Abstract
Carotenoid pigments are critical for plant survival, and carotenoid composition is tuned to the developmental stage, tissue, and to environmental stimuli. We report the cloning of the CAROTENOID CHLOROPLAST REGULATORY1 (CCR1) gene. The ccr1 mutant has increased shoot branching and altered carotenoid composition, namely, reduced lutein in leaves and accumulation of cis-carotenes in dark-grown seedlings. The CCR1 gene was previously isolated as EARLY FLOWERING IN SHORT DAYS and encodes a histone methyltransferase (SET DOMAIN GROUP 8) that methylates histone H3 on Lys 4 and/or 36 (H3K4 and H3K36). ccr1 plants show reduced trimethyl-H3K4 and increased dimethyl-H3K4 surrounding the CAROTENOID ISOMERASE (CRTISO) translation start site, which correlates with low levels of CRTISO mRNA. Microarrays of ccr1 revealed the downregulation of 85 genes, including CRTISO and genes associated with signaling and development, and upregulation of just 28 genes. The reduction in CRTISO transcript abundance explains the altered carotenoid profile. The changes in shoot branching are additive with more axillary branching mutants, but the altered carotenoid profile may partially affect shoot branching, potentially by perturbed biosynthesis of the carotenoid substrates of strigolactones. These results are consistent with SDG8 regulating shoot meristem activity and carotenoid biosynthesis by modifying the chromatin surrounding key genes, including CRTISO. Thus, the level of lutein, the most abundant carotenoid in higher plants that is critical for photosynthesis and photoprotection, appears to be regulated by a chromatin modifying enzyme in Arabidopsis thaliana.
INTRODUCTION
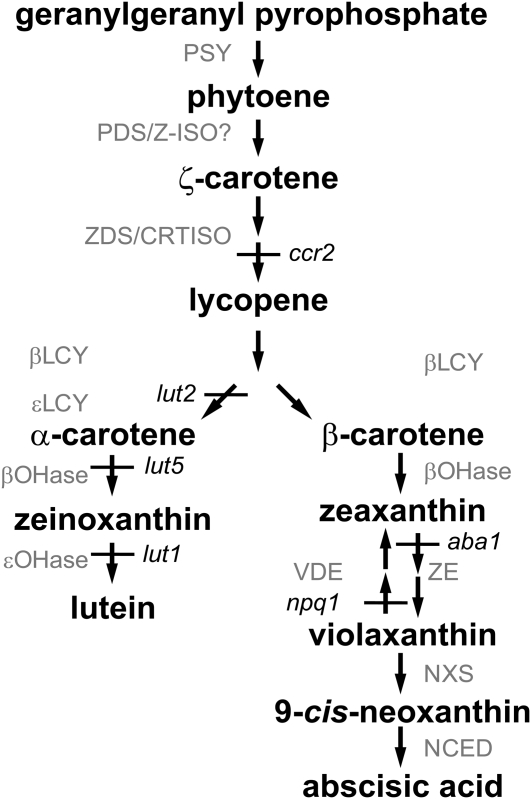
Carotenoids have a variety of crucial roles in photosynthetic organisms, including photosystem assembly, enhancing light-harvesting by absorbing a broader range of wavelengths than chlorophyll, and providing protection from excess light via energy dissipation and free radical detoxification (Niyogi, 1999; DellaPenna and Pogson, 2006; Sandmann et al., 2006; Lu and Li, 2008). Carotenoid biosynthesis in higher plants proceeds from the condensation of geranylgeranyl pyrophosphate by PHYTOENE SYNTHASE (PSY) to form phytoene, which is desaturated by PHYTOENE DESATURASE (PDS) and ZETA-CAROTENE DESATURASE (ZDS) and isomerized by CAROTENOID ISOMERASE (CRTISO) and ZETA-CAROTENE ISOMERASE (Z-ISO) to form the linear all-trans-lycopene (Figure 1) (Beyer et al., 1994; Schnurr et al., 1996; Bartley et al., 1999; Romer et al., 2000; Fraser et al., 2001; Park et al., 2002; Isaacson et al., 2004; Breitenbach and Sandmann, 2005; Li et al., 2007). The pathway branches at this point, producing α- or β-carotene. The carotenes are then subject to oxygenation reactions to produce xanthophylls, including zeaxanthin, violaxanthin, neoxanthin, and lutein, which is the most abundant carotenoid in higher plants. The major carotenoids involved in photosynthesis are β-carotene, zeaxanthin, violaxanthin, neoxanthin, and lutein. Xanthophyll composition in general and lutein content in particular can greatly affect photoprotection and plant viability (Pogson et al., 1998; Cuttriss and Pogson, 2004; Dall'Osto et al., 2006; DellaPenna and Pogson, 2006).
Figure 1.
Carotenoid Biosynthetic Pathway in Higher Plants.
The pathway shows the primary steps found in most plant species. Arabidopsis mutations, ccr2, lut1, lut2, lut5, aba1, and npq1, are shown in italics. βLCY, β-cyclase; βOH, β-hydroxylase; εLCY, ε-cyclase; εOH, ε-hydroxylase; NCED, 9-cis-epoxycarotenoid dioxygenase; NXS, neoxanthin synthase; VDE, violaxanthin deepoxidase; ZE, zeaxanthin epoxidase; Z-ISO, ζ-carotene isomerase.
Carotenoid-derived products, such as abscisic acid and β-ionone, can function as plant hormones or volatiles in plant pollinator interactions. In addition, carotenoids are precursors of signals that regulate shoot branching in Arabidopsis thaliana, pea (Pisum sativum), petunia (Petunia hybrida), and rice (Oryza sativa) (Beveridge et al., 1996, 2000; Morris et al., 2001; Stirnberg et al., 2002; Sorefan et al., 2003; Booker et al., 2004; Schwartz et al., 2004; Snowden et al., 2005; Gomez-Roldan et al., 2008; Umehara et al., 2008). Two of the genes that affect branching encode CAROTENOID CLEAVAGE-DIOXYGENASES, CCD7 and CCD8 (Johnson et al., 2002; Sorefan et al., 2003; Booker et al., 2004; Snowden et al., 2005; Zou et al., 2006; Arite et al., 2007), and appear to be essential for synthesis of a branching inhibitor hormone. Recently, this inhibitor has been revealed as a member of the strigolactone class of metabolites (Gomez-Roldan et al., 2008; Umehara et al., 2008), previously associated with functions in the rhizosphere. Root exudates stimulate germination of parasitic plant seeds, such as Striga, and influence hyphal branching in mycorrhizae (Cook et al., 1972; Akiyama et al., 2005). Now it is clear that strigolactones also act within the plant. Compounds such as 2′-epi-5-deoxystrigol in rice and orobranchyl acetate in pea are greatly reduced in ccd8 and ccd7 mutants (Gomez-Roldan et al., 2008; Umehara et al., 2008). Addition of GR24, a synthetic strigolactone analog, inhibits shoot branch outgrowth in a dose-dependent manner in mutants of both species and in the orthologous ccd8 Arabidopsis mutant. Recombinant CCD7 and CCD8 enzymes have carotenoid cleavage activities (Booker et al., 2004; Schwartz et al., 2004), and β-carotene has been proposed as an initial substrate for strigolactone biosynthesis (Matusova et al., 2005; Rani et al., 2008), but the complete biochemistry of strigolactones has not yet been described. Moreover, potential interactions between carotenoid biosynthesis, other major hormones, such as auxin, and regulation of shoot branching will likely prove interesting for future investigations.
In contrast with our understanding of the biosynthesis of carotenoids, relatively very little is known about their regulatory mechanisms (Lu and Li, 2008). Carotenoid biosynthesis appears to be tightly regulated throughout the life cycle with dynamic changes in composition matched to prevailing developmental requirements and environmental constraints, including germination, photomorphogenesis, and fruit development (Herrin et al., 1992; von Lintig et al., 1997; Cunningham and Gantt, 1998; Hoober and Eggink, 1999; Grunewald et al., 2000; Welsch et al., 2000; Hirschberg, 2001). Recent studies have linked carotenoid regulation to plastid biogenesis and morphology (Lu and Li, 2008). There are some carotenoid regulatory mutants that affect nongreen tissues; these include the orange cauliflower mutant (or) that accumulates β-carotene due to mutation of a plastid-associated DNAJ protein (Li et al., 2001; Lu et al., 2006) and the high-pigment1 tomato (Solanum lycopersicum) mutant that has increased pigmentation because of increased chromoplast compartment size (Cookson et al., 2003). In greening seedlings, PSY is strongly light induced (Welsch et al., 2000), and the transcription factor RAP2.2 (AP2/EREBP family) has been shown to bind to the PSY promoter (Welsch et al., 2007). However, modulating RAP2.2 levels resulted in only small pigment alterations in Arabidopsis root calli (Welsch et al., 2007). Overall, there are few reports describing regulatory processes that control carotenoid biosynthesis and/or transcript abundance (von Lintig et al., 1997; Cunningham and Gantt, 1998; Grunewald et al., 2000; Welsch et al., 2000; Hirschberg, 2001; Bramley, 2002). Investigations into lutein biosynthesis in Arabidopsis have yielded mutations in key biosynthetic enzymes: lut1, ε-hydroxylase (Tian et al., 2004); lut2, ε-cyclase (Cunningham et al., 1996; Pogson et al., 1996); carotenoid chloroplast regulatory2 (ccr2), carotenoid isomerase (Isaacson et al., 2002; Park et al., 2002); and lut5, an additional β-hydroxylase (Kim and DellaPenna, 2006).
Intriguingly, lutein biosynthesis can be altered by manipulating lycopene biosynthesis in higher plants (Misawa et al., 1994). This reflects the complexity of lycopene biosynthesis in higher plants that require at least four enzymes to produce all trans-lycopene, PDS, ZDS, Z-ISO, and CRTISO, in contrast with the requirement for a single desaturase in bacteria (Beyer et al., 1994; Schnurr et al., 1996; Romer et al., 2000; Fraser et al., 2001; Isaacson et al., 2002; Park et al., 2002; Isaacson et al., 2004; Breitenbach and Sandmann, 2005; Li et al., 2007). CRTISO catalyzes cis-trans reactions to reverse the four cis-bonds introduced by the desaturases (Isaacson et al., 2004). Consequently, CRTISO mutants, such as ccr2 and tangerine, result in accumulation of cis-carotenes, such as tetra-cis-lycopene, in the etioplasts (dark-grown plastids) of seedlings and chromoplasts of fruit (Isaacson et al., 2002; Park et al., 2002). Despite this block in etioplasts and chromoplasts, the biosynthetic pathway proceeds in chloroplasts of the CRTISO mutant, ccr2, via photoisomerization of the cis-bonds, but there is delayed greening and substantial reduction in lutein in Arabidopsis and varying degrees of chlorosis in tomato and rice CRTISO mutant seedlings (Isaacson et al., 2002; Park et al., 2002; Fang et al., 2008).
Carotenoid isomerization has proved intriguing with respect to regulating lutein synthesis and plastid development in etioplasts, chromoplasts, and chloroplasts. We previously identified a lutein regulatory mutant, ccr1, which exhibits a substantial decrease of lutein in leaves (with a corresponding increase in other xanthophylls) and cis-carotene accumulation in dark-grown tissue (Park et al., 2002). Additionally, ccr1, which here we demonstrate to be allelic to early flowering in short days (efs), displays increased shoot branching and early flowering (Kim et al., 2005; Zhao et al., 2005; Xu et al., 2008). Here, we demonstrate that ccr1 is a mutation in a histone methyltransferase required for CRTISO transcript accumulation, suggesting a potential role for epigenetic modification in regulating lutein content, carotenoid composition, and shoot branching.
RESULTS
Carotenoid Composition and Photosynthetic Parameters of ccr1
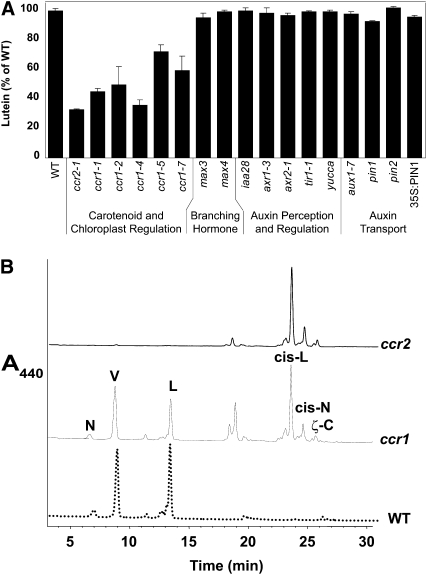
A screen for Arabidopsis mutants with reduced lutein identified six alleles of a recessive, putative regulatory locus, ccr1, that was not allelic to ccr2, lut2, or lut1 (Park et al., 2002). Lutein levels were clearly reduced in leaf tissues by 30 to 70% depending upon the allele (Figure 2A) and appear to recover during plant development as the leaf matures (see Supplemental Figure 1 online). Total carotenoid levels in ccr1 leaf tissues were similar to ccr2 and not substantially different to wild-type plants (Park et al., 2002). There was a slight decrease in chlorophyll content compared with the wild type (see Supplemental Table 1 online), which was not observed in ccr2 or lut2 but is perhaps more consistent with the chlorosis of CRTISO mutants in rice and tomato (Isaacson et al., 2002; Fang et al., 2008). There was no detectable change in chloroplast morphology, the major LHCII proteins or protochlorophyllide oxidase, maximum photosynthetic efficiency (Fv/Fm), and photosystem II operating efficiency (ΦPSII) (see Supplemental Table 1 and Supplemental Figure 2 online). Nonphotochemical quenching, a photoprotective mechanism by which excess absorbed light energy is dissipated as heat and mediated by xanthophyll pigments, was similar to other lutein-deficient mutants, lut2 and ccr2, in that it was delayed and reduced relative to the wild type (see Supplemental Figure 2 online), which is a result that has been reported previously (Pogson et al., 1998; Pogson and Rissler, 2000; Lokstein et al., 2002; Dall'Osto et al., 2006). In dark-grown seedlings, the carotenoid composition of ccr1 etioplasts was similar to ccr2, except in addition to poly-cis-carotenes, ccr1 had detectable levels of xanthophylls (Figure 2B).
Figure 2.
Carotenoid Accumulation in Arabidopsis Mutants.
(A) Lutein levels are expressed as a percentage of the total carotenoid pool relative to the wild type. Data are the average and se of three to six biological replicates of leaf tissues from 3- to 5-week-old plants.
(B) HPLC chromatogram of etiolated tissues from the wild type, ccr1, and ccr2. N, neoxanthin; V, violaxanthin; L, lutein; cis-L, cis-lycopene isomers; cis-N, cis-neurosporene isomers; ζ-C, ζ-carotene.
Phenotypic Characterization Identifies ccr1 as a Novel Branching Mutant
In addition to the carotenoid phenotype, ccr1 plants also displayed altered developmental phenotypes, most notably increased shoot branching but also early flowering and impaired fertility (Figures 3A and 3B). The increased branching cosegregated with reduced lutein accumulation in ccr1-1 through three backcrosses and in segregating F2 populations of ccr1-1 and ccr1-4. There was no change in lutein content for other mutants that show increased shoot branching, max1, max2, max3, and max4, and for auxin-related mutants, including those altered in auxin perception (axr1-3, axr2-1, and tir1-1), regulation (iaa28 and yucca), and transport (aux1-7, pin1, pin2, and 35S-PIN1) (Figure 2A).
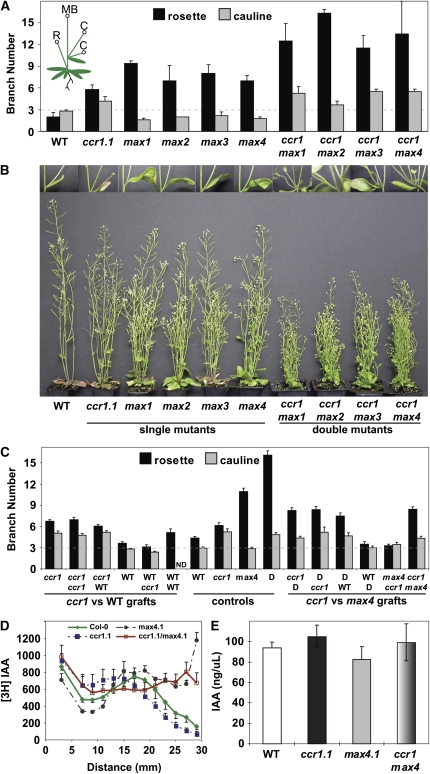
Figure 3.
Shoot Branching and Auxin Transport Are Altered in ccr1.
(A) Rosette and cauline branching. Rosette branches (R) excluding the main primary floral bolt (MB) and cauline branches (C) (see inset) were counted, and the average ± se (n = 5) are given.
(B) Representative images of single and double mutants.
(C) Rosette and cauline branching in reciprocal grafts. Genotypes are annotated as scion/rootstock with Columbia wild-type plants expressing a constitutive 35S:β-glucuronidase (GUS) marker. Averages ± se (n = 8 to 20 for grafts and 20 to 23 for control plants) are given. D refers to a double ccr1-1 × max4-1 mutant.
(D) Transport of (3H)IAA in inflorescence stem sections. Average ± se (n = 3 to 4 independent pools each of three sections) are given.
(E) IAA content in primary inflorescence stems of 30-d-old plants, including cauline leaves and branches but excluding siliques. Data are averages ± se of three pools of eight plants.
The degree of branching in ccr1 was compared with that of wild-type and max plants. On average, ccr1-1 had two- to threefold more rosette branches than the wild type but was less branched than any max mutant (Figures 3A and 3B, Table 1). Interestingly, ccr1, unlike any of the max mutants, showed a significant increase in the total number of cauline node branches (ccr1 = 1.76 ± 0.28 and wild type/max = 1 ± 0.0, P < 0.0001) (Figure 3B). Similar results were observed under both short- and long-day conditions.
Table 1.
Number of Rosette Branches in Wild Type, max4, ccr1, and ccr2
| Date | Wild Type | max4 | ccr1-1 | ccr2-1 | ccr2-3 | ccr2-4 | ccr2-5 |
|---|---|---|---|---|---|---|---|
| October 2006 | 1.5 ± 0.5 | 20.7 ± 1.5* | 5.8 ± 0.2* | 4.7 ± 0.4* | – | – | – |
| April 2007 | 0.9 ± 0.3 | 17.8 ± 2.1* | 4.8 ± 0.4* | 1.7 ± 0.3 | 4.5 ± 0.3* | 4.1 ± 0.3* | 2.6 ± 0.7* |
| October 2007 | 1.0 ± 0.3 | 8.3 ± 0.6* | 2.2 ± 1.0 | 0.8 ± 0.4 | 2.1 ± 0.4* | 2.9 ± 0.3* | 1.8 ± 0.4 |
The average and se for each experiment is given. The asterisk indicates data significantly different from the wild type (P < 0.05); n ≥ 5 per line per experiment.
To determine whether carotenoid composition has an effect on shoot branching, pea plants were treated with 10 μM norflurazon, a potent inhibitor of carotenoid biosynthesis that causes phytoene accumulation. Pea plants showed an increase in shoot branching (as determined by an increase in the length of shoot branches) in the presence of norflurazon, relative to the untreated controls (see Supplemental Figure 3A online), which is similar to the pea CCD mutants ramosus1 (rms1) and rms4 (see Supplemental Figure 3B online).
Rosette branching was also assessed in the carotenoid isomerase mutant ccr2, which displays an altered carotenoid profile (Park et al., 2002). A small but significant (P < 0.05) increase of one to two rosette branches was observed in multiple experiments with four null alleles of ccr2 (Table 1). There were slight differences in the number of shoot branches scored for the wild type and mutants from the independent experiments, and this could be the result of minor differences in conditions among multiple growth chambers. Given this more limited rosette branching of ccr2 compared with ccr1 and that strigolactones inhibit shoot branching and are synthesized from carotenoid derived intermediates, the altered ccr1 carotenoid profile is likely to be partially responsible for the increased shoot branching.
Potential links between ccr1 and the carotenoid-derived novel branching hormone were further evaluated. Branching of ccr1 max double mutants was strongly additive for both the rosette and cauline node phenotypes (Figures 3A and 3B). In grafting experiments, branching in ccr1 scions could not be rescued by wild-type, ccr1, max4, or double mutant rootstocks (Figure 3C), whereas ccr1 max4 double mutant scions on either ccr1 or wild-type rootstocks were restored to a ccr1 single mutant phenotype (Figure 3C). Similarly, the increased number of cauline branches observed in ccr1 and double mutants could not be rescued by grafting to any of the rootstocks (Figure 3C). Taken together, these data show that, unlike max4, the increased branching phenotype of ccr1 could not be suppressed by a graft-transmissible signal.
Auxin transport and endogenous auxin levels were also compared in ccr1 and max genotypes to determine if there was a correlation with increased shoot branching. Using an isolated inflorescence stem internode assay, the basipetal wave of transported labeled auxin was found to be retarded in ccr1 compared with the wild type or max4, but the ccr1 max4 double mutant exhibited a complex intermediate pattern with no clear wave/pulse of Tritium labeled Indoleacetic acid [(3H)IAA] (Figure 3D). The difference in auxin transport profile between max4 and ccr1, and the additive profile in the double mutant, are further indications of at least partly independent regulatory processes mediated by these genes. There was no significant difference between the amount of endogenous IAA in inflorescence tissues from the wild type, ccr1-1, max4-1, and double mutants (Figure 3E).
ccr1 Encodes a Histone Methyltransferase, SDG8, and Is Allelic to efs
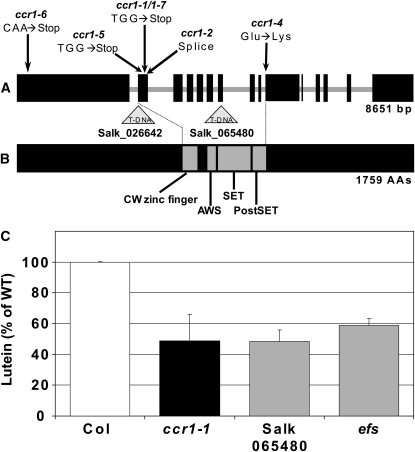
Positional cloning mapped the ccr1 mutant to a 137-kb region on chromosome 1. Scanning the genes in this interval identified a candidate gene, EFS; mutants of which have some traits similar to ccr1, specifically early flowering, increased shoot branching, and poor fertility (Soppe et al., 1999). The efs mutation has a point mutation in a SET2 domain histone methyltransferase gene (SDG8, At1g77300) (Kim et al., 2005; Zhao et al., 2005). The conserved SET domain [named from Su(var)3-9, Enhancer of zeste, and Trithorax] of histone methyltrasferases adds one, two, or three methyl groups to Lys residues of histone proteins within nucleosomes and may alter the configuration of chromatin, thereby affecting the accessibility of DNA to regulatory factors. Histone methylation is associated with promoting gene transcription (Lys [K] 4 or K36) or gene repression (K9 or K27) (Shilatifard, 2006). SET2 proteins are associated with activation of transcription. Sequence analysis of At1g77300 identified single base changes in all six ccr1 ethyl methanesulfonate–generated alleles (Figure 4A), including four lesions that introduced premature stop codons, one splice variant, and one with a Glu-to-Lys change in the conserved post-SET domain (Figure 4B). The efs mutant is allelic to ccr1-1 (data not shown) and efs and a SALK line (065480) containing a T-DNA insertion in exon 7 of SDG8 (Figure 4A), which all have reduced lutein levels (Figure 4C).
Figure 4.
Mutations in EFS/CCR1/SDG8 Impair Lutein Biosynthesis.
(A) Location of mutations in SDG8 resulting in premature stop codons (ccr1-1, 1-5, 1-6, and 1-7), a splice variant (ccr1-2), a residue change in the SET domain (ccr1-4), and T-DNA insertions (SALK_026642 and SALK_065480).
(B) SDG8 conserved domains include a Cys-rich zinc finger motif (CW domain) and the SET domain that is invariably preceded by an AWS (associated with SET) domain and followed by a Cys-rich post-SET domain.
(C) Lutein levels in leaf tissues from efs, SALK_065480, and ccr1-1 are expressed as a percentage of the total carotenoid pool relative to the wild type. The average of 3 to 10 plants and se are given.
SDG8 Regulates CRTISO Transcript Levels
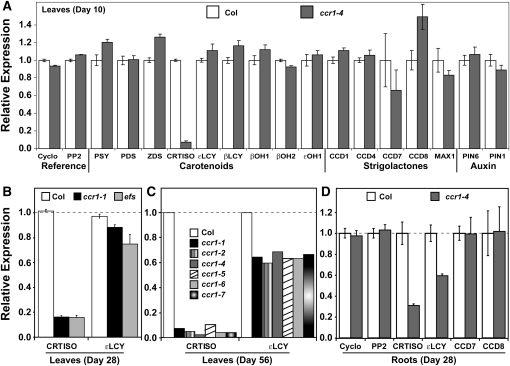
To determine how efs/ccr1 regulates carotenoid composition and shoot branching, the relative transcript abundance of key genes involved in carotenoid biosynthesis, carotenoid cleavage, and strigolactone synthesis (CCD7, CCD8, and MAX1) and auxin transport (PIN6 and PIN1) were quantified by real-time PCR. The level of CRTISO transcripts in ccr1-4 leaf tissue was only 10% that of wild-type leaves (Figure 5A). There was no change in transcript levels for any of the other genes tested (Figure 5A), and the small change in transcript abundances of CCD7 and CCD8 was not reproducible in leaf or root tissues (Figures 5A and 5D). CRTISO mRNA levels were substantially lower than the wild type in the efs mutant and five other ccr1 alleles (Figures 5B and 5C). CRTISO mRNA levels were consistently reduced by at least 90% in seedling (day 10) and mature (day 56) leaf tissues (compare Figures 5A and 5C), yet there was a substantial increase in lutein levels (from 30 to 80% of the wild type) observed during plant development (see Supplemental Figure 1 online). ccr2 does not contain any functional CRTISO, and lutein levels were observed to only increase up to 40% of wild-type levels (Park et al., 2002). It is possible that the small amount of CRTISO transcribed (<10%) in ccr1 and low turnover rate of carotenoids could be sufficient to allow a gradual accumulation of lutein in leaf tissues during plant development. CRTISO mRNA abundance was also significantly reduced in ccr1-4 roots, as was lycopene epsilon cyclase (εLCY) (Figure 5D). Transcript abundance of εLCY in ccr1 tissues was lower in mature leaf tissues and more variable in younger leaves (cf. Figures 5A and 5B with 5C).
Figure 5.
Gene Expression in ccr1 and efs.
Leaf and root issues were pooled from independent plants, and RT-PCR used to quantify gene expression levels from at least two biological replicates were determined in mutant lines and normalized to the wild type. Standard error bars are displayed (n = 4). Abbreviations are given in Supplemental Table 2 online.
(A) Gene expression of carotenoid biosynthesis, strigolactone biosynthesis, and auxin transport proteins in wild-type and ccr1-4 leaf tissues (10 d old).
(B) Relative expression levels of CRTISO and εLCY in 4-week-old leaves from the early flowering mutants efs and ccr1-1. For comparison, the dashed line indicates the level of no change in expression.
(C) Relative expression levels of CRTISO and εLCY in 8-week-old leaf tissues from six ccr1 alleles. The average transcript abundance from one biological replicate is displayed.
(D) Gene expression in roots from 4-week-old wild-type and ccr1-4 plants growing on MSO media.
Microarray Analysis of ccr1 Revealed Downregulation of a Small Set of Genes
Genome-wide transcript profiling of the ccr1-1 mutant (Affymetrix GeneChip Arrays) identified 113 differentially expressed genes (P < 0.05; see Supplemental Table 3 online). Overall there was no specific pathway, gene family, or cluster (Genevestigator analysis by anatomy, development, stimulus, and mutation) that was overrepresented in the list of differentially expressed genes downregulated in ccr1-1. The majority (75%) of genes with altered expression were downregulated (Table 2), which is consistent with the known function of SDG8, which modifies chromatin by adding marks of active transcription (Kim et al., 2005; Zhao et al., 2005; Xu et al., 2008).
Table 2.
Microarray Analysis of ccr1-1 Showing Differential Gene Expression Changes
| Differential Expression (q < 0.1)
|
Downregulated
|
Upregulated
|
|||
|---|---|---|---|---|---|
| Fold Change | Total | Total | Percentage | Total | Percentage |
| All | 113 | 85 | 75% | 28 | 25% |
| >1.5 | 86 | 77 | 90% | 9 | 10% |
The transcript profiling of the ccr1 mutant showed reduced transcript abundance of CRTISO, which concurs with quantitative RT-PCR data (Figure 5) and reduced transcript abundance of FLC, in keeping with previous findings (Kim et al., 2005) and the early flowering habit of ccr1. Microarrays previously performed on entire 6-d-old seedlings of SDG8 T-DNA insertion mutants (Xu et al., 2008) showed considerable overlap of differentially expressed genes in 10-d-old leaf tissues from ccr1 in the same direction (Table 2). Surprisingly, CRTISO expression was not altered in these previous arrays; this may reflect differences in experimental procedure, which tissues were analyzed, or the relative low abundance of CRTISO mRNA. A search for candidate genes that may be implicated in the enhanced rosette and cauline shoot branching displayed by ccr1 did not uncover obvious targets. Nonetheless, the ccr1 transcript profiling data provide a useful resource for identifying primary targets regulated by SDG8.
Chromatin Surrounding CRTISO Shows Reduced H3 Lys 4 Trimethylation
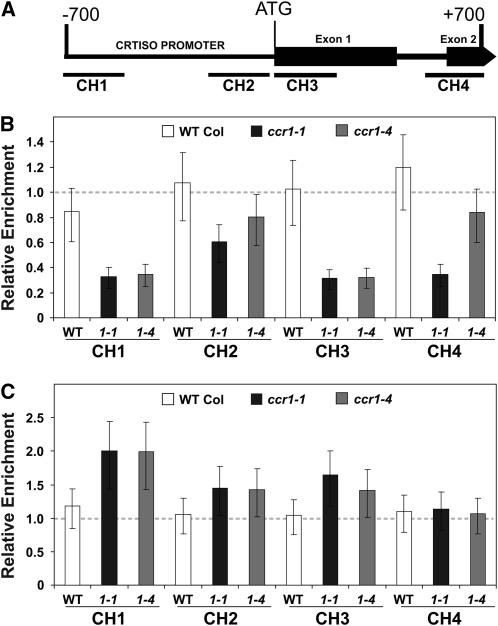
Immunoprecipitation of chromatin isolated from aerial tissue of young seedlings using antibodies against histone H3 dimethylK4 (K4me2) or H3 trimethylK4 (K4me3) was followed by quantification of precipitated DNA by real-time PCR. The analysis of two upstream (CH1 and CH2) and two downstream (CH3 and CH4) regions flanking the CRTISO translation start site (Figure 6A) was used to monitor the effect of SDG8 mutation on CRTISO chromatin. The level of H3K4me3 was 40 to 60% lower in all regions of ccr1-1 compared with the wild type (P < 0.05). While regions CH1 and CH3 showed a comparable reduction in H3K4me3 in ccr1-1 and ccr1-4, a smaller decrease in H3K4me3 was observed in regions CH2 and CH4 for ccr1-4 (Figure 6B). The smaller reduction in H3K4me3 at CH2 and CH4 in ccr1-4 compared with ccr1-1 is curious, but there is a statistically significant decrease in H3K4me3 across CRTISO chromatin in both mutant alleles. In the linear mixed model analysis of these data, the three-way interaction term, genotype by antibody by DNA region, was found to be significant (P = 0.03 on 36 residual degrees of freedom). Thus, there is a significant change in histone methylation surrounding CRTISO as the statistical analysis takes into account artifacts associated with nonspecific chromatin immunoprecipitation as well as the region of DNA targeted for histone methylation. In both ccr1 alleles, H3K4 dimethylation increased by 40 to 100% in regions CH1, CH2, and CH3 but not in CH4. Collectively, these data show that chromatin surrounding the CRTISO translation start site has altered H3K4 methylation in ccr1 alleles relative to wild-type plants, consistent with the decrease in CRTISO transcript abundance.
Figure 6.
Analysis of H3K4 Methylation of Chromatin Surrounding the Carotenoid Isomerase.
(A) The position of PCR amplicons, CH1, CH2, CH3, and CH4, used to quantify H3K4 methylation associated with CRTISO. Primer sequences are given in Supplemental Table 2 online.
(B) and (C) The level of H3K4me3 (B) and H3K4me2 (C) in CRTISO chromatin is presented as a ratio of mutant to wild type, following normalization using a region of the housekeeping gene S-ADENOSYL METHIONINE SYNTHASE. The predicted means (on the untransformed scale) of three independent experiments are given, and error bars represent the least significant difference (GenStat; analysis of variance). If mutant and wild-type error bars do not overlap, then their corresponding means can be considered as statistically significantly different at the 5% level (P < 0.05). For comparison, the dashed line indicates the level of no change in expression
The Expression of Genes Neighboring CRTISO Is Reduced in ccr1
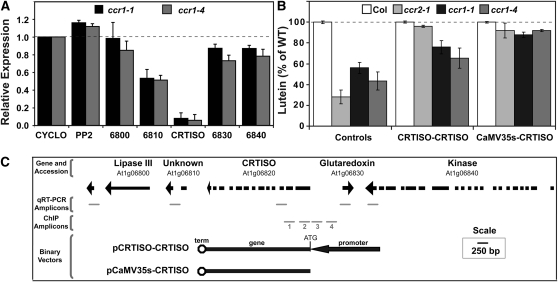
Analysis of the ccr1-1 microarrays identified one of the genes neighboring CRTISO (At1g06840) as marginally (P value < 0.01) reduced by ∼1.5-fold (see Supplemental Table 3 online). Quantitative RT-PCR analysis confirmed that neighboring genes on either side of CRTISO (Figure 7C) were downregulated (10 to 50%), but none were reduced to the same extent as CRTISO, indicating specific downregulation of CRTISO by SDG8 (Figure 7A). This suggests that transcription of CRTISO influences the activity of the adjacent genes, perhaps via changes in chromatin accessibility.
Figure 7.
SDG8 Targets the CRTISO Promoter and Alters Expression of Neighboring Genes.
(A) Expression of genes neighboring CRTISO in 4-week-old leaf tissues.
(B) Lutein levels in leaf tissues from transgenic lines (3 to 53 independent lines per transgenic) harboring pMDC32:CaMV35S-CRTISO and pPZP200:CRTISO-CRTISO are expressed as a percentage of the total carotenoid pool relative to the wild type. The average and se are given.
(C) Schematic diagram showing genes neighboring CRTISO, position of PCR amplicons, and CRTISO overexpression binary vectors.
A CRTISO Promoter-Gene Fusion Restores Lutein Levels in ccr2 but Not ccr1
The ccr1-1, ccr1-4, and ccr2-1 mutants were transformed with a genomic fragment (including the 3′ untranslated region) of the carotenoid isomerase driven by either the cauliflower mosaic virus 35S promoter (CaMV35S) or CRTISO (−1977 bp) promoters (Figures 7B and 7C). Overexpression of the carotenoid isomerase using the CaMV35S promoter was sufficient to restore 88 to 92% of wild-type lutein levels in ccr1-1 and ccr1-4 (Figure 7B) and ccr2-1 (Figure 7B) (Cuttriss et al., 2007), indicating successful complementation. However, the CRTISO promoter only partially restored lutein levels in ccr1-1 and ccr1-4 (66 to 76% of the wild type) despite completely restoring lutein levels by 96% in ccr2-1 (Figure 7B). That is, ccr1 alleles transformed with the CRTISO promoter-gene fusion showed a small increase in lutein (<20%), which was significantly lower than ccr2-1 transgenics with a 68% increase in lutein. Therefore, it seems likely that the CRTISO promoter requires SDG8 to correctly regulate CRTISO gene expression.
DISCUSSION
The ccr1 mutant is not allelic to known carotenoid structural genes (e.g., lut1, lut2, and ccr2) (Pogson et al., 1996; Park et al., 2002), yet it displays a carotenoid profile similar to that of a CRTISO mutant (ccr2) (Park et al., 2002). Together with the major changes in structure and function of the photosystems other than those attributable to decreased lutein (see Supplemental Figure 2 and Supplemental Table 1 online), the role of ccr1 is largely consistent with regulation of carotenoid biosynthesis, not accumulation. The discovery that ccr1 alters carotenoid composition by modifying the histone methylation status of chromatin surrounding the CRTISO gene, thereby reducing CRTISO transcript levels by 90%, indicates that ccr1/efs is a regulatory mutant. EFS/CCR1/SGD8 does not regulate other genes in the carotenoid biosynthetic pathway (Figure 5A). The severe reduction in CRTISO transcript levels was sufficient to cause the lutein reduction in leaves and the accumulation of cis-carotenes in etiolated tissue. This is confirmed by overexpression of CRTISO using the CaMV35S promoter, which restored wild-type lutein levels in ccr1 and ccr2 (Figure 7B). Thus, CRTISO can function as a rate-limiting step in lutein biosynthesis, altering the flux between the two branches of the pathway (Figure 1). The decrease in CRTISO transcript levels may epistatically influence εLCY expression, which was slightly (∼40%) reduced in root tissues (Figure 5D). In leaf tissues of ccr1 and efs, εLCY expression was slightly but variably reduced (cf. Figures 5A to 5C). A similar interaction between CRTISO and εLCY occurs in ccr2 etiolated tissues and therefore identifies CRTISO, and most likely εLCY, as rate-limiting steps in lutein production (Pogson et al., 1996; Cuttriss et al., 2007). The extent to which CRTISO regulates lutein via epistatic effects on εLCY mRNA abundance versus other mechanisms, such as changes in substrate preference for the two cyclases, remains an open question.
Our data demonstrate that the histone methyltransferase EFS/CCR1/SDG8 is required for expression of CRTISO mRNA in tissues tested. The ccr mutants identified to date (ccr1-1, -2, -4, -5, -6, and -7 and efs; ccr2-1, -3, and -5) have been mapped to one of two loci, namely, CCR2 that encodes the CRTISO protein itself, and the regulatory locus, EFS/CCR1. The failure to identify other loci regulating ccr2 expression is consistent with our hypothesis that SDG8 directly alters CRTISO expression through the modification of associated histones. SDG8 is developmentally regulated, increasing between 2 and 8 d after germination and has a degree of diurnal regulation (Kim et al., 2005; Zhao et al., 2005; Xu et al., 2008), and whether this modulates CRTISO expression during development is unknown.
Methylation of the N-terminal tails of histones within nucleosomes plays an important role in both promoting and repressing gene expression. In yeast, the SET1 and SET2 histone methyltransferases associate with the Polymerase Associated Factor1 and RNA Polymerase II complexes during transcription and facilitate the opening of chromatin, either enhancing transcription initiation or elongation and thereby promoting gene expression (Krogan et al., 2003). The SDG8 SET domain protein shows amino acid similarity with both SET2 (H3-K36) and SET1 (H3-K4) methyltransferases that play a key role in the methylation of Lys residues on histone H3 in animals (Zhang and Reinberg, 2001; Ng et al., 2007). Arabidopsis SDG8 is essential for the expression of FLOWERING LOCUS C (FLC) and for elevated levels of di- and trimethylation of H3K36 and trimethylation of H3K4 in FLC chromatin (Kim et al., 2005; Zhao et al., 2005; Xu et al., 2008). Recently, it was shown that sdg8 mutants have reduced global levels of H3K36me2 and H3K36me3 but unchanged global levels of H3K4me3 (Xu et al., 2008), suggesting that the main function of SDG8 is di- and trimethylation of H3K36. It is not clear whether SDG8 also trimethylates H3K4 at certain loci, such as FLC (Kim et al., 2005), or whether the decrease in H3K4me3 is a consequence of decreased H3K36 methylation. The H3K36 methylation status of CRTISO chromatin in a ccr1 backgrounds remains to be investigated, but consistent with the decrease in H3K4me3 at FLC in sdg8 mutants, ccr1 showed decreased levels of trimethyl H3K4 and slightly increased dimethyl H3K4 in chromatin surrounding the CRTISO translation start (Figure 6) (Kim et al., 2005; Zhao et al., 2005).
The expression of genes flanking CRTISO was slightly reduced in the ccr1 background, which is reminiscent of FLC, where its two flanking genes were partially repressed (Kim et al., 2005). Interestingly, the neighboring gene (At1g06830; glutaredoxin) immediately upstream of the CRTISO start of translation was only slightly repressed when compared with the gene immediately downstream (At1g06810) (Figure 7C). Our complementation studies showed that the activity of the transgenic CRTISO promoter was impaired in a ccr1 background when compared with ccr2, which further supports our hypothesis that SDG8 is required for the correct expression of the carotenoid isomerase. It seems possible that there are DNA motifs located within the CRTISO promoter that facilitate either a direct interaction with SDG8 or indirect interaction with an unknown regulatory factor that forms part of a SDG8 chromatin remodeling complex. It will be important to learn how histone modifications directed by SDG8 are targeted to specific genes like CRTISO and how these signals spread along the chromosome and affect the expression of neighboring genes.
The other phenotypes of the ccr1 mutant (e.g., low male fertility, slow germination, and increased shoot branching) indicate that CRTISO and FLC are not the only targets of SDG8 activity. Our microarray analysis of ccr1-1 identified a small number of gene expression changes (113), of which a majority was downregulated (Table 2), consistent with the proposed role of SDG8 to maintain chromatin in an active state (Kim et al., 2005; Zhao et al., 2005; Xu et al., 2008). SDG8 appears to target specific genes; however, our arrays do not distinguish between primary and secondary targets of SDG8. Future experiments will address this issue.
None of the differentially expressed genes in ccr1-1 (Table 3; see Supplemental Table 3 online) or SDG8 T-DNA insertion lines (Xu et al., 2008) readily account for the increased shoot branching. The increased branching of ccr1 may be linked to the change in carotenoid composition and CRTISO mRNA abundance. Regulation of shoot branching depends on complex relationships between perception, transport, and synthesis of auxin and a novel carotenoid-derived branching hormone, strigolactone (Gomez-Roldan et al., 2008; Ongaro and Leyser, 2008; Umehara et al., 2008). CRTISO activity may be necessary for generating the substrate(s) required for production of strigolactones. First, pea plants treated with the carotenoid biosynthesis inhibitor norflurazon showed an increase in shoot branching (see Supplemental Figure 3 online). Second, mutations in the CRTISO gene also showed a modest, yet significant, increase of one to two rosette branches (Table 1). However, this only explains a fraction of the ccr1 branching phenotype. That is, the ccr1 mutants can be distinguished from the max mutants as ccr1 has increased cauline branching that is absent in max mutants (Figure 2B), and branching in max ccr1 double mutants was additive unlike max double mutants (Figure 2A), which are indistinguishable from their single mutant parents (Stirnberg et al., 2002). Auxin transport profiles also differ between ccr1, which is slightly retarded compared with the wild type, and max4, which is enhanced (Figure 3D). Furthermore, the ccr1 phenotype is not corrected by the graft-transmissible signal, unlike max1, max3, and max4; although it should be noted that the effects of max2, an F-box protein, are also not graft transmissible (Auldridge et al., 2006). Finally, the change in lutein content is not observed in a range of max or auxin synthesis and perception mutants (Figure 2). The extent to which the observed change in auxin flux (Figure 2) could explain the rest of the branchy phenotype is worth further study.
Table 3.
Subsets of Differentially Regulated Genes in ccr1-1 Leaf Tissues
| Locus (AGI) | Locus Identifier | ccr1-1 P Value | FDR P Valuec | FC | Salkb (FC) |
|---|---|---|---|---|---|
| Downregulated | |||||
| AT5G56380 | F-box family protein | 1.42E-07 | 0.00324 | −7.7d | −2.0 |
| AT1G06820 | Carotenoid isomerase (CRTISO) | 6.32E-07 | 0.00481 | −29.2 | |
| AT5G05040;AT5G05060 | Cys protease inhibitor | 4.92E-07 | 0.00481 | −15.1 | |
| AT2G47060 | Ser/Thr protein kinase | 1.43E-06 | 0.00816 | −3.8 | −2.0 |
| AT5G44870 | Disease resistance protein (TIR-NBS-LRR class) | 3.67E-06 | 0.01065 | −25.0 | −1.9 |
| AT5G46020 | Cupin family protein | 3.74E-06 | 0.01065 | −3.0 | −2.7 |
| AT1G67620 | Unknown, Iojap-related protein | 3.28E-06 | 0.01065 | −7.0d | |
| AT3G21220 | Mitogen-activated protein kinase kinase 5 (ATMKK5) | 4.85E-06 | 0.01168 | −12.0d | −2.2 |
| AT3G04110 | Glu receptor 1 (GLR1) | 1.28E-05 | 0.01819 | −6.0 | −1.8 |
| AT4G11640 | Ser racemase (ATSR) | 1.79E-05 | 0.01942 | −5.9 | |
| AT1G65390 | Transmembrane receptor (ATPP2-A5) | 1.56E-05 | 0.01942 | −4.4 | |
| AT4G00030 | Plastid-lipid associated/fibrillin family protein | 1.48E-05 | 0.01942 | −3.7 | |
| AT5G38990;AT5G39000 | Protein kinase family protein | 1.66E-05 | 0.01942 | −3.0 | |
| AT5G24160 | Squalene monooxygenase/epoxidase (SQP1,2) | 1.87E-05 | 0.01943 | −8.3 | |
| AT3G62220 | Ser/Thr protein kinase | 2.22E-05 | 0.02036 | −14.8d | −2.5 |
| AT1G33560 | Activated disease resistance 1 (ATFUC1) | 2.32E-05 | 0.02039 | −3.0d | |
| AT3G26320 | Cytochrome P450 (CYP71B36) | 2.52E-05 | 0.02128 | −8.6d | |
| AT5G60950 | Phytochelatin synthetase-related | 3.09E-05 | 0.02521 | −5.2d | |
| AT5G65860 | Ankyrin repeat family protein | 3.43E-05 | 0.02694 | −3.7 | |
| AT5G10140 | FLOWERING LOCUS C (FLC); transcription factor | 4.87E-05 | 0.03178 | −34.7d | −6.6 |
| AT4G23570 | Pathogen resistance protein (SGT1A) | 5.65E-05 | 0.03398 | −3.1 | |
| AT1G13950 | Eukaryotic translation initiation factor (EIF-5A) | 5.66E-05 | 0.03398 | −3.7d | |
| AT2G19130 | S-locus lectin protein kinase family protein | 6.77E-05 | 0.03530 | −4.0 | |
| AT1G62290 | Aspartyl protease family protein | 6.61E-05 | 0.03530 | −9.5d | −5.1 |
| AT5G50630;AT5G50520 | Nodulin family protein | 9.03E-05 | 0.03671 | −5.1 | |
| AT5G37290 | Armadillo/β-catenin repeat family protein | 9.05E-05 | 0.03671 | −3.7 | |
| AT4G14610 | Putative disease resistance protein | 8.96E-05 | 0.03671 | −2.5d | |
| AT5G66250 | Kinectin-related/calcium-dependent protein kinase | 1.03E-04 | 0.03925 | −3.1 | |
| AT5G26110 | Ser/Thr kinase | 1.21E-04 | 0.04103 | −3.8 | |
| AT2G41100 | Calmodulin-like protein (TCH3) | 1.18E-04 | 0.04103 | −3.7 | |
| AT3G25740 | Met aminopeptidase 1C (MAP1B) | 1.19E-04 | 0.04103 | −6.2d | −2.1 |
| AT2G28100 | α-l-FUCOSIDASE 1 (ATFUC1) | 1.25E-04 | 0.04103 | −2.4d | |
| AT4G25710 | F-box family protein | 1.44E-04 | 0.04294 | −5.5 | −3.3 |
| AT1G70820 | Putative phosphoglucomutase | 1.51E-04 | 0.04294 | −2.2d | |
| AT5G57220 | Cytochrome P450 (CYP81F2) | 1.85E-04 | 0.04639 | −5.1d | −2.3 |
| AT3G20270 | Lipid binding serum glycoprotein family protein | 2.32E-04 | 0.04824 | −3.1 | |
| AT5G42670 | Agenet domain-containing protein | 2.41E-04 | 0.04960 | −3.1 | |
| Upregulated | |||||
| AT3G21720 | Isocitrate lyase | 2.75E-06 | 0.01065 | 8.1d | |
| AT5G26270 | Unknown protein | 5.12E-06 | 0.01168 | 10.7 | |
| AT3G30720 | Unknown protein | 8.94E-06 | 0.01416 | 8.8 | |
| AT1G20390 | Gypsy-like retrotransposon family protein | 6.90E-05 | 0.03530 | 3.5 | |
| AT5G04200 | Caspase/Cys-type peptidase (AMC9) | 1.12E-04 | 0.04103 | 6.6d | |
| AT5G09570 | Unknown protein | 1.56E-04 | 0.04308 | 13.0 | 2.6 |
AGI, Arabidopsis Genome Initiative.
Genes differentially regulated in seedling tissues from SDG8 T-DNA insertion lines relative to wild-type Col (Xu et al., 2008).
FDR, false discovery rate.
The actual fold change in relative expression may vary due to absent calls in ccr1 for these downregulated genes and absent calls in the wild type for these upregulated genes (see Methods).
It seems likely that ccr1 and the max mutants influence branching partly via independent pathways and partly via the ccr1-mediated change in CRTISO altering carotenoid biosynthesis, which could potentially alter the substrate for strigolactone biosynthesis. This situation will be clarified when the biosynthetic pathway for the branching hormone is completely elucidated. This is the first evidence we are aware of that targeted-chromatin modification can control carotenoid gene expression in plants and thus defines a novel mechanism for modulating carotenoid composition. The requirement of EFS/CCR1/SDG8 for CRTISO expression specifically results in changes in lutein content, a pigment critical for plants and implicated in protecting against age-related macular degeneration in humans. The extent to which SDG8 functions to regulate lutein in vegetables, fruits, and cereals and why SDG8 targets particular genes like CRTISO and FLC will be unearthed as we begin to dissect the role of SDG8 in regulating gene expression during plant development.
METHODS
Plant Growth, Grafting, and Mutants
All plants were in the Arabidopsis thaliana ecotype Columbia (Col-0) background and grown as described (Park et al., 2002) unless otherwise indicated. Germplasm used was as follows: ccr2-1, carotenoid isomerase (Park et al., 2002); ccr1, histone methyltransferase (this study); max3-9 and max4-1, carotenoid cleavage-dioxygenases (Stirnberg et al., 2002; Sorefan et al., 2003); iaa28, AUX/IAA transcription factor (Rogg et al., 2001); axr1-3, auxin-insensitive protein 1 (Lincoln et al., 1990); axr2-1, auxin-insensitive protein 2 (Nagpal et al., 2000); tir1-1, auxin receptor (Dharmasiri et al., 2005; Kepinski and Leyser, 2005); yucca (flavin monooxygenase that enhances auxin production; Zhao et al., 2001); aux1-7, auxin transporter (Lincoln et al., 1990); pin1, auxin efflux carrier (Okada et al., 1991; Galweiler et al., 1998); pin6, auxin efflux carrier (Muller et al., 1998); and 35S:PIN1, enhances polar auxin transport (Benkova et al., 2003).
Grafting studies and branching quantification were performed as described (Turnbull et al., 2002). Graft integrity was confirmed by GUS assays of root and scion shoot stocks carrying a 35S∷GUS marker. Homozygous max ccr1 double mutants were selected by visible additive phenotypes in segregating F2 populations.
Vector Constuction and Plant Transformation
Plasmid constructs were prepared using standard cloning techniques (Sambrook and Russell, 2001) and appropriate DNA segments sequenced to confirm the final structure. The pMDC32:CRTISO overexpression vector was constructed as previously described (Cuttriss et al., 2007). PCR primers CRTISO-cc1F (5′-GCTCTAGATCAACATTGCCTACGAGTC-3′) and CRTISO-cc1R (5′-ACACAAATCCATGGTTGCTCG-3′) were used to amplify the CRTISO promoter using DYNAzyme EXT DNA Polymerase (Finnzymes) and were subsequently cloned into pGEM-T (Promega) following the manufacturer's instructions. The promoter sequence contains 1977 bp of sequence upstream from the start codon, which is flanked by sequences that generate an NcoI restriction site. pGEMT:CRTISO and pTm35:FiLUC (pPZP200 binary vector harboring an intron containing firefly luciferase reporter gene under control by the minimal CaMV35S promoter) (Cazzonelli and Velten, 2008; Velten et al., 2008) were digested with XbaI/NcoI, and the CRTISO promoter was cloned upstream from the luciferase reporter, creating pTCRTISO:FiLUC. pTCRTISO:FiLUC was digested with NcoI/AflII (removes the FiLUC gene), and the entire CRTISO genomic sequence (3075 bp), including 171 bp of the 3′ untranslated region, was excised from pMDC32:CRTISO (NcoI/AflII) and cloned to create the overexpression vector pTCRTISOPromoterCRTISOGene (pTCPCG).
The binary vectors were subsequently transformed into Agrobacterium tumefaciens strain LBA4404 by electroporation followed by selection on media containing 50 μg/mL kanamycin (pMDC32:CRTISO) or 100 μg/mL spectinomycin (pTCPCG). Agrobacterium-mediated transformation of wild-type and mutant (ccr1 and ccr2 alleles) Arabidopsis plants was performed according to the floral-dip method (Clough and Bent, 1998). Transformants harboring pMDC32:CaMV35S-CRTISO were selected by plating sterilized seeds on Murashige and Skoog (MS) media (4.4 g/L MS salts, 1× MS salts, 0.5 g/L sucrose, and 0.8% agar) containing 50 μg/mL hygromycin (Invitrogen), and between 3 and 53 homozygous lines were characterized. Heterozygous lines harboring pTCPCG were selected by spraying seedlings with 50 mg/mL BASTA (glufosinate-ammonium salt; Sigma-Aldrich), and three to five independent lines were characterized. Transgenics were analyzed for lutein content by HPLC.
Photosynthetic Pigments and Auxin Assays
Carotenoid and chlorophyll measurements, electron microscopy, and photosynthetic measurements were performed as previously described (Porra et al., 1989; Pogson et al., 1998; Park et al., 2002; Cuttriss et al., 2007).
Pulse chase auxin transport experiments were performed as described (Rashotte et al., 2003). Three or four independent pools each of three basal inflorescence stem sections, mean length 34 mm, were cut from 30-d-old plants. Inverted sections were pulsed for 40 min by immersion in 20 μL 400 nM (3H)IAA, end rinsed with 400 nM cold IAA, and chased with 20 μL cold 400 nM IAA (140 min). Segments (2 mm, excluding the first 6 mm) were extracted into Ecolite scintillant and 3H content determined. For endogenous IAA measurement, three pools of eight primary inflorescence stems were cut from the base to below the silique zone, including cauline leaves and branches. Tissues were extracted in cold 80% methanol, including butylhydroxytoluene and (2H5)IAA internal standard, concentrated to aqueous phase, and partially purified by passage through C18 SPE cartridges eluted with 70% methanol. Dried eluates were converted to tetramethyl silane derivatives at 60°C using N-methyl-N-trimethylsilyltrifluoroacetamide and then analyzed by full-scan electrospray ionization–gas chromatography–mass spectrometry (EI-GC-MS) on a Hewlett Packard benchtop gas chromatography–mass spectrometry system. IAA content was calculated by isotope ratio using two ion pairs: m/z 202/207 and 319/324.
Real-Time PCR and Sequencing
RNA was extracted with the Qiagen RNeasy Plant mini kit and included an on-column DNase step using the Qiagen RNase-free DNase kit, according to the manufacturer's instructions (www.qiagen.com). First-strand cDNA synthesis was performed using oligo(dT) primer and SuperScript II reverse transcriptase according to manufacturer's instructions (Invitrogen). The relative transcript abundance was quantified using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich), and three technical replicates for each of two to three biological replicates were performed using the RotorGene 2000 (Corbett Research). Data were analyzed by relative quantification [Target Eff Ct(Wt-ccr1)/Reference Eff Ct(Wt-ccr1)] (Pfaffl, 2001) using cyclophilin (At2g29960) and Protein Phosphatase 2A (At1g13320) as housekeeper reference control genes (Czechowski et al., 2005). Primer sequences are listed in Supplemental Table 2 online.
Genomic DNA was extracted from mature leaves (DNeasy Plant mini kit; Qiagen). PCR primers (see Supplemental Table 4 online) were designed to generate overlapping DNA amplicons (400 to 650 bp) spanning the entire length of the SDG8 gene (At1g77300) and were sequenced by the Australian Genome Research Facility (www.agrf.org.au). Sequences were assembled using ContigExpress (Vector NTI).
Microarrays
Transcriptomic analysis was performed using Affymetrix GeneChip Arabidopsis Genome ATH1 Arrays (Affymetrix) as described (Rossel et al., 2007). Three biological replicates were analyzed for each treatment with each array representing a single biological replicate. For all arrays, green rosette leaves from 10-d-old seedlings were harvested from Columbia and ccr1-1 and immediately frozen in liquid nitrogen before total RNA was isolated using the RNeasy plant mini protocol (Qiagen). The quality of the RNA was verified using an Agilent Bioanalyzer (Agilent Technologies) and spectrophotometric analysis to determine concentration and the A260 to A280 ratio. Preparation of labeled cRNA from 1 to 5 μg of total RNA, target hybridization, as well as washing, staining, and scanning of the arrays were performed exactly as described in the Affymetrix GeneChip Expression Analysis Technical Manual, using the Affymetrix One-Cycle Target Labeling and Control Reagents, an Affymetrix GeneChip Hybridization Oven 640, an Affymetrix Fluidics Station 450 and an Affymetrix GeneChip Scanner 3000 7G at the appropriate steps. Data quality was assessed using GCOS 1.4 (Affymetrix) before CEL files were imported into Avadis 4.2 (Strand Genomics) for further analysis. Raw intensity data were normalized using the Guanine Cytosine Robust Multi-array Analysis (GC-RMA) algorithm (Harr and Schlotterer, 2006). Correlation plots were examined between biological replicates using the scatterplot function, in all cases r ≥ 0.981. The data were log transformed, and a list of differentially expressed genes was generated by performing t tests (unpaired with asymptotic P value computation), and P values were corrected using the Benjamini Hodgeberg false discovery rate. This produced a list of 113 genes defined as differentially expressed with P value < 0.05 (see Supplemental Table 3 online). Raw intensity data were analyzed using the MAS5 algorithm to improve quality control of the fold change values by allowing probe IDs to be called as present/absent across the arrays. A gene subset showing at least threefold changes for genes called present in all six arrays and genes called present in at least three biological replicate arrays (wild type or ccr1-1) were produced for Table 3 and ranked by false discovery rate corrected P value. All microarray data have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MEXP-1787.
Chromatin Immunoprecipitation and Statistical Analysis
Chromatin immunoprecipitation (ChIP) assays were done on 3-week-old leaf tissue as described (Johnson et al., 2002) with minor modifications. There were three biological replicates for each of the three different genotypes being compared (a randomized block design). Within a biological replicate, each of the three genotypes comprised a pool of multiple seedlings, since single seedlings would be unable to provide sufficient genetic material. The chromatin/DNA extracted from each seedling pool was spit into three aliquots, to which the two different antibodies were applied to two of the aliquots. The third aliquot was a control to which no antibody was added. Antibodies recognizing H3K4me3 and H3K4me2 were purchased from Upstate Biotechnology. The no-antibody control was included to verify that H3K4 antibodies were able to enrich ChIP DNA by at least 10-fold. ChIP DNA was resuspended in a final volume of 50 μL of Trisaminomethane-Ethylenediaminetetraacetic Acid. Sixteen microliters of ChIP DNA was diluted in 67 μL water and 5 μL used for quantitative RT-PCR. Then, each aliquot was sampled in triplicate by PCR for the presence of different regions of DNA: CH1 and CH2 (promoter regions); CH3 and CH4 (coding regions).
The relative of amounts of test and control DNA were determined using the comparative quantification analysis method described previously. DNA content was normalized to the housekeeping gene S-Adenosyl Methionine Synthase (SAM; At4g01850) (Finnegan et al., 2004), and the ratio between test and control for the gene of interest was determined. Quantitative RT-PCR was performed in triplicate, and primers used for quantitative RT-PCR analysis are given in Supplemental Table 2 online.
After normalization, the data were transformed to the log10 scale before analysis via a single linear mixed model analysis of variance, including data from all replicates. The fixed part of the statistical model was a three-way factorial structure allowing for main effects of genotype (wild type or ccr1 alleles), antibody, and DNA region, as well as their two-way and three-way interactions. Thus, using the notation as described (Wilkinson and Rogers, 1973), where A.B = interaction between A and B, A*B = A + B + A.B = “A crossed with B,” and A/B = A + A.B = “B nested within A”), the linear mixed model is then specified by: fixed equals “genotype * antibody * DNA region” and random equals “biorep/seedling pool/aliquot/triplicate,” and this term corresponds to the residual in the analysis. The random part of the model was a nested structure, triplicate reactions within aliquots, aliquots within chromatin extracts, and chromatin extracts within biological replicates. Tests for the significance of effects and Fisher's protected Least Significant Difference tests for comparisons between wild-type and ccr1 means were performed at the 5% level of significance.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following locus identifiers: At1g77300 (SDG8), At1g06810 (CYCLOPHILIN), At1g13320 (PROTEIN PHOSPHATASE 2A), At5g17230 (PSY), At4g14210 (PDS), At3g04870 (ZDS), At1g06820 (CRTISO), At5g57030 (ɛLCY), At3g10230 (βLCY), At3g53130 (ɛOH1), At4g25700 (βOH1), At5g52570 (βOH2), At3g63520 (CCD1), At4g19170 (CCD4), At2g44990 (CCD7), At4g32810 (CCD8), At2g26170 (MAX1) At1g06830 (GLUTAREDOXIN), At4g01850 (SAM), At1g06800 (LIPASEIII), At1g06810 (UNKNOWN), AT1g06840 (KINASE), At1g73590 (PIN1), and At1g77110 (PIN6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Lutein Levels Increase during Plant Development in ccr1.
Supplemental Figure 2. Photosynthetic Parameters of ccr1 Are Consistent with Reduced Lutein Content.
Supplemental Figure 3. Inhibition of Carotenoid Biosynthesis Affects Shoot Branching in Pea.
Supplemental Table 1. Pigment Content and Photosynthetic Parameters.
Supplemental Table 2. Quantitative RT-PCR Primers.
Supplemental Table 3. Microarray Analysis of ccr1-1.
Supplemental Table 4. SDG8/CCR1/EFS Sequencing Primers.
Supplementary Material
Acknowledgments
We were supported by the Australian Research Council Centre of Excellence in Plant Energy Biology (CE0561495). A.J.C. was supported by an Australian National University Endowment for Excellence PhD scholarship. We thank Christine Beveridge for rms, Candice Sheldon for efs, Jiri Friml for pin1, 35S:PIN1, and yucca, and Ottoline Leyser and Harry Klee for max lines. We thank Alec Zwart (CSIRO Math and Information Science) for the statistical analysis of the ChIP data. We thank Mathew Gordon, Sarah Kreunen, Tim Butler, Hyoungshin Park, and Alexandra Chubb (Australian National University) for valuable contributions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Barry J. Pogson (barry.pogson@anu.edu.au).
Online version contains Web-only data.
References
- Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435 824–827. [DOI] [PubMed] [Google Scholar]
- Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M., Kojima, M., Sakakibara, H., and Kyozuka, J. (2007). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51 1019–1029. [DOI] [PubMed] [Google Scholar]
- Auldridge, M.E., McCarty, D.R., and Klee, H.J. (2006). Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9 315–321. [DOI] [PubMed] [Google Scholar]
- Bartley, G.E., Scolnik, P.A., and Beyer, P. (1999). Two Arabidopsis thaliana carotene desaturases, phytoene desaturase and zeta-carotene desaturase, expressed in Escherichia coli, catalyze a poly-cis pathway to yield pro-lycopene. Eur. J. Biochem. FEMS 259 396–403. [DOI] [PubMed] [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Beveridge, C.A., Ross, J.J., and Murfet, I.C. (1996). Branching in pea (action of genes Rms3 and Rms4). Plant Physiol. 110 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge, C.A., Symons, G.M., and Turnbull, C.G. (2000). Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulated by genes Rms1 and Rms2. Plant Physiol. 123 689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, P., Nievelstein, V., Albabili, S., Bonk, M., and Kleinig, H. (1994). Biochemical aspects of carotene desaturation and cyclization in chromoplast membranes from Narcissus-pseudonarcissus. Pure Appl. Chem. 66 1047–1056. [Google Scholar]
- Booker, J., Auldridge, M., Wills, S., McCarty, D., Klee, H., and Leyser, C. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14 1232–1238. [DOI] [PubMed] [Google Scholar]
- Bramley, P.M. (2002). Regulation of carotenoid formation during tomato fruit ripening and development. J. Exp. Bot. 53 2107–2113. [DOI] [PubMed] [Google Scholar]
- Breitenbach, J., and Sandmann, G. (2005). zeta-Carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene. Planta 220 785–793. [DOI] [PubMed] [Google Scholar]
- Cazzonelli, C.I., and Velten, J. (2008). In vivo characterization of plant promoter element interaction using synthetic promoters. Transgenic Res. 17 437–457. [DOI] [PubMed] [Google Scholar]
- Clough, S., and Bent, A. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cook, C., Whichard, L., Wall, M., Egley, G., and Coggon, P. (1972). Germination stimulants. II. The structure of strigol-a potent seed germination stimulant for Witchweed (Striga lutea Lour). J. Am. Chem. Soc. 94 6198–6199. [Google Scholar]
- Cookson, P.J., Kiano, J.W., Shipton, C.A., Fraser, P.D., Römer, S., Schuch, W., Bramley, P.M., and Pyke, K.A. (2003). Increases in cell elongation, plastid compartment size and phytoene synthase activity underlie the phenotype of the high pigment-1 mutant of tomato. Planta 217 896–903. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.J., and Gantt, E. (1998). Genes and enzymes of carotenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 557–583. [DOI] [PubMed] [Google Scholar]
- Cunningham, F.X., Pogson, B., Sun, Z.R., McDonald, K.A., DellaPenna, D., and Gantt, E. (1996). Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8 1613–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttriss, A., and Pogson, B. (2004). Carotenoids. In Plant Pigments and Their Manipulation, Davies, K.M., ed. (CRC Press, Boca Raton, FL, USA, Annual Plant Reviews, Vol. 14), pp. 57–91.
- Cuttriss, A.J., Chubb, A., Alawady, A., Grimm, B., and Pogson, B. (2007). Regulation of lutein biosynthesis and prolamellar body formation in Arabidopsis. Funct. Plant Biol. 34 663–672. [DOI] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Osto, L., Lico, C., Alric, J., Giuliano, G., Havaux, M., and Bassi, R. (2006). Lutein is needed for efficient chlorophyll triplet quenching in the major LHCII antenna complex of higher plants and effective photoprotection in vivo under strong light. BMC Plant Biol. 6 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna, D., and Pogson, B.J. (2006). Vitamin synthesis in plants: Tocopherols and carotenoids. Annu. Rev. Plant Biol. 57 711–738. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Fang, J., et al. (2008). Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 54 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, E.J., Sheldon, C.C., Jardinaud, F., Peacock, W.J., and Dennis, E.S. (2004). A cluster of Arabidopsis genes with a coordinate response to an environmental stimulus. Curr. Biol. 14 911–916. [DOI] [PubMed] [Google Scholar]
- Fraser, P.D., Römer, S., Kiano, J.W., Shipton, C.A., Mills, P.B., Drake, R., Schuch, W., and Bramley, P.M. (2001). Elevation of carotenoids in tomato by genetic manipulation. J. Sci. Food Agric. 81 822–827. [Google Scholar]
- Galweiler, L., Guan, C.H., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan, V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455 189–194. [DOI] [PubMed] [Google Scholar]
- Grunewald, K., Eckert, M., Hirschberg, J., and Hagen, C. (2000). Phytoene desaturase is localized exclusively in the chloroplast and up-regulated at the mRNA level during accumulation of secondary carotenoids in Haematococcus pluvialis (Volvocales, Chlorophyceae). Plant Physiol. 122 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr, B., and Schlotterer, C. (2006). Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons. Nucleic Acids Res. 34 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrin, D.L., Battey, J.F., Greer, K., and Schmidt, G.W. (1992). Regulation of chlorophyll apoprotein expression and accumlation. Requirements for carotenoids and chlorophyll. J. Biol. Chem. 267 8260–8269. [PubMed] [Google Scholar]
- Hirschberg, J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4 210–218. [DOI] [PubMed] [Google Scholar]
- Hoober, J.K., and Eggink, L.L. (1999). Assembly of light-harvesting complex II and biogenesis of thylakoid membranes in chloroplasts. Photosynth. Res. 61 197–215. [Google Scholar]
- Isaacson, T., Ohad, I., Beyer, P., and Hirschberg, J. (2004). Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol. 136 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson, T., Ronen, G., Zamir, D., and Hirschberg, J. (2002). Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 14 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Kim, J., and DellaPenna, D. (2006). Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. USA 103 3474–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y., He, Y., Jacob, Y., Noh, Y.S., Michaels, S., and Amasino, R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., et al. (2003). Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23 4207–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Murillo, C., and Wurtzel, E.T. (2007). Maize Y9 encodes a product essential for 15-cis-zeta-carotene isomerization. Plant Physiol. 144 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Paolillo, D.J., Parthasarathy, M.V., DiMuzio, E.M., and Garvin, D.F. (2001). A novel gene mutation that confers abnormal patterns of β-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J. 26 59–67. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 11 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokstein, H., Tian, L., Polle, J.E., and DellaPenna, D. (2002). Xanthophyll biosynthetic mutants of Arabidopsis thaliana: Altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in Photosystem II antenna size and stability. Biochim. Biophys. Acta 1553 309–319. [DOI] [PubMed] [Google Scholar]
- Lu, S., and Li, L. (2008). Carotenoid metabolism: Biosynthesis, regulation, and beyond. J. Integr. Plant Biol. 50 778–785. [DOI] [PubMed] [Google Scholar]
- Lu, S., et al. (2006). The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18 3594–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusova, R., Rani, K., Verstappen, F.W., Franssen, M.C., Beale, M.H., and Bouwmeester, H.J. (2005). The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa, N., Yamano, S., Linden, H., Defelipe, M.R., Lucas, M., Ikenaga, H., and Sandmann, G. (1994). Functional expression of the Erwinia-Uredovora carotenoid biosynthesis gene CRTL in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J. 4 833–840. [DOI] [PubMed] [Google Scholar]
- Morris, S.E., Turnbull, C.G., Murfet, I.C., and Beveridge, C.A. (2001). Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 126 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, A., Guan, C.H., Galweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D.W., Wang, T., Chandrasekharan, M.B., Aramayo, R., Kertbundit, S., and Hall, T.C. (2007). Plant SET domain-containing proteins: Structure, function and regulation. Biochim. Biophys. Acta 1769 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 333–359. [DOI] [PubMed] [Google Scholar]
- Okada, K., Ueda, J., Komaki, M.K., Bell, C.J., and Shimura, Y. (1991). Requirement of the auxin polar transport-system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro, V., and Leyser, O. (2008). Hormonal control of shoot branching. J. Exp. Bot. 59 67–74. [DOI] [PubMed] [Google Scholar]
- Park, H., Kreunen, S.S., Cuttriss, A.J., DellaPenna, D., and Pogson, B.J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson, B., McDonald, K.A., Truong, M., Britton, G., and DellaPenna, D. (1996). Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson, B.J., Niyogi, K.K., Bjorkman, O., and DellaPenna, D. (1998). Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 95 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson, B.J., and Rissler, H.M. (2000). Genetic manipulation of carotenoid biosynthesis and photoprotection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Rani, K., Zwanenburg, B., Sugimoto, Y., Yoneyama, K., and Bouwmeester, H.J. (2008). Biosynthetic considerations could assist the structure elucidation of host plant produced rhizosphere signalling compounds (strigolactones) for arbuscular mycorrhizal fungi and parasitic plants. Plant Physiol. Biochem. 46 617–626. [DOI] [PubMed] [Google Scholar]
- Rashotte, A.M., Poupart, J., Waddell, C.S., and Muday, G.K. (2003). Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 133 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in iaa28 suppresses lateral root development. Plant Cell 13 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer, S., Fraser, P.D., Kiano, J.W., Shipton, C.A., Misawa, N., Schuch, W., and Bramley, P.M. (2000). Elevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 18 666–669. [DOI] [PubMed] [Google Scholar]
- Rossel, J.B., Wilson, P.B., Hussain, D., Woo, N.S., Gordon, M.J., Mewett, O.P., Howell, K.A., Whelan, J., Kazan, K., and Pogson, B.J. (2007). Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19 4091–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sandmann, G., Romer, S., and Fraser, P.D. (2006). Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab. Eng. 8 291–302. [DOI] [PubMed] [Google Scholar]
- Schnurr, G., Misawa, N., and Sandmann, G. (1996). Expression, purification and properties of lycopene cyclase from Erwinia uredovora. Biochem. J. 315 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S.H., Qin, X., and Loewen, M.C. (2004). The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J. Biol. Chem. 279 46940–46945. [DOI] [PubMed] [Google Scholar]
- Shilatifard, A. (2006). Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75 243–269. [DOI] [PubMed] [Google Scholar]
- Snowden, K.C., Simkin, A.J., Janssen, B.J., Templeton, K.R., Loucas, H.M., Simons, J.L., Karunairetnam, S., Gleave, A.P., Clark, D.G., and Klee, H.J. (2005). The Decreased apical dominance 1/Petunia hybrida carotenoid cleavage dioxygenase8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe, W.J., Bentsink, L., and Koornneef, M. (1999). The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana. Development 126 4763–4770. [DOI] [PubMed] [Google Scholar]
- Sorefan, K., Booker, J., Haurogne, K., Goussot, M., Bainbridge, K., Foo, E., Chatfield, S., Ward, S., Beveridge, C., Rameau, C., and Leyser, O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg, P., van De Sande, K., and Leyser, H.M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129 1131–1141. [DOI] [PubMed] [Google Scholar]
- Tian, L., Musetti, V., Kim, J., Magallanes-Lundback, M., and DellaPenna, D. (2004). The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc. Natl. Acad. Sci. USA 101 402–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, C.G.N., Booker, J.P., and Leyser, H.M.O. (2002). Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32 255–262. [DOI] [PubMed] [Google Scholar]
- Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., Magome, H., Kamiya, Y., Shirasu, K., Yoneyama, K., Kyozuka, J., and Yamaguchi, S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 11 195–200. [DOI] [PubMed] [Google Scholar]
- Velten, J., Pogson, B., and Cazzonelli, C.I. (2008). Luciferase as a Reporter of Gene Activity in Plants. Transgenic Plant J. 2 1–13. [Google Scholar]
- von Lintig, J., Welsch, R., Bonk, M., Giuliano, G., Batschauer, A., and Kleinig, H. (1997). Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. Plant J. 12 625–634. [DOI] [PubMed] [Google Scholar]
- Welsch, R., Beyer, P., Hugueney, P., Kleinig, H., and von Lintig, J. (2000). Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211 846–854. [DOI] [PubMed] [Google Scholar]
- Welsch, R., Maass, D., Voegel, T., Dellapenna, D., and Beyer, P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, G.N., and Rogers, C.E. (1973). Symbolic descriptions of factorial models for analysis of variance. Appl. Stat. 22 392–399. [Google Scholar]
- Xu, L., Zhao, Z., Dong, A., Soubigou-Taconnat, L., Renou, J.P., Steinmetz, A., and Shen, W.H. (2008). Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28 1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Reinberg, D. (2001). Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 15 2343–2360. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Christensen, S.K., Fankhauser, C., Cashman, J.R., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309. [DOI] [PubMed] [Google Scholar]
- Zhao, Z., Yu, Y., Meyer, D., Wu, C., and Shen, W.H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7 1256–1260. [DOI] [PubMed] [Google Scholar]
- Zou, J., Zhang, S., Zhang, W., Li, G., Chen, Z., Zhai, W., Zhao, X., Pan, X., Xie, Q., and Zhu, L. (2006). The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48 687–698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.