Abstract
Rationale: Studies show that the myosin content of the diaphragm in patients with mild to moderate chronic obstructive pulmonary disease (COPD) is reduced, compromising diaphragm contractile performance. The mechanisms for reduced contractile protein content are unknown. In the present study we hypothesized that the loss of contractile protein content is associated with activation of the ubiquitin–proteasome pathway in the diaphragm of patients with mild to moderate COPD.
Methods: Proteolytic activity of isolated 20S proteasomes was determined in diaphragm biopsies from patients with and without COPD (predicted mean FEV1, 66 and 93%, respectively). In addition, we determined 20S proteasome subunit C8 protein levels by means of Western blotting, ubiquitin-ligase mRNA levels by means of real-time polymerase chain reaction, and caspase-3 activity by determining the hydrolysis of fluorogenic substrates.
Results: The 20S proteasome activity was about threefold increased in the diaphragm of patients with COPD. C8 protein levels were not significantly different between COPD and non-COPD diaphragm, indicating increased specific activity of individual proteasomes, rather than an increased number of proteasomes. mRNA levels of the muscle-specific ubiquitin-ligase MAFbx were significantly higher in diaphragm from patients with COPD compared with patients without COPD. Caspase-3–mediated cleavage of actomyosin complexes is considered an initial step in muscle wasting, yielding fragments that can be degraded by the ubiquitin–proteasome pathway. In line with the increased ubiquitin–proteasome activity, caspase-3 activity was higher in diaphragm homogenates from patients with COPD.
Conclusions: The present study is the first to demonstrate increased activity of the ubiquitin–proteasome pathway in COPD diaphragm. Importantly, these changes occur in patients with only mild to moderate COPD (Global Initiative for Chronic Obstructive Lung Disease stage I/II).
Keywords: caspase-3, chronic obstructive pulmonary disease, diaphragm function, myosin, proteolysis
Diaphragm weakness is associated with dyspnea and increased morbidity and mortality in patients with severe chronic obstructive pulmonary disease (COPD) (1, 2). Traditionally, diaphragm weakness has been ascribed to hyperinflation-induced diaphragm shortening, which places the diaphragm at a suboptimal position on its force–length curve (3, 4). Studies show that structural, biochemical, and functional alterations may contribute to impaired diaphragm function in patients with COPD (5–12).
Force-generating capacity is strongly dependent on contractile protein content of the muscle fibers (13). Work from our laboratory demonstrated that diaphragm fibers from patients with mild to moderate COPD (Global Initiative for Chronic Obstructive Lung Disease [GOLD] stage I/II) have about 30% reduced myosin content (8, 9), which compromises diaphragm single-fiber force generation (9). This diaphragm wasting indicates an imbalance between myosin synthesis and degradation in COPD.
In animal models of muscle atrophy selective down-regulation of myosin has been found to be concurrent with accelerated protein degradation by the ubiquitin–proteasome pathway (14–18). Proteins degraded by the ubiquitin–proteasome pathway are first linked to a chain of ubiquitin molecules, which marks them for subsequent degradation by the 26S proteasome. Ubiquitination of proteins is regulated by multiple enzymes. Among these enzymes, the ubiquitin-ligases (E3-ligases) are particularly important because they account for substrate specificity of protein degradation (19). In animal studies, the expression of two newly discovered muscle-specific E3-ligases, MAFbx (atrogin-1) and MURF-1, was substantially increased in peripheral skeletal muscle during various muscle-wasting conditions (17, 20–22). Therefore, together with increased proteasome activity, elevated MAFbx and MURF-1 mRNA levels are reliable molecular markers for muscle atrophy (16, 23).
Previously, we demonstrated increased protein ubiquitination in COPD diaphragm (9). This might suggest activation of the ubiquitin–proteasome pathway, but it could also be indicative of reduced proteasome activity leading to accumulation of ubiquitinated proteins. To establish, for the first time, the activity level of the ubiquitin–proteasome pathway in COPD diaphragm, we investigated several of its key components in the diaphragm of patients with COPD. We hypothesized that proteasome activity is increased, and that E3-ligase mRNA levels are elevated, in the diaphragm of patients with mild to moderate COPD.
The sequence of events leading to muscle wasting involves additional mechanisms, because the ubiquitin–proteasome pathway does not break down intact sarcomeres (24). It has been postulated that disassociation of myosin and actin filaments from the muscle sarcomere is the rate-limiting step in muscle protein degradation. Evidence indicates that activation of caspase-3 results in cleavage of these myofilaments and yields fragments that are degraded by the ubiquitin–proteasome pathway (25). Therefore, we hypothesized that in addition to activation of the ubiquitin–proteasome pathway, caspase-3 activity is increased in COPD diaphragm.
METHODS
Subjects, Pulmonary Function Testing, and Diaphragm Biopsies
Diaphragm muscle biopsies were obtained from 15 patients with COPD and from 13 patients without COPD. Biopsies were obtained from the right anterior costal diaphragm during thoracotomy for lung cancer (stage T1–2N0–1M0 in both groups). Fresh biopsies were rapidly frozen in liquid nitrogen-cooled isopentane and stored at –80°C for later analysis. Exclusion criteria included weight loss of more than 10% in the last 6 mo before surgery, neuromuscular diseases, thyroid diseases, and chronic heart failure. None of the subjects received chemotherapy before surgery. General characteristics and pulmonary function data are shown in Table 1. Informed consent was obtained from each patient, and the study was approved by the local ethics committee.
TABLE 1.
PATIENT CHARACTERISTICS FOR PROTEASOME ACTIVITY, E3-LIGASE, CASPASE-3 ACTIVITY, AND WESTERN BLOTTING STUDIES
| Proteasome Activity
|
E3-ligase
|
Caspase-3 Activity
|
Western Blotting
|
|||||
|---|---|---|---|---|---|---|---|---|
| Non-COPD (n = 7) | COPD (n = 7) | Non-COPD (n = 10) | COPD (n = 11) | Non-COPD (n = 7) | COPD (n = 7) | Non-COPD (n = 9) | COPD (n = 9) | |
| Male/female | 6/1 | 6/1 | 9/1 | 9/2 | 6/1 | 7/0 | 8/1 | 9/0 |
| Age, yr | 64 ± 3 | 66 ± 2 | 63 ± 3 | 65 ± 3 | 66 ± 3 | 68 ± 2 | 65 ± 3 | 66 ± 2 |
| BMI, kg · m−2 | 28 ± 1 | 28 ± 2 | 28 ± 1 | 26 ± 2 | 28 ± 1 | 28 ± 2 | 28 ± 1 | 27 ± 2 |
| FEV1, % predicted | 93 ± 4 | 66 ± 7 | 98 ± 4 | 63 ± 4 | 97 ± 4 | 70 ± 7 | 96 ± 4 | 67 ± 6 |
| VC, % predicted | 91 ± 4 | 93 ± 4 | 88 ± 4 | 83 ± 6 | 95 ± 5 | 96 ± 6 | 91 ± 4 | 95 ± 5 |
| FEV1/VC, % | 77 ± 2 | 56 ± 3 | 76 ± 2 | 54 ± 2 | 74 ± 1 | 55 ± 3 | 76 ± 2 | 54 ± 3 |
| TLC, % predicted | 86 ± 3 | 111 ± 4 | 91 ± 4 | 106 ± 4 | 93 ± 5 | 99 ± 4 | 91 ± 4 | 107 ± 5 |
| RV/TLC, % | 33 ± 3 | 47 ± 2 | 38 ± 2 | 47 ± 3 | 36 ± 3 | 45 ± 4 | 37 ± 2 | 44 ± 3 |
| DlCO/Va, % predicted | 103 ± 9 | 78 ± 8 | 112 ± 5 | 76 ± 7 | 92 ± 5 | 84 ± 10 | 103 ± 6 | 79 ± 8 |
| PaO2, kPa | 11.2 ± 0.3 | 10.2 ± 0.5 | 10.2 ± 0.5 | 10.5 ± 0.5 | 10.3 ± 0.6 | 10.6 ± 0.6 | 10.2 ± 0.5 | 10.4 ± 0.3 |
| PaCO2, kPa | 4.8 ± 0.1 | 5.1 ± 0.1 | 4.7 ± 0.1 | 5.0 ± 0.2 | 4.6 ± 0.1 | 4.8 ± 0.2 | 4.7 ± 0.1 | 5.2 ± 0.2 |
Definition of abbreviations: BMI = body mass index; DlCO/Va = carbon monoxide transfer coefficient per alveolar volume; TLC = total lung capacity.
Values represent means ± SEM.
Isolation of 20S Proteasomes and Measurement of Proteolytic Activity
The 20S proteasome isolation and proteolytic activity measurements were performed according to Hobler and coworkers (26), with minor modifications. In short, the proteolytic activity of isolated 20S proteasomes was determined by measuring the activity against the fluorogenic substrates succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (LLVY) and N-carbenzoxy-Leu-Leu-Glu-7-amido-4-methylcoumarin (LLE; Sigma-Aldrich, Zwijndrecht, The Netherlands). For additional details, see the online supplement.
MAFbx and MURF-1 mRNA Determination by Real-Time Quantitative Polymerase Chain Reaction
See the online supplement for a description of the methodology for MAFbx and MURF-1 mRNA determination.
Western Blot Analysis
See the online supplement for a description of the semiquantitative determination of myosin heavy chain and 20S proteasome subunit C8 content by Western blotting.
Measurement of Caspase-3 Activity
Caspase-3 activity was determined as described by Du and coworkers (25), with minor modifications. In short, cleavage of the fluorogenic substrate N-acetyl-Asp-Glu-Val-Asp-7-amido-4-methylcoumarin (Ac-DEVD-AMC) by diaphragm homogenates was determined with a spectrophotometer. See the online supplement for details.
Statistical Methods
To evaluate the significance of differences between patients with COPD and patients without COPD, a t test was used (data were normally distributed). Because of limited diaphragm tissue available per patient, data concerning proteasome activity, caspase-3 activity, E3-ligase, and Western blotting studies are not based on the same patients, although there is extensive overlap. p < 0.05 was used as the criterion for significant significance.
RESULTS
Subject Characteristics
Patient characteristics and pulmonary function data are shown in Table 1. Patients with COPD included for proteasome activity, E3-ligase, caspase-3 activity, and Western blotting studies were classified as having mild COPD (stage I) or moderate COPD (stage II) on the basis of the GOLD classification (27).
Myosin Heavy Chain Protein Levels
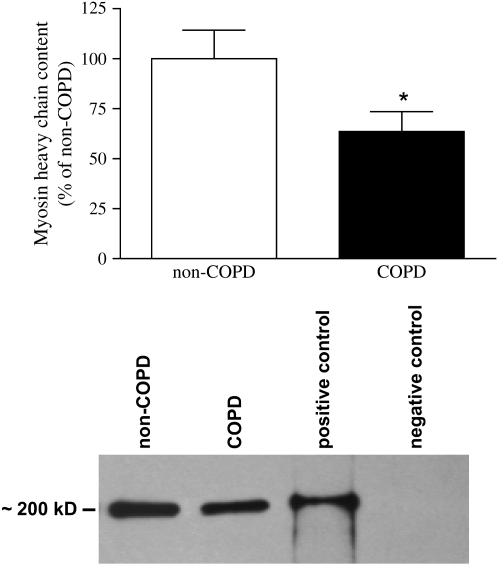
In line with previous data from our laboratory (8, 9), myosin heavy chain content was significantly lower in the diaphragm of patients with COPD compared with patients without COPD (Figure 1).
Figure 1.
Myosin heavy chain content in chronic obstructive pulmonary disease (COPD) and non-COPD diaphragm. Myosin heavy chain content was determined in diaphragm homogenates by Western blotting and subsequent densitometric quantification of protein bands. Top: Myosin heavy chain content in diaphragm homogenates from patients with COPD (solid column) versus patients without COPD (open column). Bottom: Representative myosin heavy chain immunoblot from diaphragm homogenates from a patient with COPD and a patient without COPD. For the positive control purified myosin heavy chain was used. For the negative control a homogenate from a patient without COPD was stained without addition of the primary (anti–myosin heavy chain) antibody. Data are presented as means ± SEM. * Significant difference (p < 0.05) from non-COPD.
20S Proteasome Activity
Proteasome activity against the substrate LLVY was about twofold higher, and that against LLE was about threefold higher, in COPD diaphragm compared with non-COPD diaphragm (p < 0.05; Figure 2). These data indicate that the chymotrypsin-like (LLVY) and peptidylglutamyl peptide–hydrolyzing (LLE) peptidase activities of the proteasome are increased in the diaphragm of patients with COPD. For representative proteasome activity-versus-time plots, see Figure E1 in the online supplement.
Figure 2.
Proteasome activity in diaphragm of patients with COPD and patients without COPD. Proteasome activity against fluorogenic substrates succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (LLVY) and N-carbenzoxy-Leu-Leu-Glu-7-amido-4-methylcoumarin (LLE) was higher in patients with COPD than in patients without COPD. AMC = amido-4-methylcoumarin. Data are presented as means ± SEM. * Significant difference (p < 0.05) from non-COPD.
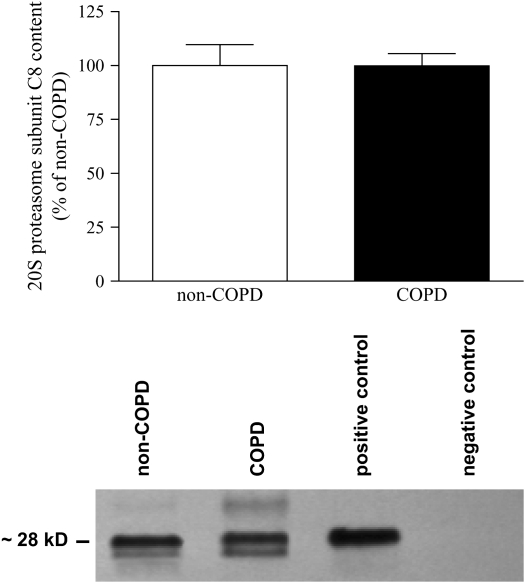
Proteasome Subunit C8 Protein Levels
To investigate whether changes in proteasome activity are the result of changes in the number of proteasomes, we determined the protein level of the C8 subunit of the 20S proteasome. Figure 3 demonstrates that the protein levels of the 20S proteasome C8 subunit were not significantly different between COPD and non-COPD diaphragm. These data suggest that the increased proteasome activity is the result of increased specific activity of the individual proteasomes, rather than of an increased number of proteasomes.
Figure 3.
20S proteasome subunit C8 protein content in diaphragm of patients with COPD and patients without COPD. Subunit C8 protein content was determined in diaphragm homogenates by Western blotting and subsequent densitometric quantification of protein bands. Top: Our analyses revealed comparable contents of subunit C8 in the diaphragm of patients with COPD (solid column) and patients without COPD (open column). Bottom: Representative C8 subunit immunoblot from diaphragm homogenates from a patient with COPD and a patient without COPD. For the positive control we used an isolated proteasome fraction from a patient without COPD (the same fraction as was used for the proteasome activity assays; see Figure 2). For the negative control a homogenate from a patient without COPD was stained without addition of the primary (anti-C8) antibody. Data are presented as means ± SEM.
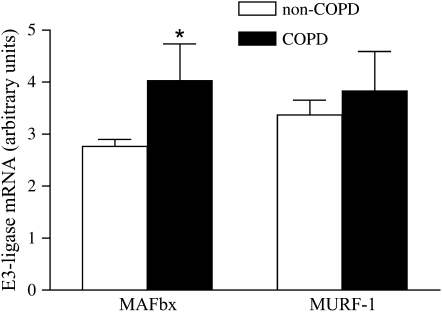
MAFbx and MURF-1 mRNA
We determined MAFbx and MURF-1 mRNA levels by means of real-time quantitative polymerase chain reaction. Compared with patients without COPD, patients with COPD had higher levels of MAFbx mRNA (p < 0.05; Figure 4). MURF-1 mRNA levels were not significantly different between patients with COPD and patients without COPD (Figure 4).
Figure 4.
E3-ligase mRNA levels in the diaphragm of patients with COPD and patients without COPD. MAFbx mRNA levels are higher in COPD versus non-COPD diaphragm. MURF-1 mRNA levels were not significantly different between COPD and non-COPD diaphragm. Data are presented as means ± SEM. * Significant difference (p < 0.05) from non-COPD.
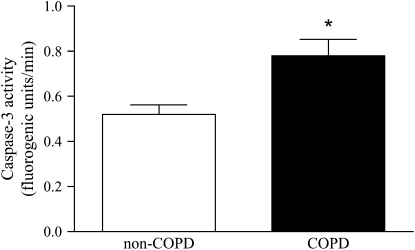
Caspase-3 Activity
Caspase-3 activity was assessed in diaphragm homogenates by determining the cleavage rate of the fluorogenic substrate Ac-DEVD-AMC. Caspase-3 activity of diaphragm homogenates was about 1.5-fold higher for patients with COPD compared with patients without COPD (p < 0.05; Figure 5). For representative caspase-3 activity-versus-time plots, see Figure E2.
Figure 5.
Caspase-3 activity in the diaphragm of patients with COPD and patients without COPD. Caspase-3 activity against the fluorogenic substrate N-acetyl-Asp-Glu-Val-Asp-7-amido-4-methylcoumarin (Ac-DEVD-AMC) was higher in diaphragm homogenates from patients with COPD (solid column) versus patients without COPD (open column). Data are presented as means ± SEM. * Significant difference (p < 0.05) from non-COPD.
DISCUSSION
This study is the first to reveal several key findings regarding diaphragm muscle proteolysis in humans, particularly in patients with COPD. We found that reduced myosin content was associated with ubiquitin–proteasome pathway activation in the diaphragm in COPD, as indicated by increased MAFbx mRNA expression and proteasome activity.
Evidence indicates that activation of caspase-3 is an initial step in myofilament proteolysis by cleavage of myosin and actin (25). In this way, activated caspase-3 yields fragments that are degradable by the ubiquitin–proteasome pathway. Indeed, we found elevated caspase-3 activity in the diaphragm of patients with COPD compared with patients without COPD. Importantly, activation of the ubiquitin–proteasome pathway and of caspase-3 activity occurred in patients with only mild to moderate COPD (GOLD stage I/II).
Increased Activity of the Ubiquitin–Proteasome Pathway in COPD Diaphragm
In line with previous work from our laboratory (8, 9), the present study confirms reduced myosin content of the diaphragm in patients with COPD. As a result, diaphragm fibers from these patients have impaired force-generating capacity (9), contributing to diaphragm weakness.
In many conditions associated with muscle wasting, such as sepsis, cancer, and chronic heart failure, loss of contractile protein occurs largely through the ubiquitin–proteasome pathway (28). This ATP-dependent proteolytic pathway can be divided into two major steps. In the first step, proteins are marked for degradation by conjugation with multiple ubiquitin molecules. This ubiquitin–protein conjugation is achieved by the combined action of ubiquitin-activating enzymes (E1), the ubiquitin-conjugating enzymes (E2), and ubiquitin-ligases (E3). In the second step, ubiquitinated proteins are recognized and subsequently degraded by the 26S proteasome, a large multicatalytic protein complex consisting of two regulatory 19S complexes and a 20S proteolytic core (29). Animal studies have shown that muscle wasting–associated conditions are characterized by a significant up-regulation of several key components of the ubiquitin–proteasome pathway (16, 24, 26, 30, 31). The present data show increased proteasome activity in the diaphragm of patients with only mild to moderate COPD. In vitro 20S proteasome activities measured under conditions similar to those reported here have been in good agreement with rates of proteolysis (32, 33).
The observed up-regulation of the E3-ligase MAFbx in COPD diaphragm strongly supports our notion that the ubiquitin–proteasome pathway is activated in these patients. Of all the proteins involved in ubiquitinating protein substrates, E3-ligases seem to have the greatest tissue and substrate specificity (19). MAFbx has been found to be markedly up-regulated during cachexia- and disuse-induced atrophy (17, 20). Moreover, when MAFbx knockout mice were subjected to an atrophic stimulus, muscle atrophy was attenuated by 50–60%, indicating an important role for this E3-ligase in atrophy (20). Previous studies in animals have shown parallel up-regulation of MAFbx and another muscle-specific E3-ligase, MURF-1, during multiple types of atrophy (16, 20). In contrast to MAFbx, diaphragmatic MURF-1 mRNA levels were not different between patients with COPD and patients without COPD. This differential response of MAFbx and MURF-1 in atrophying COPD diaphragm is in line with work on E3-ligase expression in skeletal muscle atrophy in humans (34, 35). Together, these data suggest that human skeletal muscle atrophy is regulated more by MAFbx than by MURF-1. Interestingly, MURF-1 has been suggested to target a specific set of myofibrillar proteins, including titin but not myosin (36), whereas MAFbx has been associated with specific degradation of myosin (37). Previous data from our group indicating preserved titin but reduced myosin content in COPD diaphragm (8) are in line with this concept.
Increased Caspase-3 Activity in COPD Diaphragm
Although the bulk of contractile protein degradation in several models of atrophy involves the ubiquitin–proteasome pathway, this proteolytic pathway cannot degrade intact myosin or actin (24). Evidence suggests that caspase-3 (25) and calpains (38) play a role in contractile protein degradation upstream of the ubiquitin–proteasome pathway (for a schematic representation of the proposed roles of these proteolytic systems in muscle atrophy, see Jackman and Kandarian [39]). Du and coworkers (25) have shown that caspase-3 activation is an initial step in contractile protein degradation by cleavage of actomyosin complexes, thereby rendering actin fragments. Degraded actomyosin complexes can be ubiquitinated and degraded in the 26S proteasome complex. As a result, caspase-3–mediated contractile protein cleavage has been proposed to be an early rate-limiting step in contractile protein degradation during muscle atrophy. Our data provide the first evidence of caspase-3 activation in the diaphragm of patients with COPD. The coactivation of caspase-3 and the ubiquitin–proteasome pathway in COPD diaphragm is in line with previous studies investigating mechanisms of muscle atrophy (25, 40), and supports a role for caspase-3 in the loss of diaphragmatic contractile protein content in these patients.
Potential Triggers for Activation of the Ubiquitin–Proteasome Pathway and Caspase-3 in COPD Diaphragm
Potential triggers for the observed activation of the ubiquitin–proteasome pathway and caspase-3 in COPD diaphragm might include increased local generation of proinflammatory cytokines and reactive oxygen species. Studies show increased mRNA levels of tumor necrosis factor-α and its receptors in the diaphragm of patients with moderate COPD (41). Interestingly, the selective loss of myosin during cancer cachexia appears to be mediated by cytokine-induced activation of the ubiquitin–proteasome pathway, such as induction of MAFbx (14). It could be speculated that increased local expression of proinflammatory cytokines triggers the ubiquitin–proteasome pathway and the loss of myosin in the diaphragm of patients with mild to moderate COPD. Also, oxidative stress has been shown to occur in the diaphragm of patients with severe COPD (10). Like proinflammatory cytokines, reactive oxygen species are known stimulators of the ubiquitin–proteasome pathway in muscle (42, 43) and they might therefore contribute to diaphragm wasting in these patients.
Therapeutic Implications: A Role for Inhibitors of the Ubiquitin–Proteasome Pathway or Caspase-3 Activity?
The proteasome inhibitor bortezomib, has been approved for treatment of multiple myeloma in humans (44). Animal studies have shown that in vivo administration of bortezomib in a model of disuse atrophy prevented muscle wasting by about 50% (45). Similar results were found with administration of bortezomib in a rat model of denervation atrophy (46). In both studies bortezomib was well tolerated and no signs of toxicity were observed. These findings show promise for the use of proteasome inhibitors in syndromes associated with muscle wasting, such as the diaphragm in COPD.
Theoretically, inhibition of E3-ligases rather than the proteasome provides an ideal drug target, because E3-ligases have high substrate selectivity. Therefore, a specific inhibitor of, for instance, MAFbx should be a highly specific drug with probably few side effects, and might prove beneficial in preserving contractile protein content and preventing diaphragm weakness in COPD diaphragm. Unfortunately, to date no ubiquitin-ligase inhibitor has reached the clinic.
Finally, Supinski and Callahan (47) have shown that in vivo inhibition of caspase-3 activity attenuated sepsis-induced diaphragm weakness. This new finding might provide a therapeutic target upstream of the ubiquitin–proteasome pathway.
In conclusion, the present study for the first time demonstrates increased activation of the ubiquitin–proteasome pathway and increased caspase-3 activity in the diaphragm of patients with mild to moderate COPD. These findings provide a potential molecular mechanism underlying the development of diaphragm fiber atrophy and weakness in these patients.
Supplementary Material
Acknowledgments
The authors thank Dr. A. Verhagen (Radboud University Nijmegen Medical Center, The Netherlands), Dr. F. van den Elshout, Dr. S. van Sterkenburg, Dr. W. de Vries, Dr. T. Bloemen (Rijnstate Hospital Arnhem, The Netherlands), and Dr. F. Smeenk and Dr. B. van Straten (Catharina Hospital Eindhoven, The Netherlands) for collecting diaphragm muscle biopsies, and M. Beenes, M. Linkels, and C. Pigmans (Radboud University Nijmegen Medical Center, The Netherlands) for expert technical assistance.
Supported by an unrestricted educational grant from GlaxoSmithKline, The Netherlands; the Curtis Hankamer Basic Research Fund (Y.-P.L.); and the National Institutes of Health (AR049022; Y.-P.L.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200605-721OC on August 17, 2006
Conflict of Interest Statement: C.A.C.O.'s institution has received an unrestricted educational grant from GlaxoSmithKline. L.M.A.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. Y.-P.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.J. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. H.W.H.v.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.P.N.D.'s institution has received an unrestricted educational grant from GlaxoSmithKline.
References
- 1.Laghi F, Tobin MJ. Disorders of the respiratory muscles. Am J Respir Crit Care Med 2003;168:10–48. [DOI] [PubMed] [Google Scholar]
- 2.Gray-Donald K, Gibbons L, Shapiro SH, Macklem PT, Martin JG. Nutritional status and mortality in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 1996;153:961–966. [DOI] [PubMed] [Google Scholar]
- 3.Similowski T, Yan S, Gauthier AP, Macklem PT, Bellemare F. Contractile properties of the human diaphragm during chronic hyperinflation. N Engl J Med 1991;325:917–923. [DOI] [PubMed] [Google Scholar]
- 4.Cassart M, Pettiaux N, Gevenois PA, Paiva M, Estenne M. Effect of chronic hyperinflation on diaphragm length and surface area. Am J Respir Crit Care Med 1997;156:504–508. [DOI] [PubMed] [Google Scholar]
- 5.Orozco-Levi M, Lloreta J, Minguella J, Serrano S, Broquetas JM, Gea J. Injury of the human diaphragm associated with exertion and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2001;164: 1734–1739. [DOI] [PubMed] [Google Scholar]
- 6.Levine S, Kaiser L, Leferovich J, Tikunov B. Cellular adaptations in the diaphragm in chronic obstructive pulmonary disease. N Engl J Med 1997;337:1799–1806. [DOI] [PubMed] [Google Scholar]
- 7.Levine S, Gregory C, Nguyen T, Shrager J, Kaiser L, Rubinstein N, Dudley G. Bioenergetic adaptation of individual human diaphragmatic myofibers to severe COPD. J Appl Physiol 2002;92:1205–1213. [DOI] [PubMed] [Google Scholar]
- 8.Ottenheijm CAC, Heunks LMA, Hafmans T, van der Ven PF, Benoist C, Zhou H, Labeit S, Granzier HL, Dekhuijzen PNR. Titin and diaphragm dysfunction in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;173:527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ottenheijm CAC, Heunks LMA, Sieck GC, Zhan WZ, Jansen SM, Degens H, de Boo T, Dekhuijzen PNR. Diaphragm dysfunction in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2005;172:200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barreiro E, de la Puente B, Minguella J, Corominas JM, Serrano S, Hussain SN, Gea J. Oxidative stress and respiratory muscle dysfunction in severe chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2005;171:1116–1124. [DOI] [PubMed] [Google Scholar]
- 11.Levine S, Nguyen T, Kaiser LR, Rubinstein NA, Maislin G, Gregory C, Rome LC, Dudley GA, Sieck GC, Shrager JB. Human diaphragm remodeling associated with chronic obstructive pulmonary disease: clinical implications. Am J Respir Crit Care Med 2003;168:706–713. [DOI] [PubMed] [Google Scholar]
- 12.Ribera F, N'Guessan B, Zoll J, Fortin D, Serrurier B, Mettauer B, Bigard X, Ventura-Clapier R, Lampert E. Mitochondrial electron transport chain function is enhanced in inspiratory muscles of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2003;167:873–879. [DOI] [PubMed] [Google Scholar]
- 13.Huxley AF, Simmons RM. Proposed mechanism of force generation in striated muscle. Nature 1971;233:533–538. [DOI] [PubMed] [Google Scholar]
- 14.Acharyya S, Ladner KJ, Nelsen LL, Damrauer J, Reiser PJ, Swoap S, Guttridge DC. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J Clin Invest 2004;114:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chamberlain JS. Cachexia in cancer: zeroing in on myosin. N Engl J Med 2004;351:2124–2125. [DOI] [PubMed] [Google Scholar]
- 16.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 2004;18:39–51. [DOI] [PubMed] [Google Scholar]
- 17.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 2001;98:14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanely RA, Zergeroglu MA, Lennon SL, Sugiura T, Yimlamai T, Enns D, Belcastro A, Powers SK. Mechanical ventilation–induced diaphragmatic atrophy is associated with oxidative injury and increased proteolytic activity. Am J Respir Crit Care Med 2002;166:1369–1374. [DOI] [PubMed] [Google Scholar]
- 19.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998;67:425–479. [DOI] [PubMed] [Google Scholar]
- 20.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001;294:1704–1708. [DOI] [PubMed] [Google Scholar]
- 21.Wray CJ, Mammen JM, Hershko DD, Hasselgren PO. Sepsis upregulates the gene expression of multiple ubiquitin ligases in skeletal muscle. Int J Biochem Cell Biol 2003;35:698–705. [DOI] [PubMed] [Google Scholar]
- 22.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004;117:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinnell IW, Rudnicki MA. Molecular mechanisms of muscle atrophy. Cell 2004;119:907–910. [DOI] [PubMed] [Google Scholar]
- 24.Solomon V, Baracos V, Sarraf P, Goldberg AL. Rates of ubiquitin conjugation increase when muscles atrophy, largely through activation of the N-end rule pathway. Proc Natl Acad Sci USA 1998;95:12602–12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du J, Wang X, Miereles C, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE. Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J Clin Invest 2004;113:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobler SC, Williams A, Fischer D, Wang JJ, Sun X, Fischer JE, Monaco JJ, Hasselgren PO. Activity and expression of the 20S proteasome are increased in skeletal muscle during sepsis. Am J Physiol 1999;277: R434–R440. [DOI] [PubMed] [Google Scholar]
- 27.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med 2001;163:1256–1276. [DOI] [PubMed] [Google Scholar]
- 28.Mitch WE, Goldberg AL. Mechanisms of muscle wasting: the role of the ubiquitin–proteasome pathway. N Engl J Med 1996;335:1897–1905. [DOI] [PubMed] [Google Scholar]
- 29.Glickman MH, Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82:373–428. [DOI] [PubMed] [Google Scholar]
- 30.Minnaard R, Wagenmakers AJ, Combaret L, Attaix D, Drost MR, van Kranenburg GP, Schaart G, Hesselink MK. Ubiquitin–proteasome–dependent proteolytic activity remains elevated after zymosan-induced sepsis in rats while muscle mass recovers. Int J Biochem Cell Biol 2005;37:2217–2225. [DOI] [PubMed] [Google Scholar]
- 31.Price SR. Increased transcription of ubiquitin–proteasome system components: molecular responses associated with muscle atrophy. Int J Biochem Cell Biol 2003;35:617–628. [DOI] [PubMed] [Google Scholar]
- 32.Smith HJ, Mukerji P, Tisdale MJ. Attenuation of proteasome-induced proteolysis in skeletal muscle by β-hydroxy-β-methylbutyrate in cancer-induced muscle loss. Cancer Res 2005;65:277–283. [PubMed] [Google Scholar]
- 33.Combaret L, Taillandier D, Dardevet D, Bechet D, Ralliere C, Claustre A, Grizard J, Attaix D. Glucocorticoids regulate mRNA levels for subunits of the 19 S regulatory complex of the 26 S proteasome in fast-twitch skeletal muscles. Biochem J 2004;378:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones SW, Hill RJ, Krasney PA, O'Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J 2004;18:1025–1027. [DOI] [PubMed] [Google Scholar]
- 35.Leger B, Vergani L, Soraru G, Hespel P, Derave W, Gobelet C, D'Ascenzio C, Angelini C, Russell AP. Human skeletal muscle atrophy in amyotrophic lateral sclerosis reveals a reduction in Akt and an increase in atrogin-1. FASEB J 2006;20:583–585. [DOI] [PubMed] [Google Scholar]
- 36.Witt SH, Granzier H, Witt CC, Labeit S. MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J Mol Biol 2005; 350:713–722. [DOI] [PubMed] [Google Scholar]
- 37.Schulze PC, Fang J, Kassik KA, Gannon J, Cupesi M, MacGillivray C, Lee RT, Rosenthal N. Transgenic overexpression of locally acting insulin-like growth factor-1 inhibits ubiquitin-mediated muscle atrophy in chronic left-ventricular dysfunction. Circ Res 2005;97:418–426. [DOI] [PubMed] [Google Scholar]
- 38.Williams AB, Decourten-Myers GM, Fischer JE, Luo G, Sun X, Hasselgren PO. Sepsis stimulates release of myofilaments in skeletal muscle by a calcium-dependent mechanism. FASEB J 1999;13:1435–1443. [DOI] [PubMed] [Google Scholar]
- 39.Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol 2004;287:C834–C843. [DOI] [PubMed] [Google Scholar]
- 40.Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE. Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin–proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol 2004;15:1537–1545. [DOI] [PubMed] [Google Scholar]
- 41.Casadevall C, Barreiro E, Orozco-Levi M, Minguella J, Gea J. Local expression of tumor necrosis factor-α: is it the baddy or the goody in the story of respiratory muscle adaptation occurring in COPD? Am J Respir Crit Care Med 2006;173:A26. [Google Scholar]
- 42.Gomes-Marcondes MC, Tisdale MJ. Induction of protein catabolism and the ubiquitin–proteasome pathway by mild oxidative stress. Cancer Lett 2002;180:69–74. [DOI] [PubMed] [Google Scholar]
- 43.Li YP, Chen Y, Li AS, Reid MB. Hydrogen peroxide stimulates ubiquitin-conjugating activity and expression of genes for specific E2 and E3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol 2003; 285:C806–C812. [DOI] [PubMed] [Google Scholar]
- 44.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben Yehuda D, Lonial S, Goldschmidt H, et al. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med 2005;352:2487–2498. [DOI] [PubMed] [Google Scholar]
- 45.Krawiec BJ, Frost RA, Vary TC, Jefferson LS, Lang CH. Hindlimb casting decreases muscle mass in part by proteasome-dependent proteolysis but independent of protein synthesis. Am J Physiol Endocrinol Metab 2005;289:E969–E980. [DOI] [PubMed] [Google Scholar]
- 46.Beehler BC, Sleph PG, Benmassaoud L, Grover GJ. Reduction of skeletal muscle atrophy by a proteasome inhibitor in a rat model of denervation. Exp Biol Med (Maywood) 2006;231:335–341. [DOI] [PubMed] [Google Scholar]
- 47.Supinski GS, Callahan LA. Caspase activation contributes to endotoxin induced diaphragm weakness. J Appl Physiol 2006;100:1770–1777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.