Abstract
RNA binding proteins (RBPs) are integral components of ribonucleoprotein (RNP) complexes and play a central role in RNA processing. In plants, some RBPs function in a non-cell-autonomous manner. The angiosperm phloem translocation stream contains a unique population of RBPs, but little is known regarding the nature of the proteins and mRNA species that constitute phloem-mobile RNP complexes. Here, we identified and characterized a 50-kD pumpkin (Cucurbita maxima cv Big Max) phloem RNA binding protein (RBP50) that is evolutionarily related to animal polypyrimidine tract binding proteins. In situ hybridization studies indicated a high level of RBP50 transcripts in companion cells, while immunolocalization experiments detected RBP50 in both companion cells and sieve elements. A comparison of the levels of RBP50 present in vascular bundles and phloem sap indicated that this protein is highly enriched in the phloem sap. Heterografting experiments confirmed that RBP50 is translocated from source to sink tissues. Collectively, these findings established that RBP50 functions as a non-cell-autonomous RBP. Protein overlay, coimmunoprecipitation, and cross-linking experiments identified the phloem proteins and mRNA species that constitute RBP50-based RNP complexes. Gel mobility-shift assays demonstrated that specificity, with respect to the bound mRNA, is established by the polypyrimidine tract binding motifs within such transcripts. We present a model for RBP50-based RNP complexes within the pumpkin phloem translocation stream.
INTRODUCTION
The delivery of proteins and RNA molecules through the phloem translocation of the angiosperms has recently emerged as a mechanism for long-distance signaling in plants (Jorgensen et al., 1998; Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Yoo et al., 2004; Kehr and Buhtz, 2008). Indeed, the phloem appears to contain a unique population of mobile RNA species (Lough and Lucas, 2006). Interestingly, the trafficking of such non-cell-autonomously acting RNA has been shown to play a role in the regulation of such processes as gene silencing, pathogen defense, and development (Kim et al., 2001; Yoo et al., 2004; Haywood et al., 2005; Aung et al., 2006; Bari et al., 2006; Baumberger et al., 2007; Gaupels et al., 2008).
In situ localization experiments established that SUCROSE TRANSPORTER1 mRNA is present in mature, functional sieve elements as well as within the plasmodesmata connecting the sieve elements and companion cells (Kühn et al., 1997). Proof of function for phloem-mobile transcripts has been demonstrated by grafting studies. For example, KNOTTED1-LIKE and GIBBERELLIC ACID–INSENSITIVE PHLOEM (GAIP) transcripts are translocated into heterografted scions, where they induce a phenotypic change in developing leaves (Kim et al., 2001; Haywood et al., 2005). A population of small RNAs (19 to 25 nucleotides) has also been detected in phloem sap of cucurbits, castor bean (Ricinus communis), yucca (Yucca filamentosa), and lupin (Lupinus albus), suggesting a role for small RNAs in long-distance signaling (Yoo et al., 2004).
Long-distance delivery of RNA molecules through the phloem appears to be mediated by RNA binding proteins (RBPs) (Lucas et al., 2001). The pumpkin (Cucurbita maxima; Cm) PHLOEM PROTEIN16 (Cm PP16) was the first phloem RBP to be characterized; it was identified as a functional homolog of a viral movement protein (Xoconostle-Cázares et al., 1999). Delivery of PP16 into specific target tissues appears to be regulated through its interaction with other proteins within the phloem translocation stream (Aoki et al., 2005). Phloem lectins such as Cm PP2, cucumber (Cucumis sativus; Cs) PP2, and Cucumis melo LECTIN have been shown to bind both phloem mRNA and viral RNA, and this interaction has been implicated in the control of systemic infection (Gómez and Pallás, 2004; Gómez et al., 2005). Pumpkin PHLOEM SMALL RNA BINDING PROTEIN1 binds selectively to single-stranded small RNA species and mediates their trafficking through plasmodesmata (Yoo et al., 2004).
Although ample evidence now exists that phloem-mobile RNA contributes to the integration of developmental processes at the whole plant level, little information is available on the nature of the ribonucleoprotein (RNP) complexes contained within the phloem translocation stream. In this study, we used the phloem system of pumpkin to identify and characterize the protein and RNA components of a plant phloem RNP complex. A 50-kD polypyrimidine tract binding (PTB) protein, RBP50, acts as the core of this complex; all mRNA species extracted from the RBP50 complex contained PTB motifs. The nature of the mRNA species contained within such phloem RNP complexes provides insights into the range of developmental and physiological processes likely regulated by the long-distance trafficking of information macromolecules.
RESULTS
Pumpkin Phloem Sap Contains a Spectrum of RNA Binding Proteins
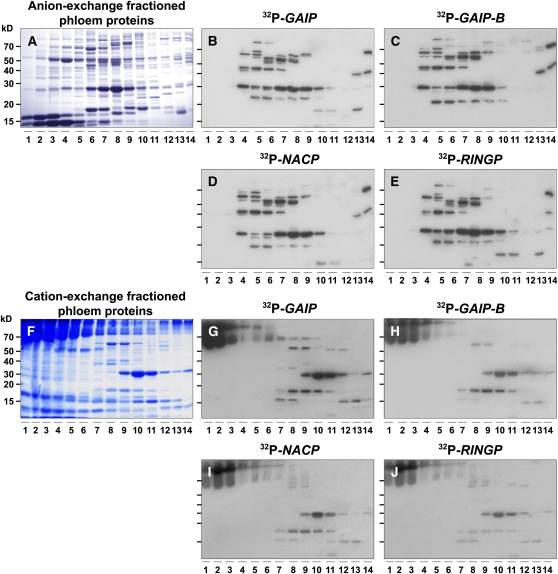
Protein and RNA gel blot overlay assays were used previously to identify potential RBPs within the pumpkin phloem sap (Xoconostle-Cázares et al., 1999; Yoo et al., 2004). In order to ascertain the complexity in terms of the number of RBPs present within the angiosperm phloem translocation stream, we first performed RNA overlay assays on phloem sap collected from stems of 6-week-old pumpkin plants. Anion- and cation-exchange fast protein liquid chromatography (FPLC) fractionated phloem proteins were separated by SDS-PAGE (Figures 1A and 1F) and then subjected to RNA overlay assays (Yoo et al., 2004) using four previously characterized phloem-mobile transcripts (Ruiz-Medrano et al., 1999; Haywood et al., 2005).
Figure 1.
Pumpkin Phloem Sap Contains a Spectrum of RNA Binding Proteins.
(A) Pumpkin phloem sap proteins separated by anion-exchange FPLC. Proteins were separated on a 13% SDS-PAGE gel and then stained with GBS reagent. Numbers represent the elution fractions from anion-exchange FPLC.
(B) to (E) RNA overlay–protein blot assays performed on FPLC-fractionated proteins from (A) using the following riboprobes specific for phloem-mobile transcripts: GAIP (B), GAIP-B (C), NACP (D), and RINGP (E).
(F) Pumpkin phloem sap proteins separated by cation-exchange FPLC. Proteins were separated on a 13% SDS-PAGE gel and then stained with GBS reagent. Numbers represent the elution fractions from cation-exchange FPLC.
(G) to (J) RNA overlay–protein blot assays performed on FPLC-fractionated proteins from (F) using the following riboprobes specific for phloem-mobile transcripts: GAIP (G), GAIP-B (H), NACP (I), and RINGP (J).
A broad range of anion-exchange fractionated phloem proteins were found to bind radioactively labeled GAIP, GAIP-B, NAM, ATAF1/2 and CUC2 DOMAIN PROTEIN PHLOEM (NACP), and RING FINGER MOTIF PROTEIN PHLOEM (RINGP) transcripts (Figures 1B to 1E); interestingly, these profiles were remarkably similar in nature, but some differences were apparent. Equivalent experiments performed on cation-exchange FPLC fractionated phloem proteins revealed that, again, quite similar RNA binding patterns were observed for the four transcripts used in these assays (Figures 1G to 1J). Based on these experiments, it appears that the pumpkin phloem sap contains a spectrum of proteins that could participate in the formation of RNP complexes with the four tested phloem-mobile transcripts.
Isolation of Candidate Phloem RBPs
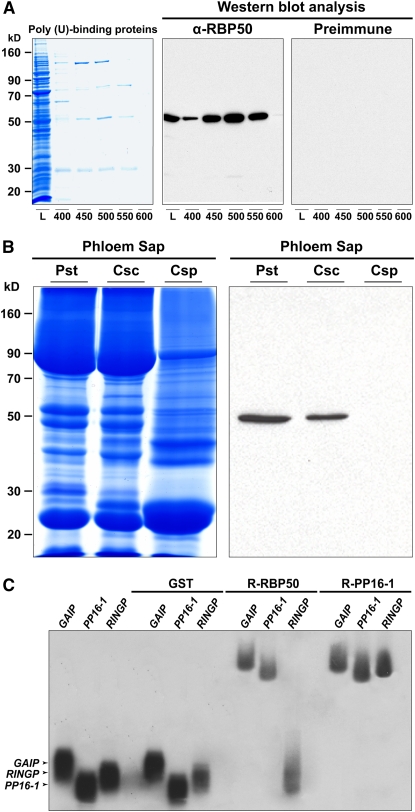
A poly(U)-affinity column was next employed to isolate and enrich for a specific set of phloem RBPs. Total phloem sap was loaded onto the poly(U) column and, following a series of washes, bound proteins were eluted using increasing salt concentrations (Figure 2A, left panel). This protocol led to the purification of four phloem RBPs. As a starting point, the phloem 50-kD RBP band was cut from the gel, trypsin-digested, and processed for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Three peptide sequences, obtained from the 50-kD band, were used to clone the encoding gene, designated RBP50. The deduced amino acid sequence for RBP50 (see Supplemental Figure 1 online) contained the four highly conserved RNA-recognition motifs present in the PTB family of proteins (Conte et al., 2000; Schmid et al., 2007). An antibody generated against RBP50 was used to confirm the identity of the 50-kD phloem protein enriched by poly(U)-affinity chromatography (Figure 2A, middle panel).
Figure 2.
RBP50, a 50-kD Poly(U) Binding Protein, Moves through the Translocation Stream and Has the Capacity to Bind, in a Sequence-Specific Manner, to Phloem-Mobile Transcripts.
(A) A poly(U)-affinity column was used to purify pumpkin phloem proteins with poly(U) binding capacity. Candidate proteins were eluted using the following step concentrations of NaCl: 400, 450, 500, 550, and 600 mM. Protein profiles (L; loading sample) were separated on a 12% SDS-PAGE gel (volumes of 20 μL were applied for each fraction) and then visualized with GBS reagent. Purified anti-RBP50 antibody (α-RBP50) specifically detected the 50-kD protein (middle panel), whereas preimmune sera did not cross-react with any protein, including those in the loading sample.
(B) Long-distance movement of RBP50 is confirmed by heterografting studies. Phloem sap was collected from pumpkin stock (Pst), cucumber scions grafted onto pumpkin stocks (Csc), and wild-type cucumber plant (Csp). Phloem sap proteins were resolved by SDS-PAGE (left panel) and subjected to protein gel blot analysis with anti-RBP50 antibody (right panel). Detection of RBP50 in phloem sap collected from cucumber scions establishes that this protein is phloem-mobile.
(C) Gel mobility-shift assays reveal that recombinant (R) RBP50 binds to phloem-mobile transcripts in a sequence-specific manner. RNA binding capacities of RBP50, PP16-1, and GST were tested against 32P-labeled in vitro–transcribed riboprobes for GAIP, PP16-1, and RINGP. Note that GST did not bind to these transcripts, RBP50 bound to GAIP and PP16-1 but not RINGP, and PP16-1 bound all three probes.
To determine whether RBP50 functions as a phloem-mobile RBP, we next performed a series of grafting experiments in which pumpkin plants were employed as the stock and cucumber as the scion. Phloem sap collected from stock and scion tissues, as well as from ungrafted cucumber plants, was used in protein gel blot analyses. The absence of signal in phloem sap collected from ungrafted cucumber plants established the specificity of the RBP50 antibody (Figure 2B). Of equal importance, the strong signal detected in the phloem sap collected from both the pumpkin stock and cucumber scion tissues (Figure 2B) established that RBP50 is a bona fide phloem-mobile protein.
To confirm the RNA binding properties of RBP50, gel mobility-shift assays were next performed using three previously identified phloem-mobile mRNAs, GAIP, PP16-1, and RINGP (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Haywood et al., 2005). Glutathione S-transferase (GST) was employed as a negative control, and, as expected, GST did not bind to these phloem transcripts (Figure 2C). As a positive control for RNA binding, we used PP16-1, which we earlier showed binds RNA in a sequence-nonspecific manner (Xoconostle-Cázares et al., 1999); this property was confirmed by our present studies (Figure 2C). Interestingly, RBP50 was found to bind GAIP and PP16-1 transcripts, but it did not appear to interact with the RNA for RINGP (Figure 2C). Taken together, these data further support the hypothesis that RBP50 is a phloem-mobile RBP that may exhibit selective binding to the population of transcripts that enter the translocation stream.
RBP50 Transcripts Are Detected in Companion Cells, Whereas RBP50 Accumulates in Sieve Elements
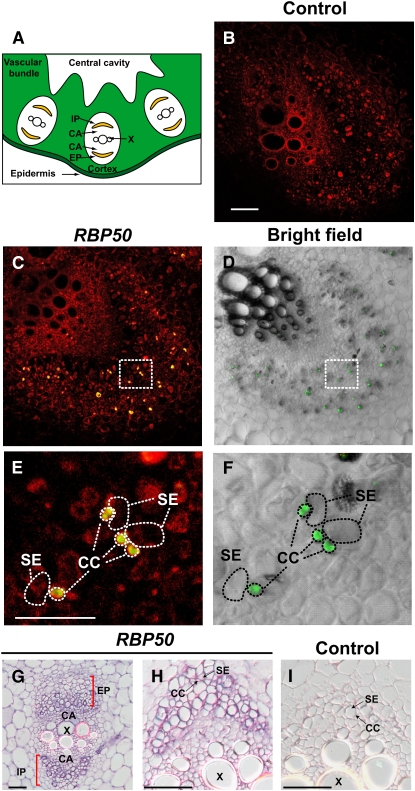
A series of in situ RT-PCR and in situ hybridization experiments was next performed to test for the expression of RBP50 in pumpkin phloem tissue. As illustrated in Figure 3, a strong RBP50 signal was detected over companion cells, whereas no signal was found over the neighboring sieve elements (Figures 3C to 3F). In situ hybridization experiments confirmed the presence of a strong RBP50 signal in companion cells of the long-distance phloem (Figures 3G and 3H). These findings are consistent with the hypothesis that RBP50 is produced in companion cells prior to its entry into the phloem translocation stream.
Figure 3.
In Situ RT-PCR– and RNA in Situ Hybridization–Based Detection of RBP50 Transcripts in Companion Cells.
(A) Schematic transverse section of a portion of the pumpkin petiole/stem. Vascular bundles are composed of internal and external phloem (IP and EP, respectively) and xylem (X), with an intervening cambium (CA).
(B) to (F) Transverse sections from pumpkin stem tissue analyzed by in situ RT-PCR using RBP50 gene-specific primers. Images were collected with a confocal laser scanning microscope; positive signal (green) represents incorporation of Alexa Fluor–labeled nucleotides.
(B) Negative control in which primers were omitted from the RT-PCR mixture. Red fluorescence represents tissue autofluorescence. Bar = 500 μm (common for [B] to [D]).
(C) Transverse section of a vascular bundle demonstrating the presence of RBP50 in phloem cells. Note that the green signal from the Alexa Fluor–labeled nucleotides appears yellow due to the red autofluorescence background.
(D) Bright-field image of (C) used to illustrate the cellular architecture of the vascular bundle. RBP50 transcripts (green fluorescent signal) accumulated in companion cells.
(E) Higher magnification of the boxed area shown in (C). CC, companion cell; SE, sieve element. Bar = 100 μm (common for [E] and [F]).
(F) Higher magnification of the boxed area shown in (D). Images presented are representative of those obtained from at least three replicate experiments.
(G) to (I) Pumpkin stem sections analyzed by in situ hybridization. Transverse sections were hybridized with an in vitro–transcribed antisense RNA probe to RBP50 ([G] and [H]); purple signal represents the presence of RBP50 transcripts in the small companion cells. Control in situ hybridization was performed with an in vitro–transcribed sense RNA probe to RBP50 (I). Bars = 500 μm.
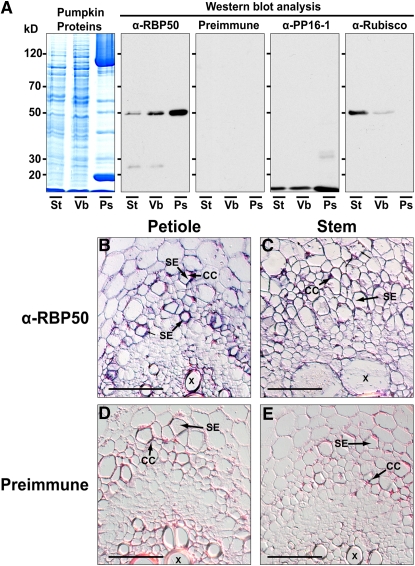
Movement of RBP50 from the companion cells into the neighboring sieve elements likely occurs through the plasmodesmata interconnecting these two cell types (Lough and Lucas, 2006). This trafficking event could result in the accumulation of RBP50 within the sieve elements. To provide a test for this hypothesis, total proteins extracted from stem tissue, vascular bundles (Lin et al., 2007), and phloem sap were separated by SDS-PAGE and then examined by protein gel blot analysis for their levels of RBP50. In comparison with the phloem sap, stem and vascular tissues were found to contain quite low amounts of RBP50 (Figure 4A). As controls for these experiments, protein gel blot analyses were performed using antibodies directed against the pumpkin PP16-1 and Ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco), which was chosen as a control to detect the presence of contamination from surrounding cell types (Sjolund, 1997; Oparka and Turgeon, 1999; Van Bel, 2003). These experiments confirmed that PP16-1 is also enriched in the phloem sap relative to its presence in the total protein population of stem or vascular tissues. Importantly, the absence of detectable amounts of Rubisco in the phloem sap suggested that the level of contaminating proteins was low; hence, the profiles observed for both RBP50 and PP16 should be reflective of the in vivo situation.
Figure 4.
RBP50 Is Synthesized in Companion Cells and Accumulates in Sieve Elements.
(A) RBP50 accumulates in the phloem. Proteins extracted from pumpkin stem (St), vascular bundles (Vb), and phloem sap (Ps) were resolved on a 12% SDS-PAGE gel and visualized with GBS reagent. Proteins were transferred to nitrocellulose membranes, and blotting was performed with anti-RBP50 antibody, preimmune serum, anti-PP16-1 antibody, or anti-Rubisco antibody. Protein gel blot analysis indicated that RBP50 and PP16-1 accumulate in sieve elements. Preimmune serum failed to detect proteins extracted from these pumpkin tissues. Anti-Rubisco antibody, which served as a control against phloem sap contamination by proteins from surrounding tissue, did not detect the presence of Rubisco in the phloem sap. Results are based on three independent replicate experiments.
(B) Immunohistochemical detection of RBP50 within companion cells (CC) and sieve elements (SE) of pumpkin petiole vascular bundles. RBP50 was detected using a combination of anti-RBP50 primary antibody followed by anti-rabbit IgG as a secondary antibody.
(C) Immunohistochemical detection of RBP50 within companion cells and sieve elements of pumpkin stem vascular bundles.
(D) and (E) Immunohistochemical controls performed on petiole and stem sections with preimmune sera. Note the absence of signal within the phloem tissues.
Bars = 500 μm for (B) to (E).
Immunolocalization studies were next performed to establish the cell types in which RBP50 accumulates. Using anti-RBP50 antibodies, signal could be detected in both the companion cells and sieve elements located in pumpkin petiole and stem tissues (Figures 4B and 4C). By contrast, no signal was detected when these tissues were treated with preimmune sera (Figures 4D and 4E). Taken together, these studies demonstrate that after translation of the RBP50 transcripts in the companion cells, RBP50 is trafficked into the translocation stream, likely through the companion cell–sieve element plasmodesmata.
RBP50 Interacts with a Subset of Phloem Proteins
The human PTB functions as a component of various RNP complexes (Reyes and Izquierdo, 2007; Auweter and Allain, 2008). If RBP50 performs a similar function in plants, then we would expect it to interact with a set of proteins contained within the pumpkin phloem translocation stream. To test this notion, anion- and cation-exchange FPLC fractionated phloem proteins were probed in protein overlay assays with RBP50. Fractionated phloem proteins were separated by SDS-PAGE (see Supplemental Figures 2A and 2B online), blotted onto nitrocellulose membranes, and then tested for the presence of RBP50 (see Supplemental Figures 2C and 2D online) or overlaid with a test protein (see Supplemental Figures 2E to 2L online). Protein gel blot analysis established that RBP50 is concentrated in anion-exchange fractions 6 and 7. Overlaid blots were performed with these RBP50-enriched phloem fractions, and we identified a range of potential interaction partners present in both the anion- and cation-exchange fractions (see Supplemental Figures 2E and 2F). By contrast, overlay assays performed with BSA failed to detect any interaction partners (see Supplemental Figures 2G and 2H online), thereby confirming the specificity of the interactions observed with RBP50.
In a parallel set of experiments performed with recombinant RBP50, produced in and purified from Escherichia coli, we observed that the ability of RBP50 to bind phloem proteins was significantly reduced (see Supplemental Figures 2I and 2J online). We recently reported a similar result for PP16-1, in which a change in the binding property of the recombinant PP16-1 was shown to be due to the absence of phosphorylation on specific residues (Taoka et al., 2007). As many phloem proteins are present in the translocation stream as phosphoproteins (Taoka et al., 2007) and RBP50 is also present as a phosphoprotein (see Supplemental Figure 3 online), we next tested whether phosphorylation status affects the capacity of RBP50 to bind its interaction partners.
Calf intestinal phosphatase (CIP) pretreatment of phloem-enriched RBP50 resulted in a significant alteration in the binding pattern when this protein was overlaid onto FPLC fractionated phloem proteins (see Supplemental Figures 2K and 2L). The similarity in binding patterns observed with recombinant (R)-RBP50 and phosphatase-pretreated phloem proteins supports the hypothesis that interaction between RBP50 and many of its phloem interaction partner proteins is likely influenced by phosphorylation status.
Identification of Phloem RBP50-Interacting Proteins
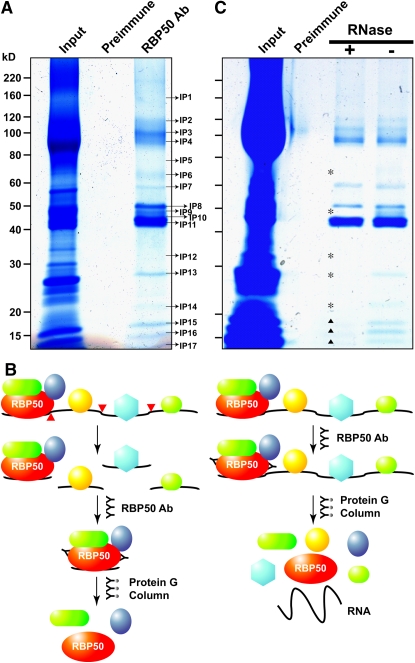
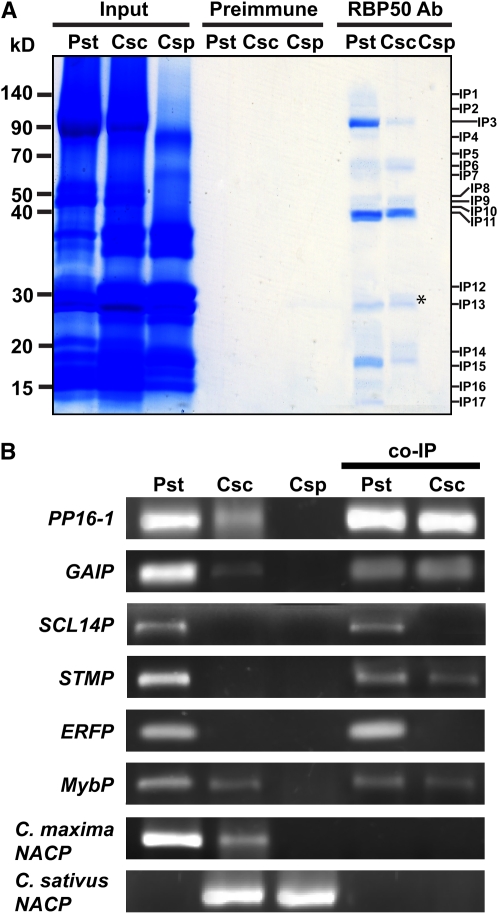
To identify the putative RBP50-interacting proteins, coimmunoprecipitation (co-IP) experiments were next performed. For these studies, unfractionated phloem sap (containing both proteins and RNA) was employed for co-IP against preimmune sera and purified RBP50 antibodies; some 17 bands were coprecipitated (Figure 5A). These RBP50-interacting proteins were identified using LC-MS/MS analysis (Table 1). Parallel co-IP experiments performed with preimmume sera yielded no positive hits when gels were subjected to LC-MS/MS analysis. This absence of proteins in our preimmume co-IP experiments confirmed the specificity of the RBP50 antibody preparation used in our co-IP experiments. Note that the 50-kD band, labeled Interacting Protein8 (IP8), was identified as RBP50. The reproducibility of these co-IP results was confirmed by replicate experiments; the same set of 17 proteins was detected in four independent experiments. Taken together, these data support the hypothesis that RBP50 can interact with a subset of pumpkin phloem proteins.
Figure 5.
Identification of the Pumpkin Phloem Proteins Coimmunoprecipitated with RBP50.
(A) Total phloem proteins (input protein) were coimmunoprecipitated using either preimmune serum or purified anti-RBP50 antibody (Ab). Proteins were visualized by GBS reagent. The co-IP complex(es) included interacting proteins IP1 to IP17; note that IP8 was confirmed to be RBP50 by LC-MS/MS analysis.
(B) Schematic illustration of phloem proteins that potentially could be bound to a phloem-mobile transcript contained within a RBP50–RNP complex. Note that transcripts bound by RBP50 may also interact with additional RNA binding proteins. RNase treatment was employed to separate a bound transcript (red darts) into individual RNP fragments, thereby allowing identification of proteins contained within the RBP50 complex.
(C) Co-IP of the RBP50 RNP complex was performed with or without RNase treatment. Asterisks indicate phloem proteins likely not directly contained within the RBP50 complex; darts indicate proteins likely bound to both the RBP50 and other protein complexes located on the same/different RNA molecule(s).
Table 1.
Potential RBP50 Phloem-Interacting Proteins Identified by Co-IP Experiments
| Interacting Protein | Protein Identity | Molecular Mass (kD) |
|---|---|---|
| IP1 | Myosin heavy chain–like protein | 149 |
| IP2 | Heat shock protein–related protein, Gly dehydrogenase | 113, 113 |
| IP3 | PP1 | 96 |
| IP4 | Expressed protein | 89 |
| IP5 | Phosphoinositide-specific phospholipase–like protein | 68 |
| IP6 | Glutathione-regulated potassium efflux system protein | 67 |
| IP7 | Glycosyl hydrolase family 1 protein | 65 |
| IP8 | RBP50 | 50 |
| IP9 | Expressed protein | 46 |
| IP10 | Putative ATP binding protein | 44 |
| IP11 | GTP binding protein | 39 |
| IP12 | Shikimate kinase precursor | 33 |
| IP13 | RBP50 cleavage product, Cys proteinase inhibitor | 26 |
| IP14 | Csf-2 related protein | 19 |
| IP15 | PP16-1 | 17 |
| IP16 | PP16-2 | 16 |
| IP17 | Cys proteinase inhibitor like protein, putative protein | 12 |
RBP50–RNP complexes were coimmunoprecipitated from total phloem sap using purified anti-RBP50 antibody. Proteins were separated by SDS-PAGE, and individual bands were excised from the gel for LC-MS/MS analysis and protein identification.
Total phloem sap contains a complex mixture of proteins and RNA, and as a number of phloem RBPs are present (Figure 1), the possibility exists that these proteins may also bind to the same transcripts as the RBP50 (Figure 5B). To test for this possibility, phloem sap was first given an RNase treatment, in order to physically separate RNP complexes located on individual transcripts (Figure 5B). This RNase pretreatment of phloem sap, prior to the RBP50 co-IP experiment, was found to significantly reduce the complexity of the co-IP protein profile (Figure 5C). Here, IP6, IP9, IP12, IP13, and IP14 were completely absent from the gel, and IP15, IP16, and IP17 were significantly reduced in their band intensities.
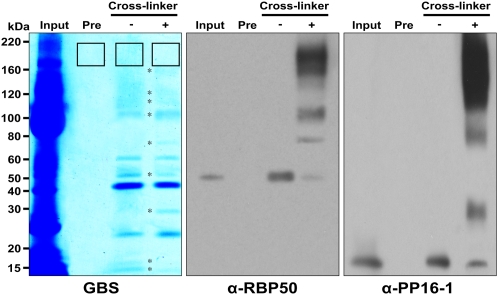
Based on the RNase treatment, it would appear that RBP50 may interact with 11 phloem proteins. To further establish the nature of the phloem proteins in this complex, we next used a chemical cross-linking method (Sinz, 2003; Koller et al., 2004) to covalently interconnect the proteins contained in the RNP complex(es) that were purified with the RBP50 antibody. The cross-linker treatment resulted in a significant change in the protein profile compared with the untreated preparation (Figure 6). Protein gel blot analysis, using anti-RBP50 antibody, allowed us to detect phloem proteins that had been covalently linked to RBP50. Here, we noted that the level of RBP50 detected at the 50-kD region of the gel, in the cross-linked sample, was significantly reduced below that observed in the control experiment; this is consistent with the presence of immunologically detected bands at higher molecular masses.
Figure 6.
Chemical Cross-Linking Identifies Proteins of the Core RBP50–RNP Complex.
Phloem proteins (Input) were coimmunoprecipitated using either preimmune serum (Pre) or purified RBP50 antibody. Isolated RBP50–RNP complexes were then treated with the chemical cross-linker, bis(sulfosuccinimidyl) suberate, and subjected to SDS-PAGE. Protein profiles were visualized using GBS reagent (left panel). To confirm the cross-linkage of RBP50 with its interacting proteins, protein gel blot assays were performed using purified anti-RBP50 antibody (middle panel) or purified anti-PP16-1 antibody (right panel). Asterisks (left panel) indicate protein bands that underwent changes in intensity after cross-linker treatment. The boxed regions on the SDS-PAGE gel were excised for protein analysis using LC-MS/MS.
As PP16-1 was identified as a putative RBP50-interacting protein (Table 1), protein gel blot analysis was also performed with the anti-PP16-1 antibody (Figure 6). Compared with the control, in which the anti-PP16-1 antibody recognized a single protein band, a range of higher molecular mass protein bands was detected with the cross-linked RBP50 coprecipitated proteins. The band in the 30-kD region of the gel represents the PP16-1 dimer, while the bands at 80, 100, and 120 to 200 kD overlap with proteins also detected by the anti-RBP50 antibody, consistent with cross-linking between PP16-1 and RBP50.
To identify the cross-linked proteins contained within the high molecular mass protein complex(es), appropriate bands were excised for trypsin in-gel digestion followed by LC-MS/MS analysis. IP1 and IP3 were detected in the same high molecular mass position on the lane loaded with no cross-linker–treated sample, suggesting that IP1 and IP3 would show nonspecific binding to RBP50 RNP complexes. As shown in Table 2, we were able to detect IP2, IP4, IP5, IP8, IP11, IP13, IP15, and IP16 in the 160- to 220-kD region of the gel. These results are consistent with the interacting proteins identified in the RNase-pretreated RBP50 coprecipitated protein complex(es) (Figure 5C). Interestingly, four additional proteins were identified as potential components of the RBP50-based phloem RNP complex (Table 2); these were eukaryotic initiation factor 5A (eIF-5A), two expressed proteins, and a molecular chaperone, Hsc70-1. Aoki et al. (2005) reported that Cm PP16 can interact with eIF-5A, so it is possible that these newly identified proteins may not interact directly with RBP50.
Table 2.
Phloem Proteins Identified within the RBP50 Cross-Linked High Molecular Mass Protein Complex(es)
| Interacting Protein | Protein Identity | Molecular Mass (kD) |
|---|---|---|
| IP2 | Heat shock protein–related protein (HSP113) | 113 |
| IP4 | Expressed protein (EP89) | 89 |
| IP5 | Phosphoinositide-specific phospholipase–like protein (PSPL) | 68 |
| IP8 | RBP50 | 50 |
| IP11 | GTP binding protein (GTPbP) | 39 |
| IP13 | Cys proteinase inhibitor (CPI) | 26 |
| IP15 | PP16-1 | 17 |
| IP16 | PP16-2 | 16 |
| CLP1 | eIF-5A | 17 |
| CLP2 | Expressed protein (EP106) | 106 |
| CLP3 | Hypothetical protein (103) | 103 |
| CLP4 | Hsc70-1 | 70 |
Coimmunoprecipitated RBP50–RNP complexes were chemically cross-linked with bis(sulfosuccinimidyl) suberate. Proteins were separated by SDS-PAGE, and high-molecular mass (160 to 220 kD) complexes were excised from the gel for LC-MS/MS analysis to identify phloem proteins cross-linked to RBP50. CLP, additional cross-linked protein not detected from analysis of proteins contained within the coimmunoprecipitated RBP50 complex(es).
Protein Phosphorylation Is Required for RNP Complex Formation
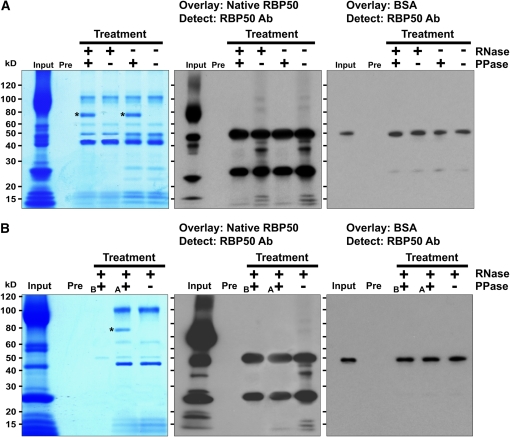
To further explore the requirement of phosphorylation for protein–protein interaction and RNP complex formation, we next performed a series of experiments involving a combination of RNase and CIP treatments. In the first experiment, total phloem sap (containing proteins and mRNA) was treated with or without RNase, followed by co-IP. Purified RBP50-based RNP complexes were then treated with or without CIP, the proteins were separated by SDS-PAGE, and their profiles were visualized by GelCode Blue Stain (GBS) reagent (Figure 6). Proteins were then blotted onto nitrocellulose membranes and overlaid with either native phloem-purified RBP50 or BSA (control for specificity of binding), and interacting proteins were then identified using anti-RBP50 antibodies (Figure 6, middle and right panels, respectively). Consistent with our earlier finding, RNase pretreatment reduced the overall complexity of the interacting proteins (Figure 7A). Importantly, CIP treatment, followed by overlay with native RBP50, caused a dramatic reduction in the capacity of RBP50 to bind the copurified phloem proteins.
Figure 7.
Protein Phosphorylation Is Required for RBP50-Based RNP Complex Formation.
(A) Total phloem sap (containing proteins and mRNA) was treated with or without RNase, followed by co-IP. Purified RBP50-based RNP complexes were then treated with or without CIP (PPase), the proteins were separated by SDS-PAGE, and their profiles were visualized by GBS reagent (left panel). Proteins were then blotted onto nitrocellulose membranes and overlaid with either native phloem-purified RBP50 or BSA, and interacting proteins were detected by anti-RBP50 antibody (Ab; middle and right panels, respectively). Asterisks indicate the bands for CIP.
(B) Total phloem sap was pretreated with RNase and CIP (PPase) before (B) the co-IP with anti-RBP50 antibodies. A second aliquot of total phloem sap was given a CIP treatment after (A) RNase pretreatment and co-IP. Proteins were separated by SDS-PAGE, and their profiles were visualized by GBS reagent (left panel). After blotting onto nitrocellulose membranes, proteins were overlaid with either native phloem-purified RBP50 or BSA, and interacting proteins were detected with anti-RBP50 antibody (middle and right panels, respectively). The asterisk indicates the band for CIP.
A second series of experiments was performed in which total phloem sap was first pretreated with RNase and CIP followed by co-IP with anti-RBP50 antibodies. As an internal control, an equal aliquot of total phloem sap was processed as in Figure 7A (i.e., pretreated with RNase, co-IP was performed, and then treated with CIP). The protein profiles shown in Figure 7B illustrate that CIP pretreatment, applied before performing the co-IP, resulted in a complete absence of the interacting proteins (left panel, lanes marked B and A). Protein overlay assays confirmed that this CIP pretreatment abolished the capacity of native RBP50 to interact with phloem proteins involved in the formation of an RNP complex (Figure 7B, middle panel). Interestingly, in its dephosphorylated state, RBP50 was still able to interact with native RBP50; note the increased signal intensity at the 50-kD region in the overlay performed with native RBP50 compared with that of the BSA control (Figure 7B, middle and right panels, respectively). The strong interacting band located at the 25-kD region of the RBP50 overlay was analyzed by mass spectrometry and shown to be a RBP50 cleavage product that was still capable of interacting with native RBP50.
Identification of the RNA Species Contained within the RBP50 Complex(es)
The mRNA species contained within the RBP50 coprecipitated protein complex(es) were next identified. To this end, the co-IP RBP50 RNP complexes were used to extract RNA and then linker-coupled reverse transcription and PCR were used to amplify these transcripts. Six genes were identified based on this approach (Table 3). The transcript displaying the highest abundance encoded PP16-1 followed by GAIP; interestingly, these two transcripts have previously been reported as phloem-mobile transcripts (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Haywood et al., 2005). Four additional phloem transcripts (denoted P) were identified: a SCARECROW-LIKE transcription factor (SCL14P), SHOOT MERISTEMLESS (STMP), an ETHYLENE RESPONSE FACTOR subfamily transcription factor (ERFP), and a MYELOBLASTOSIS (MYB) family transcription factor (MybP).
Table 3.
Phloem Transcripts Detected within RBP50-Based Ribonucleoprotein Complexes
| Phloem Transcript Identity | Transcript Abundance |
|---|---|
| PP16-1 | 15 |
| GAIP | 9 |
| SCARECROW-LIKE (SCL14P) | 7 |
| SHOOT MERISTEMLESS (STMP) | 5 |
| ETHYLENE RESPONSE FACTOR (ERFP) | 5 |
| Myb (MybP) | 4 |
Total pumpkin phloem sap was used to coimmunopurify RNP complexes. mRNA contained within these RNP complexes was extracted, cloned, and sequenced. Note that in naming these transcripts, the letter P is used to designate them as being contained within the phloem translocation stream. Transcript abundance represents the cloning results from five independent co-IP experiments in which the isolated RNA was pooled for analysis. Data presented represent the number of times each transcript was identified from all 35 amplified colonies.
To further investigate the degree to which RBP50 exhibits specificity in its binding to these identified phloem transcripts, we next performed co-IP experiments using PP2, a phloem protein previously shown to bind RNA in a non-sequence-specific manner (Gómez et al., 2005). Amplification of RNA isolated from these PP2 complexes identified a broad spectrum of mRNA species; in contrast with the six mRNA species identified from the RBP50 RNP complexes, some 75 different transcripts were cloned from the PP2 complexes (see Supplemental Table 1 online). The heterogeneous nature and high number of the transcripts identified from the PP2 co-IP, which contrasts markedly with the six transcripts identified for the RBP50 experiments, provides support for the hypothesis that RBP50 binds to a specific set of phloem-mobile mRNA species.
PTB Motifs in Phloem mRNAs Confer Specificity for RBP50 Binding
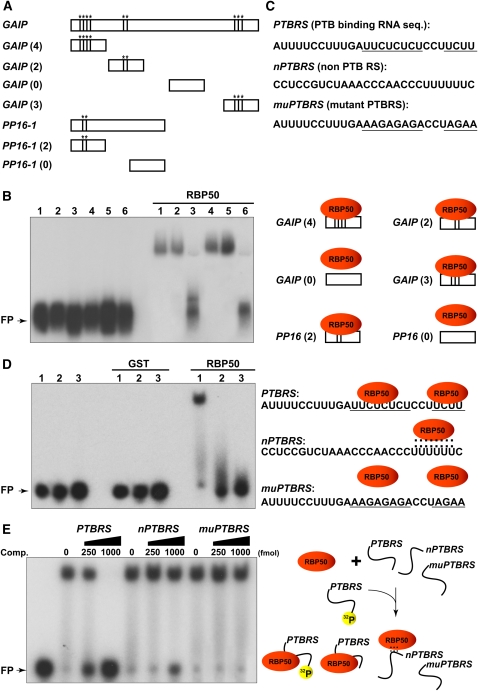
It is well established that the interaction between PTB motifs, within the mRNA, and PTB proteins confers both high binding affinity and sequence specificity (Wang and Pederson, 1990; Bothwell et al., 1991; Patton et al., 1991; Luo, 1999; Liu et al., 2002; Schmid et al., 2007). Sequence analysis of the six transcripts contained within the RBP50-based phloem RNP complexes (Table 3) revealed that they all had at least two such PTB motifs. To investigate whether these sequence motifs provided the basis for selectivity in transcript binding by RBP50, we next performed a series of gel mobility-shift assays.
For these experiments, a set of deletion mutants was engineered using GAIP and PP16-1 as representatives of the six identified phloem transcripts. Analysis of their sequences indicated that GAIP and PP16-1 have nine and two PTB motifs, respectively (Figure 8A). Based on this information, four deletion mutants were engineered for GAIP and two for PP16-1 (Figure 8A; the number of PTB motifs contained in each mutant is indicated in parentheses). In order to minimize complications associated with size-dependent effects on binding specificity (Yoo et al., 2004), all mutants were engineered to be of the same size. A gel mobility-shift assay performed with these mutant RNAs revealed that RBP50 bound RNA species containing PTB motifs (Figure 8B, lanes 1, 2, 4, and 5). RBP50 failed to bind mutant RNAs lacking any PTB motif (Figure 8B, lanes 3 and 6). These results support the hypothesis that RBP50 recognizes and binds to the isolated phloem transcripts through the presence of their PTB motifs.
Figure 8.
Selectivity of RBP50 RNA Binding Is Conferred through PTB Binding Motifs.
(A) Schematic illustration of the deletion mutant series of GAIP and PP16-1 RNA employed in gel mobility-shift assays. Asterisks indicate the locations of PTB motifs on wild-type and mutant RNAs. Values in parentheses indicate the number of PTB motifs.
(B) Gel mobility-shift assays establish that RBP50 binding to RNA is dependent on the presence of PTB motifs. Experiments were performed using recombinant RBP50 and 10 nM aliquots of the indicated 32P-labeled riboprobe. Lane 1, GAIP (4); lane 2, GAIP (2); lane 3, GAIP (0); lane 4, GAIP (3); lane 5, PP16-1 (2); lane 6, PP16-1 (0). FP, free riboprobe.
(C) Engineered 27-nucleotide PTB motif variants; PTB motifs are underlined. The two PTB motifs present in the PTBRS RNA sequence were derived from the first two motifs located in the 5′ region of the GAIP mRNA. The nPTBRS RNA represents a sequence located in the 3′ region of the GAIP mRNA devoid of PTB motifs. The muPTBRS represents PTBRS in which the two PTB motifs have been mutated.
(D) Synthetic 27-nucleotide riboprobes confirm the sequence requirement for RBP50 binding. Gel mobility-shift assays were performed using 27-nucleotide RNA probes (10 nM) and GST or recombinant RBP50. Lane 1, PTBRS; lane 2, nPTBRS; lane 3, muPTBRS. Dashed lines (right panel) indicate a weak interaction between RBP50 and nPTBRS RNA.
(E) Competition assay performed by preincubating RBP50 (250 ng) with increasing concentrations of unlabeled PTBRS, nPTBRS, or muPTBRS followed by competition with 32P-labeled synthesized PTBRS (10 nM).
To further investigate the interaction between RBP50 and the PTB motifs, we next conducted gel mobility-shift assays using 27-nucleotide RNAs with or without PTB motifs. For these studies, 27-nucleotide RNAs were designed based on the GAIP sequence; a control PTB RNA sequence (PTBRS) was synthesized using the first two PTB motifs in GAIP (Figure 8C). A 27-nucleotide probe, devoid of PTB motifs (nPTBRS), was synthesized using the 5′ region of GAIP, and a mutant PTBRS (muPTBRS) was engineered in which we replaced the PTB motifs with countermatched nucleotides. Radiolabeled 27-nucleotide RNA probes were used in gel mobility-shift assays with GST or RBP50. As expected, GST did not bind to these probes, whereas RBP50 exhibited strong binding to PTBRS (Figure 8D, lane 1). Importantly, RBP50 only weakly bound nPTBRS and failed to bind muPTBRS (Figure 8D, lanes 2 and 3). These experiments strengthened the conclusion that the RNA binding properties of RBP50 depend on the presence of PTB motifs within the target transcript.
Competition experiments were next performed by preincubating RBP50 with different concentrations of unlabeled PTBRS, nPTBRS, or muPTBRS, followed by the addition of radioactively labeled PTBRS. Only PTBRS served as a competitor to RBP50 binding to radioactively labeled PTBRS (Figure 8E). Finally, competition experiments using increasing concentrations of unlabeled PTBRS allowed us to determine the RBP50 dissociation constant for PTBRS; this has a value of 3.1 × 10−8 M (see Supplemental Figure 4 online). Taken together, these findings support the hypothesis that the PTB properties of RBP50 allow it to recognize specific phloem-mobile transcripts to form mobile RNP complexes.
RBP50 Ribonucleoprotein Complexes Move in the Phloem Translocation Stream
As RBP50 was established to be a phloem-mobile protein (Figure 2B), heterografting studies were conducted to ascertain whether the identified RBP50 complexes are similarly translocated through the phloem. Three to 4 weeks after grafting, cucumber scions were excised and phloem sap was collected for use in co-IP experiments. Here, phloem sap collected from ungrafted cucumber was employed as a negative control. Given that the anti-RBP50 antibody failed to detect the cucumber ortholog (Figure 2B), it was expected that the anti-RBP50 antibody would not result in the co-IP of cucumber phloem proteins. Our results indeed confirmed this prediction (Figure 9A).
Figure 9.
RBP50-Based RNP Complexes Move into the Cucumber Scion.
(A) Phloem sap collected from pumpkin stock (Pst), cucumber scion (Csc), and control (ungrafted) cucumber plants (Csp) was subjected to co-IP against RBP50 or purified preimmune serum. Proteins were visualized by GBS staining. Note the similarity between the protein profiles for co-IP performed with phloem sap collected from pumpkin stock and cucumber scions. The absence of proteins in the co-IP experiment performed with phloem sap collected from control cucumber plants indicates the specificity of the interaction between RBP50 and the purified anti-RBP50 antibody preparation. The asterisk indicates the presence of a 28-kD unknown cucumber phloem protein that copurified with the pumpkin RBP50-based RNP complexes.
(B) RT-PCR–based detection of phloem transcripts contained within the pumpkin stock (Pst), cucumber scion (Csc), and ungrafted cucumber stock (Csp) and RBP50 RNP complexes isolated from pumpkin stock or cucumber scion phloem sap. As controls, Cm NACP and Cs NACP transcripts were amplified using specific primer pairs. Results represent three independent RT-PCR experiments.
Parallel experiments performed on cucumber scion phloem sap, using the anti-RBP50 antibody, resulted in the co-IP of a set of proteins (Figure 9A). Inspection of this protein profile with that obtained from co-IP experiments performed on phloem sap collected from the pumpkin stock revealed a high degree of similarity. Of the 17 RBP50-interacting proteins identified from the pumpkin stock, visual inspection identified 10 from the cucumber scion co-IP experiment. LC-MS/MS analysis confirmed the identity of these proteins and led to the detection of one additional unknown cucumber protein of 28 kD. These heterografting results provide direct support for the hypothesis that the RBP50-based RNP complex functions as a phloem-mobile signaling agent. Additionally, the pumpkin proteins associated with RBP50 may represent the core of this long-distance RNP complex.
Two approaches were next employed to test for the presence of the six transcripts earlier identified as being bound in the RBP50 RNP complexes (Table 3). In the first, mRNA was isolated from phloem sap collected from the pumpkin stock, cucumber scion, and ungrafted cucumber plants. Primer pairs specific for these pumpkin transcripts were used in RT-PCR assays; all six mRNAs were detected in RNA isolated from pumpkin stock, whereas none was amplified from ungrafted cucumber (Figure 9B). Controls for these experiments were performed using primer pairs specific for Cm NACP and Cs NACP; transcripts for these pumpkin and cucumber genes were amplified from the RNA isolated from their respective phloem sap. RT-PCR analyses of the cucumber scion RNA sample detected three of the six transcripts detected in the RBP50 RNP complexes: PP16-1, GAIP, and MybP (Figure 9B). In addition, transcripts for Cm NACP as well as Cs NACP were also detected in these scion-based assays.
In the second approach, the anti-CRBP50 antibody was used in co-IP experiments performed on phloem sap collected from pumpkin stock, cucumber scion, or ungrafted cucumber plants. The RBP50 RNP complexes were then used to isolate the bound mRNA for RT-PCR analysis. Based on these experiments, all six transcripts were again amplified from the complexes isolated directly out of the pumpkin phloem sap. Analysis of the mRNA from the cucumber scion co-IP experiment gave positives with four of the six transcripts (Figure 9B). We used NACP as a negative control for these experiments, as this pumpkin mRNA was earlier shown to move into cucumber scions (Ruiz-Medrano et al., 1999) and was present in the mRNA population pulled down with PP2 (see Supplemental Table 1 online) but not RBP50. Importantly, NACP transcripts could not be amplified from RBP50 RNP complexes isolated from either pumpkin or cucumber scion phloem sap. Collectively, these results support the hypothesis that specific phloem mRNAs are carried through the phloem translocation stream within RBP50 RNP complexes.
DISCUSSION
RBP50 Is a Phloem-Mobile PTB Protein
In this study, we present evidence that pumpkin phloem sap contains a significant number of RNA binding proteins (Figure 1). This finding, along with the fact that the phloem translocation stream contains a unique subpopulation of mRNA species (Lough and Lucas, 2006; Ruiz-Medrano et al., 2007), is consistent with the pumpkin phloem serving as a conduit for the long-distance trafficking of RNP complexes.
To identify the molecular constituents of these putative RNP complexes, a biochemical approach was used to enrich and purify a 50-kD RNA binding protein, RBP50, from the pumpkin phloem sap (Figure 2). RBP50, a member of the PTB family (Schmid et al., 2007), is expressed in mature companion cells (Figure 3), gains entry into the phloem translocation stream (Figure 4), and travels to distantly located tissues and organs (Figures 2 and 9A). Of significance to the characterization of phloem RNP complexes, RBP50 displayed a degree of selectivity in terms of the phloem-mobile transcripts to which it could bind (Figure 2). This suggested that RBP50 may well function as the core protein in the formation of specific RNP complexes.
Protein Constituents of RBP50-Based Phloem-Mobile RNP Complexes
Several RNA binding proteins have previously been characterized from phloem sap (Xoconostle-Cázares et al., 1999; Lucas et al., 2001; Gómez and Pallás, 2004; Yoo et al., 2004), but as yet, the molecular constituents of these RNP complexes have remained uncharacterized. As a first step to identifying the interaction partners that constitute a RBP50-based RNP complex, we employed protein overlay methods. These studies established that RBP50 could bind to some 23 proteins present in FPLC-fractionated pumpkin phloem sap (see Supplemental Figures 2E and 2F online). This binding pattern was highly reproducible, and as the anion- and cation-fractionated pumpkin phloem sap contains >1000 proteins (Lin et al., 2008), our protein overlay assays establish that RBP50 interacts with a specific subset of phloem proteins.
Posttranslational modification of phloem non-cell-autonomous proteins plays an important role in determining their capacity to interact with NON-CELL-AUTONOMOUS PATHWAY PROTEIN1 and traffic through plasmodesmata (Taoka et al., 2007). Similarly, the animal PTB is phosphorylated, and this posttranslational modification has been shown to regulate its nucleocytoplasmic transport (Xie et al., 2003; Knoch et al., 2006). Our studies indicated that RBP50 also appears to exist in the phloem sap as a phosphoprotein (see Supplemental Figure 3 online) and, furthermore, that this posttranslational modification is required for binding to its phloem interaction partners (see Supplemental Figures 2I to 2L online). It will now be interesting to determine whether the same or different amino acid residues are involved in its trafficking through plasmodesmata and RNP complex formation. Such studies may well provide insights into the site where the RBP50-based RNP complex(es) are assembled, namely, in companion cells or the sieve elements.
Co-IP experiments performed with affinity-purified anti-RBP50 antibodies led to the identification of 16 putative interacting proteins contained within the total pumpkin phloem sap (Figure 5; Table 1). Interestingly, with the exception of IP2, a HSP113, the remaining 15 IPs did not overlap with previously reported mammalian PTB-interacting proteins (Tronchere et al., 1997; Gromak et al., 2003; Pickering et al., 2003; Hall et al., 2004; Song et al., 2005). This suggests that the function of RBP50 in the phloem may well be different from that reported for mammalian PTB family members.
Generally, RNP complex formation is mediated by the target RNA molecule establishing binding specificity for the cognate RNA binding protein(s) (MacMorris et al., 2007; Swartz et al., 2007; Galban et al., 2008). Our RNase pretreatment, applied prior to co-IP (Figure 5), allowed us to separate the 17 IPs into 11 likely contained within a RBP50-associated complex(es) and 6 that may have the capacity to bind RNA in a non-sequence-specific manner. Proteins contained within the RBP50-based complex(es) were further investigated by chemical cross-linking methods. Using this approach, in combination with LC-MS/MS analysis of the high molecular mass cross-linked proteins, we were able to further refine the nature of the RBP50 complex (Figure 6; Table 2).
Based on the consistent results obtained from three replicate cross-linker experiments, it appears that seven phloem proteins may interact directly with RBP50. Interestingly, four additional proteins not detected in RBP50 co-IP experiments were identified. This suggests that, under normal conditions, there are most likely only weak interactions between these phloem proteins and RBP50. Alternatively, these additional cross-linked proteins may bind to RBP50-interacting proteins. In the case of eIF-5A, Aoki et al. (2005) earlier demonstrated that this protein interacts with PP16-1 and PP16-2. The Hsc70-1 belongs to a subclass of non-cell-autonomous chaperones (Aoki et al., 2002) and may be involved in mediating the trafficking of the RBP50-based RNP complex, or components therein, through the companion cell–sieve element plasmodesmata.
The enucleate sieve tube system likely contains RNP complexes that function either locally, in maintenance, or in long-distance signaling. Our heterografting studies provided direct evidence that the RBP50-based RNP complexes move across the graft union into the cucumber scion (Figure 9). Thus, we conclude that the RBP50 RNP system plays a role in the delivery of specific transcripts to distantly located tissues.
RBP50 Oligomerization
Evidence that RBP50 forms homooligomers was obtained from our protein overlay assays. The band intensities, within the 50-kD region of the anion-exchange fractionated pumpkin phloem proteins and RBP50 co-IP proteins, increased significantly when overlaid with native RBP50 (Figure 7; see Supplemental Figure 2E online). The ability of RBP50 to form homodimers/oligomers is consistent with early studies showing that the mammalian PTB can exist as a dimer in solution (Perez et al., 1997; Oh et al., 1998). However, recent experiments also indicate that the animal PTB may function as a monomer (Monie et al., 2005; Auweter and Allain, 2008). Interestingly, although RBP50 phosphorylation appears necessary for an interaction with its cognate phloem protein partners, this posttranslational modification may not be required for dimer formation (Figure 7; see Supplemental Figure 2).
A 26-kD RBP50-interacting protein, identified in our overlay (Figure 7; see Supplemental Figure 2 online) and co-IP experiments (Figure 5; Table 1), was identified by mass spectrometry as a RBP50 cleavage product. This finding is consistent with an earlier report that the mammalian PTB similarly forms a 25-kD cleavage product (Venkatramana et al., 2003). Interestingly, this mammalian 25-kD PTB product was shown to prevent native PTB from binding to the 5′ untranslated region of hepatitis A viral RNA, thereby inhibiting its translation. Thus, the RBP50 cleavage product may well be involved in regulating native RBP50 binding to target transcripts (Rideau et al., 2006).
mRNA Constituents of RBP50-Based RNP Complexes in the Phloem Translocation Stream
Analysis of the RNA extracted from phloem-purified RBP50-based RNP complexes identified six different mRNA species (Table 3), and four of these identified mRNAs were delivered into the cucumber scion within RBP50 RNP complexes (Figure 9B). The presence of PP16-1 and GAIP mRNA in the cucumber scion phloem is consistent with our earlier studies (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Haywood et al., 2005). The four additional mRNA species were detected at equivalent levels and encoded transcription factors (Table 3). SCL14P and GAIP are members of the GRAS (for GAI, REPRESSOR of GA [RGA], and SCARECROW [SCR]) family (Pysh et al., 1999) involved in the regulation of meristem development (Bolle, 2004). Although GAIP mRNA was amplified from a phloem-mobile RBP50-based RNP complex, SCL14P mRNA was not detected (Figure 9B). Either the SCL14P RNP complex is present at very low levels in the cucumber scion or it may act locally; thus, it would have been removed prior to entry of the phloem sap into the cucumber scion.
STMP is a member of the KNOTTED class of homeodomain proteins (Long et al., 1996) and is required for apical meristem maintenance (Byrne et al., 2002; Cole et al., 2006). Transcripts for STMP were amplified from RBP50 RNP complexes isolated from both the pumpkin stock and cucumber scion. However, our RT-PCR assay performed on mRNA isolated from cucumber scion phloem sap failed to detect STMP transcripts. Thus, this mRNA species is likely present at low levels, in the total phloem sap, and enrichment through co-IP of the bound STMP transcripts allowed us to isolate sufficient mRNA for RT-PCR detection. This suggests that co-IP can be employed to identify low-abundance phloem-mobile mRNA species.
The ERFP and MybP transcripts both belong to large gene families that are involved in a broad spectrum of developmental, abiotic, and biotic stress signaling pathways (Riechmann et al., 2000; Ramsay and Glover, 2005; Zhao et al., 2005; Chen et al., 2006; Nurmberg et al., 2007). Interestingly, transcripts for MybP, but not ERFP, were shown to be phloem-mobile; hence, they may function to regulate gene expression in distantly located target tissues. Detailed analyses of these four transcription factors will provide insights into the roles they play in local and long-distance integration of signaling events involved in developmental and physiological processes.
All six phloem transcripts cloned from the RBP50 RNP complexes contained PTB motifs. Our gel mobility-shift assays (Figure 8) provided strong support for the hypothesis that these sequence motifs provide the foundation for transcript recognition by RBP50. These results are in agreement with earlier RNA binding studies conducted with animal PTBs (Liu et al., 2002; Oberstrass et al., 2005; Schmid et al., 2007). Indeed, the dissociation constant of the RBP50–PTB motif interaction was determined to be 30 nM, a value equivalent to that measured for the animal PTB (Song et al., 2005).
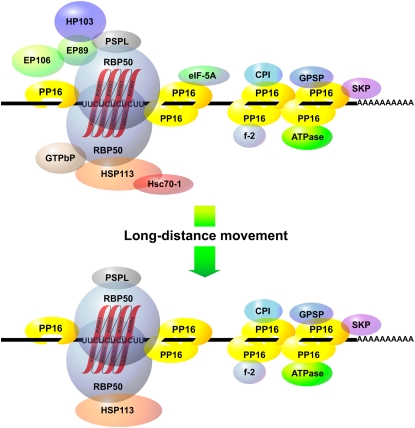
Molecular Composition of the RBP50-Based RNP Complex
A model of the RBP50-based RNP complex is illustrated in Figure 10. Some of the proteins identified from the RBP50 co-IP experiments have functions that appear consistent with their presence in a phloem RNP complex. For example, PP16-1/-2 are present in the pumpkin phloem sap, move in the translocation stream, and function as RNA binding proteins that traffic RNA through plasmodesmata (Ruiz-Medrano et al., 1999). That these proteins interact directly with RBP50 (Figures 5 to 7) implicates them in the formation of a core for the RBP50–RNP complex. As with the animal PTB, RBP50 may function as a chaperone to stabilize and deliver specific phloem-mobile transcripts to target cells for translation (Song et al., 2005).
Figure 10.
Schematic Illustration of a Phloem RBP50-Based Ribonucleoprotein Complex.
RBP50 binds to PTB motifs (UUCUCUCUCUU) present within a subclass of phloem-mobile, polyadenylated transcripts, and this interaction leads to the binding of additional RBP50 (shown as a homodimer). PP16-1/-2 interact with both the RBP50 and the target mRNA, thereby forming the core of the RNP complex. RBP50 interacts directly with a GTPbP–HSP113–Hsc70-1 complex that may chaperone the RNP complex to and through the companion cell–sieve element plasmodesmata. Another set of four proteins, composed of the 89-kD expressed protein (EP89), the 103-kD hypothetical protein (HP103), the 106-kD expressed protein, and the PSPL are shown interacting with the RBP50. eIF-5A is a component of the RBP50 core and binds directly to PP16-1/-2. Regions outside the PTB motifs are bound by PP16-1/-2 along with five additional proteins: CPI, the Csf-2 related protein (Cmf-2), the 44-kD putative ATP binding protein (ATPase), the glutathione-regulated potassium-efflux system protein (GPSP), and the shikimate kinase precursor (SKP). The bottom image shows the composition of the phloem-mobile RBP50-based RNP complex based on co-IP results obtained using cucumber scion phloem sap.
The RBP50-based RNP core also appeared to contain GTP binding protein (GTPbP), EP89, phosphoinositide-specific phospholipase-like protein (PSPL), and HSP113, and their interaction with RBP50 was dependent upon phosphorylation (Figure 7). However, Cm GTPbP and Cm EP89 were not detected in the complex isolated from cucumber scion phloem sap (Figure 9). Thus, it would appear that the core of the RBP50-based RNP complex contains five phloem proteins: RBP50, PP16-1, PP16-2, PSPL, and HSP113 (Figure 10).
GTPbP belongs to the RAB (for Ras-related in brain) GTPase superfamily (Vernoud et al., 2003; Ma, 2007); hence, one possibility is that it functions in the subcellular trafficking of the RBP50–RNP complex, perhaps for delivery to plasmodesmata (Haupt et al., 2005) for entry into or export from the phloem translocation stream. Hsc70-1 has the capacity to traffic through plasmodesmata and is present in the pumpkin phloem sap. Thus, it may interact with HSP113 to chaperone the RBP50–RNP complex through the companion cell–sieve element plasmodesmata.
An interaction between RBP50 and EP89 was indicated by the fact that this protein was isolated after RNase treatment (Figure 5) and was detected in the high molecular mass fraction from the cross-linker studies (Table 2). Hence, EP89 may also play a role in the trafficking of the RNP complex into the phloem translocation stream. Finally, both HP103 and EP106 were not detected in our initial co-IP experiments but, rather, were isolated as cross-linked components of the RBP50–RNP complex (Table 2). Thus, these proteins may be weakly bound at the periphery of the RNP complex. The functional significance of these uncharacterized proteins will await further studies.
As illustrated in Figure 5, RNase pretreatment allowed the separation of bound transcripts into a RBP50 core and RNA segments bound by other proteins. Examination of the protein profiles obtained with or without RNase treatment revealed that, of the eight proteins affected by this treatment, only PP16-1, PP16-2, and eIF-5A have the capacity to bind RNA. Furthermore, PP16-1/-2 bind RNA in a non-sequence-specific manner (Xoconostle-Cázares et al., 1999), so it would appear that these two proteins also bind along the length of the transcript to protect it from degradation.
Based on our co-IP results obtained with phloem sap collected from pumpkin stock and cucumber scion tissue, it seems that Csf-2, CPI, ATPase, GPSP, and SKP bind to PP16-1/-2. The Csf-2 was earlier identified as a ripening-induced protein from cucumber (Suyama et al., 1999) and was previously detected in the cucurbit phloem sap (Haebel and Kehr, 2001; Walz et al., 2004). The roles played by these five proteins in terms of the long-distance translocation of the RBP50-based RNP complexes must await further studies. However, their retention through the cucumber graft union indicates that they are part of a stable protein complex (Figure 10).
METHODS
Plant Materials
Pumpkin (Cucurbita maxima cv Big Max) and cucumber (Cucumis sativus cv Straight Eight) plants were grown in a special insect- and pathogen-free greenhouse under natural daylight conditions as described previously (Yoo et al., 2004; Taoka et al., 2007). Nutrients were supplied daily as described (http://greenhouse.ucdavis.edu/cef/materials/soilfert.html). Pumpkin and cucumber phloem sap were collected from well-watered plants as described previously (Yoo et al., 2004).
Grafting Protocols
Heterografting experiments were performed as described previously (Ruiz-Medrano et al., 1999), with minor modifications. Briefly, heterografts were generated between scions of 4-week-old cucumber (the vegetative apex with ∼6 cm of stem) and 4- to 5-week-old pumpkin (for heterografts) or cucumber (for homograft control) stock plants. Each excised scion was carefully inserted into a V-shaped incision made in the main stem of the stock. Graft sites were sealed with Parafilm, and then the scion was covered with a clear plastic bag that was removed 1 week later. For initial grafting experiments to test for the mobility of RBP50 in the phloem translocation stream, three independent replicates (two to three heterografts per replicate) were performed. Scions were used for phloem sap or tissue collection 3 to 4 weeks after grafting. A second, large-scale heterografting experiment was performed to test whether the RBP50-based RNP complexes move over long distances in the phloem. Here, 96 individually grafted cucumber scions were generated, and phloem sap was collected 3 to 4 weeks after grafting (see Supplemental Figure 5 online). Phloem sap was pooled and used in co-IP and RT-PCR experiments.
Phloem Sap Collection and Protein Fractionation
Phloem sap was collected from well-watered plants as described previously (Yoo et al., 2004). Anion- and cation-exchange chromatography of proteins contained within the pumpkin phloem sap was performed as described previously (Yoo et al., 2004; Taoka et al., 2007). Generally, 30 mL of pumpkin phloem sap (0.1 to 0.2 mg protein/mL) was dialyzed against buffer A (50 mM Tris, pH 7.5, 1 mM EDTA, and 30 mM 2-mercaptoethanol) and then loaded onto a buffer A–equilibrated HiTrap Q column (GE Healthcare) for anion-exchange fractionation using an FPLC system (GE Healthcare). After anion-exchange chromatography of the phloem proteins, flow-through fractions were dialyzed against buffer AC (20 mM sodium phosphate buffer, pH 7.0, 10% glycerol, 1 mM EDTA, and 1% 2-mercaptoethanol) and then loaded onto a buffer AC–equilibrated HiTrap SP column (GE Healthcare) for cation-exchange fractionation. After washing the column with buffer A or buffer AC, proteins were eluted with a linear gradient of 0 to 500 mM NaCl in buffer A or buffer AC containing 1 M NaCl. Fractionated proteins were separated on 13% SDS-PAGE gels and stained with GBS reagent (Pierce Biotechnology). Quantification of proteins was performed by the Bradford method (Bio-Rad).
Isolation of Phloem Poly(U) Binding Proteins
Anion-exchange fractioned pumpkin phloem proteins were first dialyzed against equilibration buffer (20 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 50 mM NaCl, 0.1 mM EDTA, 1.0 mM DTT, and 10% [v/v] glycerol) and then applied onto a poly(U)–Sepharose 4B matrix (GE Healthcare) column previously equilibrated with equilibration buffer. After washing with 10 column volumes of equilibration buffer, poly(U) binding proteins were eluted with elution buffer (20 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 0.1 mM EDTA, 1.0 mM DTT, and 10% [v/v] glycerol) containing, sequentially, 400, 450, 500, 550, and 600 mM NaCl. Eluted samples were separated on 12% SDS-PAGE gels and stained with GBS reagent.
Cloning of RBP50
Samples eluted from the poly(U)–Sepharose 4B column were separated by SDS-PAGE, 50 kD-bands were excised and subjected to in-gel trypsin digestion, and the proteolytic peptides were prepared for mass spectrometry as described previously (Rosenfeld et al., 1992; Lin et al., 2007). To identify the cDNA encoding RBP50, total RNA was extracted from stems of pumpkin and subjected to reverse transcription reaction using SuperScriptII (Invitrogen) according to the manufacturer's instructions. A full-length RBP50 cDNA was amplified by PCR using the following primer pair: 50-F (5′-ATGACTGAACCCTCAAAGGTTA-3′) and 50-R (5′-GAGTCAACAACAAAGCAG-3′). The amplified cDNA was confirmed by sequencing.
Pumpkin RBP50 Phylogenetic Analysis
Public databases were searched using PTB genes as queries. Their accession numbers and sequence information can be found in Supplemental Data Set 1 online. A neighbor-joining tree was constructed using MEGA 3.1 software (Lin et al., 2007), and a consensus tree was built from 1,000 bootstrap replicates (see Supplemental Figure 1C online).
In Situ mRNA Hybridization and in Situ RT-PCR Analysis
Stem and petiole tissues were excised from 6-week-old pumpkin plants and immediately fixed, dehydrated, and embedded with paraffin as described previously (Ruzin, 1999; Lin et al., 2007). A 160-bp DNA fragment for the 3′ untranslated region (UTR) of RBP50 was produced by PCR using the following primer pair: CmRBP3UF (5′-GTTAACAACGGCATTAAACGCG-3′) and CmRBP3UR (5′-GAGTCAACAACAAAGCAGAAG-3′). The PCR product (160 bp of 3′ untranslated region of RBP50) was subcloned into the TOPO II dual promoter vector (Invitrogen), and then this recombinant vector was linearized with XhoI or HindIII to generate digoxigenin-labeled sense or antisense mRNA, respectively, using the MAXIscript kit (Ambion). In situ hybridization was performed as described previously (Ruiz-Medrano et al., 1999; Lin et al., 2007). Anti-digoxigenin conjugated to alkaline phosphatase (Boehringer Mannheim) was used to detect the digoxigenin-labeled RNA. Images were analyzed with a phase-contrast microscope (Axioskop 2 Plus) using Axiovision 5 software (Zeiss).
In situ RT-PCR analyses were performed as described previously (Ruiz-Medrano et al., 1999; Haywood et al., 2005). Briefly, pumpkin stem and petiole sections were pretreated with DNaseI for 15 min to remove DNA and then subjected to in situ RT-PCR. The same primer pair was used to amplify RBP50 transcripts as that for generating the 3′ untranslated region of RBP50 described above. Negative control reactions contained all components except one or both primers. Fluorescence analysis was performed on a confocal laser scanning microscope (model DM RXE 6 TCS-SP2 AOBS; Leica Microsystems) using an Ar/ArKr laser. Autofluorescence from chlorophyll was detected using a GHeNe laser (Taoka et al., 2007). All images were processed with Adobe Photoshop version CS2.
Immunolocalization of RBP50
Petiole and stem tissues from pumpkin plants were fixed in 3.7% formaldehyde, 5% acetic acid, and 50% ethanol at 4°C for 16 h. Procedures of dehydration, paraffin-embedding, and sectioning were as described previously (Lin et al., 2007). Sections were deparaffinized in Neo-Clear (Merck) and then rehydrated. Polyclonal antibodies were produced against recombinant His-tagged RBP50 in rabbits. Immunolocalization analyses were performed as described previously (Lin et al., 2007). For immunolocalization, 1:100 diluted anti-RBP50 antibody was used as the primary antibody, and 1:300 diluted anti-rabbit IgG was employed as the secondary antibody; sections were given a 1-h incubation in both the primary and secondary antibody preparations. Slides were washed twice with 1× TTBS (1× TTBS = 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, and 0.5% Tween 20) for 5 min each and covered with diaminobenzidine substrate (Roche Applied Science). Sections were incubated until the desired color was achieved (∼5 to 15 min). After rinsing with distilled water, sections were dehydrated and mounted, and slides were examined on an Axioskop 2 Plus photomicroscope; images were collected with an Axiocam MRc digital camera (Zeiss).
Protein Gel Blot Analyses
Protein gel blot analyses were performed as described previously (Taoka et al., 2007). Briefly, nitrocellulose membrane blots were blocked for 1 h with 5% nonfat milk prepared in 1× TBS. Membrane blots were then incubated with the appropriate antibody preparation (anti-RBP50 antibody used at 1:5000 dilution, anti-PP16 antibody [Aoki et al., 2005] used at 1:10,000 dilution, and anti-Rubisco antibody used at 1:2000 dilution) along with the blocking agent. Membranes were then washed three times with 1× TTBS for 5 min each, followed by a 1-h incubation with secondary antibody (horseradish peroxidase–conjugated anti-rabbit [1:20,000 dilution]; Sigma-Aldrich). Blots were again washed three times with 1× TTBS for 5 min each and then immunodetected with chemiluminescence reagent (Perkin-Elmer Life Sciences) and film (Kodak Biomax MS; Eastman Kodak).
RNA Overlay Assays
RNA overlay assays were conducted as described previously (Yoo et al., 2004). Briefly, anion- and cation-exchange fractioned pumpkin phloem proteins were separated on 12% SDS-PAGE gels and then electrotransferred onto nitrocellulose membranes. Blots were rinsed with RNA binding buffer ([RBB]; 10 mM Tris, pH 7.0, 50 mM KCl, 1 mM EDTA, 0.02% [w/v] Ficoll, and 0.02% [w/v] polyvinylpyrrolidone) and blocked with the RBB containing 0.02% (v/v) ultrapure BSA (Ambion) and 0.1 mg/mL yeast total RNA (Sigma-Aldrich) at 25°C for 1 h. 32P-labeled GAIP (Haywood et al., 2005), GAIP-B (Haywood et al., 2005), NACP (Ruiz-Medrano et al., 1999), or RINGP (Ruiz-Medrano et al., 1999; Yoo et al., 2004) mRNA probes were synthesized using the MAXIscript kit (Ambion). Next, membranes were incubated with each of the 32P-labeled probes (2 × 105 cpm/mL) in RBB with BSA at 20°C for 1 h. Membranes were washed three times for 5 min each with RBB, air-dried, and autoradiographed.
Protein Overlay Assay and Co-IP
Protein overly assays were performed essentially as described (Lee et al., 2003; Taoka et al., 2007). Briefly, the anion- or cation-exchange fractionated pumpkin phloem proteins were separated on 13% SDS-PAGE gels and then transferred onto nitrocellulose membranes. Recombinant RBP50 in pET28a was expressed in Escherichia coli strain Rosseta2 (DE3) pLysS (Novagen); protein expression was induced with 0.5 mM isopropylthio-β-galactoside for 16 h at 28°C. Protein purification was performed as described previously (Taoka et al., 2007). Protein blots were overlaid with native or recombinant RBP50 diluted in BSA buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, and 2 mg/mL BSA) for 40 min at 20°C. Blotted nitrocellulose membranes were washed with 1× TTBS four times for 5 min each and subjected to normal protein gel blot analysis, as described above, with polyclonal RBP50 antibody and anti-rabbit horseradish peroxidase–conjugated secondary antibody (Sigma-Aldrich).
Co-IP was performed as follows. Briefly, pumpkin phloem sap proteins (2 mg protein/mL) were dialyzed in bind/wash buffer (0.14 M NaCl, 8.0 mM sodium phosphate, 2.0 mM potassium phosphate, and 11 mM KCl, pH 7.4) overnight at 4°C. Anti-RBP50 antiserum was first purified with a MAbTrap affinity column (GE Healthcare) and then used with a HiTrap N-hydroxy-succinimide (NHS)-activated HP column (GE Healthcare) with immobilized recombinant His-tagged RBP50. Antigen-specific anti-RBP50 IgG was then immobilized to ImmunoPure Plus Immobilized Protein G (Pierce Biotechnology) and immunoprecipitation was performed using the Seize X immunoprecipitation kit (Pierce Biotechnology), following the manufacturer's instructions with minor modifications (Aoki et al., 2005). Input samples (2 mg pumpkin phloem sap proteins/mL) were loaded onto an anti-RBP50 IgG spin column and incubated at 4°C for 16 h. Proteins were eluted with elution buffer supplied from the kit, and elution fractions were directly neutralized with a one-twentieth volume of 1 M Tris-HCl, pH 9.5. Elution fractions were separated on 4 to 15% gradient SDS-PAGE gels (Bio-Rad) and then stained with GBS reagent.
To identify RNA sequences required for RBP50 binding, total RNA was extracted from RBP50 coprecipitated proteins and then subjected to linker-coupled reverse transcription reaction (forward linker, 5′-AAGCAGTGGTATCAACGCAGAGTACGCGGG-3′; reverse linker, 5′-TGGTATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTVN-3′) using SuperScriptII (Invitrogen). Reverse transcription reaction samples were amplified by PCR using the following primer pair: LL (5′-AAGCAGTGGTATCAACGCAGAGT-3′) and RL (5′-TGGTATCAACGCAGAGTAC-3′). All PCR products were subcloned into the TOPO II dual promoter vector (Invitrogen) and confirmed by sequencing.
RT-PCR Analyses
RT-PCR analyses of mRNA extracted from pumpkin stock, cucumber scion, ungrafted cucumber scion phloem sap, and RBP50 co-IP complexes isolated from pumpkin stock or cucumber scion phloem sap were performed as follows. One hundred nanograms of RNA was used for the reverse transcription reaction with SuperScriptII reverse transcriptase (Invitrogen) following the manufacturer's instructions. One microliter of each reverse transcription reaction was used in the PCR with the following conditions: 30 s at 98°C, 30 s at 53°C, and 30 s at 72°C (35 cycles). PP16-1, GAIP, SCL14P, STMP, ERFP, MybP, NACP, and Cs NACP were amplified using the following gene-specific primer pairs: CmPP16-F (5′-GTGGTAAAGGACTTCAAGCCCACGACC-3′) and CmPP16-R (5′-ATGGGTTTGAAGAAGCCAAGCCACTTA-3′); CmGAIP-F (5′-GTGTCGAATAGCTTGGCGGATCTGGAC-3′) and CmGAIP-R (5′-GAGCATGCTTGCTTGCTTGAATGCATT-3′); CmSCL14P-F (5′-ACAACCTACTTGACGAAACTGTCGAA-3′) and CmSCL14P-R (5′-TCTGTCATCTGAGAGGGAACGTATTG-3′); CmSTMP-F (5′-GTGAATTGTCAGAAGGTGGGTGCACCG-3′) and CmSTMP-R (5′-CTTCCTTTGGTTAATGAACCAATTATT-3′); CmERFP-F (5′-GCAGATCATTGCATTGTCTTCAAGT-3′) and CmERFP-R (5′-CGGAAGCCAACGGAGGTG-3′); CmMybP-F (5′-ATACCATTTCTGGCAGCAAAGAGC-3′) and CmMybP-R (5′-AGTTGATGACCAAAATAGTGGGAAAACT-3′); CmNACP-F (5′-GTCATGCATGAATTTCGACTCGAACCC-3′) and CmNACP-R (5′-GCATCGCCATTGTTCGATCATAACATC-3′); and CsNACP-F (5′-CCGGGGTTTTGGCATCCGACGGACGAG-3′) and CsNACP-R (5′-GTCGGTGCCGGTGGCCTTCCAGTAGCCAT-3′) (Ruiz-Medrano et al., 1999). Amplified cDNAs were separated on 1% agarose gels, stained with ethidium bromide, and imaged using ChemiImager 5500 (Alpha Innotech).
Synthetic RNA
Chemically synthesized and HPLC-purified 27-nucleotide RNA oligonucleotides were obtained from Integrated DNA Technologies. Oligonucleotides used in gel mobility-shift experiments were as follows: PTBRS (5′-rArUrUrUrUrCrCrUrUrUrGrArUrUrCrUrCrUrCrUrCrCrUrUrCrUrU-3′), nPTBRS (5′-rCrCrUrCrCrGrUrCrUrArArArCrCrCrArArCrCrCrUrUrUrUrUrUrC-3′), and muPTBRS (5′-rArUrUrUrUrCrCrUrUrUrGrArArArGrArGrArGrArCrCrUrArGrArA-3′). These oligonucleotides were derived from GAIP. 5′ end-labeling of 27-nucleotide single-stranded RNAs with [γ-32P]ATP (20 μCi/μL) was performed with a KinaseMax kit (Ambion) following the manufacturer's instructions.
Electrophoretic Mobility-Shift Assays
Gel mobility-shift assays were performed as described (Xoconostle-Cázares et al., 1999; Yoo et al., 2004). To generate the various deletion mutants of GAIP and PP16-1, PCR was performed on corresponding cDNAs using the following primer pairs: CmGAIP(4)F (5′-GGTTCACACCATTTTCCTTTGATTC-3′) and CmGAIP(4)R (5′-CTGTTCGAGCTTCTGTGCAACTTC-3′); CmGAIP(2)F (5′-GAAGAAGCTATGTGTCAAGTTCAG-3′) and CmGAIP(2)R (5′-CCTGTAGATTCGGCGTGCTAATGC-3′); CmGAIP(N)F (5′-ATGCATTTCTACGAGAGCTGTCCC-3′) and CmGAIP(N)R (5′-CCCCCGGCCGAGCCAGTAGCTGATG-3′); CmGAIP(3)F (5′-CAAGAATGAAACAGGACTGACTGTC-3′) and CmGAIP(3)R (5′-CTTTTTATTCCCACCAATCTAATTA-3′); CmPP16-1(2)F (5′-GCACCTTTGTATTCCTTCAACTTC-3′) and CmPP16-1(2)R (5′-GATATGGAAGTCACCACCGCTC-3′); and CmPP16-1(N)F (5′-CAAGGTCATGGACCATGACGCT-3′) and CmPP16-1(N)R (5′-TTATGTTAAGTCATAGGAGTTTG-3′). All PCR products were subcloned into the TOPO II dual promoter vector (Invitrogen) and confirmed by sequencing.
All reactions were performed on ice in 20 μL of binding buffer (20 mM HEPES, pH 8.0, 50 mM KCl, 1 mM DTT, and 5% [v/v] glycerol). PP16-1, GAIP, RINGP, and various deletion mutants of PP16-1 and GAIP sense RNA probe, labeled with [α-32P]UTP (10 μCi/μL), were generated by in vitro transcription using the MAXIscript system according to the manufacturer's instructions (Ambion). An aliquot (1 nM) of 32P-labeled PP16-1, GAIP, RINGP, or various deletion mutants of PP16-1 and GAIP RNA, or 10 nM of 32P-labeled synthetic RNAs, was used for assays with 250 ng of purified GST, RBP50, or GST-PP16-1. Reaction mixtures were incubated on ice for 30 min and then separated on a 1% (v/v) nondenaturing agarose gel or a 5% (v/v) nondenaturing polyacrylamide gel. Electrophoresis was performed at 4°C and 120 V, and then gels were dried and exposed to x-ray film. Competition assays were performed as described for the electrophoretic mobility-shift assays, except that RBP50 was incubated with various amounts of unlabeled synthesized RNAs for 10 min followed by the addition of labeled synthesized RNA and a further incubation for 15 min. To measure the dissociation constant, band intensities were quantified using the ImageQuant Tools software, version 5.2 (GE Healthcare), and calculated as described previously (Haynes, 1999; Yoo et al., 2004).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU793994 (RBP50), AY325306 (GAIP), AY326307 (GAIP-B), AF079170 (PP16-1), AAC12676 (PP1), and AF527794 (Hsc70-1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Analysis Indicates That RBP50 Is a Member of the PTB Family.
Supplemental Figure 2. RBP50 Interacts with a Range of Phloem Proteins.
Supplemental Figure 3. RBP50 Is Present in the Pumpkin Phloem Translocation Stream as a Phosphoprotein.
Supplemental Figure 4. Determination of the RBP50 Dissociation Constant.
Supplemental Figure 5. Plant Heterografting System Using Pumpkin as the Stock (Pst) and Cucumber as the Scion (Csc).
Supplemental Data Set 1. Sequences Used to Generate the RBP50 Phylogeny Presented in Supplemental Figure 1C Online.
Supplementary Material
Acknowledgments
We thank Gary Thompson for providing polyclonal antisera to pumpkin PP2. This work was supported by the Department of Energy, Division of Energy Biosciences (Grant DE-FG02-94ER20134), and the National Science Foundation (Grant IOS-0444725). B.X.-C. was supported by a University of California Institute for Mexico and the U.S.–Mexico National Council for Science and Technology Sabbatical Leave Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: William J. Lucas (wjlucas@ucdavis.edu).
Online version contains Web-only data.
References
- Aoki, K., Kragler, F., Xoconostle-Cázares, B., and Lucas, W.J. (2002). A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc. Natl. Acad. Sci. USA 99 16342–16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K., Suzui, N., Fujimaki, S., Dohmae, N., Yonekura-Sakakibara, K., Fujiwara, T., Hayashi, H., Yamaya, T., and Sakakibara, H. (2005). Destination-selective long-distance movement of phloem proteins. Plant Cell 17 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung, K., Lin, S.I., Wu, C.C., Huang, Y.T., Su, C.L., and Chiou, T.J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter, S.D., and Allain, F.H.-T. (2008). Structure-function relationships of the polypyrimidine tract binding protein. Cell. Mol. Life Sci. 65 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari, R., Datt Pant, B., Stitt, M., and Scheible, W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N., Tsai, C.H., Lie, M., Havecker, E., and Baulcombe, D.C. (2007). The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17 1609–1614. [DOI] [PubMed] [Google Scholar]
- Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218 683–692. [DOI] [PubMed] [Google Scholar]
- Bothwell, A.L., Ballard, D.W., Philbrick, W.M., Lindwall, G., Maher, S.E., Bridgett, M.M., Jamison, S.F., and Garcia-Blanco, M.A. (1991). Murine polypyrimidine tract binding protein. Purification, cloning, and mapping of the RNA binding domain. J. Biol. Chem. 266 24657–24663. [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chen, Y.H., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60 107–124. [DOI] [PubMed] [Google Scholar]
- Cole, M., Nolte, C., and Werr, W. (2006). Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 34 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte, M.R., Grune, T., Ghuman, J., Kelly, G., Ladas, A., Matthews, S., and Curry, S. (2000). Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19 3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galban, S., et al. (2008). RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 28 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupels, F., Furch, A.C., Will, T., Mur, L.A., Kogel, K.H., and van Bel, A.J. (2008). Nitric oxide generation in Vicia faba phloem cells reveals them to be sensitive detectors as well as possible systemic transducers of stress signals. New Phytol. 178 634–646. [DOI] [PubMed] [Google Scholar]
- Gómez, G., and Pallás, V. (2004). A long-distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with Hop stunt viroid RNA. J. Virol. 78 10104–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G., Torres, H., and Pallás, V. (2005). Identification of translocatable RNA-binding phloem proteins from melon, potential components of the long-distance RNA transport system. Plant J. 41 107–116. [DOI] [PubMed] [Google Scholar]
- Gromak, N., Rideau, A., Southby, J., Scadden, A.D., Gooding, C., Huttelmaier, S., Singer, R.H., and Smith, C.W. (2003). The PTB interacting protein raver1 regulates alpha-tropomyosin alternative splicing. EMBO J. 22 6356–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haebel, S., and Kehr, J. (2001). Matrix-assisted laser desorption/ionization time of flight mass spectrometry peptide mass fingerprints and post source decay: a tool for the identification and analysis of phloem proteins from Cucurbita maxima Duch. separated by two-dimensional polyacrylamide gel electrophoresis. Planta 213 586–593. [DOI] [PubMed] [Google Scholar]
- Hall, M.P., Huang, S., and Black, D.L. (2004). Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell 15 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt, S., Cowan, G.H., Ziegler, A., Roberts, A.G., Oparka, K.J., and Torrance, L. (2005). Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17 164–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, S.R. (1999). RNA-Protein Interaction Protocols. (Totowa, NJ: Humana Press).
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42 49–68. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A., Atkinson, R.G., Forster, R.L., and Lucas, W.J. (1998). An RNA-based information superhighway in plants. Science 279 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kehr, J., and Buhtz, A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59 85–92. [DOI] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293 287–289. [DOI] [PubMed] [Google Scholar]
- Knoch, K.P., Meisterfeld, R., Kersting, S., Bergert, H., Altkruger, A., Wegbrod, C., Jager, M., Saeger, H.D., and Solimena, M. (2006). cAMP-dependent phosphorylation of PTB1 promotes the expression of insulin secretory granule proteins in beta cells. Cell Metab. 3 123–134. [DOI] [PubMed] [Google Scholar]
- Koller, D., Ittner, L.M., Muff, R., Husmann, K., Fischer, J.A., and Born, W. (2004). Selective inactivation of adrenomedullin over calcitonin gene-related peptide receptor function by the deletion of amino acids 14–20 of the mouse calcitonin-like receptor. J. Biol. Chem. 279 20387–20391. [DOI] [PubMed] [Google Scholar]
- Kühn, C., Franceschi, V.R., Schulz, A., Lemoine, R., and Frommer, W.B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275 1298–1300. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Yoo, B.C., Rojas, M.R., Gomez-Ospina, N., Staehelin, L.A., and Lucas, W.J. (2003). Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPPl. Science 299 392–396. [DOI] [PubMed] [Google Scholar]
- Lin, M.K., Belanger, H., Lee, Y.J., Varkonyi-Gasic, E., Taoka, K., Miura, E., Xoconostle-Cázares, B., Gendler, K., Jorgensen, R.A., Phinney, B., Lough, T.J., and Lucas, W.J. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19 1488–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M.K., Lee, Y.J., Lough, T.J., Phinney, B.S., and Lucas, W.J. (2008). Analysis of the pumpkin phloem proteome provides functional insights into angiosperm sieve tube function. Mol. Cell. Proteomics, in press. http://dx.doi.org/10.1000/123456/10.1074/mcp.M800420-MCP200. [DOI] [PubMed]
- Liu, H., Zhang, W., Reed, R.B., Liu, W., and Grabowski, P.J. (2002). Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA 8 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Lough, T.J., and Lucas, W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57 203–232. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Yoo, B.C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2 849–857. [DOI] [PubMed] [Google Scholar]
- Luo, G. (1999). Cellular proteins bind to the poly(U) tract of the 3′ untranslated region of hepatitis C virus RNA genome. Virology 256 105–118. [DOI] [PubMed] [Google Scholar]
- Ma, Q.H. (2007). Small GTP-binding proteins and their functions in plants. J. Plant Growth Regul. 26 369–388. [Google Scholar]
- MacMorris, M., Kumar, M., Lasda, E., Larsen, A., Kraemer, B., and Blumenthal, T. (2007). A novel family of C. elegans snRNPs contains proteins associated with trans-splicing. RNA 13 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie, T.P., Hernandez, H., Robinson, C.V., Simpson, P., Matthews, S., and Curry, S. (2005). The polypyrimidine tract binding protein is a monomer. RNA 11 1803–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmberg, P.L., Knox, K.A., Yun, B.W., Morris, P.C., Shafiei, R., Hudson, A., and Loake, G.J. (2007). The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc. Natl. Acad. Sci. USA 104 18795–18800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass, F.C., Auweter, S.D., Erat, M., Hargous, Y., Henning, A., Wenter, P., Reymond, L., Amir-Ahmady, B., Pitsch, S., Black, D.L., and Allain, F.H. (2005). Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309 2054–2057. [DOI] [PubMed] [Google Scholar]
- Oh, Y.L., Hahm, B., Kim, Y.K., Lee, H.K., Lee, J.W., Song, O., Tsukiyama-Kohara, K., Kohara, M., Nomoto, A., and Jang, S.K. (1998). Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 331 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J., and Turgeon, R. (1999). Sieve elements and companion cells—Traffic control centers of the phloem. Plant Cell 11 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.G., Mayer, S.A., Tempst, P., and Nadal-Ginard, B. (1991). Characterization and molecular cloning of polypyrimidine tract-binding protein: A component of a complex necessary for pre-mRNA splicing. Genes Dev. 5 1237–1251. [DOI] [PubMed] [Google Scholar]
- Perez, I., Lin, C.H., McAfee, J.G., and Patton, J.G. (1997). Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3 764–778. [PMC free article] [PubMed] [Google Scholar]
- Pickering, B.M., Mitchell, S.A., Evans, J.R., and Willis, A.E. (2003). Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 31 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18 111–119. [DOI] [PubMed] [Google Scholar]
- Ramsay, N.A., and Glover, B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. [DOI] [PubMed] [Google Scholar]
- Reyes, R., and Izquierdo, J.M. (2007). The RNA-binding protein PTB exerts translational control on 3′-untranslated region of the mRNA for the ATP synthase beta-subunit. Biochem. Biophys. Res. Commun. 357 1107–1112. [DOI] [PubMed] [Google Scholar]
- Rideau, A.P., Gooding, C., Simpson, P.J., Monie, T.P., Lorenz, M., Huttelmaier, S., Singer, R.H., Matthews, S., Curry, S., and Smith, C.W. (2006). A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat. Struct. Mol. Biol. 13 839–848. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290 2105–2110. [DOI] [PubMed] [Google Scholar]
- Rosenfeld, J., Capdevielle, J., Guillemot, J.C., and Ferrara, P. (1992). In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203 173–179. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Moya, J.H., Xoconostle-Cázares, B., and Lucas, W.J. (2007). Influence of cucumber mosaic virus infection on the mRNA population present in the phloem translocation stream of pumpkin plants. Funct. Plant Biol. 34 292–301. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cázares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126 4405–4419. [DOI] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999). Plant Microtechnique and Microscopy. (Oxford: Oxford University Press).
- Schmid, N., Zagrovic, B., and van Gunsteren, W.F. (2007). Mechanism and thermodynamics of binding of the polypyrimidine tract binding protein to RNA. Biochemistry 46 6500–6512. [DOI] [PubMed] [Google Scholar]
- Sinz, A. (2003). Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 38 1225–1237. [DOI] [PubMed] [Google Scholar]
- Sjolund, R.D. (1997). The phloem sieve element: A river runs through it. Plant Cell 9 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., Tzima, E., Ochs, K., Bassili, G., Trusheim, H., Linder, M., Preissner, K.T., and Niepmann, M. (2005). Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 11 1809–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama, T., Yamada, K., Mori, H., Takeno, K., and Yamaki, S. (1999). Cloning cDNAs for genes preferentially expressed during fruit growth in cucumber. J. Am. Soc. Hortic. Sci. 124 136–139. [Google Scholar]
- Swartz, J.E., Bor, Y.C., Misawa, Y., Rekosh, D., and Hammarskjold, M.L. (2007). The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J. Biol. Chem. 282 19844–19853. [DOI] [PubMed] [Google Scholar]
- Taoka, K., Ham, B.K., Xoconostle-Cázares, B., Rojas, M.R., and Lucas, W.J. (2007). Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell 19 1866–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronchere, H., Wang, J., and Fu, X.D. (1997). A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 388 397–400. [DOI] [PubMed] [Google Scholar]
- Van Bel, A.J.E. (2003). The phloem, a miracle of ingenuity. Plant Cell Environ. 26 125–149. [Google Scholar]
- Venkatramana, M., Ray, P.S., Chadda, A., and Das, S. (2003). A 25 kDa cleavage product of polypyrimidine tract binding protein (PTB) present in mouse tissues prevents PTB binding to the 5′ untranslated region and inhibits translation of hepatitis A virus RNA. Virus Res. 98 141–149. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A.C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz, C., Giavalisco, P., Schad, M., Juenger, M., Klose, J., and Kehr, J. (2004). Proteomics of cucurbit phloem exudate reveals a network of defence proteins. Phytochemistry 65 1795–1804. [DOI] [PubMed] [Google Scholar]
- Wang, J., and Pederson, T. (1990). A 62,000 molecular weight spliceosome protein crosslinks to the intron polypyrimidine tract. Nucleic Acids Res. 18 5995–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J., Lee, J.A., Kress, T.L., Mowry, K.L., and Black, D.L. (2003). Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 100 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, F.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98. [DOI] [PubMed] [Google Scholar]
- Yoo, B.C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C.S., Craig, J.C., Petzold, H.E., Dickerman, A.W., and Beers, E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138 803–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.