and Leister, D. (2003). Plastocyanin is indispensable for photosynthetic electron flow in Arabidopsis thaliana. J. Biol. Chem. 278 31286–31289. [DOI] [PubMed] [Google Scholar]
- Wintz, H., Fox, T., Wu, Y.Y., Feng, V., Chen, W., Chang, H.S., Zhu, T., and Vulpe, C. (2003). Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278 47644–47653. [DOI] [PubMed] [Google Scholar]
- Witte, C.P., Rosso, M.G., and Romeis, T. (2005). Identification of three urease accessory proteins that are required for urease activation in Arabidopsis. Plant Physiol. 139 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S.A., and Carrington, J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, H., Abdel-Ghany, S.E., Cohu, C.M., Kobayashi, Y., Shikanai, T., and Pilon, M. (2007). Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282 16369–16378. [DOI] [PubMed] [Google Scholar]
- Yamasaki, H., Pilon, M., and Shikanai, T. (2008). How do plants respond to copper deficiency? Plant Signal. Behav. 3 231–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Wang, W., Gu, S., Hu, Z., Xu, H., and Xu, C. (2008). Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407 1–11. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Schwarz, S., Saedler, H., and Huijser, P. (2007). SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 63 429–439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.