Abstract
In the wild tomato Solanum habrochaites, the Sst2 locus on chromosome 8 is responsible for the biosynthesis of several class II sesquiterpene olefins by glandular trichomes. Analysis of a trichome-specific EST collection from S. habrochaites revealed two candidate genes for the synthesis of Sst2-associated sesquiterpenes. zFPS encodes a protein with homology to Z-isoprenyl pyrophosphate synthases and SBS (for Santalene and Bergamotene Synthase) encodes a terpene synthase with homology to kaurene synthases. Both genes were found to cosegregate with the Sst2 locus. Recombinant zFPS protein catalyzed the synthesis of Z,Z-FPP from isopentenylpyrophosphate (IPP) and dimethylallylpyrophosphate (DMAPP), while coincubation of zFPS and SBS with the same substrates yielded a mixture of olefins identical to the Sst2-associated sesquiterpenes, including (+)-α-santalene, (+)-endo-β-bergamotene, and (−)-endo-α-bergamotene. In addition, headspace analysis of tobacco (Nicotiana sylvestris) plants expressing zFPS and SBS in glandular trichomes afforded the same mix of sesquiterpenes. Each of these proteins contains a putative plastid targeting sequence that mediates transport of a fused green fluorescent protein to the chloroplasts, suggesting that the biosynthesis of these sesquiterpenes uses IPP and DMAPP from the plastidic DXP pathway. These results provide novel insights into sesquiterpene biosynthesis and have general implications concerning sesquiterpene engineering in plants.
INTRODUCTION
Isoprenoids constitute a large class of natural compounds made of multiples of the five-carbon isoprene unit. Among isoprenoids, terpenoids are low molecular weight compounds composed of two to eight isoprene units (10 to 40 carbon atoms) that include monoterpenoids (C10), sesquiterpenoids (C15), diterpenoids (C20), triterpenoids (C30), and tetraterpenoids (C40). The diterpenoid gibberellins and the triterpenoid brassinosteroids are examples of plant hormones playing a crucial role in the regulation of development. There are also numerous terpenoid products of secondary metabolism that have been exploited by humans in different ways, for example, as drugs such as paclitaxel, an anticancer diterpenoid, and artemisinin, an antimalarial sesquiterpenoid, or as fragrance and aroma ingredients (menthol, patchoulol, nootkatone, vetiverol, and santalols, to cite a few).
Our understanding of the biosynthesis of terpenoids has made important progress in the last 15 years with the discovery of genes coding for many of the pathway steps all the way from the precursors to the final product. Isopentenylpyrophosphate (IPP) and dimethylallylpyrophosphate (DMAPP) are the precursors of all isoprenoids. Chain elongation of DMAPP by successive head-to-tail addition of IPP units generates linear isoprenylpyrophosphates (IPrPs). These head-to-tail condensations are catalyzed by isoprenyl pyrophosphate synthases (IPPS) with two possible configurations for the double bonds in the resulting isoprenyl chains, either cis (Z) or trans (E) (Kellog and Poulter, 1997; Koyama, 1999). Although the reaction mechanism of both types of IPPS is similar, they share little similarity at the sequence level and they constitute two separate classes of enzymes (Koyama, 1999). It has been found to date that the biosynthesis of terpenoids proceeds from IPrPs in the E configuration via the action of a conserved family of enzymes, the terpene synthases. Thus, 2E-geranylpyrophosphate (GPP; C10) is the precursor of monoterpenoids, 2E,6E-farnesylpyrophosphate (E,E-FPP; C15) of sesquiterpenoids and triterpenoids, and 2E,6E,10E-geranylgeranylpyrophosphate (E,E,E-GGPP; C20) of diterpenoids and tetraterpenoids (Wendt and Schulz, 1998; Tholl, 2006).
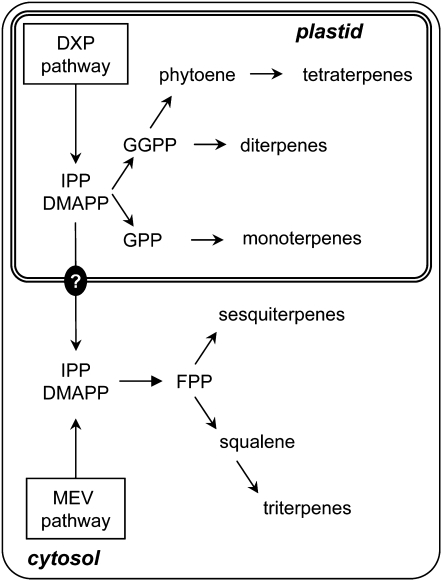
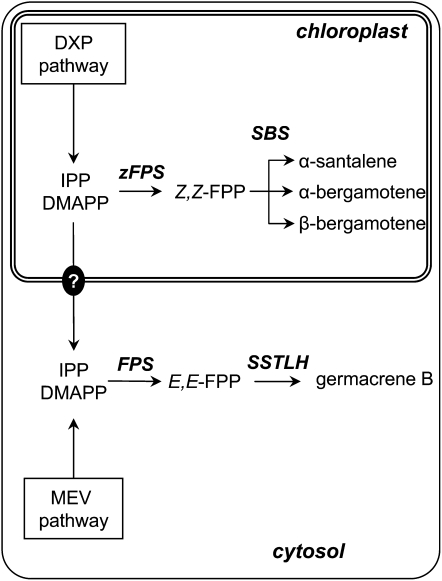
In plants, DMAPP and IPP can be produced by two pathways that are localized in different subcellular compartments. The mevalonate (MEV) pathway, common to all eukaryotes, takes place in the cytosol, whereas the deoxyxylulose 5-phosphate (DXP) pathway proceeds in the plastids (Eisenreich et al., 1998; Kuzuyama, 2002). Accordingly, the monoterpenoids, diterpenoids, and tetraterpenoids are produced in the plastids and made of IPrPs derived from the DXP pathway, whereas the sesquiterpenoids and triterpenoids are synthesized in the cytosol from IPrPs of the MEV pathway (Tholl, 2006) (Figure 1). However, in some cases, it has been shown that sesquiterpenes are derived from IPP and DMAPP of plastidic origin, suggesting that a transport of these compounds from the plastids to the cytosol might occur (Adam et al., 1999; Dudareva et al., 2005).
Figure 1.
Overview of Terpene Biosynthesis Pathways and Their Subcellular Localizations in Plant Cells.
Monoterpenes, diterpenes, and tetraterpenes are derived from IPP and DMAPP of the plastidic DXP pathway. Sesquiterpene and triterpene biosyntheses are localized to the cytosol and use IPP and DMAPP from the MEV pathway. However, in some cases, sesquiterpenes have been shown to originate from IPP and DMAPP of the DXP pathway (e.g., Dudareva et al., 2005), suggesting that transport of IPP could occur from the plastids to the cytosol, represented in the figure by the black circle with a question mark. FPP, E,E-farnesylpyrophosphate; GGPP, E,E,E-geranylgeranylpyrophosphate.
Wild tomato species (Solanum section Lycopersicon) are native to western South America and are important sources of insect- and pathogen-resistant germplasm. In several cases, the diverse phytochemicals responsible for the resistance are produced by glandular trichomes. Methyl ketones from Solanum habrochaites f. sp glabratum, sesquiterpene carboxylic acids from S. habrochaites (accession LA1777 from the Tomato Genetics Resource Center), and acyl-glucose esters from Solanum pennellii are examples of such compounds that possess toxic properties against Lepidoptera or aphids (Williams et al., 1980; Goffreda et al., 1990; Juvik et al., 1994; Frelichowski and Juvik, 2001). By contrast, the cultivated tomato S. lycopersicum does not produce these compounds, indicating a loss of expression or genomic information critical to their biosynthesis upon evolution and selection. Interspecific crosses between S. lycopersicum and its wild relatives are viable and have been used to generate populations of introgression lines (ILs) (Monforte and Tanksley, 2000). Each IL carries a single or a limited number of chromosome segments from one of the wild relatives in a S. lycopersicum genome background. These populations provide a convenient material to map the genetic loci responsible for various traits, including pathogen resistance or secondary metabolite production (Young et al., 1988; van Der Hoeven et al., 2000).
Glandular trichomes of S. habrochaites accession LA1777 produce sesquiterpene olefins of two different classes. Class I is represented mainly by germacrene B, whereas class II consists of a mix of α-santalene and related sesquiterpenes, which are the likely precursors of the sesquiterpene carboxylic acids (Coates et al., 1988). Using the ILs resulting from a cross between S. lycopersicum and S. habrochaites, Van Der Hoeven et al. (2000) showed that the biosynthesis of class I and II sesquiterpene olefins is effected by two independent loci, Sst1 and Sst2, respectively. The Sst1 locus on chromosome 6 is composed of two gene clusters 24 centimorgans apart, Sst1-A and Sst1-B, but only the Sst1-A genes are responsible for the accumulation of class I sesquiterpenes. At this locus, the S. lycopersicum allele is associated with the biosynthesis of β-caryophyllene and α-humulene, whereas the S. habrochaites allele is associated with the biosynthesis of germacrene B and D and an unidentified sesquiterpene. The Sst2 allele of S. habrochaites on chromosome 8 is responsible for the accumulation of class II sesquiterpenes, including α-santalene, α-bergamotene, and β-bergamotene and several unidentified sesquiterpenes. To date, the genes at the Sst2 locus of S. habrochaites have not been identified or characterized.
Here, we present the characterization of two genes from S. habrochaites that are responsible for the biosynthesis of all chromosome 8–associated class II sesquiterpenes. Using a S. habrochaites trichome EST database, two candidate genes that are highly and specifically expressed in trichome cells and that mapped to the Sst2 locus on chromosome 8 were identified. A first gene encoding a putative terpene synthase (Santalene and Bergamotene Synthase [SBS]) with homology to kaurene synthases was found to cosegregate with a second gene encoding a putative Z-prenyltransferase (zFPS). In vitro experiments with both recombinant proteins produced in Escherichia coli allowed us to demonstrate that (1) zFPS encodes a Z,Z-farnesyl pyrophosphate synthase, and (2) Z,Z-FPP is the substrate of the SBS enzyme that is responsible for the biosynthesis of type II sesquiterpenes. In addition, transgenic tobacco (Nicotiana sylvestris) plants having both genes were able to produce all type II sesquiterpenes, confirming in vivo the function of these two genes.
RESULTS
Identification of Two Candidate Genes for the Biosynthesis of Class II Sesquiterpenes
Class II sesquiterpenes are produced specifically in S. habrochaites trichomes, and their occurrence is restricted to particular accessions (Tomato Genetics Resource Center accession LA1777; van Der Hoeven et al., 2000). By contrast, accessions of the closely related S. habrochaites f. glabratum (accession PI26449) and S. pennellii (accession LA716) do not produce any class II sesquiterpenes. Trichome-specific ESTs from these accessions with highly distinct trichome metabolic profiles are publicly available (e.g., Tomato Expression Database, http://ted.bti.cornell.edu/) and were recently used successfully to identify genes involved in methyl ketone biosynthesis in S. habrochaites f. glabratum (Fridman et al., 2005). The same approach was applied here to identify candidate genes for the biosynthesis of class II sesquiterpenes, and a S. habrochaites EST collection (cLHT; accessible online at http://www.sgn.cornell.edu/index.pl) was screened for highly expressed genes. Besides the contig for the highly expressed SSTLH1 gene responsible for the biosynthesis of class I sesquiterpene (van Der Hoeven et al., 2000), two EST contigs, SGN-U313980 and SGN-U315327, were singled out as potentially involved in isoprenoid metabolism, although not a priori in sesquiterpene biosynthesis.
General Features of SBS
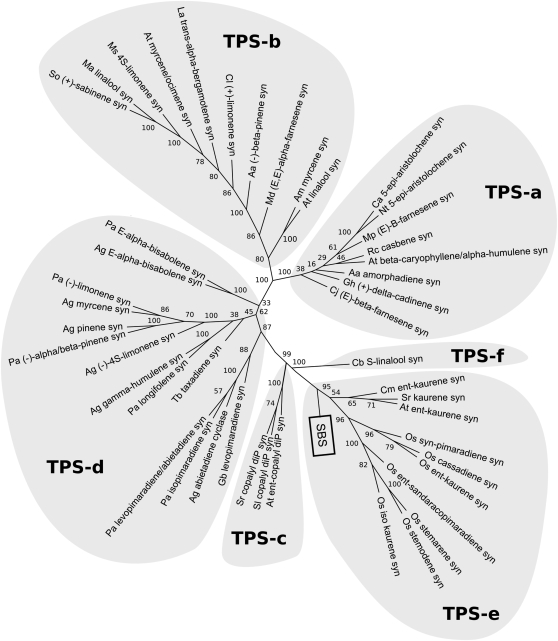
The first contig, unigene SGN-U313980, with 35 ESTs (1.42% of the library), was found to encode a protein of 778 amino acids with significant similarity to members of a group of diterpene synthases known as the kaurene synthase like (KSL) family. The corresponding gene was named SBS. SBS contains a putative chloroplast targeting signal peptide in the N-terminal position as predicted by the ChloroP program (Emanuelsson et al., 1999) with a cleavage site located at position 36. A phylogenetic tree with SBS and plant terpene synthases representative of the different classes identified so far is presented in Figure 2. The grouping of terpene synthases, or Tps, follows the nomenclature proposed by Bohlmann et al. (1998) and updated in Martin et al. (2004). It shows that SBS is clustered with members of the Tps-e group that includes the kaurene synthases involved in gibberellin biosynthesis and other KSL proteins. A multiple alignment with other members of the KSL family shows that SBS contains the catalytic motif DDXXD characteristic of type A terpene cyclases (MacMillan and Beale, 1999), but nothing may be inferred regarding its putative function (see Supplemental Figure 1 online).
Figure 2.
Unrooted Phylogenetic Tree of Plant Terpene Synthases, Including SBS.
Numbers at each root branch indicate the number of times the underlying descendants were found grouped together among the 1000 most parsimonious trees, expressed as a percentage. The complete list of the plant terpene synthases in the tree is supplied in Supplemental Table 3 online, and the alignment used to generate the tree is available as Supplemental Data Set 1 online.
General Features of zFPS
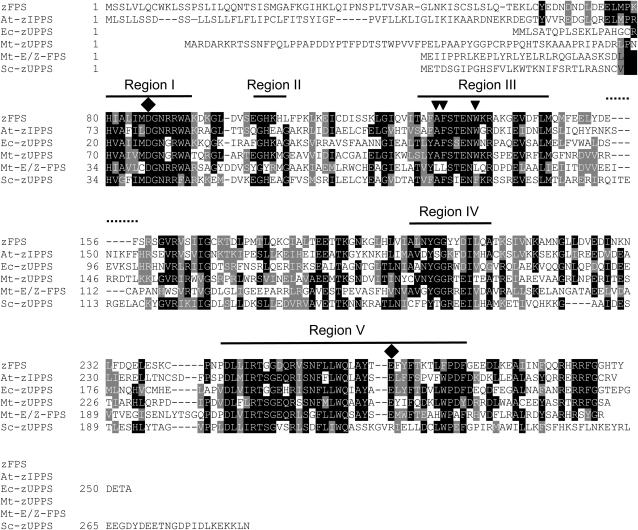
The second contig, unigene SGN-U315327, with 17 ESTs from the trichome library (0.69% of the library) was found to encode a protein of 303 amino acids with a predicted molecular mass of 34 kD. This protein has homology to Z-isoprenyl pyrophosphate synthases and was named zFPS. The ChloroP 1.1 program (Emanuelsson et al., 1999) gives a positive score (0.510) for the presence of a chloroplast targeting sequence in zFPS with a cleavage site localized after the Ser residue at position 45. Figure 3 shows an alignment of the zFPS protein with other functionally characterized Z-isoprenyl pyrophosphate synthases (Z-IPPS). The five conserved regions (I through V) of Z-IPPS described by Kharel and Koyama (2003) are also present in zFPS, as are the conserved Asp at position 86 in region I and the Glu residue at position 268 in region V. Among the functionally characterized proteins, the most closely related sequence is a polyprenyl pyrophosphate synthase from Arabidopsis thaliana encoded by the locus At2g23410 and named ACPT (for Arabidopsis cis-prenyl transferase) (Cunillera et al., 2000; Oh et al., 2000).
Figure 3.
Amino Acid Sequence Alignment of zFPS with Functionally Characterized Z-Isoprenyl Transferases.
Identical residues are shaded in black, and similar residues are shaded in gray. Dashes are gaps introduced to maximize alignment. In the N-terminal regions before the conserved domains, the identical amino acids were not highlighted because of the low complexity of the sequences. The sequences from top to bottom of the alignment are as follows: zFPS, S. habrochaites Z,Z-farnesyl pyrophosphate synthase; At-zIPPS, Arabidopsis dehydrodolichyl diphosphate synthase; Ec-zUPPS, E. coli undecaprenyl pyrophosphate synthetase; Mt-zUPPS, Mycobacterium tuberculosis Z-decaprenyl diphosphate synthase; Mt-E/Z-FPS, M. tuberculosis short-chain Z-isoprenyl pyrophosphate synthase; Sc-zUPPS, Saccharomyces cerevisiae dehydrodolichyl diphosphate synthase. The five conserved regions (I to V) and the conserved Asp and Glu residues (black filled lozenges) are indicated. The Leu residues highlighted by black inverted triangles in Mt-E,Z-FPS are those that could play a role in determining isoprenyl chain length according to Kharel et al. (2006) but are absent in zFPS. The dotted horizontal line between regions III and IV indicates a loop with a potential role in determining the length of the isoprenyl chain.
Genetic Mapping of zFPS and SBS
To ascertain their status as candidate genes for class II sesquiterpene biosynthesis, SBS and zFPS were mapped on the tomato genome by restriction fragment length polymorphism (RFLP) using a population of ILs from a S. lycopersicum × S. pennellii cross. Both genes were localized at the same position on ILs 8-1, 8-1-1, and 8-1-5, which contain the chromosome fragment bin-8A localized at the top of the chromosome 8 (see Supplemental Figure 2 online). This position concurs with the genetic mapping of the Sst2 locus (van Der Hoeven et al., 2000). PCR amplification of genomic DNA with gene-specific primers of zFPS and SBS resulted in distinct band profiles between S. lycopersicum and S. habrochaites. Line TA517 from the set of ILs of the S. lycopersicum × S. habrochaites cross was previously shown to support class II sesquiterpene biosynthesis (van Der Hoeven et al., 2000), indicating that one of the chromosome fragments from S. habrochaites introgressed in S. lycopersicum carries the Sst2 locus. When genomic DNA from TA517 was used to amplify zFPS and SBS fragments, the profile matched that of S. habrochaites, confirming the association of the zFPS and SBS genes with the Sst2 locus (see Supplemental Figure 3 online). Consequently, further characterization of the zFPS and SBS proteins was undertaken to verify whether they were actually involved in the biosynthesis of tomato class II sesquiterpenes.
The Tomato zFPS Protein Is a Z,Z-FPP Synthase
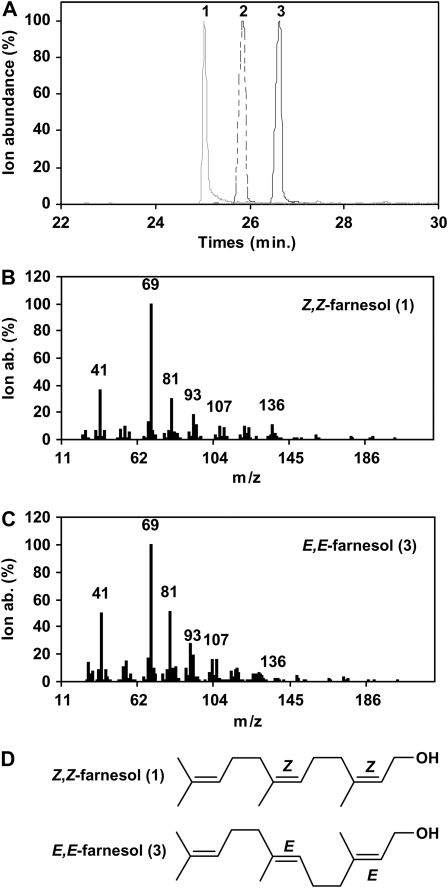
To test the catalytic activity of zFPS, its full-length coding cDNA was cloned in a bacterial expression vector with a poly-histidine tag located at the C terminus of the protein. The apparent molecular mass of this recombinant enzyme in nondenaturing conditions determined by gel filtration chromatography is 28 kD, which is consistent with a monomeric state. The recombinant protein was purified on a nickel column and was incubated with various isoprenyl pyrophosphate substrates. The assay mixture was treated with alkaline phosphatase to dephosphorylate the products and then analyzed by gas chromatography coupled with mass spectrometry (GC-MS). Incubation of zFPS with IPP and DMAPP gave a single compound (Figure 4A). Its retention time was compared with that of E,E-farnesol, which contains small detectable amounts of two other farnesol isomers (E,Z-farnesol and Z,E-farnesol; Figure 4A). The unknown molecule had a shorter retention time than the three other farnesol isomers. In addition, the mass spectrum of this dephosphorylated compound (Figure 4B) was found to be highly similar to that of a commercial E,E-farnesol standard (Figure 4C). This, together with the homology of zFPS to Z-IPPS, suggested that the dephosphorylated product could be Z,Z-farnesol. To unambiguously establish this fact, sufficient quantities of the compound, in the range of hundreds of micrograms, were produced to determine its structure. 1H and 13C nuclear magnetic resonance (NMR) chemical shifts and 2D correlation data from the NOESY spectrum (see Supplemental Figure 4 online) are reported in Supplemental Table 1 online. Significantly, the CH-2 and CH-6 methines showed a cross-peak with the CH3-15 and CH3-14 methyls, respectively. These correlations demonstrate that the molecule is (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, also known as Z,Z-farnesol. These results show that zFPS catalyzes the synthesis of Z,Z-FPP from IPP and DMAPP.
Figure 4.
GC-MS Analysis of the zFPS Enzymatic Reaction Products.
(A) Superimposed chromatograms of the dephosphorylated product obtained in vitro from zFPS recombinant protein incubated with DMAPP and IPP (gray line, peak 1), the mixture of the E,Z- and Z,E-farnesol isomers from a commercial source (dashed line, peak 2), and a pure E,E-farnesol sample (black line, peak 3).
(B) and (C) Mass spectra of (Z,Z)-farnesol and (E,E)-farnesol corresponding to peaks 1 and 3, respectively.
(D) Structures of (2Z,6Z)- and (2E,6E)-farnesol.
Enzymatic Characterization of zFPS
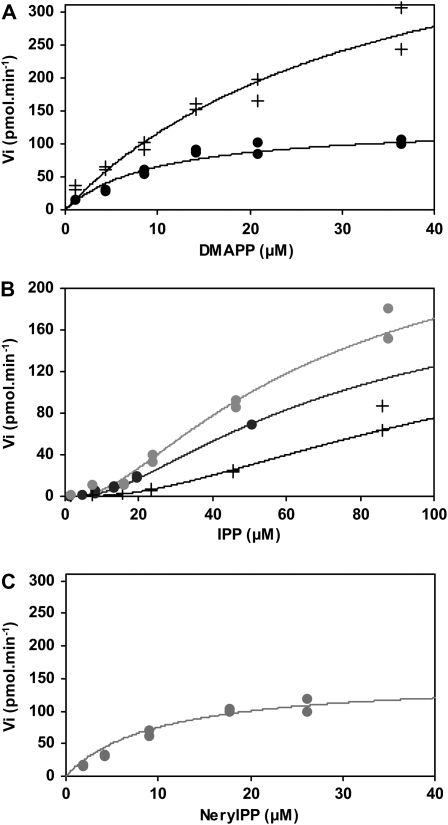
zFPS uses two substrates: IPP and DMAPP. Therefore, the affinity constant for each substrate was determined by varying the concentration of the first substrate, while the second was maintained constant. For these assays, [1-14C]IPP was used as a labeled cosubstrate with unlabeled DMAPP. Reactions were stopped by high concentration of salt, and the product was extracted using butanol and counted. The affinity of zFPS for DMAPP was studied first, with two concentrations of IPP, 35 and 210 μM. In these conditions, the variation of Vi(DMAPP) followed the equation of Michaelis-Menten. Km(DMAPP) values were 10 ± 2 μM and 35 ± 5 μM, respectively (Figure 5A). This showed that increasing concentrations of IPP decreased the affinity of zFPS for DMAPP and increased the Vmax (Figure 5A). Next, the affinity of zFPS for IPP was measured at DMAPP concentrations of 46 and 210 μM. Vi(IPP) followed the equation of Hill. Mathematical fits allowed the determination of values for the IPP Hill constant of 16 and 36 μM for DMAPP at 46 and 210 μM, respectively (Figure 5B). This indicated that increasing the concentration of DMAPP decreased the affinity of zFPS for IPP. The Hill number was 4 for both concentrations of DMAPP. This value is indicative of a positive cooperativity for the binding of zFPS to IPP. These results suggested that zFPS activity is tightly regulated in a complex manner first by IPP, both as a substrate and as a positive effector, and second by DMAPP.
Figure 5.
Kinetic Analysis of zFPS.
(A) Affinity measurements of zFPS for DMAPP at IPP 35 μM (black bullet) and 210 μM (black cross). Fits are hyperbolic according to the Michaelis-Menten equation.
(B) Affinity measurements of zFPS for IPP at DMAPP 46 μM (black bullet) and 210 μM (black cross), and for IPP at NerylPP 180 μM (gray bullet). Data are mathematically fitted according to the Hill equation.
(C) Affinity measurements of zFPS for NPP at IPP 180 μM (gray bullet). Fits are hyperbolic according to the Michaelis-Menten equation. Each measure was done in duplicate.
Because zFPS makes two sequential additions of IPP, the putative intermediate compound nerylpyrophosphate (NPP) may also be used to produce Z,Z-FPP. Therefore, NPP was tested as an alternate cosubstrate of IPP. zFPS was able to use NPP as a starter of the reaction and a Km of 10 ± 2 μM was measured at 180 μM of IPP (Figure 5C). As for DMAPP, the sigmoid distribution (17 μM and 4 for Hill constant and number) of Vi confirmed the role of IPP as activator of its own catalysis (Figure 5B).
SBS Catalyzes the Formation of Class II Sesquiterpene Olefins from Z,Z-FPP
The existence of a Z,Z-FPP synthase in tomato trichomes suggested that class II sesquiterpene biosynthesis could proceed from Z,Z-FPP. However, there has been no description to date of sesquiterpene synthases using Z,Z-FPP as a substrate. SBS, the other gene we had identified as comapping with zFPS at the Sst2 locus, was a potential candidate. As for zFPS, the SBS cDNA was cloned in a bacterial expression vector with a poly-histidine tag located at the C terminus of the protein. The SBS protein was purified, and we first tested the E-IPrP as substrates, namely, GPP, E,E-FPP, and E,E,E-GGPP. None of these gave a product (see Table 1). Because Z,Z-FPP is not commercially available, in vitro enzymatic assays with both zFPS and SBS recombinant proteins were conducted using IPP and DMAP as substrates. The products of the reaction were analyzed by GC-MS (Figure 6A). Remarkably, the profile of the chromatogram showed the presence of several peaks corresponding to sesquiterpene olefins and matched the profile of the chromatogram obtained from leaves of TA517 (Figure 6B). Incubation of zFPS and SBS with IPP and GPP did not yield any product. Interestingly, SBS was able to use NPP and transform it into a product whose mass spectrum matched that of α-terpineol in the National Institute of Standards and Technology database (Table 1). Small amounts of α-terpineol could also be detected when NPP and IPP were used in the combined assay with zFPS and SBS. However, no terpineol could be detected with IPP and DMAPP as substrates with zFPS and SBS combined. The coupled assays are not adequate for a kinetic analysis; thus, we were unable to gather kinetic data for SBS. Altogether, these results show that SBS is specific for Z-IPrPs versus E-IPrPs substrates but that it is able to use NPP in addition to Z,Z-FPP to make a monoterpene.
Table 1.
Substrate Specificity and Reaction Products of zFPS and SBS Enzymes
| Enz 1 | Enz 2 | Substrate 1 | Substrate 2 | Product |
|---|---|---|---|---|
| zFPS | DMAPP | IPP | Z,Z-FPP | |
| zFPS | GPP | IPP | ND | |
| zFPS | NPP | IPP | Z,Z-FPP | |
| SBS | DMAPP | None | ND | |
| SBS | GPP | None | ND | |
| SBS | E,E-FPP | None | ND | |
| SBS | E,E,E-GGPP | None | ND | |
| SBS | NPP | None | Terpineol | |
| zFPS | SBS | DMAPP | IPP | S |
| zFPS | SBS | GPP | IPP | ND |
| zFPS | SBS | NPP | None | Terpineol |
| zFPS | SBS | NPP | IPP | S + terpineol |
In vitro enzymatic assays of recombinant SBS and zFPS, either alone or in combination, were done with various isoprenyl pyrophosphate substrates. Products of the reaction were analyzed by GC-MS. ND, no product detected. NPP, nerylpyrophosphate. S, class II sesquiterpenes mixture comprising (+)-α-santalene, (−)-endo-α-bergamotene, (−)-exo-α-bergamotene, (−)-epi-β-santalene, and (+)-endo-β-bergamotene.
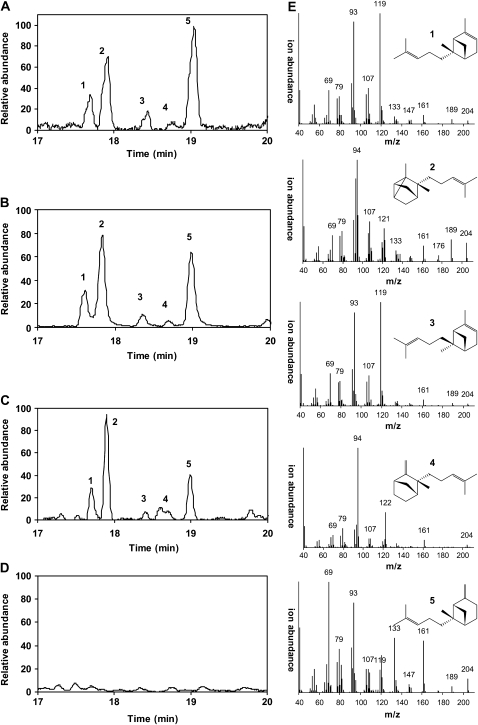
Figure 6.
Characterization of the Products of SBS by GC-MS.
(A) Chromatogram of the products obtained in vitro with the zFPS and SBS recombinant proteins incubated with DMAPP and IPP.
(B) Chromatogram of the NIL TA517 leaf exudates.
(C) Chromatogram of volatiles emitted by the transgenic line 6009 carrying zFPS and SBS transgenes.
(D) Chromatogram of tobacco line 1223, which is the background line in which the zFPS and SBS were expressed.
(E) Structure and mass spectra of the five identified sesquiterpenes, numbered according to the peaks in the chromatograms: 1, (−)-endo-α-bergamotene; 2, (+)-α-santalene; 3, (−)-exo-α-bergamotene; 4, (−)-epi-β-santalene; 5, (+)-endo-β-bergamotene.
Analysis of the Sesquiterpene Products of SBS
Of the five peaks we could detect either in S. habrochaites, in IL TA517, or from the in vitro assays, three of them have been identified previously as (−)-endo-α-bergamotene (peak 1), (+)-α-santalene (peak 2), and (+)-endo-β-bergamotene (peak 5) (Figure 6) (Coates et al., 1988; van Der Hoeven et al., 2000). Two minor peaks, 3 and 4, could also be consistently resolved. These had not been assigned to specific sesquiterpenes to date. Sandalwood oil (from Santalum album) contains several sesquiterpene olefins related to tomato class II sesquiterpenes, including (+)-α-santalene, (−)-exo-α-bergamotene, (+)-epi-β-santalene, and (−)-β-santalene (Kawamura et al., 1995; Jones et al., 2006). Comparison with a GC-MS chromatogram of an authentic sandalwood oil sample showed that peaks 3 and 4 had identical retention times and mass spectra highly similar to (−)-exo-α-bergamotene and (+)-epi-β-santalene, respectively (see Supplemental Figure 5 online). When the samples were compared on a chiral column, peak 3 still coeluted with (−)-exo-α-bergamotene, but peak 4 had a different retention time than (+)-epi-β-santalene of sandalwood oil (see Supplemental Figure 5 online). This led us to conclude that peak 3 corresponded to (−)-exo-α-bergamotene, while peak 4 was (−)-epi-β-santalene, of the opposite stereochemistry of the (+)-epi-β-santalene from sandalwood.
Expression of zFPS and SBS in Tobacco Trichomes Leads to the Production of Tomato Class II Sesquiterpenes
Glandular trichomes of the wild tobacco Nicotiana sylvestris were chosen as an in planta expression system for terpenoid biosynthesis pathway genes. In a previous study, this system was used successfully to produce novel diterpenoids (Rontein et al., 2008). To facilitate the identification of the novel terpenoids produced, the endogenous tobacco diterpene synthase, cembratrien-ol synthase (CBTS) (Wang and Wagner, 2003), was silenced by an intron hairpin construct. A single transgenic line (number 1223) with this construct was used here. As in the previous study, the CBTS trichome-specific promoter was used to direct the expression of the cDNAs of zFPS and SBS. Both transgenes under the control of the CBTS trichome-specific promoter were cloned within the same T-DNA vector. This T-DNA was introduced into N. sylvestris line number 1223 by Agrobacterium tumefaciens–mediated transformation. Thirty transgenic T0 lines were recovered, and 16 of them presented a high level of transgene expression, as verified by quantitative real-time PCR (see Supplemental Figure 6 online).
The presence of novel compounds produced by the glandular trichomes may be detected by washing the leaves with a solvent (e.g., pentane for terpene olefins) followed by GC-MS analysis. Leaf exudates of the transgenic lines expressing zFPS and SBS did not reveal the presence of new sesquiterpenoid compounds. This could be explained by the fact that tobacco plants have secretory trichomes that are devoid of specialized structures to retain volatile compounds, such as sesquiterpene olefins. Thus, to detect sesquiterpene olefins produced by these transgenic plants, the volatile compounds were collected from the headspace and analyzed by GC-MS. The chromatogram (Figure 6C) indicated that the transgenic plants harboring both genes were able to produce a mixture of new sesquiterpene olefins identical to the mixture obtained from both TA517 tomato plants and also the in vitro assay with the recombinant enzymes. In particular, the proportion of each major sesquiterpene, such as endo-α-bergamotene, α-santalene, and endo-β-bergamotene, was maintained. By contrast, the headspace of plants from line 1223 showed no sesquiterpenes in the same region of the chromatogram (Figure 6D). Also, plants expressing SBS only did not produce any detectable sesquiterpene, indicating that zFPS was necessary for the biosynthesis of class II sesquiterpenes in planta (data not shown).
zFPS and SBS Are Targeted to the Plastid Compartment
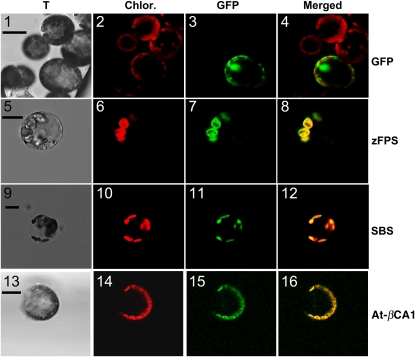
Sesquiterpene biosynthesis in higher plants usually takes place in the cytosol and uses IPP and DMAPP, which are produced from the cytosolic MEV pathway. The zFPS and SBS proteins, however, contain putative chloroplast targeting sequences of 45 and 36 amino acids, respectively, as predicted by the ChloroP software. Thus, determining whether they are actually targeted to the chloroplasts would provide novel insight into sesquiterpene biosynthesis in plants. Expression of fusions to a reporter such as green fluorescent protein (GFP) provides a convenient method to test the functionality of subcellular targeting sequences. To ensure that no amino acid residue critical to the proper function of the targeting signal peptide was missed, the fused fragments were chosen to span the predicted targeting peptide and extend to the beginning of the first conserved region with other related proteins. Thus, cDNA fragments encoding the first 58 and 63 amino acids of zFPS and SBS, respectively, were fused to the GFP coding sequence under the control of a constitutive promoter. The resulting plasmids were introduced into tobacco protoplasts for transient expression, and GFP transient expression was monitored by confocal laser scanning microscopy. Chlorophyll and GFP fluorescences were recorded at distinct wavelengths, thus allowing the separate recording of their signals. Chlorophyll is an indicator of chloroplasts, and colocalization of chlorophyll and GFP signals in merged images indicates that the GFP fusion protein is targeted to plastids. The construct with GFP alone showed nonoverlapping signals for chlorophyll and GFP fluorescences, confirming previous observations that GFP was targeted to the cytosol (Figure 7, panels 1 through 4). By contrast, the constructs with the zFPS and SBS signal peptides fused to GFP gave signals overlapping with the chloroplasts' fluorescence, indicating that the fused peptides were able to direct the GFP to the plastids (Figure 7, panels 5 through 12). A GFP fusion construct with the Arabidopsis β-carbonic anhydrase 1 (At-βCA1) protein that was previously shown to be targeted to the chloroplasts (Fabre et al., 2007) gave a similar fluorescence profile (Figure 7, panels 13 through 16). These results provided evidence that the biosynthesis of type II sesquiterpenes in tomato takes place in the chloroplasts of the trichome secretory cells.
Figure 7.
Transit Peptides of zFPS and SBS Direct GFP Fusions to the Chloroplasts.
Mesophyll tobacco protoplasts transfected with the control pENP50PMA4-GFP plasmid (GFP, panels 1 through 4), with pENP50PMA4-zFPS-GFP (zFPS, panels 5 through 8), with pENP50PMA4-SBS-GFP (SBS, panels 9 through 12), or with At-αCA1-GFP (At-αCA1, panels 13 through 16). Images were taken with a confocal laser scanning microscope. T, transmitted light images; Chlor., chlorophyll fluorescence images false-colored in red; GFP, GFP fluorescence images false-colored in green; Merged, merged of chlorophyll and GFP fluorescence images. Bars = 10 μm.
DISCUSSION
Using a candidate gene approach based on the in silico analysis of trichome ESTs, we have identified two genes (zFPS and SBS) associated with the biosynthesis of tomato class II sesquiterpenes, which comprise (−)-endo-α-bergamotene, (+)-α-santalene, (−)-exo-α-bergamotene, (−)-epi-β-santalene, and (+)-endo-β-bergamotene. Both genes were mapped to the top of chromosome 8 of the tomato genome, a position consistent with that of the Sst2 locus responsible for class II sesquiterpene biosynthesis. The functional characterization of these two genes has led to the discovery of several unusual features concerning the biosynthesis of sesquiterpenes that will be discussed below.
Z,Z-FPP Is a Novel and Atypical Substrate for Sesquiterpene Synthases
The data presented here demonstrate that the tomato zFPS protein is a short-chain Z-IPPS, that its main product is Z,Z-FPP, and that the SBS protein uses Z,Z-FPP as a substrate to produce the tomato class II sesquiterpenes.
Most Z-IPPS discovered to date have been shown to catalyze the conversion of E,E-FPP to medium- and long-chain isoprenyl diphosphates, which are not substrates for terpene synthases. Rather, for example, the medium-chain Z-isoprenyl diphosphates are biosynthetic precursors of C55-C100 dolichols, themselves involved in the biosynthesis of glycoproteins (Sato et al., 1999), whereas long-chain Z-isoprenyl pyrophosphates are biosynthetic precursors of polymers like rubber (Asawatreratanakul et al., 2003). A short-chain Z-isoprenyl pyrophosphate synthase (Rv1086) was discovered in Mycobacterium tuberculosis, but it catalyzes the synthesis of E,Z-FPP from GPP. E,Z-FPP is then metabolized to decaprenyl pyrophosphate, which plays a key role in the biosynthesis of the arabinogalactan-peptidoglycan complex of the bacterial cell wall (Crick et al., 2001). In a study to identify the amino acids involved in chain length determination, Kharel et al. (2006) found three Leu residues (Leu-84, Leu-85, and Leu-90) and one His residue (His-237) (see Figure 3) that, when introduced in long-chain Z-IPPS, reduced the chain length of the isoprenyl pyrophosphate synthesized. However, the tomato zFPS does not contain any of these residues and in that respect is very similar to medium- or long-chain Z-IPPS. Thus, it may be concluded that amino acid residues at these positions are unlikely to affect chain length determination by zFPS. We noted, however, that zFPS and Rv1086 present similarities in another region potentially involved in chain length determination (Kharel et al., 2006), a loop situated between the conserved regions III and IV (Figure 3). Both short-chain Z-IPPS have a loop two or three amino acids shorter than that of medium-length Z-IPPS and five or seven amino acids shorter than that of a long-chain Z-IPPS, the yeast UPPS. This could be a region to modify to manipulate the length of the Z-isoprenyl length synthesized by zFPS.
Could Z-Isoprenylpyrophosphates Be Widespread Substrates for Terpene Synthases in Plants?
Despite the fact that all FPP synthases involved in sesquiterpene biosynthesis found so far are of the E type, the implication of Z-isoprenyl pyrophosphates in sesquiterpene biosynthesis has been hinted at in previous work. For example, cotton (Gossypium hirsutum) enzymatic extracts converted Z,Z-FPP to δ-cadinene much more efficiently than they converted E,E-FPP (Heinstein et al., 1970). In subsequent work, it was also found that a Z,Z-FPP synthase could be identified from cotton extracts, although it could not be separated from the E,E-FPP synthase (Adams and Heinstein, 1973). More recently it was suggested that cyclization of the class II sesquiterpenes from tomato, the objects of this study, would be favored from E,Z-FPP compared with the typical E,E-FPP substrate because of stereochemical constraints (Coates et al., 1988). Also, the proposed cyclization mechanism for a number of sesquiterpenes from E,E-FPP involves an initial isomerization step leading to the formation of nerolidyl pyrophosphate and allowing the conversion of the pyrophosphate proximal double bond from the E to the Z configuration (for example, see Bohlmann et al., 1998; Benedict et al., 2001). This initial isomerization could be bypassed with Z,E-FPP as a substrate rather than with E,E-FPP. The existence of a bona fide Z,E-FPP synthase in plants remains to be shown, however, and zFPS, the tomato enzyme described here, failed to yield Z,E-FPP when supplied with GPP and IPP. Examination of complete plant genome sequences may help identify putative candidates. The Arabidopsis genome, for example, contains nine putative Z-prenyl pyrophosphate synthases (see Supplemental Table 2 online), and only one of them, ACPT encoded by the locus At2g23410, has been characterized as a polyprenyl pyrophosphate synthase (Cunillera et al., 2000; Oh et al., 2000). Depending on the microorganism in which it was expressed, the chain length of the products synthesized by ACPT varied from 75 to 85 carbon atoms in yeast (Cunillera et al., 2000) to ∼120 in E. coli (Oh et al., 2000). The actual length of the products of ACPT in Arabidopsis plants remains to be determined, but it seems unlikely that it produces short-chain Z-IPrPs. Because the amino acids involved in determining chain length elongation of Z-IPrPs are not known with confidence, it is not possible to predict the chain length of the product of the remaining Arabidopsis Z-IPPS. Thus, it remains to be seen whether any of these putative Z-IPPS actually encodes a Z,Z-FPP or a Z,E-FPP synthase. According to the TargetP 1.1 software (Emanuelsson et al., 2000), none of the Arabidopsis proteins of the Z-IPPS family is targeted to the chloroplast, whereas two of them are presumably targeted to the mitochondria and four are predicted to be secreted, including ACPT (see Supplemental Table 2 online). This would suggest that, unlike tomato, Arabidopsis does not possess a chloroplastic Z,Z-FPS.
More generally, the existence of Z,Z-FPP as a substrate for sesquiterpene synthases and the activity of SBS on NPP suggest that Z-IPrPs of different lengths could be the substrates of terpene synthases. For example, nerylpyrophosphate (C10) could be the substrate of monoterpene synthases to give products such as neral, which is present in the essential oil of a number of fragrant species. Similarly, Z,Z,Z-GGPP could be the substrate of diterpene synthases.
Proposed Mechanism for the Conversion of Z,Z-FPP to Class II Sesquiterpenes
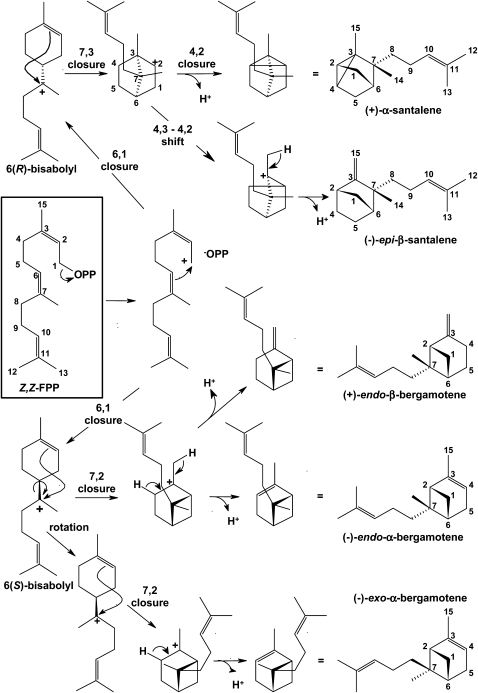
A cyclization pathway is proposed to account for the multiple sesquiterpenes arising from the action of SBS on Z,Z-FPP (Figure 8). In this scheme, the first step is the formation of the Z,Z-farnesyl C1 carbocation, which, because of the Z configuration of the C2-C3 bond, is conveniently converted to a bisabolyl carbocation by a C6-C1 closure. Similarly to what has been shown for monoterpene synthases (Wise et al., 1998), the stereochemistry of this closure, however, will determine which specific stereoisomers are produced. Earlier work demonstrated that the main class II sesquiterpenes have the following absolute stereochemistry: (+)-α-santalene, (+)-endo-β-bergamotene, and (−)-endo-α-bergamotene (Coates et al., 1988). The opposite stereochemistry at the bridging carbon to which the side chain is attached between (+)-α-santalene on one hand and (+)-endo-β-bergamotene and (−)-endo-α-bergamotene on the other implies that (+)-α-santalene arises from the 6(R)-bisabolyl cation, whereas (+)-endo-β-bergamotene and (−)-endo-α-bergamotene arise from the 6(S)-bisabolyl cation. A C4-C2 closure on the 6(R)-bisabolyl cation followed by proton loss yields (+)-α-santalene, whereas a C4-C3 shift to a C4-C2 bond followed by deprotonation leads to (−)-epi-β-santalene. Similarly, a C7-C2 closure on the 6(S)-bisabolyl cation followed by an endocyclic or an exocyclic deprotonation leads to either (+)-endo-β-bergamotene or (−)-endo-α-bergamotene, respectively. The opposed stereochemistry at the C-7 in exo-α-bergamotene imposes a rotation of the C6-C7 bond of the 6(S)-bisabolyl cation. The cyclization events that follow are analogous to those for the formation of endo-α-bergamotene. This multiproduct cyclization pathway is yet another example of the plasticity of terpene synthases, with the added novelty that it uses the atypical Z,Z-FPP as a substrate.
Figure 8.
Pathway for the Cyclization of Z,Z-FPP to Class II Sesquiterpenes by SBS.
Following dephosphorylation of Z,Z-FPP (boxed), the Z,Z-farnesyl carbocation is converted to either the 6(R)- or the 6(S)-bisabolyl cations depending on the stereochemistry of the 6,1 closure. The 6(R)-bisabolyl cation will then lead to the formation of (+)-α-santalene and (−)-epi-β-santalene, while the 6(S)-bisabolyl cation will give rise to (−)-exo-α-bergamotene, (−)-endo-α-bergamotene, and (+)-endo-β-bergamotene.
SBS Is an Atypical Sesquiterpene Synthase
Another unexpected finding of this work is the sequence similarity of the SBS enzyme to terpene synthases of the kaurene synthase group. This group, Tps-e according to the classification proposed previously (Bohlmann et al., 1998), is typically involved in the biosynthesis of polycyclic diterpenes, both of primary (e.g., gibberellins) but also of secondary metabolites like the rice (Oryza sativa) diterpenoid phytoalexins (kaurenoids, phytocassanes, and momilactones) (Sakamoto et al., 2004). All angiosperm sesquiterpene synthases described so far, including the tomato SSTLH1 responsible for the synthesis of germacrene B in S. habrochaites, belong to either the Tps-a group or the more recently defined Tps-g group (Dudareva et al., 2003; Aharoni et al., 2004). SBS has a low percentage of sequence similarity with SSTLH1 (16%) but is most closely related to a tobacco protein of unknown function (PID: AAS98912). It shares 39% sequence identity and 57% sequence similarity with the Arabidopsis kaurene synthase, and 31% sequence identity and 50% sequence similarity with the maize (Zea mays) kaurene synthase. Enzymatic assays with various IPrP substrates demonstrated that SBS is highly specific for Z-IPrPs versus E-IPrP. On the other hand, SBS exhibited some flexibility toward the Z-IPrP substrate because it was able to transform NPP to a monoterpene, α-terpineol. The absence of α-terpineol in S. habrochaites LA 1777 extracts suggests that NPP does not accumulate to significant levels in planta, most likely because of the complete conversion of IPP and DMAPP to Z,Z-FPP by zFPS. The absence of commercially available Z,Z,Z-GGPP did not allow the testing of this substrate with SBS, but it would certainly be worth investigating this further.
Cellular Localization of Sesquiterpene Biosynthesis in Tomato Trichomes
Terpene synthases have been shown to localize to different cellular compartments. Typically, monoterpene and diterpene synthases are targeted to the chloroplasts and sesquiterpene synthases to the cytosol. There is also one example of a terpene synthase localized to the mitochondrion (Aharoni et al., 2004). This cellular localization of terpene synthases is coupled with the cellular localization of the two pathways for the universal terpenoid precursors, IPP and DMAPP. The MEV pathway is cytosolic, whereas DXP is plastidic. As a result, plant monoterpenes and diterpenes are typically synthesized from precursors of the DXP pathway and sesquiterpenes from precursors of the MEV pathway. This rule suffers some exceptions, however, with, for example, a reported chloroplastic E,E-FPP synthase in rice (Sanmiya et al., 1999). In addition, labeling experiments in chamomile (Matricaria recutita) and snapdragon (Antirrhinum majus) showed that sesquiterpenes produced by these plants were derived from IPP and DMAPP of the DXP pathway rather than the cytosolic MEV pathway (Adam et al., 1999; Dudareva et al., 2005). Assuming that the sesquiterpene synthase involved was cytosolic, Dudareva et al. (2005) inferred that the IPP and DMAPP were transported from the plastids and transformed into E,E-FPP by a cytosolic FPS. The authors were supported in this hypothesis by a prior report describing the existence of a transport of isoprenyl diphosphates, particularly IPP and GPP, from the chloroplast to the cytosol (Bick and Lange, 2003). A recent report shows that there are two very closely related terpene synthases in snapdragon, both able in vitro to produce a monoterpene, linalool, and a sesquiterpene, nerolidol (Nagegowda et al., 2008). One of these isoforms is targeted to the plastids and the other to the cytosol. These results seem to confirm the hypothesis of a transport of IPP and DMAPP from the plastids to the cytosol, thus accounting for the incorporation of precursors from the DXP pathway into sesquiterpenes in snapdragron flowers. However, the existence of a plastidic E,E-FPP synthase in rice (Sanmiya et al., 1999) indicates that sesquiterpene biosynthesis taking place directly in the plastids is also a possibility, albeit in a different plant species.
The results of GFP fusion experiments with both zFPS and SBS described here demonstrate that both an FPS and a sesquiterpene synthase are localized in the chloroplasts, thus providing an explanation for the existence of sesquiterpenes synthesized from precursors of the DXP pathway. Interestingly, the cytosolic SSTLH1 (van Der Hoeven et al., 2000) and the plastidic SBS (this study) coexist in the trichomes of S. habrochaites (Figure 9). It is currently not known whether SSTLH1 uses IPP and DMAPP from the MEV or the DXP pathway, and labeling experiments will have to be performed to address this issue. Altogether, results from this and previous studies show that the subcellular localization of sesquiterpene biosynthesis in plants is subject to a high level of variation in a species-specific fashion. Cytosolic E,E-FPP synthesis in plants is likely to be under strong regulatory constraints because of its central role in signaling via protein farnesylation but also in the biosynthesis of brassinosteroid hormones and membrane sterols. The variations in sesquiterpene biosynthesis observed in tomato, snapdragon, chamomile, and rice may thus be seen as different ways to circumvent these regulatory constraints to achieve higher levels of production.
Figure 9.
Cellular Organization of Sesquiterpene Biosynthesis Pathways in S. habrochaites LA1777 Trichomes.
Two pathways coexist. The first, in the chloroplasts, leads to class II sesquiterpenes, including α-santalene, α-bergamotenes, and β-bergamotene. The second, in the cytosol, produces germacrene B.
In conclusion, this work describes a novel pathway for the biosynthesis of sesquiterpenes from Z,Z-FPP in plants. The availability of Z,Z-FPP now makes it possible to test sesquiterpene synthases that might use this substrate. This could have broad implications, both for engineering of terpene biosynthesis and also for gene function discovery.
METHODS
Oligonucleotide Primers and Probes
A complete list of oligonucleotide primers and probes used in this study is available in Supplemental Table 4 online.
Plant Material
Solanum habrochaites LA1777, Solanum pennelli LA716, Solanum lycopersicum E6203, and NIL TA517 seeds were provided by the Tomato Genetics Resource Center (University of California, Davis, CA). Seeds were germinated directly in soil at 25°C with 14-h light/day in a growth chamber for 2 weeks. Individual plants were then transferred to pots and grown in a greenhouse supplied with 14 h light/day (400-W high-pressure sodium lights).
The tobacco line 1223 used for transformation in this study was a transgenic Nicotiana sylvestris line containing an ihpRNAi construct to silence the CBT-ol synthase gene (CBTS) used previously (Rontein et al. 2008).
Isolation of Nucleic Acid, Gene Detection, and Expression Analysis of Tobacco Transgenics Lines
Total cellular DNA isolation was performed from 50 mg of leaves using a MATAB procedure modified as follows (Sallaud et al., 2003): After chloroform separation, the supernatant (250 μL) was loaded on a 96-well filter plate (Acroprep omega 100K; PALL). The plate was then centrifuged at 3000g for 20 min, washed with 250 μL of sterile water, and centrifuged again at 3000g for 20 min to remove remaining traces of water on the membrane. The DNA was eluted from the filter plate by applying 150 μL of sterile water on the membrane, and after 30 min of gentle agitation, the DNA solution was recovered by pipetting. RNA isolation was achieved from 50 mg of leaves using the RNeasy kit (Qiagen) according to the manufacturer's instructions. To remove traces of genomic DNA contamination, RNA samples were treated with a DNase (Turbo DNA-free; Ambion) according to the manufacturer's instructions. For RT-PCR, cDNAs were synthesized from 1 μg of total RNA with the Transcriptor reverse transcriptase kit (Roche) using oligo(dT) or random primers. Transgene expression was quantified by real-time quantitative RT-PCR in duplex experiments on an ABI7000 instrument (Applied Biosystems). As we routinely saw good coexpression of both transgenes on the T-DNA, only SBS expression was recorded. This was done by amplification with primers SBS-For and SBS-Rev and the TaqMan probe SBS-Pr. The reference signal was provided by amplifying a tobacco (Nicotiana sylvestris) Actin gene with primers Actin-For and Actin-Rev and monitoring amplification with the TaqMan probe Actin-Pr (see Supplemental Figure 6 online). The SBS-specific TaqMan probe was labeled with a FAM fluorescent dye (Eurogentec), and the Actin TaqMan probe with a VIC fluorescent dye (Applied Biosystems). Single-copy lines with the best expression levels were selected for volatile analysis by GC-MS.
Cloning of Full-Length zFPS and SBS cDNAs
zFPS and SBS full-length cDNAs were amplified by PCR from cDNA obtained from mRNA of S. habrochaites leaves using primers flanked with convenient restriction sites, subcloned into pGEM-T vectors (Promega), and verified by sequencing. RNA isolation was achieved from 50 mg of leaves using the RNeasy kit (Qiagen) according to the manufacturer's instructions. To remove traces of genomic DNA contamination, RNA samples were treated with a DNase (Turbo DNA-free; Ambion) according to the manufacturer's instructions. For RT-PCR, cDNAs were synthesized from 1 μg of total RNA with the Transcriptor reverse transcriptase kit (Roche) using oligo(dT) or random primers. Forward primer 10D01 and reverse primer 10D02 (sequences provided in Supplementable Table 4 online) for zFPS, and forward primer 7F05 and reverse primer 8D05 for SBS were used to amplify the full-length cDNAs. Sequence information was obtained from the Tomato Gene Index Database (Dana Farber Cancer Institute, Boston, MA 02115; http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=tomato). A NdeI site within the SBS cDNA that interfered with subsequent cloning was eliminated by site-directed mutagenesis (QuickChange XL kit; Stratagene) using primers 8H08 and 8H09, without modification of its primary amino acid sequence.
An Agrobacterium tumefaciens transformation binary vector was made to directly clone transgene expression cassettes with Gateway recombination technology (Invitrogen). The destination binary vector for LR recombination (Invitrogen) was created from pCAMBIA2300 (Genbank accession number AF234315) as follows: The AttR4/AttR3 DNA fragment was isolated from pDEST R4R3 plasmid (Invitrogen) by PCR using primers 7E06 and 7E07 both containing a XbaI site and cloned into the pGEM-T vector at the XbaI site. The internal XbaI site was removed by site-directed mutagenesis (QuickChange XL kit; Stratagene) using primers 7I06 and 7I07. The AttR4/AttR3 DNA fragment was then cloned into the unique XbaI site of pCAMBIA2300 to give R3R4-pC2300. For the selection of the recombined clones, the bla gene was isolated from pUC18 and inserted outside the T-DNA region into the unique ScaI restriction site of R3R4-pC2300 to give the R3R4-pC2300-Amp destination vector.
The enhancer 35S-CBTS 1.0-kb promoter (BamHI, NcoI), the cDNA (zFPS or SBS) (NcoI, XhoI), and the terminator (NOS or CBTS) (XhoI, XbaI) were inserted into a PUC19 cloning vector by regular ligation procedure to create pUC19-zFPS and pUC19-SBS. These expression cassettes containing zFPS or SBS were amplified with a high-fidelity enzyme (Pwo, Roche) using primers with 5′ extensions containing the recombination sites AttB4 (primer 7C06) and AttB1 (primer 7C07) for zFPS, and AttB1 (primer 7C08) and AttB2 (primer 7C09) for SBS. The zFPS and SBS expression cassettes were introduced into pDONR-P4-P1R and pDONR 221(Invitrogen), respectively, by recombination with BP clonase (Invitrogen) to give the vectors pENTR-L4R1-zFPS and pENTR-L1R2-SBS. Two LR recombination reactions with LR clonase (Invitrogen) into R3R4-pC2300-Amp destination vector were done as follows: (1) pENTR-L4R1-SBS with two empty pENTR-L1R2 and pENTR-R2L3 that gave rise to the pC2300-SBS cassette; (2) pENTR-L4R1-zFPS, pENTR-L1R2-SBS, and the empty vector pENTR-R2L3 that gave rise to pC2300-zFPS-SBS double expression cassettes. The empty pENTR vectors were first created by amplification of PUC19-zFPS with primers 7C06 and 7C07, or 7D01 and 7D02, for subsequent cloning in pDON221 or pDONR-P2R-P3, respectively, and then the resulting pENTR vectors were digested with BamHI and XbaI to remove the cassettes.
GFP Fusion Experiments
The nucleic acid sequences corresponding to the first 58 and 63 amino acids of zFPS and SBS, respectively, were fused upstream of and in frame with the GFP coding sequence, at the KpnI site in the plasmid EN50PMA4-KGFP (Hosy et al., 2005). The primers used to amplify the corresponding coding region were 14F02 and 14F03 for zFPS, and 14F04 and 14F05 for SBS. In this plasmid, the GFP coding sequence is under the control of a strong constitutive promoter EN50PMA4, which consists of the pma4 (for plasma membrane H+-ATPase isoform #4 gene) promoter in which the cauliflower mosaic virus 35S enhancer is inserted at position −50 upstream from the transcription start of the pma4 gene (Zhao et al., 1999). The plasmid with the fusion with Arabidopsis thaliana At-βCA1 and GFP was as described by Fabre et al. (2007) and was a gift from D. Rumeau.
Thirty to fifty micrograms of plasmid was used to transform tobacco and Arabidopsis protoplasts according to Verret et al. (2004). Transient expression experiments were repeated three times. Images were taken on a Leica SP2 AOBS inverted confocal microscope (Leica Microsystems) equipped with an argon ion laser. Chlorophyll fluorescence was measured with wavelengths ranging from 620 to 750 nm, and GFP fluorescence was measured with wavelengths ranging from 500 to 530 nm.
Gene Mapping
The IL population used for mapping the zFPS and SBS genes was that from the S. pennellii × S. lycopersicum cross because the IL population from the S. habrochaites × S. lycopersicum was not available in the laboratory at the time of the experiments. However, because of the extensive colinearity of the genomes of S. habrochaites, S. pennellii, and S. lycopersicum, the mapping can be transposed from one species to the other. In addition, the mapping was confirmed by screening the individual IL lines of the S. habrochaites × S. lycopersicum cross carrying the appropriate chromosome fragments, thus validating the initial mapping with the S. pennellii × S. lycopersicum IL lines. The IL population is composed of 75 lines, each containing a single introgression from S. pennellii (LA716) in the genetic background of the processing tomato S. lycopersicum variety M82 (Pan et al., 2000). The IL population was connected to the high-density map of tomato (Tanksley et al., 1992) by probing all the ILs with the RFLP markers from the framework F2 map (Pan et al., 2000). The clones were mapped by RFLP after screening for polymorphism using four restriction enzymes (EcoRI, EcoRV, HindIII, and XbaI). Genomic DNA extraction, digestion, and hybridization were as previously described (Saliba-Colombani et al., 2000). In addition, PCR on genomic DNA with primers 9I01 and 9I02 for zFPS and 8D08 and 8G01 for SBS produced a different band profile for S. esculentum and S. habrochaites (see Supplemental Figure 3 online). The profile for IL TA517 that carries the S. habrochaites Sst2 allele was the same as that of S. habrochaites (see Supplemental Figure 3 online).
Phylogenetic Analysis and Prediction Program
BLAST searches were performed against the nonredundant protein database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) using default parameters. Protein sequences were aligned using the ClustalW2 program at the European Bioinformatics Institute (http://www.ebi.ac.uk/tools/clustalw2) with the default matrix. The alignment is available as Supplemental Data Set 1 online. Phylogeny was inferred using the maximum likelihood method from the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html), where a consensus tree was derived from the most parsimonious trees out of 1000 data sets.
Analysis of amino acids sequence of SBS and zFPS for prediction of transit peptide occurrence and length was done with the ChloroP program at http://www.cbs.dtu.dk/services/ (Emanuelsson et al., 1999).
Expression of Recombinant zFPS and SBS Proteins in Escherichia coli Cells
The full-length coding sequences of zFPS and SBS, including the putative transit peptides, were amplified with primers 9I01/9I02 and 8D08/8D06, respectively. The PCR products were digested with NdeI and XhoI for cloning into the pET30b expression vector (Novagen) to generate fusions with 6-His-tags at the C terminus. The vectors were then introduced by electroporation into E. coli cells [BL21-CodonPlus (DE3)-RIL strain; Stratagene]. For protein production, the culture (500 mL) was incubated at 37°C to OD 0.5 at 600 nm. The temperature was then decreased to 16°C, and ethanol (1%, v/v) was added to the medium. After 1 h incubation, isopropylthio-β-galactoside was added to a final concentration of 1 mM to induce the expression of the protein. The culture was incubated for 18 h and then centrifuged at 4°C. The pellet was then resuspended in phosphate buffer (200 mM, pH 7), and the cells were lysed by a one-shot depression (ΔP = 19 bar). After centrifugation at 15,000g, the supernatant containing soluble proteins was desalted on a PD-10 column (General Electric Healthcare) and then equilibrated with a phosphate buffer (50 mM, pH 7) before being loaded on a nickel-nitrilotriacetate column (Ni-NTA; Qiagen) and eluted according to the manufacturer's instructions. The enriched fractions were then identified by SDS-PAGE gel, and the elution buffer was exchanged using a PD-10 column equilibrated with the activity buffer (50 mM HEPES, pH 7.8, 100 mM KCl, 1 mM DTT, and glycerol 5% [w/v]).
In Vitro Enzymatic Assays with the Recombinant Enzymes zFPS and SBS
In vitro enzymatic assays were done in the following buffer: 50 mM HEPES, pH 7.8, 100 mM KCl, 7.5 mM MgCl2, 5% (w/v) glycerol, and 5 mM DTT. For each assay, 25 μg of protein were incubated with IPP, DMAPP, GPP, FPP, GGPP (Sigma-Aldrich), or NPP (a gift from Skip Waechter), alone or in combination at 65 μM each and incubated at 32°C for 2 h. For zFPS assay, the reaction products were dephosphorylated by adding 20 units of alkaline phosphatase (intestinal calf; New England Biolabs) for 1 h at 37°C. Terpene alcohols produced by dephosphorylation were then extracted with a mix of chloroform:methanol:water (0.5:1:0.4, v:v:v) (Bligh and Dyer, 1959). Water and organic phases were separated by adding 0.5 volume each of water and chloroform. The organic phase was then dried under nitrogen, resuspended in pentane, and analyzed by GC-MS (Agilent Technologies). Terpene alcohols were identified by comparison of their mass spectra with those of the National Institute of Standards and Technology database and of their retention times with known standards, such as geraniol, farnesol, and geranylgeraniol (Fluka) or essential oils in which compounds have been well characterized. For the SBS assay, the reaction product was directly extracted three times with pentane (v:v) before direct injection on GC-MS.
Determination of Enzymatic Parameters of zFPS
Assays for the kinetic analysis were performed using unlabeled DMAPP (Sigma-Aldrich) and [1-14C]IPP (59.0 mCi/mmol; General Electric Healthcare) used as a tracer, with unlabeled IPP (Sigma-Aldrich) added to get the required final concentration in the assay. Each assay was performed in 50 μL contained in glass tubes. DMAPP and zFPS (6 μg enriched on Ni-NTA resin) were preincubated at 4°C. Reactions were started by adding IPP and by placing the glass tube in a 30°C water bath. Reactions were performed and shown to be linear over a period of 4 min. Assays were stopped by adding 10 volumes of water saturated with NaCl. Z,Z-FPP was extracted with 10 volumes of butanol (Schulbach et al., 2000). The radioactivity in the solvent phases was estimated by a scintillation counter. As a control for the specificity of the butanol extraction toward FPP, DMAPP and [1-14C]IPP were extracted from an assay without enzyme, but still with NaCl. In this case, the butanol phase was not significantly labeled compared with a sample without labeling and never exceeded 1% of the label extracted from the assay in the presence of enzyme at the tested concentrations of IPP.
Mathematic fits were determined using Kaleidagraph software and by entering the Michaelis or Hill equation and approximate values for Km, Vm, and Hill number when required. The fit to the experimental data is automatically done by the software according to the least square method.
Analysis of Tobacco Volatiles
The headspace of transgenic tobacco lines was analyzed in a glass chamber in which the air composition (CO2 and O2) was maintained. An airflow system containing a pump and a cartridge filled with SuperQ resin (Alltech) trapped the volatiles emitted by the plant. The airflow rate was ∼20 mL·min−1. Volatiles were sampled over 48 h, eluted from the cartridge with 1 mL of pentane, and analyzed by GC-MS.
GC-MS Analysis
Pentane samples were injected directly into GC-MS for analysis in a 6890N gas chromatograph coupled to a 5973N mass spectrometer (Agilent Technologies). Separation was assured with a 30 m × 0.25-mm diameter with 0.25-μm film of HP-1ms (Agilent Technologies) or a 30 m × 0.25-mm diameter MEGADEX DETTBS fused silica column (Mega). Samples were injected using a cool on-column injector at 40°C and then increased by 10°C/min to 150°C, with a 2-min hold. The oven was programmed to start at 40°C and then increased by 10°C/min to 100°C, followed by 3°C/min to 280°C, with a 5-min hold. Electronic impact was recorded at 70 eV.
Purification of Z,Z-Farnesol
A three-liter culture of the E. coli strain expressing zFPS was grown. An in vitro reaction with the resulting recombinant zFPS and its substrates (IPP and DMAPP) was performed. Following dephosphorylation, ∼600 μg of the compound was recovered. The pentane extract containing the Z,Z-farnesol was loaded on HPLC (Agilent Technologies 1100 series equipped with a quaternary pump) using a reverse phase column (UP5-ODB 15QS, 150 × 2 mm; Interchrom). Solvents used for separation were water, acetonitrile, and a ternary blend of dichloromethane:hexane:methanol (66:33.5:0.5, v:v:v). Gradient program was as follows: 0 to 15 min from 60% acetonitrile in water to 100%; 15 to 18 min from 0 to 100% of ternary blend; 18 to 22 min from 100% of ternary blend; flow rate of 0.5 mL/min. The HPLC effluent was ionized in the atmospheric pressure chemical ionization source using a nitrogen nebulizing gas in the positive-ion mode. The atmospheric pressure chemical ionization vaporizer and capillary temperature were 300°C and 200°C, respectively. The instrument was operated to select the Z,Z-farnesol peak at a mass-to-charge ratio of 203 [M – 17]+, which was collected in a tube containing 1 mL of pentane. Due to the volatily of the dephosphorylated compound, the yield to concentrate the HPLC fractions was low and only 150 μg of pure product was obtained. However, it was sufficient for NMR analysis and structural elucidation.
NMR of Z,Z-Farnesol
Analyses were done on a Bruker Avance DRX500 spectrometer (1H-500.13 MHz) equipped with a 5 mm inverse cryoprobe TXI triple resonance (1H-13C-15N), with z gradient. Spectra were recorded with a 1.7 mm NMR capillary tube (150 μg of sample) in 50 μL of CDCl3 solvent (δ1H 7.26 ppm - δ13C 77.16 ppm) at 300° K. Pulse programs for all 1D/2D experiments (1H, 13C-DEPTQ, NOESY, HMQC) were taken from the Bruker standard software library.
Accession Numbers
Sequences data from this article can be found in GenBank under the following accession numbers: At-GA2 (Arabidopsis ent-kaurene synthase) AAC39443; Sr-KS (Stevia rebaudiana ent-kaurene synthase) AAD34295; Cm-KS-B (Cucurbita maxima ent-kaurene synthase) AAB39482; Nt-KSL (N. tabacum terpenoid cyclase, unknown function) AAS98912; Hv-KS (Hordeum vulgare ent-kaurene synthase-like protein 1) AAT49066; Os-KS (Oryza sativa cv japonica ent-kaurene synthase 1A) AAQ72559; Os-KS4i (O. sativa cv indica syn-pimara-7,15-diene synthase) AAU05906; Os-KSL7j (O. sativa cv japonica ent-cassadiene synthase) BAC56714; Os-KSL10j (O. sativa cv japonica ent-sandaracopipmaradiene synthase) ABH10735; Os-KSL11j (O. sativa cv japonica stemodene synthase) AAZ76733; Os-KSL8j (O. sativa cv japonica ent-kaurene synthase like protein) NP_001067887; Os-KSL5i (O. sativa cv indica isokaurene synthase) ABH10732; Os-KSL6j (O. sativa cv japonica isokaurene synthase) ABH10733; Mt-zUPPS (Mycobacterium tuberculosis Z-decaprenyl diphosphate synthase) NP_216877; Ec-zUPPS (E. coli undecaprenyl pyrophosphate synthetase) P60472; Mt-E/Z-FPS (M. tuberculosis short-chain Z-isoprenyl diphosphate synthetase) O53434; At-zIPPS (Arabidopsis dehydrodolichyl diphosphate synthase) O80458; Sc-zUPPS (Saccharomyces cerevisiae dehydrodolichyl diphosphate synthase) P35196; zFPS (S. habrochaites Z,Z-FPP synthase, this study, ACJ38408); SBS (S. habrochaites Santalene and bergamotene synthase, this study, ACJ38409).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SBS Homology with Proteins of the Kaurene Synthase-Like Family.
Supplemental Figure 2. Chromosome Localization of the SBS and zFPS Genes on the Tomato IL Map.
Supplemental Figure 3. PCR Polymorphism of the zFPS and SBS Genes.
Supplemental Figure 4. 1H/1H NMR 2D-NOESY Spectrum of Z,Z-Farnesol.
Supplemental Figure 5. GC-MS Comparison of a Leaf Exudate Extract from S. habrochaites and Santalum album Essential Oil.
Supplemental Figure 6. Quantification of SBS Transgene Expression in Tobacco.
Supplemental Table 1. Complete 1H and 13C NMR Assignments for Z,Z-Farnesol and Correlation Table (CDCl3).
Supplemental Table 2. Arabidopsis Genes and Proteins Presenting Similarity to Z-IPPS.
Supplemental Table 3. List of Plant Terpene Synthases Used to Generate the Phylogenetic Tree in Figure 2.
Supplemental Table 4. Complete List of Oligonucleotide Primers and Probes Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree in Figure 2.
Supplementary Material
Acknowledgments
We thank the Tomato Genetics Resource Center (University of California, Davis, CA) for providing tomato seeds, the Groupe de Recherche Appliquées en Phytotechnologie of the Institut de Biologie Environnementale et de Biotechnologie at the Commissariat à l'Energie Atomique Cadarache for assistance in plant cultivation, Gérard Audran for supplying a chiral GC column, Jean-Marc Adriano and David Pignol for assistance in protein preparation, Dominique Rumeau for gift of the At-βCA1:GFP fusion plasmid, and Skip Waechter for supplying nerylpyrophosphate. We also thank Ethan Signer for critical reading of the manuscript. This research was supported in part by the French Agence Nationale de la Recherche Biotech program Grant ANR05PRIB02101 to Librophyt.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alain Tissier (alain.tissier@librophyt.com).
Online version contains Web-only data.
References
- Adam, K.P., Thiel, R., and Zapp, J. (1999). Incorporation of 1-[1-(13)C]Deoxy-D-xylulose in chamomile sesquiterpenes. Arch. Biochem. Biophys. 369 127–132. [DOI] [PubMed] [Google Scholar]
- Adams, S.R., and Heinstein, P.F. (1973). Evidence for trans-trans and cis-cis farnesyl pyrophosphate synthesis in Gossypium hirsutum. Phytochemistry 12 2167–2172. [Google Scholar]
- Aharoni, A., Giri, A.P., Verstappen, F.W., Bertea, C.M., Sevenier, R., Sun, Z., Jongsma, M.A., Schwab, W., and Bouwmeester, H.J. (2004). Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16 3110–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asawatreratanakul, K., Zhang, Y.-W., Wititsuwannakul, D., Wititsuwannakul, R., Takahashi, S., Rattanapittayaporn, A., and Koyama, T. (2003). Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis. A key factor participating in natural rubber biosynthesis. Eur. J. Biochem. 270 4671–4680. [DOI] [PubMed] [Google Scholar]
- Benedict, C.R., Lu, J.-L., Pettigrew, D.W., Liu, J., Stipanovic, R.D., and Williams, H.J. (2001). The cyclization of farnesyl diphosphate and nerolidyl diphosphate by a purified recombinant δ-cadinene synthase. Plant Physiol. 125 1754–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick, J.A., and Lange, B.M. (2003). Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: Unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 415 146–154. [DOI] [PubMed] [Google Scholar]
- Bligh, E.G., and Dyer, W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37 911–917. [DOI] [PubMed] [Google Scholar]
- Bohlmann, J., Crock, J., Jetter, R., and Croteau, R. (1998). Terpenoid-based defenses in conifers: cDNA cloning, characterization, and functional expression of wound-inducible (E)-alpha-bisabolene synthase from grand fir (Abies grandis). Proc. Natl. Acad. Sci. USA 95 6756–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates, R.M., Denissen, J.F., Juvik, J.A., and Babka, B.A. (1988). Identification of alpha-santalenoic and endo-beta-bergamotenoic acids as moth oviposition stimulants from wild tomato leaves. J. Org. Chem. 53 2186–2192. [Google Scholar]
- Crick, D.C., Mahapatra, S., and Brennan, P.J. (2001). Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11 107R–118R. [DOI] [PubMed] [Google Scholar]
- Cunillera, N., Arro, M., Fores, O., Manzano, D., and Ferrer, A. (2000). Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett. 477 170–174. [DOI] [PubMed] [Google Scholar]
- Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., Boland, W., and Gershenzon, J. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 102 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., Martin, D., Kish, C.M., Kolosova, N., Gorenstein, N., Faldt, J., Miller, B., and Bohlmann, J. (2003). (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15 1227–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich, W., Schwarz, M., Cartayrade, A., Arigoni, D., Zenk, M.H., and Bacher, A. (1998). The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol. 5 R221–R233. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, N., Reiter, I.M., Becuwe-Linka, N., Genty, B., and Rumeau, D. (2007). Characterization and expression analysis of genes encoding α and β carbonic anhydrases in Arabidopsis. Plant Cell Environ. 30 617–629. [DOI] [PubMed] [Google Scholar]
- Frelichowski, J.E., and Juvik, J.A. (2001). Sesquiterpene carboxylic acids from a wild tomato species affect larval feeding behavior and survival of Helicoverpa zea and Spodoptera exigua (Lepidoptera: Noctuidae). J. Econ. Entomol. 94 1249–1259. [DOI] [PubMed] [Google Scholar]
- Fridman, E., Wang, J., Iijima, Y., Froehlich, J.E., Gang, D.R., Ohlrogge, J., and Pichersky, E. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffreda, J.C., Szymkowiak, E.J., Sussex, I.M., and Mutschler, M.A. (1990). Chimeric tomato plants show that aphid resistance and triacylglucose production are epidermal autonomous characters. Plant Cell 2 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinstein, P.F., Herman, D.L., Tove, S.B., and Smith, F.H. (1970). Biosynthesis of gossypol. Incorporation of mevalonate-2-14C and isoprenyl pyrophosphates. J. Biol. Chem. 245 4658–4665. [PubMed] [Google Scholar]
- Hosy, E., Duby, G., Véry, A.A., Costa, A., Sentenac, H., and Thibaud, J.B. (2005). A procedure for localisation and electrophysiological characterisation of ion channels heterologously expressed in a plant context. Plant Methods 1 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C.G., Ghisalberti, E.L., Plummer, J.A., and Barbour, E.L. (2006). Quantitative co-occurrence of sesquiterpenes; a tool for elucidating their biosynthesis in Indian sandalwood, Santalum album. Phytochemistry 67 2463–2468. [DOI] [PubMed] [Google Scholar]
- Juvik, J.A., Shapiro, J.A., Young, T.E., and Mutschler, M.A. (1994). Acylglucoses of the wild tomato Lycopersicon pennellii (Corr.) D'Arcy alter behavior and reduce growth and survival of Helicoverpa zea (Boddie) and Spodoptera exigua (Hubner). J. Econ. Entomol. 87 482–492. [Google Scholar]
- Kawamura, M., Ogasawara, K., and Saito, M. (1995). Diastereocontrolled and enantiocontrolled synthesis of sandalwood constituents (-)-beta-santalene and (+)-epi-beta-santalene starting from the same (+)-norcamphor. Tetrahedron Lett. 49 9003–9006. [Google Scholar]
- Kellog, B.A., and Poulter, C.D. (1997). Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol. 1 570–578. [DOI] [PubMed] [Google Scholar]
- Kharel, Y., and Koyama, T. (2003). Molecular analysis of cis-prenyl chain elongating enzymes. Nat. Prod. Rep. 20 111–118. [DOI] [PubMed] [Google Scholar]
- Kharel, Y., Takahashi, S., Yamashita, S., and Koyama, T. (2006). Manipulation of prenyl chain length determination mechanism of cis-prenyltransferases. FEBS J. 273 647–657. [DOI] [PubMed] [Google Scholar]
- Koyama, T. (1999). Molecular analysis of prenyl chain elongation enzymes. Biosci. Biotechnol. Biochem. 63 1671–1676. [DOI] [PubMed] [Google Scholar]
- Kuzuyama, T. (2002). Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotechnol. Biochem. 66 1619–1627. [DOI] [PubMed] [Google Scholar]
- MacMillan, J., and Beale, M.H. (1999). Diterpene biosynthesis. In Comprehensive Natural Products Chemistry No. 2, Isoprenoid Including Carotenoids and Steroids, S.D. Barton and K. Nakanishi, eds (Oxford, UK: Elsevier Science), pp. 217–243.
- Martin, D.M., Fäldt, J., and Bohlmann, J. (2004). Functional characterization of nine norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 135 1908–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monforte, A.J., and Tanksley, S.D. (2000). Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in an L. esculentum background: A tool for gene mapping and gene discovery. Genome 43 803–813. [PubMed] [Google Scholar]
- Nagegowda, D.A., Gutensohn, M., Wilkerson, C.G., and Dudareva, N. (2008). Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 55 224–239. [DOI] [PubMed] [Google Scholar]
- Oh, S.K., Han, K.H., Ryu, S.B., and Kang, H. (2000). Molecular cloning, expression, and functional analysis of a cis-prenyltransferase from Arabidopsis thaliana. Implications in rubber biosynthesis. J. Biol. Chem. 275 18482–18488. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Liu, Y.-S., Budai-Hadrian, O., Sela, M., Carmel-Goren, L., Zamir, D., and Fluhr, R. (2000). Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rontein, D., Onillon, S., Herbette, G., Lesot, A., Werck-Reichhart, D., Sallaud, C., and Tissier, A. (2008). CYP725A4 from yew catalyzes complex structural rearrangement of taxa-4(5),11(12)-diene into the cyclic ether 5(12)-oxa-3(11)-cyclotaxane. J. Biol. Chem. 283 6067–6075. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 34 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba-Colombani, V., Causse, M., Gervais, L., and Philouze, J. (2000). Efficiency of AFLP, RAPD and RFLP markers for the construction of an intraspecific map of the tomato genome. Genome 43 29–40. [PubMed] [Google Scholar]
- Sallaud, C., et al. (2003). Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor. Appl. Genet. 106 1396–1408. [DOI] [PubMed] [Google Scholar]
- Sanmiya, K., Ueno, O., Matsuoka, M., and Yamamoto, N. (1999). Localization of farnesyl diphosphate synthase in chloroplasts. Plant Cell Physiol. 40 348–354. [DOI] [PubMed] [Google Scholar]
- Sato, M., Sato, K., Nishikawa, S.-I., Hirata, A., Kato, J.-I., and Nakano, A. (1999). The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes a cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulbach, M.C., Brennan, P.J., and Crick, D.C. (2000). Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J. Biol. Chem. 275 22876–22881. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl, D. (2006). Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 9 297–304. [DOI] [PubMed] [Google Scholar]
- van Der Hoeven, R.S., Monforte, A.J., Breeden, D., Tanksley, S.D., and Steffens, J.C. (2000). Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell 12 2283–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret, F., Gravot, A., Auroy, P., Leonhardt, N., David, P., Nussaume, L., Vavasseur, A., and Richaud, P. (2004). Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 576 306–312. [DOI] [PubMed] [Google Scholar]
- Wang, E., and Wagner, G.J. (2003). Elucidation of the functions of genes central to diterpene metabolism in tobacco trichomes using posttranscriptional gene silencing. Planta 216 686–691. [DOI] [PubMed] [Google Scholar]
- Wendt, K.U., and Schulz, D.E. (1998). Isoprenoid biosynthesis: Manifold chemistry catalyzed by similar enzymes. Structure 6 127–133. [DOI] [PubMed] [Google Scholar]
- Williams, W.G., Kennedy, G.G., Yamamoto, R.T., Thacker, J.D., and Bordner, J. (1980). 2-Tridecanone: A naturally occurring insecticide from the wild tomato Lycopersicon hirsutum f. glabratum. Science 207 888–889. [DOI] [PubMed] [Google Scholar]
- Wise, M.L., Savage, T.J., Katahira, E., and Croteau, R. (1998). Monoterpene synthases from common sage (Salvia officinalis). cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1,8-cineole synthase, and (+)-bornyl diphosphate synthase. J. Biol. Chem. 273 14891–14899. [DOI] [PubMed] [Google Scholar]
- Young, N.D., Zamir, D., Ganal, M.W., and Tanksley, S.D. (1988). Use of isogenic lines and simultaneous probing to identify DNA markers tightly linked to the Tm-2a gene in tomato. Genetics 120 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R., Moriau, L., and Boutry, M. (1999). Expression analysis of the plasma membrane H+-ATPase pma4 transcription promoter from Nicotiana plumbaginifolia activated by the CaMV 35S promoter enhancer. Plant Sci. 149 157–165. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.