Abstract
Seed development in Arabidopsis thaliana undergoes an initial phase of endosperm proliferation followed by a second phase in which the embryo grows at the expense of the endosperm. As mature seed size is largely attained during the initial phase, seed size is coordinately determined by the growth of the maternal ovule, endosperm, and embryo. Here, we identify SHORT HYPOCOTYL UNDER BLUE1 (SHB1) as a positive regulator of Arabidopsis seed development that affects both cell size and cell number. shb1-D, a gain-of-function overexpression allele, increases seed size, and shb1, a loss-of-function allele, reduces seed size. SHB1 is transmitted zygotically. The increase in shb1-D seed size is associated with endosperm cellurization, chalazal endosperm enlargement, and embryo development. SHB1 is required for the proper expression of two other genes that affect endosperm development, MINISEED3 (MINI3) and HAIKU2 (IKU2), a WRKY transcription factor gene and a leucine-rich repeat receptor kinase gene. SHB1 associates with both MINI3 and IKU2 promoters in vivo. SHB1 may act with other proteins that bind to MINI3 and IKU2 promoters to promote a large seed cavity and endosperm growth in the early phase of seed development. In the second phase, SHB1 enhances embryo cell proliferation and expansion through a yet unknown IKU2-independent pathway.
INTRODUCTION
In angiosperms, double fertilization leads to the formation of a diploid embryo and a triploid endosperm, as the endosperm arises from the central cell that contains two identical haploid genomes. The endosperm constitutes the major volume of the mature seed in monocots and some dicots. In Arabidopsis thaliana and many other dicots, seed development is marked by two distinct phases (Sundaresan, 2005). In an initial phase, rapid growth and proliferation of the endosperm results in a large increase in size (Boisnard-Lorig et al., 2001). Then, embryo growth takes place at the expense of endosperm during the second phase. At maturity, the seed contains only a single layer of endosperm cells, and the maternal integument ultimately becomes the seed coat (Scott et al., 1998; Garcia et al., 2003). Seed coat formation and endosperm growth precede embryo growth in Arabidopsis, and the seed reaches almost its final size before the enlargement of the embryo. Therefore, seed size is determined by the coordinated growth of the diploid embryo, the triploid endosperm, and the diploid maternal ovule.
Both maternal and nonmaternal factors are involved in seed size regulation (Garcia et al., 2005). In Arabidopsis, an increased dosage of the paternal genome in the endosperm increases seed size, whereas an increased dosage of the maternal genome reduces seed size, with delayed cellularization of the peripheral endosperm and hypertrophy of the chalazal endosperm and associated nodules (Scott et al., 1998). Mutations in DNA METHYLTRANSFERASE1 (MET1) and DECREASE IN DNA METHYLATION1 (DDM1) dramatically reduce DNA methylation and cause parent-of-origin effects on F1 seed size (Xiao et al., 2006). Pollination of met1-6 or ddm1-2 pistils with wild-type pollen produced larger F1 seeds with a hypomethylated maternal genome, delayed endosperm development, and a larger endosperm volume than the wild type. A reciprocal cross led to small F1 seeds with a hypomethylated paternal genome, early endosperm cellularization, and, ultimately, a smaller endosperm volume than the wild type.
The seeds of the triple cytokinin receptor mutant are twice the size of wild-type seeds, and cytokinin may regulate embryo size via a maternal and/or endospermal mechanism (Hutchison et al., 2006; Riefler et al., 2006). Mutations in the transcription factor APETALA2 (AP2) increase seed size due to an increase in both embryonic cell number and cell size, and the seed trait is passed through the maternal sporophyte and the endosperm genomes (Jofuku et al., 2005; Ohto et al., 2005). The integument also plays a role in seed size determination, and the Arabidopsis megaintegumenta/auxin response factor2 mutant causes extra cell division in the integuments surrounding the mutant ovules, enlarges seed coats, and increases seed size (Schruff et al., 2006).
Mutations in either HAIKU2 (IKU2) or MINISEED3 (MINI3) reduce seed size, and the mutant seed phenotypes depend on the genotype of both the embryo and the endosperm (Luo et al., 2005). MINI3 encodes a WRKY family transcription factor and is expressed in both the endosperm and the embryo. IKU2 encodes a leucine-rich repeat (LRR) receptor kinase and is expressed in the endosperm but not in the embryo or elsewhere in the plant. Interestingly, IKU2 expression is downregulated in mini3, and MINI3 may act upstream of IKU2. The reduced seed size of the iku mutants is closely associated with a reduced growth of endosperm, a premature cellularization of the endosperm, and a reduced proliferation of the embryo after the early torpedo stage (Garcia et al., 2003). AGAMOUS-like 62 (AGL62) is another regulator of endosperm cellularization, as the endosperm cellularizes prematurely in agl62 seeds (Kang et al., 2008).
SHORT HYPOCOTYL UNDER BLUE1 (SHB1) was initially isolated from a gain-of-function mutant, shb1-D or short hypocotyl under blue 1-Dominant, with a long hypocotyl phenotype under red, far-red, and blue light (Kang and Ni, 2006). Its recessive T-DNA knockout allele, shb1, exhibits a short hypocotyl phenotype under blue light. In this study, we show that SHB1 also has specific functions in Arabidopsis seed development, regulating endosperm proliferation and the timing of endosperm cellularization. SHB1 associates with MINI3 and IKU2 promoters, as demonstrated by chromatin immunoprecipitation (ChIP) analysis, and SHB1 may be recruited by other transcription factors that specifically recognize MINI3 and IKU2 promoter sequences.
RESULTS
shb1-D Produces Significantly Enlarged Seeds, and shb1 Notably Reduces Seed Size
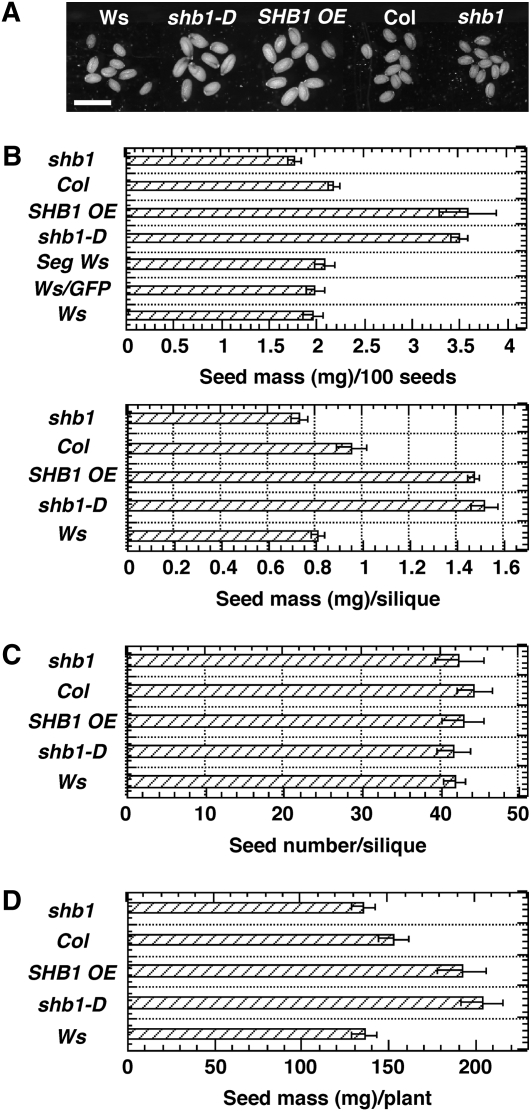
We determined the average seed mass by weighing mature dry Arabidopsis seeds in batches of 100 for seed lots from independently propagated Wassilewskija (Ws), shb1-D, Columbia (Col), and shb1 lines or independent transgenic lines that overexpress either green fluorescent protein (GFP) or full-length SHB1:GFP. shb1-D and the SHB1:GFP overexpression (SHB1 OE) plants had seeds that were 1.5- to 1.7-fold heavier per 100 seeds or per silique than the Ws wild type, whereas the shb1 mutation reduced seed mass by 15% compared with the Col wild type (P < 0.01, two-tailed Student's t tests) (Figures 1A and 1B). By contrast, the number of seeds per silique and the number of siliques per plant were not changed for Ws and shb1-D (Figure 1C; see Supplemental Figure 1A online). We harvested all seeds from each plant, and, as shown in Figure 1D, the seed mass of shb1-D plants was increased compared with Ws wild type, and the seed mass of shb1 was notably reduced compared with Col (P < 0.001, two-tailed Student's t tests).
Figure 1.
SHB1 Regulates Seed Size.
(A) Mature seeds of Ws, shb1-D, SHB1 OE, Col, and shb1. Bar = 0.5 mm.
(B) Average seed mass per 100 seeds (top) or per silique (bottom) for Ws, shb1-D, SHB1 OE, Col, and shb1. Data from Ws plants that segregated from the original SHB1 OE lines or Ws plants that carry a GFP transgene are also shown as controls.
(C) Seed number per silique for Ws, shb1-D, SHB1 OE, Col, and shb1.
(D) Total seed mass (mg) for Ws, shb1-D, SHB1 OE, Col, and shb1. Data are means ± se from at least six independently propagated wild-type and mutant lines or independent transgenic lines.
Seed Enlargement of shb1-D Is Due to Increases in Cell Size and Cell Number
We isolated and visualized embryos from mature seeds and found that the cotyledon area of shb1-D was 1.5-fold larger than that of the Ws wild type (Figures 2A and 2B). The embryonic shoot or hypocotyl and embryonic root apex of shb1-D also appeared larger than those of the Ws wild type (Figure 2A). We further examined whether the seed size increase of shb1-D is due to an increase in cell division or cell expansion. The average size of shb1-D cotyledon cells is 1.2-fold that of the wild type (Figure 2B). The size of shb1-D hypocotyl cells also appeared larger than that of the Ws wild type (Figure 2C). We then monitored the numbers of epidermal cells in mature embryos under scanning electron microscopy and counted the cell number from three columns in the central region of cotyledons or in the hypocotyl plus the embryonic root of Ws and shb1-D. The average number of cells in the cotyledon and in the hypocotyl plus the embryonic root was 1.23- and 1.29-fold greater, respectively, in shb1-D than in the Ws wild type (Figure 2D). Interestingly, the 1.2-fold increase in cell size (Figure 2B) and 1.23- to 1.29-fold increase in cell number accounted for a 1.44- to 1.56-fold increase in embryo size of shb1-D. Therefore, both increased cell size and cell number contribute to the overall enlargement of mature embryo in the gain-of-function mutant shb1-D.
Figure 2.
The Increase in Seed Size of shb1-D Is Due to an Increase in Both Cell Size and Cell Number.
(A) Mature embryos dissected from imbibed seeds of Ws (top left) and shb1-D (top right) plants. Epidermal cell layer from the central region of Ws (bottom left) and shb1-D (bottom right) cotyledons. Bars = 100 μm in top panels and 10 μm in bottom panels.
(B) Cotyledon area (top) and cotyledon cell size (bottom) of Ws and shb1-D plants.
(C) Epidermal cell layer from the central region of Ws (left) and shb1-D (right) hypocotyls. Bars = 50 μm.
(D) Average cell numbers from three columns in the central region of the cotyledon (left) and the hypocotyl plus the embryonic (Em) root (right) in Ws and shb1-D plants. Data in (B) and (D) are means ± se from at least 20 independently propagated Ws or mutant lines.
shb1-D Seeds Accumulate More Proteins and Fatty Acids Than Do Wild-Type Seeds
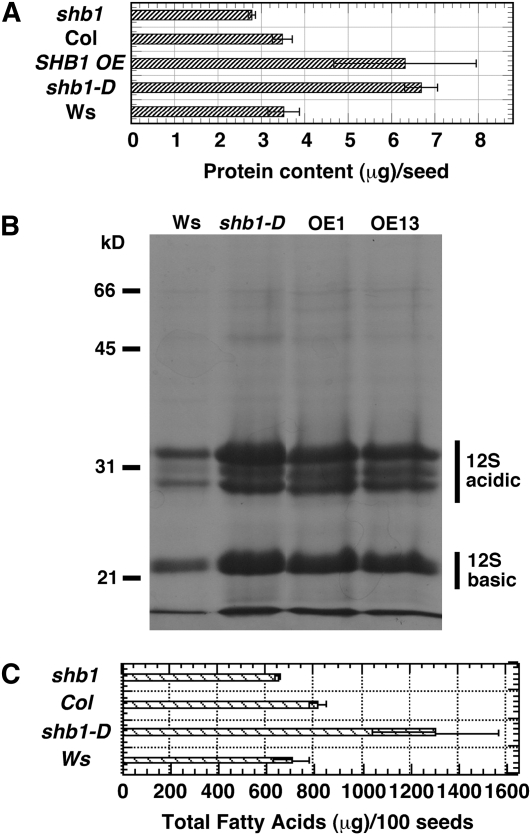
The differences in seed mass between shb1-D or shb1 plants and wild-type plants may be due in part to an increase or a decrease in seed reserves, respectively. We prepared protein extracts from 200 seeds each of Ws wild type or shb1-D and examined the content and spectrum of soluble proteins. Two major classes of Arabidopsis seed storage proteins include the 12S cruciferins and the 2S albumins (Heath et al., 1986; Pang et al., 1988). The seeds of shb1-D or the SHB1 OE plants accumulated 1.5-fold more proteins, when expressed as mg/100 seeds, than those of the Ws wild type (Figure 3A). By contrast, the total protein content was reduced by >20% in shb1 plants relative to the Col wild type (P < 0.01, two-tailed Student's t tests). No such difference in protein content was detected when expressed on the basis of seed weight, and the increase in protein content is apparently due to the larger seeds and not a higher concentration of protein in the seeds (see Supplemental Figure 3A online). The spectrum of the seed storage proteins or the relative proportion of 12S storage proteins to other proteins was not selectively affected by the shb1-D mutation, whereas the 2S albumin proteins were not retained in the 12% SDS-PAGE gel (Figure 3B). We also measured the content of reducing sugars and total soluble sugars in wild-type and shb1-D mutant seeds. shb1-D and SHB1 OE lines accumulated more reducing sugars and total soluble sugars per seed than did Ws, and the shb1 mutant accumulated less sugars per seed than did Col (see Supplemental Figure 4 online).
Figure 3.
shb1-D Seeds Have Higher Protein and Fatty Acid Content Than Do Ws Seeds.
(A) Total protein content per seed for Ws, shb1-D, SHB1 OE, Col, and shb1.
(B) Spectrum of seed storage proteins in Ws, shb1-D, and SHB1 OE. Molecular standard ladder is shown to the left of the gel.
(C) Total fatty acid methyl-ester content per 100 seeds of Ws, shb1-D, Col, and shb1. Data are means ± se from at least six independently propagated Ws, mutant lines, or independent transgenic lines.
We next measured the content and composition of total fatty acid methyl-ester for Ws wild-type and shb1-D mutant seeds. Like Brassica napus, Arabidopsis seeds contain a small number of fatty acid species that represent ∼80% of the total fatty acid content in seeds (Downey 1983; James and Dooner, 1990; Taylor et al., 1995). As shown in Figure 3C, shb1-D accumulated more total fatty acids per seed due to increased seed size than did Ws, and a 20% reduction in total fatty acid content per seed was observed in shb1 relative to Col (P < 0.01, two-tailed Student's t tests). By contrast, the fatty acid composition, including palmitic (C16:0), palmitoleic (C16: 1), stearic (C18:0), oleic (C18: 1), linoleic (C18: 2), linolenic (C18: 3), arachidic (C20:0), eicosenoic (C20: 1), behenic (C22:0), and erucic (C22:1) acids, was not significantly altered between shb1-D and Ws or shb1 and Col plants (see Supplemental Figure 3B online).
Activation of SHB1 Enlarges the Seed Cavity and Delays Endosperm Cellularization and Embryo Development
To explore the developmental alterations of shb1 mutants, cleared seeds of shb1-D or shb1, and also of their corresponding wild types, were compared at different developmental days after pollination (DAP) (Figure 4). The embryo sac or seed cavity in the shb1-D mutant was significantly larger than that of the Ws wild type at 4 DAP and remained larger throughout embryogenesis. By contrast, shb1-D embryos were developmentally delayed compared with those of Ws at the early phase of seed development but gradually caught up at the later phase of seed development to obtain a larger size than those of the wild type at maturity (Figures 1 and 4). By contrast, the shb1 embryo sac was similar in size or slightly smaller than that of Col from 4 to 9 DAP, and the embryo of shb1 was smaller than that of Col at 9 DAP (Figure 4). Seeds of shb1-D were longer, wider, and more pointed at the micropylar pole than those of Col (Figure 5A). By 5 DAP, wild-type seeds had reached the late heart stage, the chalazal endosperm formed a compact rounded cyst, and the peripheral endosperm was cellularizing from the micropylar pole (Figures 4 and 5A). By contrast, shb1-D seeds of the same age had globular to early heart stage embryos and long and enlarged chalazal endosperms, and the peripheral endosperm had not undergone substantial cellulerization by this stage, as that seen in the Ws wild type (Figure 5A).
Figure 4.
Seed Development in Wild-Type and shb1 Mutant Plants.
Cleared Ws, shb1-D, Col, and shb1 seeds were imaged with differential contrast optics at 3, 4, 5, 6, 7, and 9 DAP. Bars = 50 μm.
Figure 5.
The shb1-D Mutation Affects Endosperm Development, and SHB1 Is Transmitted Zygotically.
(A) Five-day-old Ws seed at the heart stage, with cellularized peripheral endosperm, and shb1-D seed at the late globular stage, with uncellularized endosperm. PEN (arrow), peripheral endosperm; CZE (asterisk), chalazal endosperm. Bars = 50 μm.
(B) F1 and F2 seed mass from reciprocal crosses between Ws and shb1-D. Ws shb1-D indicates Ws pollen crossed to a shb1-D pistil.
(C) F1 seed mass from reciprocal crosses between Col and shb1. Data are presented as means ± se from at least six independently propagated Ws, Col, or mutant lines.
SHB1 Acts Zygotically to Regulate Seed Size
To determine the mode of transmission of parental SHB1, we performed reciprocal crosses between Ws and shb1-D or between Col and shb1 using flowers at identical positions (11th to 14th flowers) on secondary inflorescences. Seed masses were determined from five inflorescences per plant and four to five plants in total. If SHB1 acted through the sporophyte genome to regulate seed mass, the effect of the dominant shb1-D mutation on seed mass would be observed only when maternal plants are homozygous or heterozygous in the shb1-D locus. As shown in Figure 5B, the dominant shb1-D allele regulates seed mass through the embryo genome in these reciprocal-cross experiments. The shb1-D (+/−) F1 seeds of shb1-D pistils pollinated with Ws wild-type pollen were heavier than wild-type seeds but were very close in weight to shb1-D (−/−) seeds produced when shb1-D pistils were pollinated with shb1-D pollen. The shb1-D (+/−) F1 seeds resulting from Ws wild-type pistils being pollinated with shb1-D pollen were also heavier than those of the wild type but were 12% lighter than shb1-D (−/−) seeds (Figure 5B). Apparently, maternal homozygous shb1-D mutants produced seeds that were consistently heavier than those from maternal wild-type plants due to a gene dosage effect from the triploid endosperm, and SHB1 may also partially act through the endosperm genome to regulate seed mass.
The large seed phenotype was still visible in a segregating F2 population for the two different reciprocal crosses performed. The F2 seeds from selfed shb1-D (+/−), regardless of maternal or paternal origin, all showed a 1.4-fold increase in seed mass compared with the Ws wild type (Figure 5B). The shb1 (−/−) F1 seeds of shb1 pistils pollinated with shb1 pollen were, on average, 15% lighter than those of the Col wild type (P < 0.01, two-tailed Student's t tests) (Figure 5C). As shb1 is recessive and has a relatively weak seed phenotype, shb1 (+/−) F1 seeds formed in pistils either of shb1 or of the Col wild-type produced seeds of the same mass as Col wild-type seeds (Figure 5C).
SHB1 Regulates the Expression of MINI3 and IKU2
Through real-time RT-PCR analysis, we found that the shb1-D mutation enhanced, and the shb1 mutation reduced, the expression of MINI3 and IKU2, a WRKY family transcription factor gene and an LRR receptor kinase gene, but not that of AP2, a transcription factor gene involved in floral organ and seed development and expressed in the embryo but not in the endosperm (P < 0.001, two-tailed Student's t tests) (Figure 6A). The expression of MINI3 in shb1-D green siliques was three times higher than in the Ws wild type, and the expression of MINI3 in shb1 was reduced by 40% compared with Col (P < 0.001, two-tailed Student's t tests) (Figure 6A). IKU2 expression was 10-fold greater in shb1-D mutants and six-fold less in shb1 mutants than in the Ws or Col wild type (Figure 6A). By contrast, the expression of SHB1 relative to UBQ10 in Col, mini3, iku2, and ap2 was not changed (Figure 6B). In shb1-D, the expression of IKU2 was enhanced by more than that of MINI3, and in the shb1 mutant, the expression of IKU2 was reduced by more than that of MINI3. IKU2 is specifically expressed in the endosperm (Luo et al., 2005). To test if the altered expression of IKU2 in shb1-D is caused by increased endosperm volume, we examined the expression of another endosperm-specific gene, HEAT SHOCK FACTOR15 (HSF15) (Winter et al., 2007; http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi) in the wild type and shb1 mutants. No difference in HSF15 expression was found between Ws and shb1-D or between Col and shb1 (see Supplemental Figure 5A online).
Figure 6.
SHB1 Regulates the Expression of MINI3 and IKU2.
(A) Expression of MINI3, IKU2, and AP2 in Ws, shb1-D, Col, and shb1 as determined by real-time RT-PCR analysis.
(B) Expression of SHB1 in Col, mini3 (mini3-2), iku2 (iku2-4), and ap2 as determined by real-time RT-PCR analysis.
(C) Genetic interactions of SHB1 with MINI3 and IKU2. All genotypes are in a mixed Ws/Col background identified from a F2 segregation population, and at least eight individuals were included in data calculation. Bar = 0.5 mm
(D) Expression of IKU2 in Ws Col, mini3, shb1-D, and shb1-D mini3 as determined by real-time RT-PCR analysis. Means were calculated from three biological samples, and each biological sample was examined in triplicate. The calculated se includes technical error and biological error.
SHB1 Acts Upstream of MINI3 and IKU2
To test whether SHB1 acts upstream of MINI3 and IKU2, we performed double mutant analysis, combining shb1-D, which produces large seeds, with either mini3-2 (SALK_050364) or iku2-4 (SALK_073260), which produce small seeds. As shb1-D (Ws) and mini3-2 or iku2-4 (Col) are in different genetic backgrounds, we performed the genetic analysis in a mixed Ws and Col background after a cross of shb1-D to either mini3-2 or iku2-4. We selected the genotypes from the segregating F2 population by PCR genotyping, and at least 10 individuals of each genotype were identified and used in the analysis. As shown in Figure 6C, a mutation in IKU2 diminished the large seed phenotype of shb1-D. Since the shb1-D iku2-4 double mutant phenotypically resembled iku2-4, IKU2 appears to act downstream of SHB1. The mini3-2 mutation also significantly suppressed the large seed phenotype of shb1-D to that of the mini3-2 single mutant (Figure 6C). We then examined the expression of IKU2 in the shb1-D mini3-2 double mutant. The mutation in MINI3 completely blocked the expression of IKU2 and reduced the expression of IKU2 from a relative level of 10, as seen in the shb1-D single mutant, to a relative level of 0.6 (Figure 6D).
SHB1 Has an Overlapping Expression Pattern with MINI3 and IKU2
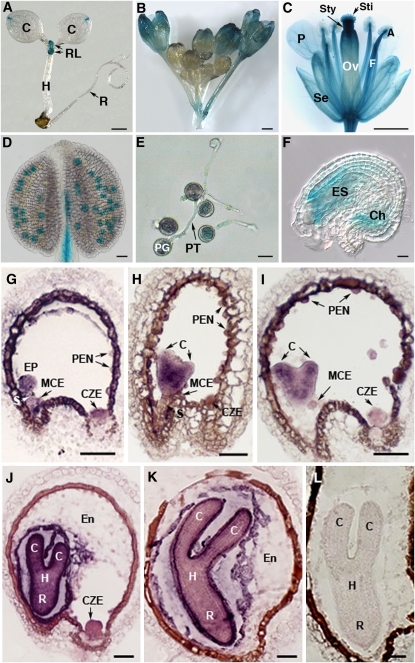
To explore SHB1 function, we examined the expression pattern of a β-glucuronidase (GUS) reporter gene driven by a native SHB1 promoter. GUS activity was observed in the apical meristem of young seedlings, rosette leaves, inflorescence, floral organs, mature pollen grains, germinating pollens, and mature unfertilized ovules (Figures 7A to 7F). The expression of SHB1 overlaps with that of MINI3 in floral buds, pollen grains, pollen tubes, and ovules (Figure 7; Luo et al., 2005). The expression of SHB1 in developing seeds was also monitored by in situ hybridization. Within the developing siliques, SHB1 has an overlapping expression pattern with MINI3 in developing and cellularized endosperm and in the globular and early heart stage embryo 12 to 96 h after fertilization (Figures 7G to 7K; Luo et al., 2005). SHB1 also has an overlapping expression pattern with IKU2 in endosperm, and the expression of IKU2 was only detected in the endosperm, but not in the embryo or elsewhere in the plant (Luo et al., 2005). In addition, SHB1 expression was also detected in the chalazal endosperm or posterior cyst and in the late heart-stage and torpedo-stage embryos, suggesting a role for SHB1 in promoting embryonic development in the second phase of seed development (Figures 7G to 7J).
Figure 7.
SHB1 Is Expressed in Endosperm and Embryo.
Expression of ProSHB1:GUS in eight-day old seedling (A), inflorescence (B), flower (C), mature pollen (D), in vitro germinating pollen (E), and unfertilized ovule (F). In situ hybridization of Ws with SHB1 antisense probe at early globular stage (G), early heart stage (H), heart stage (I), torpedo stage (J), and late torpedo stage (K) or with SHB1 sense probe at torpedo stage (L). A, anther; C, cotyledon; Ch, chalazal end; CZE, chalazal endosperm; En, endosperm; EP, embryo proper; ES, embryo sac; F, filament; H, hypocotyl; MCE, micropylar endosperm; Ov, ovule; P, petal; PEN, peripheral endosperm; PG, pollen grain; PT, pollen tube; R, embryonic root; RL, rosette leaf; S, suspensor; Se, sepal; Sti, stigma; Sty, style. Bars = 1 mm for (A) to (C), 25 μm for (D) to (F), and 50 μm for (G) to (L).
SHB1 Associates with MINI3 and IKU2 Promoters
To investigate how SHB1 regulates the expression of MINI3 and IKU2, we performed a ChIP assay with either anti-SHB1 or anti-GFP antibodies followed by quantitative PCR analysis. Figure 8A shows the various amplicons in MINI3 and IKU2 promoters used for ChIP analyses. Amplicon M3, which spans from −986 to −1216 in the MINI3 promoter, was highly enriched using either anti-SHB1 or anti-GFP antibodies, and amplicon M1, which spans from +3 to −349 in the MINI3 promoter, was also significantly enriched (Figures 8B and 8C). By contrast, amplicon M2, which spans from −399 to −620 in the MINI3 promoter, was not enriched (Figures 8B and 8C). Amplicon 1, which spans from −3 to −277 in the IKU2 promoter, was moderately enriched, and amplicon 2, which spans from −296 to −665 in the IKU2 promoter was significantly enriched (Figures 8B and 8C). We also performed ChIP analysis of the promoter sequences of HSF15, an endosperm-specific gene, and LEAF COTYLEDON2 (LEC2; Stone et al., 2001), an embryo-specific gene, and neither of the promoter sequences was significantly enriched (see Supplemental Figures 5B and 5C online). In addition, SHB1 was unable to bind directly to either MINI3 or IKU2 promoter sequences in a gel mobility shift assay (Y. Zhou and M. Ni, unpublished data).
Figure 8.
SHB1 Associates with MINI3 and IKU2 Promoters in Vivo.
(A) Schematic diagram of the MINI3 and IKU2 loci and the five amplicons (i.e., M1, M2, M3, IK1, and IK2) used for ChIP analysis.
(B) and (C) Enrichment of particular MINI3 and IKU2 chromatin regions with anti-SHB1 antibody in wild-type plants (B) or with anti-GFP antibody in SHB1:GFP transgenic plants (C) as detected by real-time PCR analysis. Preimmune serum was used as mock control. Means were calculated from three biological samples, and each biological sample was examined with three PCR technical replicates. The calculated se includes technical error and biological error.
DISCUSSION
The Level of SHB1 Is Limiting during Seed Development
SHB1 has a specific function in endosperm and embryo development, as the sizes of many other organs, such as leaves, stems, roots, carpel, petals, and sepals, were not affected by either the shb1-D or the shb1 mutation. SHB1 does not influence general plant growth and development, and mature shb1-D or shb1 plants were indistinguishable from those of the Ws or Col wild type (see Supplemental Figures 2A and 2B online). The seed phenotype is not tightly related to its function in light signaling since there was no particular enhancement of shb1-D or shb1 seed size by red, far-red enriched, or blue light compared with white light (see Supplemental Figure 1B online). Although SHB1 gain- or loss-of-function of mutants had altered seed size, shb1 had a relatively weak phenotype, with a 15% reduction in seed mass, compared with a 1.5- to 1.6-fold increase in seed mass of the shb1-D lines (Figure 1). The changes in protein or fatty acid content were also less dramatic in shb1 than in shb1-D (Figure 3). The development of embryo sac and endosperm was not affected by the shb1 mutation, but the embryo of shb1 appeared smaller at 9 DAP than that of the wild type (Figure 4). One possibility is that the level of SHB1 is limiting for endosperm and seed enlargement since we were unable to detect SHB1 message in regular RNA gel blot of total RNA prepared from rosette leaves, which show a strong SHB1 expression compared with other organ types by GUS reporter assay (Kang and Ni, 2006; Figure 7A). Therefore, SHB1 activation has a stronger effect on seed development than does the loss of SHB1. By contrast, the weak seed phenotype in shb1 may be due to redundant SHB1 homologs or developmental pathways that regulate seed size. For example, one of the SHB1 homologs, gi 3548806 or At2g03240, is expressed at the globular and heart stages of the Arabidopsis embryo and has detectable expression levels in developing siliques (Winter et al., 2007; http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). The expression pattern of another SHB1 homolog, gi 3548805 or At2g03250, is not available in the public domain and merits further studies.
SHB1 Promotes Endosperm Enlargement at the Early Phase of Seed Development
The early phase of endosperm development in most species is marked by cell proliferation and growth to generate a large multinucleate cell and is defined as the syncytial phase (Olsen, 2001; Berger, 2003). This syncytium is then partitioned into individual cells by a specific type of cytokinesis called cellularization. Cellularization is initiated in the micropylar pole and occurs after the eighth mitotic cycle in the peripheral endosperm (Garcia et al., 2003). Endosperm development is essential for providing nutrients to the developing embryo and for enforcing a space limitation (Brink and Cooper, 1947). As the initial endosperm enlargement affects the final seed size, the endosperm may initiate a signal to regulate the subsequent embryo development (Miller and Chourey, 1992; Hong et al., 1996). The mini3 and iku mutations cause premature cellularization of the endosperm and reduce the proliferation of the cellularized endosperm (Garcia et al., 2003; Luo et al., 2005). IKU and MINI3 may regulate endosperm size through the timing of endosperm cellularization (Luo et al., 2005). We demonstrated in this study that SHB1 gain-of-function regulates a similar set of cellular responses to MINI3 and IKU2, such as endosperm proliferation and the timing of endosperm cellularization (Figure 5A). More significantly, the activation of SHB1 significantly enlarges the embryo sac at 4 DAP, and the endosperm remains larger throughout the subsequent stages of seed development (Figure 4).
The posterior pole of the endosperm does not undergo cellularization and contains a multinucleate pool of cytoplasm, chalazal endosperm, or cyst. The cyst is located above the placentochalazal area of the seed integument where vascular elements terminate and is of potential importance for transfer of maternal nutrients to the seed (Schultz and Jensen, 1971; Otegui et al., 2002). The overall size of the posterior pole in iku seeds is reduced relative to that of the wild type, and cellularization reaches the posterior pole in a few iku seeds, suggesting that iku mutations cause a posterior displacement of the boundary between the peripheral endosperm and the posterior pole (Garcia et al., 2003). In the shb1-D mutant, the posterior cyst is significantly enlarged. This may result in an excess of nutrients from the maternal parent, which may contribute to the subsequent development of the embryo (Figure 5A).
SHB1 Regulates Embryo Cell Proliferation and Elongation at the Second Phase of Seed Development
The mini3 and iku mutations also retard embryo proliferation after the early torpedo stage and reduce the total number of cells, but not the cell size, in the mature embryo (Garcia et al., 2003; Luo et al., 2005). Similarly, mutations in EXTRA SPOROGENOUS CELLS/EXCESS MICROSPOROCYTES1 (EXS/EMS) also produced smaller seeds due to a reduction in cell size, but not in cell number (Canales et al., 2002; Zhao et al., 2002). EXS/EMS encodes a putative LRR receptor kinase and is expressed in both the embryo and the endosperm. The effect of the iku2 mutation on embryo development may be indirect as a consequence of the effects of this mutation on the endosperm development at the initial phase of seed development. In addition, the expression of IKU2 is limited to the endosperm (Luo et al., 2005). Although the endosperm may initiate a signal to regulate the subsequent embryo development, this hypothesis requires further validation. For example, it is still unknown if overexpression of IKU2 promotes embryo development and leads to enlarged seeds. Although MINI3 message can be detected in both the endosperm and embryo, it remains unknown if MINI3 functions in embryonic development in addition to its role in endosperm proliferation (Luo et al., 2005). By contrast, SHB1 activation causes active endosperm proliferation in the initial phase of seed development at the expense of embryonic development, but the embryo subsequently reaches its full mature size 2 to 3 d later than does the wild type. At maturity, the shb1-D mutation increases both embryo cell size and cell number (Figure 2). Therefore, SHB1 may activate other developmental pathways that regulate embryo cell division and cell expansion in addition to the MINI3-IKU2 pathway that regulates endosperm proliferation.
SHB1 Regulates the Expression of Both MINI3 and IKU2
SHB1 is required for the proper expression of both MINI3 and IKU2 (Figure 6A). Double mutant analyses indicate that the effect of shb1 mutations on seed development was largely dependent on MINI3 and IKU2 function (Figure 6C). SHB1 contains an N-terminal SPX (for SYG1, PHOSPHATE TRANSPORTER81, and XENOTROPIC AND POLYTROPIC MURINE LEUKEMIA VIRUSES RECEPTOR1) domain and a C-terminal EXS (for ER RETENSION-DEFECTIVE1, XENOTROPIC AND POLYTROPIC MURINE LEUKEMIA VIRUSES RECEPTOR1, and SYG1) domain (Kang and Ni, 2006). A major function of the SPX domain in yeast SYG1 protein is to mediate protein–protein interactions (Spain et al., 1995). The EXS domain contains several predicted transmembrane helices; however, SHB1 is localized to the nucleus (Kang and Ni, 2006). Thus, the EXS domain in SHB1 may have a distinct function from those in SYG1-like proteins. SHB1 protein does not contain DNA binding motifs that recognize specific elements in MINI3 and IKU2 promoters, and SHB1 did not directly bind to either the MINI3 or IKU2 promoter in a gel mobility shift assay (Y. Zhou and M. Ni, unpublished data). ChIP experiments indicate that SHB1 associates with MINI3 and IKU2 promoters in vivo (Figure 8). A similar function has been discovered for numerous other proteins, such as FLOWERING LOCUS T (FT), GIGANTEA (GI), and FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1) (Wigge et al., 2005; Sawa et al., 2007).
FT does not have DNA binding activity but associates with AP1 promoters through a direct interaction with FLOWERING LOCUS D, a bZIP transcription factor that directly binds to the AP1 promoter (Wigge et al., 2005). GI and FKF1 also associate with CONSTANS promoters through a direct interaction with CYCLING DOF FACTOR1, a Dof transcription factor, in a blue light–dependent manner (Sawa et al., 2007). Similarly, SHB1 may interact with other proteins that are specific transcription factors for MINI3 and IKU2 promoters and achieve its regulation over the transcription of MINI3 and IKU2 genes during the initial phase of seed development (Figure 9A). It remains unclear if SHB1 acts in a similar way to regulate embryo cell proliferation and elongation during the second phase of seed development (Figure 9B). Future studies will be directed to identify the protein factors that may interact with SHB1 in the regulation of either endosperm development or embryo development.
Figure 9.
A Working Hypothesis Is Proposed for SHB1 Function in Arabidopsis Seed Development.
(A) SHB1 is recruited to the promoters of MINI3 and IKU2 by protein X and regulates the expression of key genes such as MINI3 and IKU2 that are required for endosperm proliferation and cellularization.
(B) It remains unknown if SHB1 deploys a similar mechanism to present itself through protein Y to the promoters of unknown targets such as Z gene or MINI3 that are essential for embryo cell proliferation and elongation. IKU2 is expressed uniquely in the endosperm, whereas MINI3 is expressed in both the endosperm and the embryo. MINI3 may play additional roles in embryonic development.
METHODS
Plant Materials and Growth Conditions
shb1 (SALK_128406), mini3-2 (SALK_050364), iku2-4 (SALK_073260), and ap2 (SALK_071140) are in the Col background and were obtained from the ABRC (Ohio State University, Columbus). shb1-D was isolated in the Ws background, and the SHB1 overexpression lines in the Ws or Col background were generated as described previously (Kang and Ni, 2006). shb1-D and shb1 mutants were backcrossed twice to the wild type before phenotypic analysis. Plants were grown at 23°C, and all seeds from a single plant were harvested when the plant was mature and the last siliques on the inflorescence had elongated, turned brown, and dried. At the four-leaf stage, the younger plants were also fitted with a plastic well to catch all seeds released by dehiscing siliques in later developmental stages. These hand-harvested seeds were combined with those caught in the well, dried at 25°C for 7 d, and weighed.
Cytological Experiments
Mature seeds of Ws, shb1-D, SHB1 OE, Col, shb1, Ws/Col, mini3-2, iku2-4, mini3-2 shb1-D, and iku2-4 shb1-D were imaged with a Cool Cam color CCD camera (Cool Camera) coupled to a Nikon Eclipse E800 microscope, using Imago Pro Plus version 3.0 software (Media Cybernetics). Mature dried seeds of the wild type and shb1 mutants were also imbibed for 1 h and dissected under a microscope to isolate mature embryos. The embryos was incubated for 12 h in buffer that contains 50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 1% Triton X-100, and 1% DMSO at 37°C, fixed for 45 min in FAA (formalin, acetic acid, and ethanol) that contains 10% formalin, 5% acetic acid, 45% ethanol, and 0.01% Triton X-100, and dehydrated through an ethanol series (Ohto et al., 2005). The embryos were then treated for 1 h in Hoyer's solution that contains 3:0.8:0.4 of chloral hydrate:water:glycerol. Observations were made with a Zeiss 510 Meta confocal laser scanning microscope. Cotyledon area and average epidermal cell size from the central region of cotyledons were determined using NIH IMAGE analysis software as described (Ohto et al., 2005). The area of the cotyledon was calculated from 50 mature embryos. The cell size in the middle of the cotyledon was also determined from 20 mature embryos by dividing 130 μm2 image area by the number of cells. Data are presented as the average of two independent experiments.
Scanning Electron Microscopy
Mature embryos from the mutant and the wild type were released from the seed coats by applying gentle pressure and fixed in FAA at 4°C for 2 h. After fixation, the samples were dehydrated through an ethanol series (70, 85, 95, and 100%), each for 30 min. The embryos were then dried using the Autosamdri-814 Critical Point Dryer. The individual embryos were mounted on scanning electron microscopy stubs, sputter-coated with gold with a Fullam Sputter-coater, and examined under a Hitachi S3500N variable pressure scanning electron microscope at an accelerating voltage of 5 k.
Protein, Carbohydrate, and Fatty Acid Analysis
Total proteins were extracted from the same number of wild-type and mutant seeds as described (Heath et al., 1986). Protein content was determined using the Bio-Rad DC protein assay kit with BSA as standard. Total protein extracts from 200 seeds of the wild type and mutants was also fractionated on 12% SDS-PAGE and stained with Coomassie Brilliant Blue as described (Ohto et al., 2005).
To extract total fatty acids, 100 seeds from wild-type and mutant plants were homogenized in 500 μL 1 n HCl-methanol, and 200 mL hexane was added to each vial and heated for 2 h at 85°C (Larson and Graham, 2001). After heating, the solutions were cooled and 250 μL 0.9% KCl was added to partition the hexane phase that contained the fatty acid methyl esters. A 1-μL aliquot of the hexane solution was then analyzed by gas chromatography with flame ionization detection. Free fatty acid C21:0 (Sigma-Aldrich) was added as an internal control. Total fatty acid content was estimated by comparing the total fatty acid methyl ester peak area to that of the C21:0 internal standard.
One hundred seeds each from the wild type and the shb1 mutant were also homogenized in 500 mL of 80% ethanol, and the extracts were incubated at 70°C for 90 min (Focks and Benning, 1998). Following centrifugation, the supernatant was saved and the pellet was extracted twice again with 500 μL of 80% ethanol. The combined supernatants were evaporated, and the residue was dissolved in 100 μL of deionized water and measured for reducing sugars by the Nelson-Somogyi assay (Nelson, 1944; Somogyi, 1952). The total sugar content was measured using the phenol sulfuric acid carbohydrate assay and interpreted against a 1-mg/mL glucose-generated standard curve (Dubois et al., 1956).
Developmental Alteration Analysis
To examine the developmental alterations of shb1 mutants, wild-type and shb1 mutant flowers were hand-pollinated, and the developing seeds were harvested at different days after pollination. The siliques were then cut into 2-mm-long fragments, fixed, and dehydrated as described above. The segments were next cleared in Hoyer's solution overnight and examined using an Olympus BX51 with differential interference contrast optics. Data after 10 DAP were not collected as siliques were mature and preparation of clear seeds at these late developmental stages becomes difficult. To investigate endosperm development, the siliques were fixed in FAA solution overnight at 4°C, dehydrated as described above, and embedded in EPON 812 (SERVA). Sections were stained with 0.1% (w/v) toluidine blue O in distilled water. Stained sections were photographed using a Cool Cam color CCD camera (Cool Camera) coupled to an Olympus BH-2 microscope.
Real-Time RT-PCR Analysis
For quantitative RT-PCR analysis, total RNAs were isolated from green siliques 4 to 5 d after hand-pollination (i.e., early to late heart stage) using the SV Total RNA Isolation Kit (Promega). SuperScript III reverse transcriptase (Invitrogen) was used to synthesize the first-strand cDNA with oligo(dT) primer and 1 μg of total RNA at 50°C for 1 h. Quantitative PCR was then performed with the Platinum SYBR Green qRT-PCR Kit (Invitrogen) on an Applied Biosystems 7500 Real-Time PCR machine. The thermal cycling program was 50°C for 10 min and 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 1 min, and a one-cycle dissociation stage at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The primers used in quantitative RT-PCR were MINI3, 5′-TTTGATGATATTGCAACGGAA-3′ and 5′-GATCCTTTGTGTCTTGCTTGT-3′; IKU2, 5′-CGTGTGAGACAAGCGTTAGC-3′ and 5′-GAGGAGACTTGTCCGTGCAT-3′; AP2, 5′-ATTCGGCTAATTCGAAGCATAA-3′ and 5′-AGAGGAGGTTGGAAGCCATT-3′; SHB1, 5′-CAGGTTCAAGCACTGAGGAGT-3′ and 5′-TGCTTCCTCGGTTTAGAGTA-3′; HSF15, 5′-CAACCTCAAAACTTCCTTTC-3′ and 5′-TAATGATTTCCACAAGGTGA-3′; and UBQ10, 5′-AGGTACAGCGAGAGAAAGTAGCA-3′ and 5′-TAGGCATAGCGGCGAGGCGT-3′. Data were calculated from three biological replicates, and each biological replicate was examined in triplicate.
Reciprocal Crosses and Double Mutant Analysis
The reciprocal crosses between Ws and shb1-D and between Col and shb1 were made with flowers at identical positions (11th to 14th flowers) on secondary inflorescences. Seed weight and seed number per silique were determined from four separate inflorescences per plant and from four to five plants in total. A series of double mutants of shb1-D with small seed size mutants mini3-2 and iku2-4 were made and PCR genotyped. Gene-specific primers for MINI3 are 5′-TGTCGTTGCAATCTCTCCA-3′ and 5′-GATCCTTTGTGTCTTGCTTGT-3′. Gene-specific primers for IKU2 are 5′-TCTTTTTAATGCCACTAGCTT-3′ and 5′-CGAACATTCCAAGAGACACA-3′. Gene-specific primers for SHB1 are 5′-TAAGCAGCACGAGCTCAAAT-3′ and 5′-TGCTTCCTCGGTTTAGAGTA-3′. Gene-specific primers for AP2 are 5′-TAGGTGGATTTGACACTGCT-3′ and 5′-AGAGGAGGTTGGAAGCCATT-3′. The T-DNA–specific primer for mini3-2, iku2-4, shb1, and ap2 is 5′-GGAACCACCATCAAACAGGAT-3′. For shb1-D genotyping, gene-specific primers are 5′-GAAGATACGGGTTTTGCAT-3′ and 5′-GGGAAGCTTGGATGTCTTGAA-3′, and the T-DNA–specific primer is 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′.
ProSHB1:GUS Construct and GUS Staining
A 2886-bp SHB1 promoter region was amplified by PCR using forward primer 5′-CTAAGCTTCTTCGCGTTATATGACACAACG-3′ and reverse primer 5′-GCGTCGACTGAATCTTGGCCTCTCTCTGTC-3′, which contain a HindIII and a SalI restriction site (underlined), respectively. The PCR fragment was digested with HindIII and SalI and inserted in frame in front of the GUS gene in the pCAMBIA1391 plasmid. The constructs were transformed into Ws by the floral dip method using Agrobacterium tumefaciens GV3101 (Clough and Bent, 1998). Transgenic plants were selected on 25 mg/L of hygromycin B medium (Roche). GUS staining was performed using the method of Sieburth and Meyerowitz (1997). Briefly, tissue was gently fixed by incubation in 90% acetone on ice for 15 to 20 min and rinsed with buffer that contains 50 mM NaPO4, pH 7.2, 0.5 mM K3Fe(CN)6, and 0.5 mM K4Fe(CN)6. Tissue was then transferred to staining solution that contains 50 mM NaPO4, pH 7.2, 2 mM X-gluc (Sigma-Aldrich), 0.5 mM K3Fe(CN)6, and 0.5 mM K4Fe(CN)6, vacuum infiltrated for 10 min, and incubated at 37°C overnight. After staining, tissue was hydrated with 70% ethanol for 1 h and cleared in Hoyer's solution. Ovules were observed using an Olympus BX51 microscope with differential interference contrast optics. Other tissues were photographed using a Cool Cam color CCD camera coupled to an Olympus BH-2 microscope.
In Situ Hybridization
Ws wild-type flowers were hand-pollinated, and the developing seeds were harvested at different days after pollination. The harvested siliques were fixed in FAA overnight at 4°C, embedded in paraplast (Sigma-Aldrich) after dehydration, and sectioned at 8 μm. A 379-bp fragment specific to the 5′ end of the SHB1 gene was amplified with forward primer 5′-GAGGTTTGGGAAAGAGTTTGTGTC-3′ and reverse primer 5′-AGCTGTCTCCACTTTCAGCCTG-3′ and cloned into the pGEM-T easy vector. Antisense and sense RNA probes were synthesized in vitro with digoxigenin-UTP by SP6 and T7 RNA polymerases (Digoxigenin RNA labeling kit; Boehringer Mannheim). Sections were hybridized with 200 ng/mL probes at 42°C overnight in hybridization solution that contains 50% formamide. Hybridization signals were detected using antidigoxigenin antibody conjugated to alkaline phosphatase (DIG nucleic acid detection kit; Boehringer Mannheim). Photographs were taken using a Cool Cam color CCD camera coupled to an Olympus BH-2 microscope.
Anti-SHB1 Antiserum and ChIP Assays
A PCR-generated 2-kb SHB1 C-terminal fragment was subcloned into the EcoRI and SalI sites of the pET-30a expression vector. Approximately 3 mg of SHB1 protein was purified using a His-tag affinity column (Novagen) and injected into rabbits for polyclonal antibody production (Cocalico Biologicals). The specificity of the antibody was verified using preimmunoserum and the fourth bleeding serum in protein gel blot analysis.
ChIP assays were performed as described (Bowler et al., 2004; Sawa et al., 2007) with the following minor modifications. Briefly, developing siliques were harvested 4 to 5 d after hand pollination (early to late heart stage) and fixed with 1% formaldehyde under vacuum. Chromatins were incubated with either anti-SHB1 antibody or anti-GFP antibody for 3 h at 4°C and then incubated with protein A/G agarose beads that had been preequilibrated with salmon sperm DNA for another 2 h at 4°C. Preimmune serum was used as the negative control. After washing twice with low salt buffer and high salt buffer that contains 0.05% SDS and 0.5% Triton X-100, the immunocomplexes were eluted from the agarose beads twice using freshly prepared elution buffer at 65°C (Bowler et al., 2004). DNA was purified by phenol/chloroform extraction and recovered by ethanol precipitation in the presence of glycogen (Sawa et al., 2007). The DNA pellet was resuspended in 150 μL of water, and 3-μL aliquots were used for quantitative PCR analysis. Quantitative PCR was performed as described for real-time PCR analysis. The fold enrichment of the specific chromatin fragment was normalized to the expression level of the UBQ10 amplicon and calculated for each amplicon using the following equation: 2(Ct MINI3-M1 MOCK-Ct MINI3-M1 ChIP)/2(Ct UBQ10 MOCK-Ct UBQ10 ChIP). The primer pairs used in real-time PCR experiments were as follows: MINI3/M1, 5′-CTCATTTTTGACAAATCCTT-3′ and 5′-AACCGAAGTAGAAACCTAAA-3′; MINI3/M2, 5′-TTGTTTTCAATTTTTCACAT-3′ and 5′-AACAAACGGTTATTTTTCTT-3′; MINI3/M3, 5′-TGATGGTATAGTACCGTGATCG-3′ and 5′-TTGGTTGGACTGGTAGAGTCAG-3′; IKU2/IK1, 5′-TCTCCGGTCTCTCTTGATAA-3′ and 5′-GTAGGCGAAACAACATAAGG-3′; IKU2/IK2, 5′-CCTTATGTTGTTTCGCCTAC-3′ and 5′-TCTCTACGTCGGAAGGATTA-3′; LEC2/L1, 5′-CACAAAAGAAAACGAGTCAA-3′ and 5′-TATAAGGAAGAGAGCCAAGG-3′; LEC2/L2, 5′-CTAAGTTAGTCGTTTTTCCA-3′ and 5′-ACTCGATTTTCACAAAACTA-3′; HSF15/H1, 5′-AGATACAATGTTGTGGACTT-3′ and 5′-ATTTTGGATCAAAAGCACTC-3′; HSF15/H2, 5′-GAGTCTTTTGGGTCTTTAAT-3′ and 5′-CATCTTTTGGTAAACAATCT-3′; UBQ10, 5′-TCCAGGACAAGGAGGTATTCCTCCG-3′ and 5′-CCACCAAAGTTTTACATGAAACGAA-3′.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers UBQ10 (At4G05320), SHB1 (At4G25350), AP2 (At4G36920), MINI3 (At1G55600), and IKU2 (At3G19700).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Silique Number of Wild-Type and shb1 Mutants and Seed Mass per Wild-Type and shb1-D Plant under Various Light Conditions.
Supplemental Figure 2. Adult Vegetative and Reproducing Wild-Type and shb1 Mutant Plants.
Supplemental Figure 3. Protein Content on the Basis of Seed Mass and Fatty Acid Composition of Wild-Type and shb1 Mutants.
Supplemental Figure 4. Reducing and Total Soluble Sugar Content of Wild-Type and shb1 Mutants.
Supplemental Figure 5. Expression and ChIP Analysis of SHF15 and LEC2.
Supplementary Material
Acknowledgments
We thank David Marks and Changbin Chen for help and suggestions on microscopy experiments. We thank Gilbert Ahlstrand from the College of Biological Science imaging center for help with scanning electron microscopy. We appreciate the help from Tom Krick with fatty acid analysis in the Center for Mass Spectrometry and Proteomics. We also thank the Ohio State Stock Center for Arabidopsis T-DNA insertion collections and mutant seeds. This work was supported by a grant from the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (2004-35304-14939 to M.N.) and by a grant from the program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, IRT0635 to X.Z.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Min Ni (nixxx008@tc.umn.edu).
Online version contains Web-only data.
References
- Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A.V., Tariq, M., and Paszkowski, J. (2004). Chromatin techniques for plant cells. Plant J. 39 776–789. [DOI] [PubMed] [Google Scholar]
- Berger, F. (2003). Endosperm, the crossroad of seed development. Curr. Opin. Plant Biol. 6 42–50. [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the histone:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R.A., and Cooper, D.C. (1947). The endosperm in seed development. Bot. Rev. 13 423–541. [Google Scholar]
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12 1718–1727. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Downey, R.K. (1983). High and low erucic acid rapeseed oils. In Biochemical Aspects of Crop Improvement, J. Kamer, F. Sauer, and W. Pigden, eds (New York: Academic Press), pp. 253–292.
- Dubois, M., Gilles, K.A., Hamilyon, J.K., Rebers, P.A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28 350–356. [Google Scholar]
- Focks, N., and Benning, C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Fitz Gerald, J.N., and Berger, F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jurgens, G., and Berger, F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, J.D., Weldon, R., Monnot, C., and Meinke, D.W. (1986). Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169 304–312. [DOI] [PubMed] [Google Scholar]
- Hong, S.K., Kitano, H., Satoh, H., and Nagato, Y. (1996). How is embryo size genetically regulated in rice? Development 122 2051–2058. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.E., Li, J., Argueso, C., Gonzalez, M., Lee, E., Lewis, M.W., Maxwell, B.B., Perdue, T.D., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18 3073–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, D.W., and Dooner, H.K. (1990). Isolation of EMS-induced mutants in Arabidopsis altered in seed fatty-acid composition. Theor. Appl. Genet. 80 241–245. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., Omidyar, P.K., Gee, Z., and Okamuro, J.K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 102 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, I.H., Steffen, J.G., Portereiko, M.F., Lloyd, A., and Drews, G.N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, X., and Ni, M. (2006). Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, T.R., and Graham, I.A. (2001). A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J. 25 115–125. [DOI] [PubMed] [Google Scholar]
- Luo, M., Dennis, E.S., Berger, F., Peacock, W.J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102 17531–17536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.E., and Chourey, P.S. (1992). The maize invertase-deficient miniature-1 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell 4 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, N. (1944). A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 153 375–380. [Google Scholar]
- Ohto, M., Fischer, R.L., Goldberg, R.B., Nakamura, K., and Harada, J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102 3123–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.A. (2001). Endosperm development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 233–267. [DOI] [PubMed] [Google Scholar]
- Otegui, M.S., Capp, R., and Staehelin, L.A. (2002). Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 14 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, P.P., Pruitt, R.E., and Meyerowitz, E.M. (1988). Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 11 805–820. [DOI] [PubMed] [Google Scholar]
- Riefler, M., Novak, O., Strnad, M., and Schmülling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, M., Nusinow, D.A., Kay, S.A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff, M.C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., and Scott, R.J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133 251–261. [DOI] [PubMed] [Google Scholar]
- Schultz, P., and Jensen, W.A. (1971). Capsella embryogenesis: The chalazal proliferating tissue. J. Cell Sci. 8 201–227. [DOI] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi, M. (1952). Notes on sugar determination. J. Biol. Chem. 195 19–23. [PubMed] [Google Scholar]
- Spain, B.H., Koo, D., Ramakrishnan, M., Dzudzor, B., and Colicelli, J. (1995). Truncated forms of a novel yeast protein suppress the lethality of a G protein subunit deficiency by interacting with the subunit. J Biol. Chem. 270 25435–25444. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V. (2005). Control of seed size in plants. Proc. Natl. Acad. Sci. USA 102 17887–17888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98 11806–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D.C., Giblin, E.M., Reed, D.W., Hogge, L.R., Olson, D.J., and MacKenzie, S.L. (1995). Stereospecific analysis and mass-spectrometry of triacylglycerols from Arabidopsis thaliana (L) heynh Columbia seed. J. Am. Oil Chem. Soc. 72 305–308. [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G.V., and Provart, N.J. (2007). An electronic fluorescent pictograph browser for exploring and analyzing large-scale biological data sets. PLoS One 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., Brown, R.C., Lemmon, B.E., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2006). Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 142 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D.Z., Wang, G.F., Speal, B., and Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.