Abstract
Infection thread–dependent invasion of legume roots by rhizobia leads to internalization of bacteria into the plant cells, which is one of the salient features of root nodule symbiosis. We found that two genes, Nap1 (for Nck-associated protein 1) and Pir1 (for 121F-specific p53 inducible RNA), involved in actin rearrangements were essential for infection thread formation and colonization of Lotus japonicus roots by its natural microsymbiont, Mesorhizobium loti. nap1 and pir1 mutants developed an excess of uncolonized nodule primordia, indicating that these two genes were not essential for the initiation of nodule organogenesis per se. However, both the formation and subsequent progression of infection threads into the root cortex were significantly impaired in these mutants. We demonstrate that these infection defects were due to disturbed actin cytoskeleton organization. Short root hairs of the mutants had mostly transverse or web-like actin filaments, while bundles of actin filaments in wild-type root hairs were predominantly longitudinal. Corroborating these observations, temporal and spatial differences in actin filament organization between wild-type and mutant root hairs were also observed after Nod factor treatment, while calcium influx and spiking appeared unperturbed. Together with various effects on plant growth and seed formation, the nap1 and pir1 alleles also conferred a characteristic distorted trichome phenotype, suggesting a more general role for Nap1 and Pir1 in processes establishing cell polarity or polar growth in L. japonicus.
INTRODUCTION
Like in many legume species, invasion of lotus (Lotus japonicus) roots by Mesorhizobium loti occurs via root hairs. The rhizobial bacteria gain access to the interior of the root and, subsequently, to a nodule primordium located within the underlying root cortex through tubular structures called infection threads (ITs). ITs are initiated from infection pockets, rhizobial microcolonies that have been entrapped within a shepherds-crook structure of curled root hairs (Schauser et al., 1998, 1999; Szczyglowski et al., 1998; Esseling et al., 2003). Upon formation of infection pockets, the root hair cell wall dissolves and an IT is initiated by invagination and subsequent polar extension of the plasma membrane, which is accompanied by the deposition of new cell wall material (for a review, see Gage, 2004). Inward-growing ITs progress through the root hair and ramify by a cell autonomous mechanism within cells of the growing nodule primordia. The emergent IT network acts as conduit to deliver the bacteria to a subset of these cells. Eventually, an endocytotic release of bacteria from ITs into the plant cytoplasm of this subset establishes the infected cells containing bacteria in membrane bound symbiosome organelles. These cells enlarge and become the nitrogen-fixing cells of legume root nodules.
Several classes of L. japonicus mutants arrested in either the early or the late stages of the infection process have been isolated and characterized (Kistner et al., 2005; Krusell et al., 2005; Kumagai et al., 2007). Perception of the bacterially produced Nod factor signal is mediated by the Lys motif (LysM)–containing NOD FACTOR RECEPTORS, NFR1 and NFR5, and both receptors are required for the host plant to initiate infection and nodule organogenesis (Madsen et al., 2003; Radutoiu et al., 2003, 2007). The pathway shared with mycorrhizal symbiosis is involved in Nod factor signal transduction downstream from the receptors (Kistner et al., 2005; Yano et al., 2006). In the model legumes lotus and Medicago truncatula, an overlapping set of gene products contribute to the shared pathway (Kistner et al., 2005; Oldroyd and Downie 2008). The Leu-rich repeat Lj SYMBIOSIS RECEPTOR-LIKE KINASE/Mt DOES NOT MAKE INFECTIONS2 (DMI2) (Endre et al., 2002; Stracke et al., 2002), along with the predicted cation channel(s) Lj CASTOR and Lj POLLUX/Mt DMI1 (Ané et al., 2004; Imaizumi-Anraku et al., 2005) and the nucleoporins Lj NUP133 and Lj NUP85 (Kanamori et al., 2006; Saito et al., 2007), are all required for induction of calcium spiking, a rapid physiological response that is detectable in root hairs of the susceptible host plant within 5 to 10 min after Nod factor application (Ehrhardt et al., 1996; Miwa et al., 2006). The calcium spiking is believed to be interpreted by a calcium calmodulin-dependent kinase, CCAMK, which acts together with the CYCLOPS protein to mediate downstream responses (Lévy et al., 2004; Mitra et al., 2004; Yano et al., 2008; Tirichine et al., 2006). Downstream from the shared pathway, the putative transcription regulators NODULE INCEPTION (NIN), NODULATION SIGNALING PATHWAY1 (NSP1), and NSP2 specifically mediate bacterial infection at the root epidermis and nodule organogenesis in the root cortex (Schauser et al., 1999; Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006; Marsh et al., 2007).
In addition to Ca2+ spiking, Nod factor signaling elicits other epidermal responses in compatible host roots, such as membrane depolarization, ion fluxes across the membrane, and cytoskeleton rearrangements (Allen et al., 1994; Cárdenas et al., 1998; Felle et al., 1998; Miller et al., 2000; Weerasinghe et al., 2005). In fact, one of the first cellular changes observed in root hairs responding to rhizobial inoculation or Nod factor application is an alteration in actin and microtubule organization (Cárdenas et al., 1998; de Ruijter et al., 1999; Weerasinghe et al., 2003, 2005; Vassileva et al., 2005).
Treatment of bean (Phaseolus vulgaris), alfalfa (Medicago sativa), common vetch (Vicia sativa), or lotus root hairs with Nod factor leads to rapid, within 3 to 5 min, changes in the polymerization pattern of actin filaments (Allen et al., 1994; Cárdenas et al., 1998; Weerasinghe et al., 2005). The long actin bundles extending into the root hair apical tips were observed to undergo fragmentation, and fine bundles of filaments accumulated in the apical/subapical region of the responding root hairs (Cárdenas et al., 1998; de Ruijter et al., 1999; Weerasinghe et al., 2005). These fine bundles of actin filaments were proposed to originate from fragmentation or unbundling of thick actin filaments or from new nucleation and polymerization (Cárdenas et al., 1998; de Ruijter et al., 1999). Similarly, significant changes in the dynamic behavior of cortical and endoplasmic microtubules preceding nodule development and root hair curling were reported. These were found to be associated with all early steps during symbiotic interaction, including preinfection thread formation, initiation and polar growth of ITs, as well as the activation of root pericycle and cortical cells that initiates nodule primordia organogenesis (van Spronsen et al., 1995; Timmers et al., 1999; Vassileva et al., 2005). Given a well-recognized role of the cytoskeleton in mediating cell divisions and cell growth, including directionality and rate of cell expansion (Bannigan and Baskin, 2005), it was proposed that the observed Nod factor–dependent reorganization of microtubules and actin filaments might be a prerequisite for successful symbiotic interaction (van Spronsen et al., 1995; Timmers et al., 1999; Vassileva et al., 2005). However, until now, there has been no genetic evidence to support this notion and none of the components regulating the process have been identified.
Although all early root responses to bacterial signaling, including formation of nodule primordia and ITs, are either drastically impaired or absent in the receptor, shared pathway, and the transcriptional regulator mutants (Madsen et al., 2003; Radutoiu et al., 2003; Kistner et al., 2005; Heckmann et al., 2006; Murakami et al., 2006), none of the underlying genes have been directly linked to regulatory components that are known to participate in cytoskeleton rearrangements. Interestingly, a class of mutants characterized by a lack of or impairment in IT progression and a concomitant formation of uncolonized empty nodules was identified (Schauser et al., 1998; Lombardo et al., 2006; Murray et al., 2006; Yano et al., 2006). The defective phenotypes typically observed in these mutant lines were characterized by ITs that were arrested either within root hairs/epidermis or within the first cortical cell layers. Furthermore, mutant lines were also identified in which IT formation was not followed by the release of bacteria into cells of the nodule primordium (Imaizumi-Anraku et al., 1997).
By surveying mutant lines with a defective infection process, we have selected a subclass, which in addition to aberrant infections had distorted trichomes, a phenotype often associated with defects in endomembrane, microtubules, or actin-dependent morphogenesis (Szymanski, 2005). We demonstrate here that the deleterious mutations in two L. japonicus genes, Nap1 (for Nck-associated protein 1; see Eden et al., 2002) and Pir1 (for 121F-specific p53 inducible RNA; see Saller et al., 1999), were responsible for the observed aberrant trichome phenotype. Furthermore, we show that the actin rearrangement preceding root hair deformation, root hair curling, and IT initiation, as mediated by these genes, is essential for efficient root invasion by the symbiotic bacteria and also for the subsequent controlled enlargement of infected cells.
RESULTS
Nodulation Phenotype of nap1 and pir1 Mutants
Screening of mutagenized lotus populations has identified an unusual class of mutants showing both aberrant symbiotic and trichome phenotypes (Tansengco et al., 2003; Ooki et al., 2005; this article). We have characterized the nap1 and pir1 mutants that belong to this category and cloned the corresponding genes that encode proteins that are most likely involved in the assembly of F-actin filaments (see below). Eight monogenic recessive mutant alleles defined below as nap1-1, nap1-2, and nap1-3, and pir1-1, pir1-2, pir1-3, pir1-4, and pir1-5 (Table 1),corresponding to two lotus loci, Nap1 and Pir1, respectively, were found in three independent screens for symbiotic mutants. nap1-1 and pir1-1 were isolated from a mutant population that was originally established to identify transposon-tagged mutants (Thykjær et al., 1995; Schauser et al., 1998), while the remaining alleles were found in two independent ethyl methanesulfonate (EMS) populations (Murray et al., 2006; Yano et al., 2006).
Table 1.
Lotus pir and nap Mutant Alleles
| Allele (Previous Name) | Mutation | Reading Frame Change |
|---|---|---|
| pir1-1 (sym40) | 414-bp deletion (position 12847–13261) | IT945-946 → VV L947→ stop |
| pir1-2 (S14-3) | C6934 → T | Q470 → stop |
| pir1-3 (sym80) | C11514 → T | Q854 → stop |
| pir1-4 (S57-F) | C3123 → T | Q136 → stop |
| pir1-5 (B31-C) | G11051 → A | Splice site mutation |
| nap1-1 (sym67) | LORE-1 insertion in exon 18 (position 17216) | Insertion of 15 amino acids after P692 → premature stop |
| nap1-2 (S12-5A) | 3574-bp deletion, affecting exons 2 to 5, (position 3393–6966) | Possibly cryptic start at M205 |
| nap1-3 (S90-D) | C6473 → T | Q177 → stop |
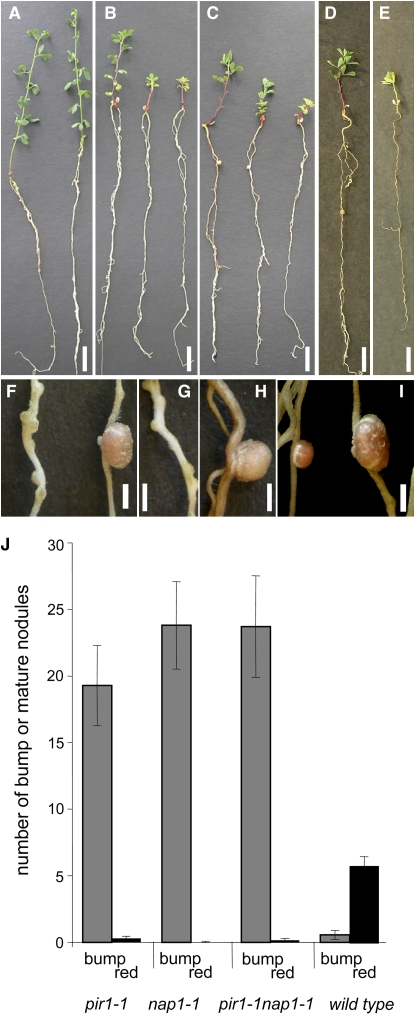
nap1 and pir1 mutants were identified as small nitrogen-starved plants when grown on nitrogen-deficient nutrient medium. As exemplified by nap1-1 and pir1-1 (Figure 1),all mutant alleles identified had a comparable effect on nodulation and all mutants show the red stem pigmentation characteristic for nitrogen-starved Lotus plants. Inoculation of plants carrying any of the above-mentioned alleles with M. loti triggered the formation of supernumerary small white nodules, hereafter called bumps. This contrasted significantly with the symbiotic phenotype of the corresponding wild-type plants (Figures 1A to 1E and 1J), which developed on average five pink nodules at the comparable time point after inoculation with M. loti (Figure 1J). In six-week experiments, nap1-1 and pir1-1 mutants developed an average of 23 and 19 bumps per plant, respectively. In nap1-1, one pink, wild-type-looking nodule, developed per 881 bumps, while in pir1-1 this frequency was one pink nodule per 69 bumps (Figure 1J). Compared with the wild type, the rare pink nodules that formed on nap1-1 and pir1-1 roots were grossly enlarged (Figures 1F to 1I). Greenhouse-grown nap1-1 and pir1-1 mutants were also severely nitrogen starved, and, on average, one small pink nodule was observed on 25% of the plants cultivated for an extended period of 11 weeks. Thus, compared with the wild type, a drastic reduction in the frequency of fully developed nodules was evident in nap1 and pir1 mutants regardless of the growth conditions tested (Figure 1J).
Figure 1.
Whole Plant and Nodulation Phenotype of L. japonicus Wild-Type, pir1-1, nap1-1, and pir1-1 nap1-1 Mutant Plants.
(A) Wild-type L. japonicus.
(B) pir1-1.
(C) nap1-1.
(D) and (E) pir1-1 nap1-1 double mutant.
(F) pir1-1 Fix− bump (left) and one of the rare oversized red nitrogen-fixing nodules (right).
(G) nap1-1 Fix− nodules.
(H) One of the rare nap1-1 oversized red nitrogen-fixing nodules.
(I) Size comparison between L. japonicus wild-type (left) and an oversized pir1-1 nodule (right).
Bars = 1 cm in (A) to (E) and 2 mm in (F) to (I).
(J) Nodule numbers on 6-week-old nap1-1, pir1-1, pir1-1 nap1-1, and wild-type plants inoculated with M. loti NZP2235. Black columns: mature, red nodules (red); gray columns: white immature, Fix− nodules (bump). Bars indicate 95% confidence intervals.
Microscopy of Nodules from nap1 and pir1 Mutants
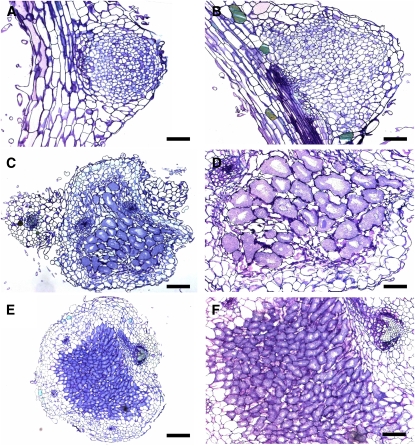
The bumps that developed on nap1 and pir1 roots were further characterized by microscopy (Figure 2). Light microscopy showed that mutant nodules consisted of uniformly sized cells, an anatomy characteristic of developmentally arrested empty nodules (Figures 2A and 2B). Large infected cells or ITs were absent, which was further confirmed using electron microscopy (data not shown). Furthermore, while infected cells were not observed, the presence of bacteria in the intracellular spaces was occasionally detected. By contrast, enlarged infected cells were clearly visible in sections of wild-type nodules (Figures 2E and 2F). Microscopy of large pink nodules that occasionally developed on nap1-1 and pir1-1 roots showed that bacteria were successfully endocytosed into the nodule cells. However, the infected cells of the nap1-1 and pir1-1 mutants differed from the corresponding cells of wild-type nodules. They appeared enlarged and more irregular (cf. Figures 2C and 2D to 2E and 2F). Furthermore, they also showed enlarged vacuoles (Figure 2D). These larger than normal infected cells were independently observed in pink nodules of pir1-3 mutants (Yano et al., 2006).
Figure 2.
Light Microscopy of Nodule Sections.
(A) Thin section of pir1-1 noninfected white bump.
(B) Close-up of nap1-1 noninfected bump.
(C) Thin section of large, red, infected nap1-1 nodule.
(D) Close-up of bacteroid-containing cells in large, red nap1-1 nodule. Note the enlarged size of the cells.
(E) Wild-type nodule.
(F) Wild-type nodule; close-up of the bacteroid-containing cells.
Bars = 100 μm in (A), (C), and (E) and 50 μm in (B), (D), and (F). [See online article for color version of this figure.]
Root Hair Response and IT Formation in nap1 and pir1 Mutants
The preponderance of empty nodules formed on nap1 and pir1 roots suggested that the infection process might be perturbed in these mutants. To investigate this possibility, we examined nap1-1, nap1-2, and pir1-1 roots following inoculation with a M. loti strain constitutively expressing a β-galactosidase (lacZ) reporter gene or a green fluorescent protein (GFP) reporter (Figure 3).
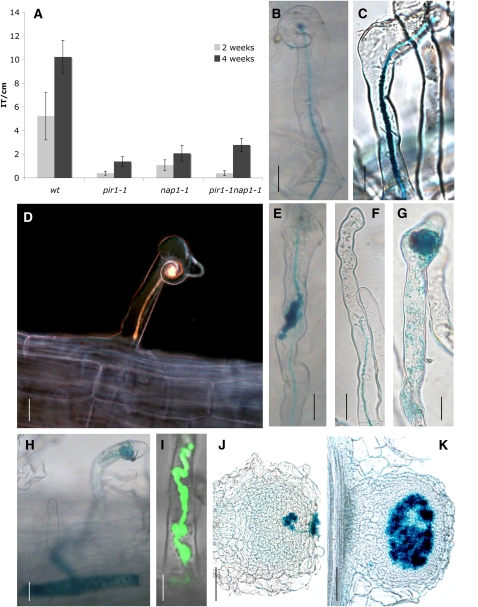
Figure 3.
IT Formation in Wild-Type, pir1-1, nap1-1, nap1-2, and pir1-1 nap1-1 Mutants.
(A) Number of ITs per centimeter in wild-type, pir1-1, nap1-1, and pir1-1 nap1-1. The roots were harvested at 2 and 4 weeks after inoculation with rhizobia. The values are presented with 95% confidence intervals.
(B) IT (blue color) in wild-type plant 2 weeks after inoculation with M. loti expressing lacZ. The root hair is curling in response to rhizobia, and the IT develops reaching the base of the root hair.
(C) IT in the nap1-1 mutant, wild-type-like.
(D) nap1-2 IT stopping in the first epidermis cell.
(E) IT in nap1-1 with inflated sac-like structure on the IT containing rhizobia.
(F) to (I) Various stages of IT damage, from partially burst IT releasing rhizobia into root hair to totally disrupted ITs. pir1-1 nap1-1 double mutant (F), nap1-1 ([G] and [H]), and inflated sac-like structure on a nap1-1 IT and release of rhizobia expressing GFP (I).
(J) Colonization attempts from rhizobial patch on a nap1-2 bump.
(K) Wild-type nodule showing cells colonized with rhizobia.
For (B) to (H), (J), and (K), inoculation was with an M. loti–expressing lacZ. Bars = 20 μm, except for (J) and (K), where bars = 100 μm.
When examined 2 and 4 weeks after inoculation, a significantly decreased number of ITs was observed in all mutants tested compared with wild-type roots (Figure 3A). Scanning the whole root 4 weeks after inoculation, we detected ∼10 ITs on average per centimeter of wild-type roots, while only two or fewer ITs were observed per centimeter of the nap1-1 and pir1-1 roots (Figure 3A). A similarly reduced level of ITs was found on roots of the mutants 6 weeks after inoculation, supporting the notion that the IT formation was significantly impaired rather than delayed.
Like the wild type, nap1 and pir1 mutants were capable of forming ITs extending through the root hair (Figures 3B and 3C). Nevertheless, wild-type-looking ITs were very rarely observed. Instead, deformed ITs containing inflated sac–like structures filled with rhizobia were frequently found (Figures 3E and 3I). In many cases, enlarged infection pockets with no or only very short ITs were formed (Figure 3G). Some ITs appeared to burst, releasing rhizobia into the root hair cell (Figures 3F, 3H, and 3I). In contrast with wild-type plants, all ITs that formed on nap1-1, nap1-2, and pir1-1 roots were arrested either within the root hairs or within the base of the corresponding epidermal cells (Figure 3D).
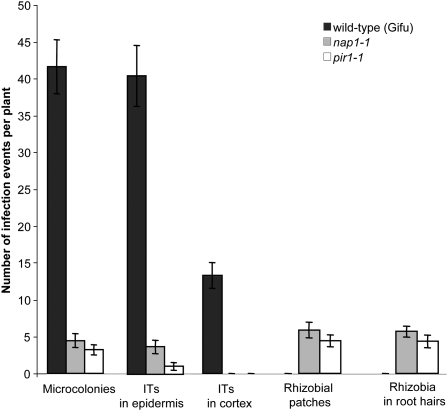
To gain further insight into the role of Nap1 and Pir1 in the infection process, the ability of the mutant plants to respond to inoculation with rhizobia by root hair curling and the formation of infection pockets in the root zone susceptible for bacterial invasion was investigated. Root hair curling and subsequent infections were scattered along the mutant roots, in comparison with wild-type controls in which curling and infection were localized to the invasion zone. In addition, a detailed analysis of the early infection events in the pir1-1, nap1-1, and nap1-2 mutants 10 d after inoculation with M. loti clearly demonstrated that all phases, including formation of infection pockets and initiation and subsequent progression of ITs into the root epidermal base, were significantly restricted in all three mutants and that this was correlated with the appearance of rhizobia within some root hairs in the absence of ITs. The results for pir1-1 and nap1-1 are shown in Figure 4.
Figure 4.
Early Infection Events in lotus Wild-Type, nap1-1, and pir1-1 Mutants.
Microcolonies and ITs were scored 10 d after inoculation with the M. loti strain expressing lacZ. The ITs were grouped into two categories: those present only within the epidermis (ITs in epidermis) and those that managed to penetrate the root cortex (ITs in cortex). In addition, the presence of M. loti at the surface of nodule primordia (rhizobial patches), or within root hairs in the absence of accompanying IT structures, were quantified and compared between wild-type and mutant plants. Mean values ± 95% confidence intervals are given for each genotype and category (n = 20).
Later in the developmental process, patches of rhizobia were visible on top of the numerous empty nodules that formed on the mutant roots, from which colonization attempts similar to those observed in L. japonicus root-hairless mutant background (Karas et al., 2005) were occasionally launched (Figure 3J).
Cloning of the Nap1 Gene
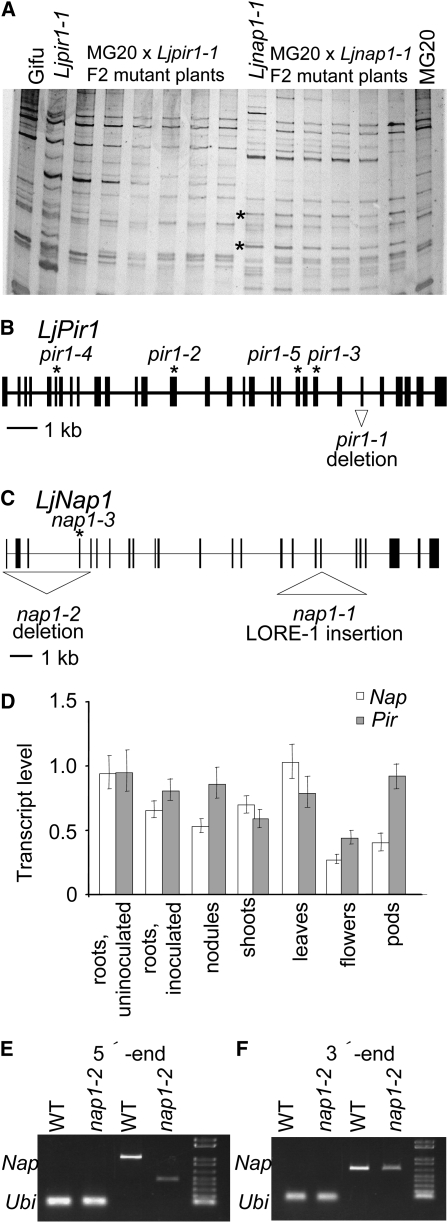
Positional cloning was initiated to identify and characterize the Nap1 gene. The Nap1 locus was mapped to the L. japonicus linkage group 4 flanked by the TM0227 and TM0347 markers (Sandal et al., 2006). Fine mapping and subsequent genotyping of 1077 nap1-1 mutant plants identified markers TM1846 and TM0229, which delimited the Nap1 locus to a 0.4-centimorgan (cM) region, as defined by 12 and 6 recombinations on each side, respectively (see Supplemental Figure 1 online).
In a parallel approach, recombinant plants delimiting the Nap1 and Pir1 loci were tested for insertion of the LORE1 and LORE2 retroelements previously found to be active in lotus (Madsen et al., 2005; Fukai et al., 2008). The sequence-specific amplification polymorphism (SSAP) technique for amplifying genomic DNA flanking the retrotransposon's long terminal repeats (Madsen et al., 2005) was used to investigate whether the nap1-1 or pir1-1 alleles were caused by insertion of LORE1 or LORE2 (see also below). Two LORE1 SSAP fragments were specifically amplified from five nap1-1 individuals carrying recombination end points defining the Nap1 locus (Figure 5A). Sequencing of the two SSAP fragments produced the same stretch of 140 nucleotides, which was identical to a sequence in exon 18 of the Nap1 gene found by searching the lotus genome sequence. Subsequent mapping of a Nap1 gene–specific marker showed that the nap1-1 LORE1 insertion and the mutant phenotype cosegregated in the 1077 mutant plants derived from the F2 mapping population. The nap1-2 allele was located within a 0.4-cM segment of linkage group 4 flanked by the TM1846 and TM0229 markers (see Supplemental Figure 1 online), which overlapped the LORE1 insertion site in nap1-1. Cloning of the Nap1 locus was further confirmed by the identification of a large 3.6-kb deletion spanning the first four exons of this gene in the nap1-2 mutant (Figure 5C) and a premature stop codon in exon 3 of the nap1-3 allele (Table 1).
Figure 5.
Cloning, Gene Structure, and Expression Patterns of Nap1 and Pir1 Genes.
(A) SSAP detection of LORE-1 element integrations in pir1-1 and nap1-1 mutant plants compared with parental lines L. japonicus Gifu and Miyakojima MG20. Asterisk indicates new integration in the nap1-1 allele. Both bands represent the same integration event.
(B) Pir1 gene structure. Positions of mutations in the pir1-1, pir1-2, pir1-3, pir1-4, and pir1-5 alleles are indicated.
(C) Nap1 gene structure. Positions of the mutations in the nap1-1, nap1-2, and nap1-3 alleles are indicated. In (B) and (C), filled rectangles indicate exons, and thin lines indicate introns.
(D) Relative levels of Nap1 and Pir1 mRNA in different wild-type lotus organs. The level in uninoculated roots is set to 1 for each gene. Bars represent 95% confidence intervals.
(E) and (F) RT-PCR detection of 5′- and 3′-ends of the Nap1 transcript in leaves of the nap1-2 mutant and wild-type plants. In each panel: lanes 1 and 2 are wild-type and nap1-2 ubiquitin controls; lane 3 and 4 are wild-type and nap1-2 Nap1 transcripts; lane 5 is the size marker.
Sequencing of a full-length cDNA determined the transcription start site at least 58 bp upstream from the predicted start codon and a 3′ untranslated region of 236 or 440 nucleotides in length, corresponding to at least two alternative polyA addition sites (Figure 5C). Alignment of genomic and cDNA sequences defined 24 exons in the Nap1 gene, 23 of which made up the coding region, while one noncoding exon was in the predicted 5′ untranslated region (Figure 5C). The fifth intron of Nap1 contained a rare noncanonical splice donor-acceptor pair, GC-AG. In Arabidopsis thaliana, GC-AG donor-acceptor sequences were found in 0.8% of the introns (Sheth et al., 2006).
The lotus Nap1 cDNA was shown to encode a conceptual protein of 1395 amino acids, corresponding to ∼156 kD. In the fully sequenced Arabidopsis and rice (Oryza sativa) genomes, Nap1 homologs are single genes in each species. The predicted Arabidopsis (NP_181056) and rice (NP_001062406) proteins are 77 and 69% identical to lotus NAP1, respectively (see Supplemental Figure 2 online). Less similar, with 22% global identity, is the human NAP protein. To our knowledge, no function or structure has been assigned to any particular domains of animal-, protist-, or plant-derived NAP proteins.
Cloning of the Pir1 Gene
The Pir locus was positioned within 4 cM on linkage group 1 flanked by TM0109 and TM0105 markers (Sandal et al., 2006). Additional fine mapping of an F2 population by genotyping of 822 pir1-1 mutant plants delimited the Pir locus to a 0.12-cM region (see Supplemental Figure 3 online). In contrast with nap1-1, the pir1-1 allele was not generated by the insertion of LORE1 or LORE2 (Figure 5A); therefore, a direct, candidate gene approach was used. The BAC and TAC genome sequences covering the Pir locus, as defined by recombination end points, were searched for gene content (see Supplemental Figure 3 online), and a gene encoding a presumed lotus PIR component of the SCAR/WAVE (suppressor of cAMP receptor defect/WASP family verpolin homologous protein) complex, which is known to participate in the regulation of actin cytoskeleton (see below), stood out as a likely candidate. Amplification and sequencing of this gene from the pir1-1 mutant revealed a short 414-nucleotide deletion encompassing exon 25 (Table 1, Figure 5B). Subsequent mapping of a Pir gene–specific marker showed that the pir1-1 deletion and the mutant phenotype cosegregated in the 822 mutant plants. Four independent mapping projects positioned pir1-2, pir1-3, pir1-4, and pir1-5 within an ∼5-cM region overlapping the Pir locus on linkage group 1 (see Supplemental Figure 3 online). In addition to the similar trichome and symbiotic phenotypes, this genetic evidence suggested that they were allelic to pir1-1. Subsequent sequencing of these alleles identified stop codons in exon 14 of pir1-2, exon 23 of pir1-3, exon 6 of pir1-4, and a donor splice site mutation in pir1-5 (Table 1, Figure 5B). Genotyping of these additional alleles confirmed the mapping and identification of the Pir1 gene.
Sequencing of full-length cDNAs determined the transcription start site at least 99 bp upstream of the start codon and a 3′ untranslated region of 232 nucleotides (Figure 5B). Alignment of genomic and cDNA sequences defined 30 exons in Pir (Figure 5B). The twelfth intron of Pir1 contains a highly unusual splice donor-acceptor pair, AT-AC, that is conserved in intron 12 of the Arabidopsis Pir gene (Basu et al., 2004). The donor sites of these unusual introns contain a highly conserved ATATCCTT sequence (Wu and Krainer, 1999) that is also conserved in the lotus and Arabidopsis Pir genes.
The lotus Pir1 cDNA encodes a conceptual protein of 1277 amino acids, corresponding to ∼144.9 kD. In the fully sequenced Arabidopsis and rice genomes, Pir1 homologs are single genes in each species, and their predicted proteins are 83 and 68% identical to the predicted lotus PIR1 protein, respectively (see Supplemental Figure 4 online). Less similar, with 30% overall identity, is the human PIR protein (NP_055191). To our knowledge, no function has been assigned to any particular domains of animal, protist, or plant PIR proteins, and domains involved in Rac GTP interaction have not yet been defined.
Expression of the Nap1 and Pir1 Genes
The mutant symbiotic phenotypes suggest a function for Pir1 and Nap1 genes in root and shoot tissues. To test this prediction, their expressions in different lotus organs were determined by quantitative RT-PCR analysis. Nap1 and Pir1 were expressed in roots and in all other organs tested (Figure 5D). No significant changes in the steady state level of Pir1 and Nap1 mRNA were detected in roots after inoculation with M. loti. Thus, both Nap1 and Pir1 appear to be ubiquitously expressed in lotus.
The effect of the 3.6-kb deletion in the nap1-2 allele on the transcript was also investigated. The products of RT-PCR, using cDNA templates from wild-type and nap1-2 leaves, were compared. A reduced size of Nap1 mRNA was detected using the RT-PCR amplification of the 5′ region across the deletion boundaries. In the 3′ region, the amplification across exon 28 produced products of similar size in nap1-2 compared with the wild-type control (Figures 5E and 5F). The sequencing and subsequent analysis of the shorter, 5′-derived product predicted an open reading frame that could give raise to a truncated NAP1 protein, with the Met205 codon as a potential new translational start site.
The Trichome, Root Hair, and Growth Phenotypes of nap1 and pir1 Mutants
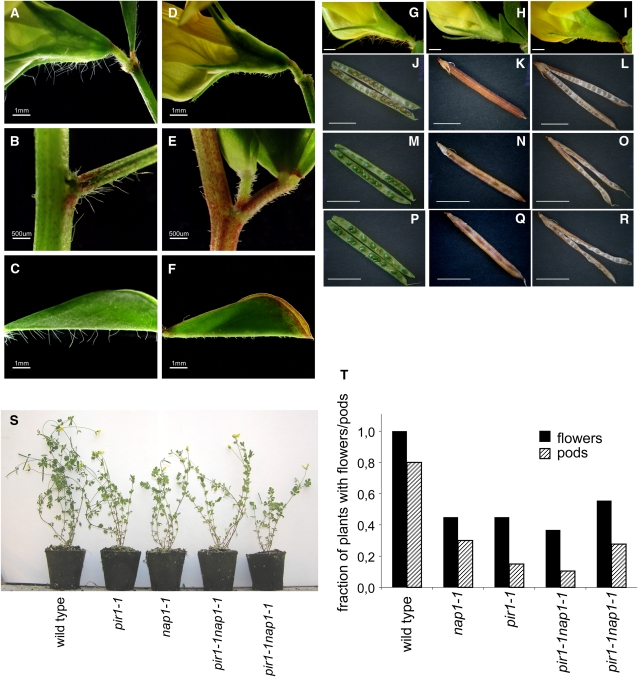
As indicated above, one of the common phenotypic characteristics of lotus plants carrying any of the nap1 or pir1 mutant alleles was the arrested trichome development. The elongated, filamentous trichomes were visible on the sepals, the abaxial midribs of leaves, and leaf stalks of wild-type plants (Figures 6A to 6C and 6G).By contrast, trichomes formed on nap1 and pir1 mutants were distinctively shorter and deformed (Figures 6D to 6F, 6H, and 6I).
Figure 6.
Trichome and Growth Phenotype of L. japonicus Wild-Type Gifu, nap1-1, pir1-1, and pir1-1 nap1-1 Double Mutant Plants.
(A) to (F) Trichomes on the wild type ([A] to [C]) and the nap1-2 mutant ([D] to [F]). Images of trichomes formed on fully expanded flowers ([A] and [D]), stem internodes ([B] and [E]), and the abaxial surface of the leaf midvein ([C] and [F]) are shown. Bars = 1 mm in (A), (C), (D), and (F) and 500 μm in (B) and (E).
(G) to (I) Flower of wild-type (G), pir1-1 (H), and nap1-1 (I) showing the trichome phenotype of the sepals. Bars = 5 mm.
(J) to (R) Pod and seed phenotype of wild-type ([J] to [L]), pir1-1 ([M] to [O]), and nap1-1 ([P] to [R]). Bars = 1 cm.
(S) and (T) Shoot growth phenotype and developmental status of 9-week-old soil-grown wild-type, pir1-1, nap1-1, and pir1-1 nap1-1 plants. The fractions of plants that reached the stages of flowering (black bars) and pod development (cross hatched bars) are shown. Results are shown for two different pir1-1 nap1-1 lines.
A distorted trichome phenotype has been described in Arabidopsis nap and pir mutants, and this was correlated with a disorganized structure of F-actin bundles (Li et al., 2004). In spite of several independent attempts, we were unable to visualize actin filaments in lotus trichomes using Alexa-phalloidin staining, a method successfully applied to Arabidopsis. Perhaps cell walls of lotus trichomes are thicker or less penetrable to the stain used than those in Arabidopsis trichomes. Nevertheless, the altered trichome phenotype of lotus mutants further suggested the importance of NAP1 and PIR1 in lotus cells with polar growth pattern, such as those participating in IT or trichome formation.
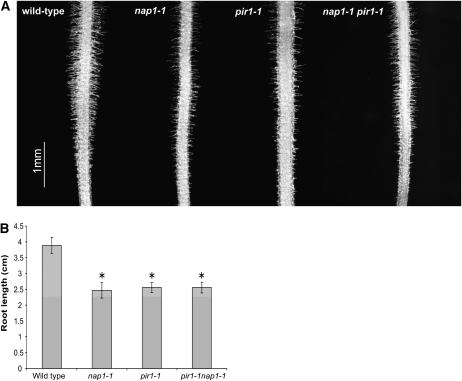
Two other types of polarized growth, namely, pollen tubes and root hair development were also evaluated. In vitro pollen germination experiments revealed about half the germination frequency in nap1-1, nap1-2, pir1-1, pir1-2, and pir1-3 mutant pollen (10 to 42%) compared with the wild type (57 to 82%). Likewise, when grown on the surface of vertically positioned agar plates (Karas et al., 2005), root hair formation appeared adversely affected in nap1-1, pir1-1, and pir1-1 nap1-1 double mutant roots when compared with wild-type control roots (Figure 7A). Although consistent across all mutant genotypes and growth conditions tested, this phenotype was rather subtle. Furthermore, its severity (in terms of the length and number of root hairs formed) varied substantially even between individuals of the same genotype, precluding quantification. Like the corresponding Arabidopsis mutants, the lotus nap1 and pir1 were affected in various growth characteristics. Thus, root elongation of the mutant roots was diminished in comparison to wild-type plants grown under the same experimental conditions (Figure 7B). Furthermore, comparing soil-grown plants, the nap1 and pir1 mutants were generally less vigorous than the wild type, reflected in the reduced size of mature plant shoots (Figure 6S). Flowering and seedpod formation was also less prolific in nap1 and pir1 mutants in comparison with wild-type plants. The fraction of soil-grown plants setting flowers and developing seedpods within the first 9 weeks after sowing was reduced to ∼50 and 30% in nap1-1 and pir1-1 mutant populations, respectively (Figure 6T). Nevertheless, the mutants could complete their life cycle and produce viable seeds. Seed formation was also affected, and both nap1 and pir1 mutants displayed a shrunken pod phenotype due to reduced seed set (Figures 6J to 6R). The shrunken seedpod phenotype, the nodulation phenotype, and the distorted trichomes cosegregated in 20 pir1-1 mutants selected from an F2 population of a backcross to wild-type lotus. Furthermore, all mutant phenotypic features described above were common to all nap1 and pir1 alleles, suggesting the involvement of Nap1 and Pir1 in the same actin-regulating pathway within different cell types.
Figure 7.
Root Development in L. japonicus Wild-Type Gifu, nap1-1, pir1-1, and pir1-1 nap1-1 Mutants.
(A) Light microscopy images of root segments (root tip toward bottom) showing diminished root hair development in the mutant plants compared with the wild-type control. The depicted differences in root hair density between individual mutant lines reflect the range of variation observed independently in each mutant genetic background tested.
(B) Root length of 7-d-old seedlings, showing significantly (*P < 0.05 in a t test) reduced elongation of the mutant roots. Mean values ± 95% confidence intervals are given for each genotype (n = 20).
The Role of Nap1 and Pir1 in the Infection Process
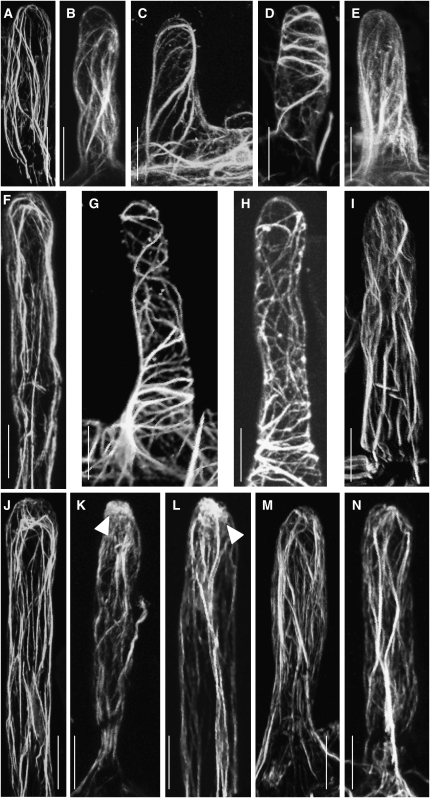
The function of NAP and PIR proteins has been investigated in detail in animals, protists, and Arabidopsis. Both proteins are part of the SCAR/WAVE complex that mediates actin dynamics. To investigate the role of the lotus NAP1 and PIR1 proteins during infection, we analyzed the actin structure in root hairs at three developmental stages. Two different techniques were used: Alexa-phalloidin staining of unfixed root hairs (Van Gestel et al., 2001) and imaging of live root hairs of transgenic roots using the 35S:GFP-ABD2-GFP F-actin reporter (Wang et al., 2007). Actin filaments in short (40 to 50 μm) wild-type root hairs visualized with Alexa-phalloidin (n = 25) show a characteristic arrangement with long cables of aligned actin filaments running longitudinally (Figure 8A). In the equivalent root hairs of pir1-1 and in nap1-1 mutants (n = 25), actin filaments were significantly more transverse and less longitudinally aligned (Figures 8C and 8E). Comparable stuctural differences were observed in short root hairs (n = 36) and in medium length root hairs (80 to 100 μm, n = 20) using the 35S:GFP-ABD2-GFP F-actin reporter (Figures 8B, 8D, and 8F to 8I). In the longer (>120 μm, n = 20) mutant roots hairs, an actin structure more comparable to the long bundles of actin filaments running longitudinally in wild-type root hairs (n = 39) was observed (Figures 8J, 8K, 8M, and 8N).
Figure 8.
Actin Cytoskeleton of Root Hairs, Visualized by Alexa-Phalloidin Staining or Expression of the 35S:GFP-ABD2-GFP F-Actin Reporter in Transgenic Roots.
(A) to (E) Short root hairs (40 to 50 μm).
(F) to (I) Medium length root hairs (80 to 100 μm).
(J) to (N) Long root hairs (>120 μm).
(A) Wild-type root hair, phalloidin-stained actin.
(B) Wild-type root hair; actin visualized using the 35S:GFP-ABD2-GFP F-actin reporter.
(C) Actin filaments in pir1-1 root hairs, phalloidin.
(D) Actin filaments in pir1-1 root hairs, F-actin reporter.
(E) Actin filaments in nap1-1 root hairs, phalloidin.
(F) Wild-type root hair, F-actin reporter.
(G) and (H) Examples of actin filaments in pir1-1 root hairs, F-actin reporter.
(I) Actin filaments in nap1-1 root hairs, F-actin reporter.
(J) Wild-type root hair before Nod factor application, phalloidin.
(K) Wild-type root hair 30 min after Nod factor application. Note the zone of diffuse actin accumulation at the tip of the root hair (arrowhead), phalloidin.
(L) Wild-type root hair 30 min after M. loti inoculation. Note the zone of diffuse actin accumulation at the tip of the root hair (arrowhead), phalloidin.
(M) pir1-1 root hair 30 min after application of Nod factor, phalloidin.
(N) nap1-1 root hair 30 min after application of Nod factor, phalloidin.
Bars = 20 μm.
Application of purified Nod factor to wild-type root hairs caused a rapid rearrangement of actin filaments visualized using Alexa-phalloidin. The longitudinally oriented actin bundles became thinner and a regional accumulation of more diffuse actin, close to the root hair tip, was observed in 160/260 root hairs (Figures 8J and 8K). In the pir1-1 mutants, no distinct alteration in actin cytoskeleton was observed in 130 root hairs examined after application of Nod factor (Figure 8M). In the nap1-1 mutants, accumulation of diffuse actin was observed in only 10 out of 350 mutant root hairs, and no accumulation was detected in 340/350 root hairs (Figure 8N). M. loti inoculation also induced accumulation of diffuse actin, in 5 out 10 roots hairs, of the wild type (Figure 8L), but no alteration in actin cytoskeleton was observed in root hairs of pir1-1 and nap1-1 root hairs (0/10 each; data not shown) following inoculation.
The root hair deformation assay showed a drastically attenuated response of mutant root hairs to external application of Nod factor. nap1-1 and pir1-1 showed only sporadic and limited root hair deformations. These were characterized by some root hair swelling with a markedly reduced size of the root hair tip polar outgrowth as compared with the wild-type control (see Supplemental Figure 5 online). Prolonged incubation for an additional 16 h in the presence of Nod factor did not increase the size of the polar outgrowth, suggesting that this defect was not due to a delayed or slower response of mutant root hairs. By contrast, wild-type roots showed abundant root hair deformations and a significant polar outgrowth of root hair tips was reproducibly observed after overnight incubation. The apparent failure of mutant root hairs to properly respond to Nod factor application could explain the strong impairment in the ability of nap1 and pir1 mutant plants to support root colonization by M. loti. In addition, these results provide further support for the crucial role of Nap1 and Pir1 in the dynamic actin reorganization that mediates polar cell growth in response to Nod factor signaling.
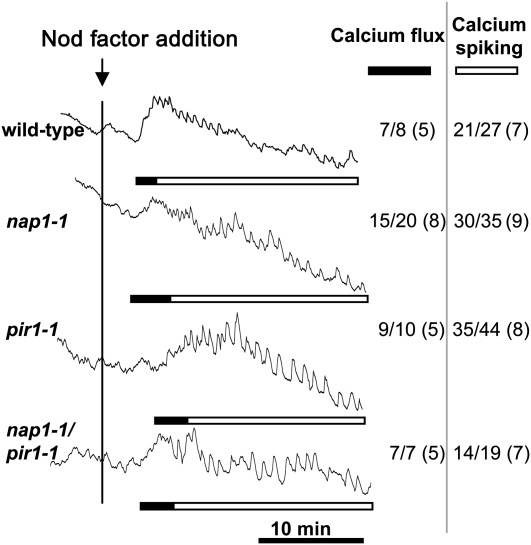
Nod factor–induced calcium influx coincides temporally and spatially with actin rearrangement and was proposed to be involved in the rapid reorganization of actin in responding root hairs (Sanchez et al., 1991; de Ruijter et al., 1999). Calcium influx and calcium spiking was therefore measured in pir1-1 and nap1-1 mutants to investigate the relationship between the early electrophysiological and cellular changes observed in root hairs. Ratios of fluorescence of Oregon Green to Texas Red were determined after Nod factor application (Miwa et al., 2006), and as seen in Figure 9, nosignificant differences in calcium influx or calcium spiking were observed between the wild type and the pir1-1 or nap1-1 mutants. This suggests that Nod factor–induced actin rearrangement mediated by Nap and Pir acts downstream or in parallel to calcium influx and spiking.
Figure 9.
Nod Factor–Induced Calcium Flux and Calcium Spiking in L. japonicus nap1-1, pir1-1, and pir1-1 nap1-1 Mutants.
Calcium levels were monitored in individual root hairs of the wild type and nap1-1, pir1-1, and pir1-1 nap1-1 mutants following addition of 100 nM M. loti Nod factor (black vertical line). The ratios (arbitrary units) of fluorescence of Oregon Green (calcium sensitive) to Texas Red (calcium insensitive) were recorded every 5 s for >30 min. The number of cells showing calcium spiking or calcium flux is shown in the inset table as a fraction of the total number of cells analyzed (with the total number of plants tested in parentheses). Solid bars indicate region of the trace where there is a significant transient increase in cellular calcium particularly at the root tip compared with other cytoplasmic or nuclear regions, and the open bars indicate the parts of the traces showing nuclear-associated calcium spiking.
Phenotype of nap1-1 pir1-1 Double Mutants
A double mutant that combines the lotus nap1-1 and pir1-1 alleles was constructed. The resulting nap1-1 pir1-1 homozygous double mutant was viable and showed essentially the same mutant phenotypes as observed in the corresponding single mutants (Figures 1, 6S, and 6T). Thus, the double mutant developed empty nodules upon inoculation with M. loti, and only occasionally a few pink nodules were found (Figure 1A). This was associated with the impairment in the initiation and/or progression of ITs (Figures 3A and 3F). The double mutant also showed the trichome and shrunken pod phenotypes, its overall growth was affected to the similar extent as observed in the corresponding single mutants, and no differences were seen in calcium spiking or calcium flux (Figure 9). Phalloidin visualization of actin in root hairs of double mutants revealed a filament structure that was similar to the milder perturbations observed also in single mutants.
DISCUSSION
The cytoskeleton plays a central role in regulating plant cell growth. Fundamental processes, such as cell division, cell expansion, organelle movement, stomatal closure, and cell morphogenesis, depend on dynamic rearrangements of the microtubule and actin networks of the cytoskeleton (Wasteneys and Galway, 2003; Ketelaar et al., 2003; Takemoto and Hardham, 2004; Bannigan and Baskin, 2005; Hussey et al., 2006). Since polarized cell growth has been particularly amenable to investigations, processes such as pollen tube elongation, root hair development, trichome morphogenesis, and lobe formation in leaf epidermal cells have been used as convenient models for describing the mechanisms underlying the dynamic reorganization of the cytoskeleton in expanding plant cells (Ketelaar et al., 2003; Smith and Oppenheimer, 2005; Samaj et al., 2006). It has become clear that in spite of traditionally assigned functions, with microtubules determining the directionality and actin mediating the rate and overall extent of cell expansion, a significant cooperation between these two cytoskeletal arrays exist, and this is required for a proper cell morphogenesis (Bannigan and Baskin, 2005).
Rearrangements of microtubule and actin cytoskeleton were observed in root hair cells reacting to rhizobial signaling by initiation of a variety of growth responses. The initial swelling and subsequent re-initiation of polar root hair tip growth as well as the initiation and subsequent inward oriented extension of ITs have all been shown to be preceded by and associated with significant cytoskeletal reorganization. It was postulated that Nod factor–induced actin rearrangements in root hairs are a prerequisite for IT formation (Cárdenas et al., 1998). Our data provide strong genetic and experimental support for this hypothesis by showing that L. japonicus Nap1 and Pir1 genes are essential for establishing the actin organization in lotus root hairs and that they are required for IT-dependent root colonization by M. loti. Observation of both mild and severe distortion of actin filaments in nap1 and pir1 mutant root hairs emphasizes the dynamics of actin rearrangements. The lotus nap1 and pir1 mutants showed a defective symbiotic phenotype. The collective evidence obtained by performing phenotypic and genetic analyses, physical mapping, and map-based cloning and sequencing of mutant alleles identified lotus Pir1 and Nap1 as essential genes for IT-dependent infection. The corresponding conceptual proteins, NAP1 and PIR1, showed amino acid sequence homology with Arabidopsis NAP125 and PIR121, respectively, which are components of the SCAR/WAVE complex that regulates actin cytoskeleton (Brembu et al., 2004; Deeks et al., 2004; Li et al., 2004).
NAP and PIR Regulation of Actin Dynamics
The actin cytoskeleton is formed by polymerization of G-actin monomers. Its dynamic reorganization is mediated by specialized proteins that can nucleate, stabilize, cross-link, cap, separate, and degrade F-actin polymers. Although species-specific differences at the cellular level have been documented, both the protein components and composition of the protein complexes involved were found to be conserved (Deeks and Hussey, 2005). Thus, actin polymerization was shown to be mediated by the actin-related protein ARP2/3 complex, which binds preexisting actin polymers and nucleates new filaments (Le et al., 2003; Li et al., 2003; Mathur et al., 2003; El-Din El-Assal et al., 2004; Smith and Oppenheimer, 2005). The ARP2/3 complex is activated by the SCAR/WAVE complex, which in Arabidopsis consists of the PIR121, NAP125, ABI, HSPC300, and SCAR proteins (Deeks et al., 2004; Gautreau et al., 2004). Activation of the SCAR/WAVE complex was most extensively characterized in mammalian cells (Eden et al., 2002). Binding of Rac GTPase to PIR121 activates the SCAR complex linking actin polymerization to endogenous or external signaling events.
In agreement with earlier observations, we showed that signaling by Nod factor–producing M. loti leads to a rapid and transient rearrangement of actin cytoskeleton in lotus root hairs. By contrast, root hairs of the nap1 and pir1 mutants had a disorganized actin cytoskeleton that was not responsive to bacterial inoculation. Therefore, these results indicate that a key function of NAP1 and PIR1 proteins is to mediate the organization of actin polymers in lotus root hairs. They also suggest that the dynamic reorganization of the actin cytoskeleton in response to bacterial signaling requires Nap1 and Pir1. The attenuated responses of nap1 and pir1 mutant root hairs to external application of Nod factor and the markedly reduced size of the root hair tip polar outgrowth compared with the wild type indicate a role in the earliest cellular responses of root hairs. An attractive explanation for the root hair deformation response is a NAP1- and PIR1-dependent role for the SCAR/WAVE complex in activation of the nucleation and polymerization that has been suggested to cause the subapical accumulation of fine actin filaments in response to Nod factor (Cárdenas et al., 1998; de Ruijter et al., 1999). Nod factor–induced root hair deformation, Ca2+ influx, and Ca2+ spiking all appear to branch from the initial perception event (Miwa et al., 2006). Ca2+ spiking is not necessary for root hair deformation (Miwa et al., 2006), and although the Ca2+ influx has been proposed to be required for root hair deformation (Esseling et al., 2003), this seems unlikely, because at least three orders of magnitude less Nod factor is sufficient to induce root hair deformation compared with that required for the Ca2+ influx, as measured using a Ca2+-sensitive dye (Miwa et al., 2006) or the Ca2+ influx–associated alkalinization measured using a microelectrode (Felle et al., 1998; Radutoiu et al., 2003). Our observations are consistent with the Ca2+ responses occurring in parallel with the NAP1- and PIR1-induced cytoskeletal changes, although we cannot exclude the possibility of a low level or localized change in Ca2+ that cannot be measured in root hairs using existing techniques.
The Nonsymbiotic Phenotypes of nap and pir Mutants
Deleterious mutations in the genes encoding the components of the SCAR/WAVE complex, such as NAP and PIR, have been reported to cause the so-called distorted mutant phenotypes in Arabidopsis, including distortion of trichome branching. The observation that lotus nap1 and pir1 mutants have an aberrant trichome phenotype further supports the function of the NAP1 and PIR1 proteins in a presumed L. japonicus SCAR/WAVE complex. This notion is also consistent with the disorganized and unresponsive structure of the actin cytoskeleton observed in the mutant root hairs, as described above.
Inactivation of the SCAR/WAVE complex in animal cells causes a lethal phenotype, but lotus nap1 and pir1 mutants were fully viable. In line with the latter observation, the corresponding Arabidopsis mutants were also viable. In fact, the lethality has not been observed in mutant plants carrying deleterious mutations in genes known to regulate the actin cytoskeleton. Nevertheless, more general effects on plant growth and development, including reduced chlorophyll content, shortened siliques, reduced seed production, and substantially increased length of dark-grown roots in comparison with the wild-type control, were reported in Arabidopsis nap-1 and pir-1 mutants (Brembu et al., 2004; Deeks et al., 2004; Li et al., 2004). Similarly, lotus nap1 and pir1 showed various growth aberrations, further substantiating the general cell biological effects of the mutations. Unlike in Arabidopsis, however, the overall development of roots, including their elongation and the formation of root hairs, was diminished in lotus nap1 and pir1.
Actin Rearrangement and the Bacterial Infection Process
L. japonicus plants carrying a mutant nap1 or pir1 allele had a significantly diminished ability to respond to inoculation by capturing bacteria within curled root hairs and initiating ITs. The rare ITs that formed underwent rapid disintegration, and only sporadically ITs that extended to the base of the epidermal cell were observed. Thus, NAP1 and PIR1 were required during all stages of the infection process, including the initiation and maintenance of IT integrity. A total lack of IT formation within the root cortex further suggested that NAP1 and PIR1 were essential for the progression of the infection process beyond the root epidermis. By contrast, nap1 and pir1 mutants appeared fully competent to initiate nodule primordia, which culminated with the organogenesis of empty nodule structures, indicating that NAP1 and PIR1 were not required for this process. A significantly increased number of nodule formation events in the nap1 and pir1 mutants in comparison to wild-type plants likely reflects the functioning of a homeostatic mechanism. Such a regulatory mechanism could operate to allow the host plant to monitor the progression of infection events and to respond appropriately by adjusting the competency of the root to subsequent bacterial signaling.
Considering all of the growth defects observed in lotus nap1 and pir1 mutants, the most dramatic effect was associated with the infection process. Therefore, the molecular characterization of lotus nap1 and pir1 mutants provides a new model with which several fundamental questions in the nodulation process could be answered while furthering the overall understanding of the cellular processes that regulate polarized cell growth in response to external stimuli. Signaling by Nod factor likely activates small GTPases that, in turn, participate in rapid cellular responses, including the activation of SCAR/WAVE-dependent reorganization of the actin cytoskeleton. In this context, a comprehensive understanding of the interrelationship between Nod factor–induced dynamic actin reorganization and calcium signaling, processes that are central to symbiosis, will be essential and should be facilitated by the availability of lotus nap1 and pir1 mutants.
We have demonstrated the essential role of Nap1 and Pir1 genes in the dynamic reorganization of the actin cytoskeleton and showed that they are required for IT and trichome formation in L. japonicus. Based on our data, we predict that other symbiotic legume mutants with trichome and IT defects, such as crinkle and lot1, might define additional components influencing cytoskeleton dynamics. Together, this should improve our understanding of the cellular mechanisms that mediate IT-dependent root colonization of legume roots by symbiotic bacteria.
METHODS
Plant Material
The eight nap1 and pir1 mutants were isolated as symbiotic mutants, and they are all in an ecotype Gifu B-129 background (Handberg and Stougaard, 1992). nap1-1 previously called sym67 was found in a population screened for Ac-tagged mutants (Thykjær et al., 1995; Sandal et al., 2006), nap1-2 and nap1-3 mutants previously called S12-5A and S90-D originate from an EMS-mutagenized population (Murray et al., 2006). pir1-1 previously called sym40 was found in a population screened for Ac-tagged mutants (Thykjær et al., 1995; Sandal et al., 2006), pir1-2, pir1-3, pir1-4, and pir1-5 originate from EMS populations and were previously called sym80 (Kawaguchi et al., 2002), S14-3, S57-F, and B31-C (Murray et al., 2006), respectively. Seeds of the wild type, nap1-1, pir1-1, and nap1-1pir1-1 double mutant were surface sterilized as described previously (Handberg and Stougaard, 1992) and grown for 6 weeks in Magenta boxes or 5 weeks on solid quarter-strength B&D (Broughton and Dilworth, 1971) slants with 1 mM KNO3. Plants were grown with or without Mesorhizobium loti strains NZP2235 or R7A (Sullivan et al., 1995).
Phalloidin Staining and Fluorescence Microscopy
A method modified after Van Gestel et al. (2001) was used for phalloidin staining of unfixed root material. Roots from young seedlings, 5 to 6 d old, were carefully excised to avoid damaging the root hairs and placed in actin stabilizing buffer (ASB) containing 100 mM PIPES, 10 mM EGTA, and 5 mM MgSO4, pH 6.8, supplemented with 1.5% glycerol, 0,1% Triton X-100, and 2 units of AlexaFluor 488–conjugated phalloidin (Invitrogen). The roots were incubated in 100 μL ASB buffer in the dark overnight at room temperature, washed three times in ASB, and mounted in ASB on glass slides for confocal fluorescence microscopy. The stained material was analyzed with a Zeiss LSM 510 Meta confocal microscope. The AlexaFluor 488 dye was excited with the 488-nm line of the Ar laser, and emission was captured with the 505- to 530-nm band-pass filters. The same settings, with an additional bright-field channel, were used for GFP visualization. Transgenic root hairs expressing the 35S:GFP-ABD2-GFP F-actin reporter (Wang et al., 2007) were generated and analyzed as described by Radutoiu et al. (2003).
IT Analysis and Root Hair Deformation
To visualize ITs, roots were inoculated with M. loti NZP2235 expressing the lacZ reporter gene. Plants were grown at 21/16°C and a 16/8 h day/night regime on a substrate of Leca (Optiroc) and Vermiculite (3:1 mixture) and supplemented with 50 mL of quarter-strength B&D medium with 1 mM KNO3 in Magenta containers (Sigma-Aldrich). Each seedling was inoculated with 400 μL (OD600 ∼0.01 to 0.02) of M. loti NZP2235 expressing the lacZ reporter gene strain, and ITs were visualized 2, 4, and 6 weeks after inoculation by staining for β-galactosidase activity, as described by Boivin et al. (1990). ITs were observed using Zeiss fluorescence microscope under bright-field illumination. Alternatively, roots from plants inoculated as described above with M. loti R7A expressing GFP were harvested at 7, 14, and 21 d after inoculation, mounted on slides in deionized water, and observed directly. The number of ITs was recorded for each plant. Nod factor isolation and root hair deformation assays were performed as described (Miwa et al., 2006). At least 20 roots were scored for each genotype in three independent experiments.
Pollen Germination and Root Hair Assays
Pollen released from mutant and wild-type flowers was germinated in 100 μL of a filter-sterilized medium consisting of 1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, 0.01% H3BO3, and 18% sucrose, pH 7.0. Pollen was germinated at room temperature in the dark for 3 to 4 h before 30 μL was spotted onto a glass slide and observed at ×20 magnification (Li et al., 1999).
For the analysis of root growth and root hair phenotypes of nap1-1 and pir1-1 mutants, as well as the nap1-1 pir1-1 double mutant, seeds were germinated on Whatman filter paper for 2 d in the dark. These were then carefully transferred to plates containing half-strength Gamborg's B5 with minimal organics, 2.5 mM MES hydrate, 4.5% sucrose, and 0.8% phytagel, where they were grown and analyzed as described (Karas et al., 2005).
Microscopy
Sectioning of nodules for light microscopy was performed as described previously (van Spronsen et al., 2001). Nodules and bumps from nap1-1 and pir1-1 mutant plants grown for 6 weeks were fixed in 1.5% paraformaldehyde and 2.5% glutaraldehyde. The samples were dehydrated by two washes of distilled water and three washes in 50 mL of 2.2 dimethoxypropane to which 100 μL of 10% HCl had been added prior to embedding in Technovite 7100 (according to the manufacturer Haereus Kluwer) for sectioning. Thin sections (5 μm) were stained in 0.1% Toluidine blue for 1 min and rinsed in water before the light microscopy.
Calcium spiking and calcium flux were analyzed essentially as described previously (Miwa et al., 2006) using the calcium-sensitive dye Oregon Green-dextran 10,000 MW and the reference dye Texas Red-dextran 10,000 MW (Molecular Probes). Nod factor was added directly to an incubation chamber containing 100 μL of Fahraeus nitrogen-free plant medium (Fahraeus, 1957) to give an estimated final concentration of 100 nM. For calcium flux analysis, fluorescence was imaged over the tip and over the entire root hair cell. Only those cells showing a significant transient increase in both tip calcium and in the entire root hair cell were considered positive for calcium flux.
SSAP Analyses and Positional Cloning
Nap1 and Pir1 were mapped with two and four independent F2 mapping populations, resulting from crosses between each mutant and MG-20 Miyakojima (Kawaguchi et al., 2001). Plants were grown in a greenhouse in pots containing Leca (Optiroc) as described previously (Radutoiu et al., 2003).
F2 plants homozygous for the nap1 and pir1 mutants were screened by white bump phenotype 4 weeks after inoculation with M. loti NZP2235. In total, 1077 and 822 homozygous F2 mutant plants of nap1-1 and pir1-1 were analyzed. Microsatellite markers developed from BAC and TAC clones anchored to the general genetic map of the region were used for fine mapping and for building the physical BAC/TAC contig. The SSAP analyses were performed as described previously (Madsen et al., 2005). Protein sequences were aligned using ClustalX set with a gap opening of 25, and the percentage identities were determined using DNAsis. DNA sequences of wild-type and mutant alleles were processed using the Sequencher software, which was also used to delimit reading frames.
Gene Expression Analyses
Total RNA was isolated using Trizol (Sigma-Aldrich), and RNA was treated with RQ1-DNase (Promega). Transcript levels were determined by quantitative real-time RT-PCR (Radutoiu et al., 2003). All cDNA samples were tested for contaminating DNA using PCR primers specific for the Nin gene promoter (Radutoiu et al., 2003). Primers for transcript amplification were as follows: PIR-fw, 5′-CACGCACCTCCCTGTTCAGGGATG-3′, and PIR-rev, 5′-TGGAGCACCACTTTGCTTAATAGC-3′; NAP-fw, 5′-CTTGGAGTGAAACACAAAGAGCTC-3′, and NAP-rev, 5′-CAATGGAGTGGATGGAGTGTCTGC-3′; TIP41-fw, 5′-TCAAGCTTTGTCTGCGAAAGG-3′, and TIP41-rev, 5′-ATCAATTTCACTTTCTGCATTAAGG-3′. For each sample normalized, relative ratios of analyzed genes and three independent housekeeping genes (Protein phosphatase2A, TIP41, and Tubulin β-chain [Czechowski et al., 2005; Tirichine et al., 2007]) were calculated using Relative Quantification software (Roche). The geometric mean of relative expression ratios for three biological and three technical repetitions and corresponding upper and lower 95% confidence intervals were calculated (Vandesompele et al., 2002).
For comparing the nap1-2 and wild-type transcripts shown in Figure 5, total RNA was extracted from leaf tissue using the RNeasy Plant Mini Kit (Qiagen) and treated with DNaseI. Random hexamer-primed cDNA was synthesized using the Thermoscript RT-PCR system (Invitrogen) in a total reaction volume of 20 μL, and a control reaction for each RNA sample was included to which no reverse transcriptase was added. A 5′ portion of the Lj NAP gene spanning the presumed deletion in nap1-2 mutants was amplified (wild-type product size = 1039 bp) using the following conditions: 5 min denature at 94°C, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, 68°C for 2 min, followed by a 7-min soak at 68°C. For the comparative analysis of Lj NAP transcripts produced by wild-type Gifu and the nap1-2 mutant, a 664-bp fragment from exon 28 of the Lj NAP gene was amplified (5 min denature at 94°C, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, 68°C for 1 min, followed by a 7 min soak at 68°C). All RT-PCR reactions were conducted using 1 μL of cDNA template and High Fidelity Platinum Taq DNA Polymerase (Invitrogen) and a GeneAmp 9700 series thermocycler. Primers used are as follows: (Ubiquitin reference gene) Ubi-F, 5′-TTCACCTTGTGCTCCGTCTTC-3′, Ubi-R, 5′-AACAACAGAACACACAGACAATCC-3′; (Lj NAP RT-PCR 3′ region) Lj NAP Exon28, F 5′-AAACACGAAGCACCCACTCT-3′, Lj NAP Exon28, R1 5′-GGATACCGAGGGTGATATGG-3′; (Lj NAP RT-PCR 5′ region) Lj NAP 5′ RT F1, 5′-AAACACGAAGCACCCACTCT-3′, Lj NAP 5′ RT R2 5′-GGATACCGAGGGTGATATGG-3′.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Nap1 gene (AM946362), Pir1 gene (AM946363), Pir1 mRNA (AM946364), Nap1 mRNA (AM946365); BAC and TAC clones: LjB386A21 (BM2150), AP009620; LjT39G23 (TM2115), AP009621; LjT61G21 (TM2120), AP009622; and LjB309G02 (BM2152), AP009623.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Map-Based Cloning of Lj Nap1.
Supplemental Figure 2. Alignment of NAP1 Proteins.
Supplemental Figure 3. Map-Based Cloning of Lj Pir1.
Supplemental Figure 4. Alignment of PIR1 Proteins.
Supplemental Figure 5. Root Hair Deformation after Nod Factor Treatment.
Supplementary Material
Acknowledgments
We thank Makoto Hayashi and Masayoshi Kawaguchi for making the sym80 mutant line available and Elison Blancaflor for providing the GFP-Fimbrin-F-actin reporter. This work was supported by the Danish National Research Foundation, H.O. and K.Y. were supported by grants from the Bio-oriented Technology Research Advancement Institution of Japan, J.A.D. and G.E.D.O. were supported by the Biotechnology and Biological Sciences Research Council and a European Union grant (MRTN-CT-2006-035546) to support G.M. within the “Nodperception” network.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jens Stougaard (stougaard@mb.au.dk).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Allen, N.S., Bennett, M.N., Cox, D.N., Shipley, A., Ehrhardt, D.W., and Long, S.R. (1994). Effects of Nod factors on alfalfa root hair Ca++ and H+ currents and on cytoskeletal behavior. In Advances in Molecular Genetics of Plant-Microbe Interactions, Vol. 3. M.J. Daniels, J.A. Downie, and A.E. Osbourn, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 107–113.
- Ané, J.-M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Bannigan, A., and Baskin, T.I. (2005). Directional cell expansion – Turning toward actin. Curr. Opin. Plant Biol. 8 619–624. [DOI] [PubMed] [Google Scholar]
- Basu, D., El-Din El-Assl, S., Le, J., Mallery, E.L., and Szymanski, D.B. (2004). Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA1 during leaf epidermal development. Development 131 4345–4355. [DOI] [PubMed] [Google Scholar]
- Boivin, C., Camut, S., Malpica, C.A., Truchet, G., and Rosenberg, C. (1990). Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembu, T., Winge, P., Seem, M., and Bones, A.M. (2004). NAPP and PIRP encode subunits of a putative Wave regulatory protein complex involved in plant cell morphogenesis. Plant Cell 16 2335–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton, W.J., and Dilworth, M. (1971). Control of leghemoglobin synthesis in snake beans. Biochem. J. 125 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas, L., Vidali, L., Domínguez, J., Pérez, H., Sánchez, F., Hepler, P.K., and Quinto, C. (1998). Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etli nodulation signals. Plant Physiol. 116 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, M.J., and Hussey, P.J. (2005). Arp2/3 and SCAR: Plants move to the fore. Nat. Rev. Mol. Cell Biol. 6 954–964. [DOI] [PubMed] [Google Scholar]
- Deeks, M.J., Kaloriti, D., Davies, B., Malhó, R., and Hussey, P.J. (2004). Arabidopsis NAP1 is essential for Arp2/3-dependent trichome morphogenesis. Curr. Biol. 14 1410–1414. [DOI] [PubMed] [Google Scholar]
- de Ruijter, N.C.A., Bisseling, T., and Emons, A.M.C. (1999). Rhizobium Nod factors induce an increase in sub-apical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol. Plant Microbe Interact. 12 829–832. [Google Scholar]
- Eden, S., Rohatgi, R., Podtelejnikov, A.V., Mann, M., and Kirschner, M.W. (2002). Mechanism of regulation of WAVE-induced actin nucleation by Rac1 and Nck. Nature 418 790–793. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal, S., Le, J., Basu, D., Mallery, E.L., and Szymanski, D.B. (2004). DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J. 38 526–538. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kaló, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Esseling, J.J., Lhuissier, F.G.P., and Emons, A.M.C. (2003). Nod factor-induced root hair curling: Continous polar growth towards the point of Nod factor application. Plant Physiol. 132 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus, G. (1957). The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 16 374–381. [DOI] [PubMed] [Google Scholar]
- Felle, H.H., Kondorosi, E., Kondorosi, A., and Schultze, M. (1998). The role of ion fluxes in Nod factor signalling in Medicago sativa. Plant J. 13 455–463. [Google Scholar]
- Fukai, E., Dobrowolska, A.D., Madsen, L.H., Madsen, E.B., Umehara, Y., Kouchi, H., Hirochika, H., and Stougaard, J. (2008). Transposition of a 600 thousand year old LTR retrotransposon in the model legume Lotus japonicus. Plant Mol. Biol. 68 653–663. [DOI] [PubMed] [Google Scholar]
- Gage, D.J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau, A., Ho, H.H., Steen, J., Gygi, S.P., and Kirschner, M.W. (2004). Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA 101 4379–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg, K., and Stougaard, J. (1992). Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 2 487–496. [Google Scholar]
- Heckmann, A.B., Lombardo, F., Miwa, H., Perry, J.A., Bunnewell, S., Parniske, M., Wang, T.L., and Downie, J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in no-legume. Plant Physiol. 142 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey, P.J., Ketelaar, T., and Deeks, M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57 109–125. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku, H., Kawaguchi, M., Koiwa, H., Akao, S., and Syono, K. (1997). Two ineffective-nodulating mutants of Lotus japonicus – Different phenotypes caused by the blockage of endocytotic bacterial release and nodule maturation. Plant Cell Physiol. 38 871–881. [Google Scholar]
- Imaizumi-Anraku, H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531. [DOI] [PubMed] [Google Scholar]
- Kaló, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kanamori, N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas, B., Murray, J., Gorzelak, M., Smith, A., Sato, S., Tabata, S., and Szczyglowski, K. (2005). Invasion of Lotus japonicus root hairless 1 by Mesorhizobium loti involves the nodulation factor-dependent induction of root hairs. Plant Physiol. 137 1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, M., Imaizumi-Anraku, H., Koiwa, H., Niwa, S., Ikuta, A., Syono, K., and Akao, S. (2002). Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 15 17–26. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, M., Motomura, T., Imaizumi-Anraku, H., Akao, S., and Kawasaki, S. (2001). Providing the basis for genomics in Lotus japonicus: The accessions Miyakojima and Gifu are appropriate crossing partners for genetic analyses. Mol. Genet. Genomics 266 157–166. [DOI] [PubMed] [Google Scholar]
- Ketelaar, T., de Ruijter, N.C.A., and Emons, A.M.C. (2003). Unstable F-actin specifies the area and microtubule direction of cell expansin in Arabidopsis root hairs. Plant Cell 15 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner, C., Winzer, T., Pitzschke, A., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Webb, K.J., Szczyglowski, K., and Parniske, M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17 2217–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell, L., et al. (2005). The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, H., Hakoyama, T., Umehara, Y., Sato, S., Kaneko, T., Tabata, S., and Kouchi, H. (2007). A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus. Plant Physiol. 143 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, J., El-Din El-Assal, S., Basu, D., Saad, M.E., and Szymanski, D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13 1341–1347. [DOI] [PubMed] [Google Scholar]
- Lévy, J., Bres, C., Geurts, R., Chalhoub, B., Kulikova, O., Duc, G., Journet, E.P., Ané, J.M., Lauber, E., Bisseling, T., Dénarié, J., Rosenberg, C., and Debellé, F. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364. [DOI] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R., Zhu, M., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Blanchoin, L., Yang, Z., and Lord, E.M. (2003). The putative Arabidopsis Arp2/3 complex controls leaf cell morphogenesis. Plant Physiol. 132 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Sorefan, K., Hemmann, G., and Bevan, M.W. (2004). Arabidopsis NAP and PIR regulate actin-based cell morphogenesis and multiple developmental processes. Plant Physiol. 136 3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo, F., Heckmann, A.B., Miwa, H., Perry, J.A., Yano, K., Hayashi, M., Parniske, M., Wang, T.L., and Downie, J.A. (2006). Identification of symbiotically defective mutants of Lotus japonicus affected in infection thread growth. Mol. Plant Microbe Interact. 19 1444–1450. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- Madsen, L.H., Fukai, E., Radutoiu, S., Yost, Ch.K., Sandal, N., Schauser, L., and Stougaard, J. (2005). LORE1, an active low-copy-number TY3-gypsy retrotransposon family in the model legume Lotus japonicus. Plant J. 44 372–381. [DOI] [PubMed] [Google Scholar]
- Marsh, J.F., Rakocevic, A., Mitra, R.M., Brocard, L., Sun, J., Eschstruth, A., Long, S.R., Schultze, M., Ratet, P., and Oldroyd, G.E.D. (2007). Medicago truncatula NIN is essential for Rhizobium-independent nodule organogenesis induced by autoactive CCaMK. Plant Physiol. 144 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B.P., and Hülskamp, M. (2003). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region-specific fine F-actin formation. Development 130 3137–3146. [DOI] [PubMed] [Google Scholar]
- Miller, D.D., Klooster, H.B.L., and Emons, A.M.C. (2000). Lipochito-oligosaccharide nodulation factors stimulate cytoplasmic polarity with longitudinal endoplasmatic reticulum and vesicles at the tip in vetch root hairs. Mol. Plant Microbe Interact. 13 1385–1390. [DOI] [PubMed] [Google Scholar]
- Mitra, R.M., Gleason, C.A., Edwards, A., Hadfield, J., Downie, J.A., Oldroyd, G.E., and Long, S.R. (2004). A Ca2+ calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, H., Sun, J., Oldroyd, G.E., and Downie, J.A. (2006). Analysis of nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19 914–923. [DOI] [PubMed] [Google Scholar]
- Murakami, Y., Miwa, H., Imaizumi-Anraku, H., Kouchi, H., Downie, J.A., Kawaguchi, M., and Kawasaki, S. (2006). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 13 255–265. [DOI] [PubMed] [Google Scholar]
- Murray, J., et al. (2006). Genetic suppressors of the Lotus japonicus har1-1 hypernodulation phenotype. Mol. Plant Microbe Interact. 19 1082–1091. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Downie, J.A. (2008). Coordination of nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59 519–546. [DOI] [PubMed] [Google Scholar]
- Ooki, Y., Banba, M., Yano, K., Maruya, J., Sato, S., Tabata, S., Saeki, K., Hayashi, M., Kawaguchi, M., Izui, K., and Hata, S. (2005). Characterization of the Lotus japonicus symbiotic mutant lot1 that shows a reduced nodule number and distorted trichomes. Plant Physiol. 137 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Grønlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Jurkiewicz, A., Fukai, E., Quistgaard, E.M.H., Albrektsen, A.S., James, E.K., Thirup, S., and Stougaard, J. (2007). LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26 3923–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saller, E., Tom, E., Brunori, M., Otter, M., Estreicher, A., Mack, D.H., and Iggo, R. (1999). Increased apoptosis induction by 121F mutant p53. EMBO J. 18 4424–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaj, J., Müller, J., Beck, M., Böhm, N., and Menzel, D. (2006). Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci. 11 594–600. [DOI] [PubMed] [Google Scholar]
- Sanchez, F., Padilla, J.E., Perez, H.E., and Lara, M. (1991). Control of nodulin genes in root-nodule development and metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 507–528. [Google Scholar]
- Sandal, N., et al. (2006). Genetics of symbiosis in Lotus japonicus: Recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol. Plant Microbe Interact. 19 80–91. [DOI] [PubMed] [Google Scholar]
- Schauser, L., Handberg, K., Sandal, N., Stiller, J., Thykjær, T., Pajuelo, E., Nielsen, A., and Stougaard, J. (1998). Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol. Gen. Genet. 259 414–423. [DOI] [PubMed] [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Sheth, N., Roca, X., Hastings, M.L., Roeder, T., Krainer, A.R., and Sachidandam, R. (2006). Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 34 3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., and Geurts, R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791. [DOI] [PubMed] [Google Scholar]
- Smith, L.G., and Oppenheimer, D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21 271–295. [DOI] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Sullivan, J.T., Patrick, H.N., Lowther, W.L., Scott, D.B., and Ronson, C.W. (1995). Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 92 8985–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski, K., Shaw, R.S., Wopereis, J., Copeland, J., Hamburger, D., Kasiborski, B., Dazzo, F.B., and de Bruijn, F.J. (1998). Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 11 684–697. [Google Scholar]
- Szymanski, D.B. (2005). Breaking the WAVE complex: The point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 8 103–112. [DOI] [PubMed] [Google Scholar]
- Takemoto, D., and Hardham, A.R. (2004). The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physiol. 136 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansengco, M.L., Hayashi, M., Kawaguchi, M., Imaizumi-Anraku, H., and Murooka, Y. (2003). Crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome and seed development in Lotus japonicus. Plant Physiol. 131 1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thykjær, T., Stiller, J., Handberg, K., Jones, J., and Stougaard, J. (1995). The maize transposable element Ac is mobile in the legume Lotus japonicus. Plant Mol. Biol. 27 981–993. [DOI] [PubMed] [Google Scholar]
- Timmers, A.C.J., Auriac, M.-C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., et al. (2006). Deregulation of a Ca2+ calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., Sandal, N., Madsen, L.H., Radutoiu, S., Albrektsen, A.S., Sato, S., Asamizu, E., Tabata, S., and Stougaard, J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107. [DOI] [PubMed] [Google Scholar]
- Van Gestel, K., Le, J., and Verbelen, J.P. (2001). A comparison of F-actin labeling methods for light microscopy in different plant specimens: Multiple techniques complement each other. Micron 32 571–578. [DOI] [PubMed] [Google Scholar]
- van Spronsen, P.C., Grønlund, M., Pacios Bras, C., Spaink, H., and Kijne, J.W. (2001). Cell biological changes of outer cortical root cells in early determinate nodulation. Mol. Plant Microbe Interact. 14 839–847. [DOI] [PubMed] [Google Scholar]
- van Spronsen, P.C., van Brussel, A.A., and Kijne, J.W. (1995). Nod factors produced by Rhizobium leguminosarum biovar viciae induce ethylene-related changes in root cortical cells of Vicia sativa ssp.nigra. Eur. J. Cell Biol. 68 463–469. [PubMed] [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., de Paepe, A., and Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by genomic averaging of multiple internal control genes. Genome Biol. 3 RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva, V.N., Kouchi, H., and Ridge, R.W. (2005). Microtubule dynamics in living root hairs: Transient slowing by lipochitin oligosaccharide nodulation signals. Plant Cell 17 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.-S., Yoo, C.-M., and Blancaflor, E.B. (2007). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177 525–536. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., and Galway, M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54 691–722. [DOI] [PubMed] [Google Scholar]
- Weerasinghe, R.R., Bird, D.McK., and Allen, N.S. (2005). Root-knot nematodes and bacterial Nod factors elicit common signal transduction events in Lotus japonicus. Proc. Natl. Acad. Sci. USA 102 3147–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe, R.R., Collings, D.A., Johannes, E., and Allen, N.S. (2003). The distributional changes and role of microtubules in Nod-factor challenged Medicago sativa root hairs. Planta 218 276–287. [DOI] [PubMed] [Google Scholar]
- Wu, Q., and Krainer, A.R. (1999). AT.AC pre-mRNA splicing mechanisms and conservation of minor introns in volate-gated ion channel genes. Mol. Cell. Biol. 19 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, K., Tansengco, M.L., Hio, T., Higashi, K., Murooka, Y., Imaizumi-Anraku, H., Kawaguchi, M., and Hayashi, M. (2006). New nodulation mutants responsible for infection thread development in Lotus japonicus. Mol. Plant Microbe Interact. 19 801–810. [DOI] [PubMed] [Google Scholar]
- Yano, K., Yoshida, S., Müller, J., Singh, S., Banba, M., Vickers, K., Markmann, K., White, C., Schuller, B., Sato, S., Asamizu, E., Tabata, S., Murooka, Y., Perry, J., Wang, T.L., Kawaguchi, M., Imaizumi-Anraku, H., Hayashi, M., and Parniske, M. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105 20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.