Abstract
Escherichia coli O157:H7 is a leading cause of food-borne illness. This human pathogen produces Shiga toxins (Stx1 and Stx2) which inhibit protein synthesis by inactivating ribosome function. The present study describes a novel cell-based assay to detect Stx2 and inhibitors of toxin activity. A Vero cell line harboring a destabilized variant (half-life, 2 h) of the enhanced green fluorescent protein (d2EGFP) was used to monitor the toxin-induced inhibition of protein synthesis. This Vero-d2EGFP cell line produced a fluorescent signal which could be detected by microscopy or with a plate reader. However, a greatly attenuated fluorescent signal was detected in Vero-d2EGFP cells that had been incubated overnight with either purified Stx2 or a cell-free culture supernatant from Stx1- and Stx2-producing E. coli O157:H7. Dose-response curves demonstrated that the Stx2-induced inhibition of enhanced green fluorescent protein fluorescence mirrored the Stx2-induced inhibition of overall protein synthesis and identified a picogram-per-milliliter threshold for toxin detection. To establish our Vero-d2EGFP assay as a useful tool for the identification of toxin inhibitors, we screened a panel of plant compounds for antitoxin activities. Fluorescent signals were maintained when Vero-d2EGFP cells were exposed to Stx1- and Stx2-containing medium in the presence of either grape seed or grape pomace extract. The antitoxin properties of the grape extracts were confirmed with an independent toxicity assay that monitored the overall level of protein synthesis in cells treated with purified Stx2. These results indicate that the Vero-d2EGFP fluorescence assay is an accurate and sensitive method to detect Stx2 activity and can be utilized to identify toxin inhibitors.
Shiga toxin-producing Escherichia coli, with E. coli O157:H7 as the most common serotype, is an enteric pathogen known to cause human gastrointestinal illnesses ranging from bloody diarrhea and hemorrhagic colitis to life-threatening hemolytic-uremic syndrome (HUS) (1, 20). It has been estimated that E. coli O157 causes approximately 73,000 cases of illness per year in the United States from food- and waterborne sources. Shiga toxins (Stx1 and Stx2) are major virulence factors in E. coli O157 pathogenicity. These toxins inhibit protein synthesis by inactivating the ribosome and are thought to contribute to the development of HUS, a potentially fatal disease for which treatment is currently limited to supportive care (13, 14, 26). Toxin inactivation would prevent the development of HUS, but antitoxin therapeutics are not currently available (26). Detection methods to prevent the distribution of E. coli O157 in foods are thus an important component of food safety programs.
The rise in food-related outbreaks of E. coli O157 infection has heightened the importance of developing better methods to rapidly detect and characterize Stxs from E. coli O157 strains (26). Several methods have been developed to examine Stx activity against mammalian cells. Current assays that measure the viability of intoxicated Vero cells require several days of incubation and often produce poor quantitative data (5, 9, 19). Other methods that are more quantitative and sensitive measure the incorporation of radioactive amino acids into newly synthesized proteins (6, 15). However, these radioactivity assays are complex and laborious and allow only a limited number of conditions to be examined. A quantitative luciferase-based assay was recently developed to measure Stx toxicity in a high-throughput format (31), but this system requires several preparatory and processing steps to detect luciferase expression.
In the present study, we describe a simple cell-based assay for the detection of Stx2 and inhibitors of toxin activity by using a Vero cell line that expresses a destabilized variant (half-life, 2 h) of the enhanced green fluorescent protein (d2EGFP) to monitor the Stx2-induced inhibition of protein synthesis. This cell-based Vero-d2EGFP assay was used to screen a panel of natural compounds for anti-Stx activities, and we found that grape seed and grape pomace extracts both provided strong cellular protection against Stx2.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli O157:H7 strain RM1697 (environmental isolate 42 [stx1+ stx2+]) was isolated from beef cattle feed yards in Kansas (8). E. coli O157:H7 strain RM4876 (stx1 and stx2 negative) was isolated from a watershed sample collected in the Salinas Valley region in California (4). Multiplex PCR assays showed that both E. coli O157 strains RM1697 and RM4876 possessed the virulence genes fliC (for flagellin), eae (for the intimin adherence protein), and hly (for hemolysin); however, only strain RM1697 possessed stx1 and stx2 genes (data not shown). Nonpathogenic E. coli K-12 strain 5034 (ATCC 29425) was obtained from the American Type Culture Collection (Manassas, VA). Bacterial cultures were propagated in Luria-Bertani agar (Difco, Detroit, MI) or grown aerobically with constant shaking (200 rpm) in Luria-Bertani broth at 37°C.
Plant compounds.
Gold grape seed extract, grape pomace (skin) extract, and red wine concentrate were obtained from Polyphenolics (Madera, CA). Caffeic acid (3,4-dihydoxy-cinnamic acid) was purchased from Sigma-Aldrich (St. Louis, MO) and recrystallized from 95% ethanol before use. All tested plant compounds were used at nontoxic concentrations as assessed by a colorimetric cell viability assay using the cell proliferation reagent WST-1 (Roche Applied Science, Indianapolis, IN). Before each experiment, the plant compounds were prepared fresh from powdered stocks. All compounds, soluble in aqueous solutions, were readily dissolved at 10-mg/ml working concentrations in Ham's F-12 complete growth medium. The plant compounds and toxins were combined, and this mixture was immediately added to the cultured mammalian cells.
Culture and generation of the Vero-d2EGFP cell line.
The Vero CCL-81 cell line (American Type Culture Collection, Manassas, VA) was grown to 80% confluence in a six-well plate and then transfected with the pd2EGFP-N1 plasmid (BD Biosciences, Palo Alto, CA) by using Lipofectamine according to the instructions of the manufacturer (Invitrogen, Carlsbad, CA). After an overnight incubation, 20% of the cells were transferred into a 10-cm dish. After another overnight incubation, the cells were challenged with Geneticin at a final concentration of 1 mg/ml. The drug-containing medium was replaced every 3 days until colonies were visible. Individual colonies were isolated and serially passaged under continued selective pressure until they reached confluence in a 10-cm dish. At this point, a FACSCalibur cell sorter (BD, Franklin Lakes, NJ) was used to isolate the subpopulation of cells in each colony with the strongest fluorescence output. One of these subclones, the Vero-d2EGFP cell line, was used for all experimental studies. No appreciable loss of fluorescence from the Vero-d2EGFP cell line was detected over 5 months of continual passage. Both Vero and Vero-d2EGFP cells were grown at 5% CO2 and 37°C under humidified conditions in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Vero-d2EGFP cells were propagated in Dulbecco's modified Eagle medium containing 1 mg/ml Geneticin. All cell culture reagents were purchased from Gibco BRL (Grand Island, NY), with the exception of fetal bovine serum, which was purchased from Atlanta Biologicals (Lawrenceville, GA).
Microscopy analysis of Stx-treated Vero-d2EGFP cells.
Vero-d2EGFP cells were seeded at 5,000 cells per well onto Lab-Tek chambered cover glass (Nalge Nunc Corp., Naperville, IL). After 1 day, Vero-d2EGFP cells were treated for 16 h with Ham's F-12 complete growth medium containing 10-fold dilutions of cell-free supernatant from a stationary-phase culture of E. coli strain RM1697. Digital phase-contrast and fluorescence images were captured using a Phoenix digital frame grabber (Active Silicon, Chelmsford, MA) attached to a DMR light microscope featuring a PL Fluotar 40× lens objective with a numerical aperture of 0.7 and a green fluorescent protein filter cube (Leica Microsystems, Inc., Bannockburn, IL).
Immunoblot analysis of Stx2.
Various amounts of purified Stx2 (List Biological Laboratories, Campbell, CA) and protein samples in cell-free culture supernatants from E. coli strains RM1697, RM4876, and RM5034 were separated on a sodium dodecyl sulfate-10 to 20% polyacrylamide gradient gel (Bio-Rad, Hercules, CA) and then transferred onto a Whatman Schleicher and Schuell Protran (BA-83) nitrocellulose membrane. Immunoblot analysis was performed with a 1:200 dilution of the mouse monoclonal anti-Stx2A antibody VT 135/6-B9 (Sifin GmbH, Berlin, Germany). After subsequent incubation with a horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA), immunoreactive proteins were detected by chemiluminescence with the SuperSignal West Dura extended-duration substrate (ThermoScientific, Rockford, IL). AlphaEaseFC image analysis software (Alpha Innotech Corp., San Leandro, CA) was used for protein quantification by spot densitometry.
Fluorescence-based toxicity assay.
One day before intoxication, Vero-d2EGFP cells were seeded into Ham's F-12 complete growth medium at 10,000 cells per well in Greiner black 96-well microplates with clear bottoms (VWR International, Aurora, CO). The Vero-d2EGFP cells (∼80% confluence) were then exposed to Ham's F-12 medium containing either purified Stx2 or 10-fold serial dilutions of cell-free supernatants from stationary-phase cultures of E. coli strains RM1697, RM4876, and RM5034. Toxin-treated cells were grown for 16 h at 37°C in a 5% CO2 humidified incubator before enhanced green fluorescent protein (EGFP) fluorescence was measured on a Synergy HT multidetection microplate reader (BioTek, Winooski, VT) with the 485/20-nm excitation filter and the 528/20-nm emission filter. Results for toxin-treated cells were expressed as percentages of the values obtained for control cells incubated without toxin. When additional reagents were added to the intoxicated cells, a corresponding control condition with that reagent was used for the control cells incubated without toxin.
Toxicity assay based on overall protein synthesis.
Vero cells were seeded into a 24-well plate and grown to 80% confluence. The medium was then removed and replaced with serum-free medium containing the toxin concentrations stated in the figures. When indicated, grape seed or grape pomace extract was also present in the serum-free medium. After 2 or 16 h of incubation, the cells were washed with phosphate-buffered saline and incubated in methionine-free medium for 30 min at 37°C. The cells were subsequently exposed to 0.25 ml of methionine-free medium supplemented with 2.5 μCi of [35S]methionine (PerkinElmer, Waltham, MA). Following 15 min of radiolabeling at 37°C, the cells were bathed at 4°C in phosphate-buffered saline containing 10% trichloroacetic acid in sequential 30- and 15-min washes. Cell lysates generated in 0.2 N NaOH were loaded into scintillation vials, and the acid-precipitated radiolabel was quantified with an LS 6500 multipurpose scintillation counter (Beckman Coulter, Fullerton, CA). Results for toxin-treated cells were expressed as percentages of the values obtained for control cells incubated without toxin. When additional reagents were added to the intoxicated cells, a corresponding control condition with that reagent was used for the control cells incubated without toxin. All measurements were performed in triplicate.
RESULTS
Cell-based fluorescence assay to monitor Stx activity.
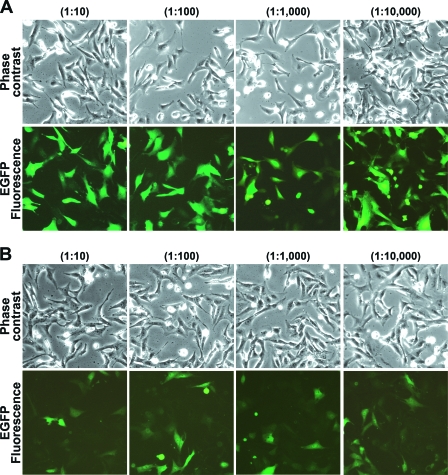
The Vero-d2EGFP cell line was generated as described in Materials and Methods. Vero cells were chosen for this study because they are highly sensitive to Stxs (9). Fluorescence microscopy demonstrated that EGFP expression was readily apparent in the Vero-d2EGFP cell line (Fig. 1A), whereas no fluorescence was observed in the parental Vero cells under identical imaging conditions (data not shown). Exposure to 10-fold dilutions of cell-free culture supernatant from non-Stx-producing E. coli O157 strain RM4876 did not affect Vero-d2EGFP fluorescence (Fig. 1A), but an obvious overall decrease in the fluorescence intensity of Vero-d2EGFP cells occurred after the cells were incubated with culture supernatant from Stx1- and Stx2-producing E. coli O157 strain RM1697 (Fig. 1B). Vero-d2EGFP cells in Ham's F-12 growth medium exhibited the same level of fluorescence as Vero-d2EGFP cells cultivated in the same growth medium supplemented with the culture supernatant from non-Stx-producing E. coli O157 strain RM4876 (data not shown). Thus, exposure to the culture supernatant from an Stx1- and Stx2-producing strain specifically eliminated the fluorescent signal from Vero-d2EGFP cells.
FIG. 1.
Fluorescence intensity of Vero-d2EGFP cells in the absence or presence of Stx. Vero-d2EGFP cells were incubated for 16 h with the indicated 10-fold dilutions of stationary-phase culture supernatant from non-Stx-producing E. coli O157 strain RM4876 (A) or Stx1- and Stx2-producing E. coli O157:H7 strain RM1697 (B). Digital phase-contrast and fluorescence images are shown.
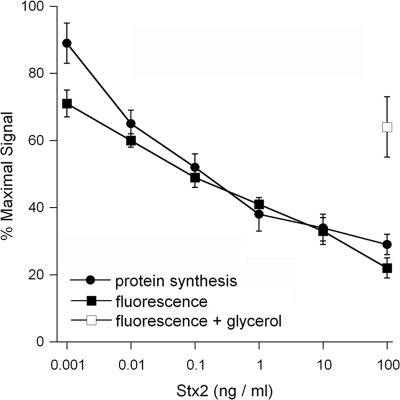
Epidemiological studies suggest that Stx2 may be more virulent than Stx1 since Stx2-expressing strains appear to be more frequently associated with HUS than strains harboring Stx1 or both Stx1 and Stx2 (3, 7, 17, 25). Therefore, we further examined the quantitative loss of EGFP fluorescence in toxin-treated cells by using a plate reader to perform the Vero-d2EGFP assay with purified Stx2 (Fig. 2). Vero-d2EGFP cells were treated with various concentrations of Stx2 for 16 h before toxicity was assessed with our fluorescence-based assay or with a standard assay to monitor the overall level of protein synthesis. Both assays recorded a dose-dependent loss of the signal, and the Stx-induced inhibition of EGFP fluorescence mirrored the Stx-induced inhibition of overall protein synthesis. A 50% effective concentration (EC50) of Stx2 of 100 pg/ml was calculated with both the Vero-d2EGFP assay and the overall protein synthesis assay. In addition, both assays could reproducibly detect the biological activity of Stx2 at a concentration of just 10 pg/ml. Taken together, these results confirmed that our assay could be used to quantify the extent of intoxication. Our assay could also be used to identify toxin inhibitors, as a substantial fluorescent signal was detected when Vero-d2EGFP cells were coincubated with 100 ng/ml of Stx2 and 10% glycerol (Fig. 2). Previous studies have demonstrated that glycerol treatment confers cellular resistance to the plant toxin ricin (23) and to the plasmid-encoded toxin of enteroaggregative E. coli (24). The results of our assay showed that glycerol treatment confers resistance to Stx2 as well.
FIG. 2.
Effect of Stx2 on Vero-d2EGFP fluorescence and overall protein synthesis. Protein synthesis and fluorescence were measured in separate samples of Vero-d2EGFP cells after a 16-h incubation with the indicated concentrations of Stx2. The means ± standard errors of the means of at least three independent experiments with triplicate samples for each condition are shown. The open square represents the average ± the range of two experiments with cells that were coincubated with 100 ng/ml of Stx2 and 10% glycerol.
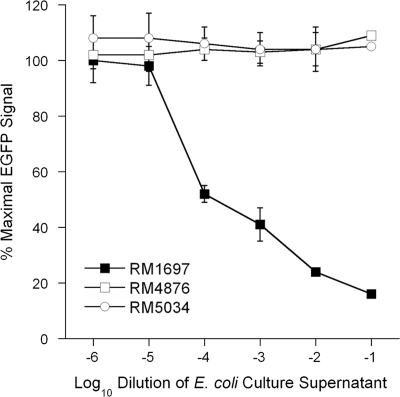
A dose-response curve was also generated with cell-free culture supernatant from Stx1- and Stx2-producing E. coli O157 strain RM1697 (Fig. 3). Vero-d2EGFP cells were exposed to 10-fold dilutions of cell-free culture supernatant from strain RM1697, and the toxin-induced loss of EGFP fluorescence was detected with a plate reader. As negative controls, Vero-d2EGFP cells were treated with dilutions of cell-free culture supernatant from non-Stx-producing E. coli O157 strain RM4876 or from nonpathogenic E. coli strain RM5034. A dose-dependent inhibition of EGFP fluorescence was documented for the toxin-containing supernatant from strain RM1697, whereas no loss of EGFP fluorescence was observed in the cells treated with supernatant from strain RM4876 or RM5034. The loss of the fluorescent signal was therefore directly correlated to the presence of Stx in the stationary-phase culture supernatant from strain RM1697. This finding was consistent with the fluorescent images presented in Fig. 1. Visualization of the toxic effect by fluorescence microscopy suggested that the EGFP signal had been substantially eliminated by a 1:10,000 dilution of the culture supernatant from strain RM1697 (Fig. 1B), but the plate reader indicated that this dilution only reduced the fluorescent signal to 50% of the control value for unintoxicated cells. The plate reader was obviously more sensitive to subtle changes in EGFP fluorescence than visual inspection. These results demonstrated that the qualitative toxic effect documented by microscopy could be quantified with a plate reader.
FIG. 3.
Effect of E. coli culture supernatants on Vero-d2EGFP fluorescence. Vero-d2EGFP cells were exposed to Ham's F-12 medium containing 10-fold dilutions of culture supernatant from Stx1- and Stx2-producing E. coli O157 strain RM1697, non-Stx-producing E. coli O157 strain RM4876, or nonpathogenic E. coli strain RM5034. After 16 h of incubation, the fluorescence output was recorded with a plate reader. The averages ± standard deviations of three independent experiments with duplicate samples for each condition are shown.
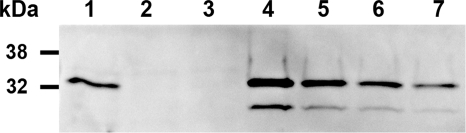
Immnunoblot analysis was used to quantify the amount of Stx2 in the cell-free culture supernatant from strain RM1697 (Fig. 4). By comparing the immunoblot signal intensities for known concentrations of purified Stx2, we estimated that the supernatant from strain RM1697 contained about 1.5 μg/ml of Stx2. No Stx2 was detected in the culture supernatants from strains RM4876 and RM5034 (Fig. 4). Based upon our quantitative assessment, the amounts of Stx2 used for the dose-response curve in Fig. 3 for cells exposed to strain RM1697 culture supernatant ranged from approximately 1.5 pg/ml to 150 ng/ml, which was similar to the range of toxin concentrations used for the dose-response curve for purified Stx2 (Fig. 2). In fact, the toxicity assays with RM1697 culture supernatant and with purified Stx2 generated similar dose-response curves and nearly identical EC50s of ∼100 pg of Stx2/ml. These comparisons do not account for the presence of Stx1 in the RM1697 culture supernatant, but our preliminary analysis of other Stx-producing O157 strains has indicated that Stx2 is more potent than Stx1 in our Vero-d2EGFP assay (data not shown). The sensitive and reproducible detection of Stxs in a cell-free culture supernatant suggested that our Vero-d2EGFP assay can be used to screen E. coli isolates for Stx production.
FIG. 4.
Immunoblot analysis of Stx2 expression in E. coli culture supernatants. Samples of cell-free E. coli culture supernatants (35 μl) and various amounts of purified Stx2 were separated on a sodium dodecyl sulfate-10 to 20% polyacrylamide gradient gel, transferred onto nitrocellulose, and probed with the mouse monoclonal anti-Stx2 antibody VT 135/6-B9 (Sifin GmbH, Berlin, Germany), which recognizes the 32-kDa A subunit and the 27-kDa A1 fragment of Stx2 (10). Molecular masses of the protein standards are shown to the left. Lanes: 1, Stx1- and Stx2-producing E. coli O157 strain RM1697; 2, non-Stx-producing E. coli O157 strain RM4876; 3, nonpathogenic E. coli strain RM5034; 4, 0.25 μg of Stx2; 5, 0.125 μg of Stx2; 6, 0.06 μg of Stx2; and 7, 0.03 μg of Stx2.
The Vero-d2EGFP assay can be used to identify Stx inhibitors.
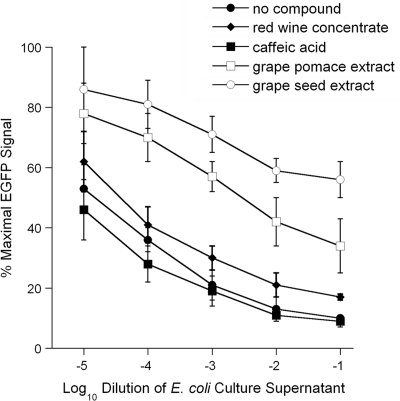
Previous work has shown that certain plant compounds inhibit the cytopathic action of cholera toxin (12, 16, 22). We accordingly used our Vero-d2EGFP system to screen a panel of plant compounds for anti-Stx activities. Cells were exposed to various dilutions of Stx1- and Stx2-containing medium and a single concentration of a plant compound. A preliminary screen of apple skin extract, chlorogenic acid, N-acetyl-l-cysteine, epigallocatechin gallate, theaflavin-3-gallate, trans-cinnamaldehyde, green tea polyphenols, and oregano oil did not identify any Stx inhibitors (data not shown). However, a second round of screening isolated two promising plant compounds: grape seed extract and grape pomace extract (Fig. 5). In contrast, caffeic acid and red wine concentrate failed to inhibit Stxs present in the culture supernatant from strain RM1697 (Fig. 5). The concentration of Stx1- and Stx2-containing medium required to reach the EC50 for cells exposed to grape seed extract was at least 6,700-fold greater than that required for the control cells intoxicated in the absence of plant compounds. Likewise, the concentration of Stx1- and Stx2-containing medium required to reach the EC50 for cells exposed to grape pomace extract was 200-fold greater than that required for the control cells intoxicated in the absence of plant compounds (Fig. 5). Thus, both grape seed and grape pomace extracts strongly inhibited intoxication of the Vero-d2EGFP cell line.
FIG. 5.
Effect of plant compounds on fluorescence output from Stx-treated Vero-d2EGFP cells. Vero-d2EGFP cells were incubated for 16 h with plant polyphenolic compounds and various dilutions of a cell-free culture supernatant from E. coli O157:H7 strain RM1697. The fluorescence output was then recorded with a plate reader. Cells were coincubated with no plant compound, 0.5 mg/ml of caffeic acid, 1 mg/ml of red wine concentrate, 0.5 mg/ml of grape pomace extract, or 0.5 mg/ml of grape seed extract. The averages ± standard deviations of three independent experiments with triplicate samples for each condition are shown.
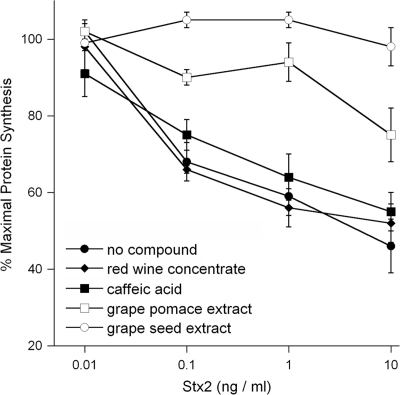
The antitoxin properties of our plant compounds were verified by an independent toxicity assay that monitored the overall level of protein synthesis in cells exposed to purified Stx2 (Fig. 6). A short, 2-h intoxication was used for this experiment in order to detect any subtle antitoxin effects that might have been missed by the 16-h intoxication in the Vero-d2EGFP fluorescence assay. Identical Stx2 dose-response curves were obtained for untreated Vero-d2EGFP cells, Vero-d2EGFP cells treated with caffeic acid, and Vero-d2EGFP cells treated with red wine concentrate. Thus, caffeic acid and red wine concentrate did not inhibit the cytotoxic effects of Stx2. In contrast, Vero-d2EGFP cells incubated with grape seed extract or grape pomace extract were strongly protected from Stx2 intoxication. Consistent with the results of the Vero-d2EGFP fluorescence assay, our protein synthesis assay found that grape seed extract provided greater protection against Stx2 than grape pomace extract. Collectively, these data demonstrated that both grape seed extract and grape pomace extract were indeed Stx inhibitors. Therefore, the Vero-d2EGFP assay can be used to detect both Stx2 and inhibitors of toxin activity.
FIG. 6.
Effect of plant compounds on protein synthesis levels in Stx2-treated Vero-d2EGFP cells. Protein synthesis in Vero-d2EGFP cells was measured after a 2-h coincubation with plant polyphenolic compounds and Stx2. Cells were coincubated with no plant compound, 1 mg/ml of caffeic acid, 1 mg/ml of red wine concentrate, 0.5 mg/ml of grape pomace extract, or 0.1 mg/ml of grape seed extract. The means ± standard errors of the means of at least four independent experiments with triplicate samples for each condition are shown.
DISCUSSION
The recent food-borne outbreaks of E. coli O157:H7 infection make it a public health problem of serious concern. Numerous other food-borne diseases also result from ingesting foods that are contaminated with bacterial toxins (1, 20). Thus, in order to improve food safety, there is a need for technologies to detect and/or inactivate bacterial toxins. The objective of this study was to develop and validate a simple yet quantitative method for the detection of Stx2 from E. coli O157. In addition, we were able to adapt our method for the identification of toxin inhibitors.
A Vero-d2EGFP cell line which expresses a destabilized variant (half-life, 2 h) of EGFP was used to detect Stx activity. Since Stxs inhibit protein synthesis, toxin-susceptible cells will degrade d2EGFP and will not produce more of the protein. Exposure to Stx will consequently eliminate the fluorescent signal from Vero-d2EGFP cells. This simple and quantitative assay has a number of advantages over current technologies for toxin detection. Monitoring Stx activity as a function of cell viability requires several days of incubation and often produces poor quantitative data (2, 19). Other assays that are more quantitative and sensitive measure the incorporation of radioactive amino acids into newly synthesized proteins (6, 15). Yet these assays are laborious, require the use of radioisotopes, and allow only a limited number of conditions to be examined. Nonradioactive cell-free assays have recently been described; however, these assays require high concentrations of toxin and/or commercially purchased kits to monitor the toxin-induced inhibition of protein synthesis (18, 27). A more recent cell-free method that utilizes an immuno-PCR assay for toxin detection appears to exhibit the same general level of sensitivity as our Vero-d2EGFP assay (30). Finally, Zhao and Haslam (31) have described a system similar to our Vero-d2EGFP assay in which the toxin-induced loss of luciferase expression (and therefore light output) is used to monitor intoxication. A high-throughput screen of toxin inhibitors has been executed with this system (21), but in comparison to our assay, the luciferase system requires additional preparatory steps, time, and end point sample processing. Our assay requires only three basic steps: (i) seeding of cells into a 96-well plate, (ii) the addition of culture supernatants or purified toxin, and (iii) reading of the fluorescence output with a plate reader after 16 h of intoxication. This procedure does not require radioisotopes, commercial kits, or additional processing steps. Since the assay is performed in a 96-well format, it is also possible to screen large numbers of samples at once.
Dose-response curves for purified Stx2 demonstrated that the toxin-induced inhibition of EGFP fluorescence mirrored the toxin-induced inhibition of overall protein synthesis. The Vero-d2EGFP assay thus produced reliable, quantitative results with picogram-per-milliliter quantities of either purified toxin or toxin present in cell-free bacterial culture supernatants. The sensitive, quantitative nature of our assay suggests that it may be useful for detecting differences in E. coli O157 strain virulence related to toxigenic potential or for detecting differences in the biological activities of Stx2 variants. However, the Vero-d2EGFP assay is not specific for Stx but can instead be used to detect any toxin that inhibits protein synthesis. This limitation on specificity also applies to the standard toxicity assays which measure cell viability or the incorporation of radiolabeled amino acids into newly synthesized proteins. Additional confirmatory procedures would therefore be required if the Vero-d2EGFP assay was used in a clinical application to screen specifically for Stx2. Our present study focused on assay development and the quantification of Stx2, but future studies should validate the Vero-d2EGFP assay as a tool to detect Stx1 as well.
As seen by microscopy, a small number of Vero-d2EGFP cells were still fluorescent after exposure to all the dilutions of toxin-containing medium. Likewise, the plate reader detected some residual fluorescence from Vero-d2EGFP cells challenged with the highest Stx2 concentration of 100 ng/ml. Vero cells express the Gb3 receptor of Stx in a cell cycle-dependent manner, so the subpopulation of apparently toxin-resistant cells most likely represented those cells that did not bind Stx during G0/G1 and S phases of the cell cycle (11).
The Vero-d2EGFP assay can also be used to identify toxin inhibitors. Daio (Rhei rhizoma), apple, hop bract, and green tea extracts have been shown previously to inhibit cholera intoxication or the release of Stx from E. coli O157 (12, 16, 22, 28). This evidence suggests that plant polyphenols and other plant compounds are potential therapeutic agents that can be used to protect consumers against food-borne, toxin-mediated disease. We accordingly tested a select panel of plant compounds for anti-Stx activities and identified two grape extracts with putative anti-Stx properties. The grape seed and grape pomace extracts were bona fide Stx2 inhibitors as determined by an independent assay based upon the overall levels of protein synthesis in toxin-treated cells. Yu and Haslam have also used a destabilized EGFP construct to identify an endoplasmic reticulum-localized Hsp40 protein that, when expressed as a dominant negative truncation, protects Vero cells from Stx1 (29). However, this work did not further characterize the EGFP system with toxin dose-response curves, direct comparisons to alternative toxicity assays, or the use of Stx-containing culture supernatants. Collectively, our results and the work of Yu and Haslam demonstrate the versatility of the Vero-d2EGFP assay as a tool that can be used for toxin detection, the isolation of toxin inhibitors, and the identification of host factors involved in the intoxication process.
The active antitoxin component(s) in the grape seed and grape pomace extracts has yet to be identified, as has the molecular basis for the antitoxin effect. The grape extracts and toxins were applied simultaneously to the cells, so toxin resistance may result from a number of possible mechanisms: toxin neutralization before cell contact, the inhibition of toxin binding to the host cell, alterations to toxin trafficking or translocation, and the disruption of toxin-target interactions may each account for the extract-induced toxin resistance. A comprehensive set of future studies with the Vero-d2EGFP assay will distinguish among these possibilities and provide a molecular basis for cellular protection against Stx2. Identification of the antitoxin compound(s) will also be accomplished by using the Vero-d2EGFP assay to screen individual constituents of the grape extracts for anti-Stx2 activity. Since the active antitoxin component(s) of the grape extracts likely constitutes a small fraction of the total extract, it must be functional at much lower quantities than the 0.1- to 0.5-mg/ml extract concentrations used in our work. This suggests that the active antitoxin component(s) of the grape extracts may function at the low concentrations required for medicinal applications. Identification and further characterization of the anti-Stx2 component(s) in the grape extracts may thus lead to the development of new antitoxin therapeutics or prophylactics.
In summary, we have developed and validated a novel technique for the detection of Stx2 and inhibitors of toxin activity. The assay, which requires minimal labor, is simple and quantitative and provides reproducible results. It requires only the use of a fluorescence plate reader and black 96-well microplates with clear bottoms. The Vero-d2EGFP assay can also serve as a general platform for toxin detection: Vero cells are highly sensitive to many toxins, so the method can be used in conjunction with a range of toxins that inhibit protein synthesis. Finally, the 96-well format of the assay is amenable to high-throughput operations involving toxin detection or the identification of toxin inhibitors.
Acknowledgments
This work was supported by the USDA Agricultural Research Service through CRIS project number 5325-42000-045.
We thank Rommel D. Alfonso, Carol Levin, and Anna Bates for technical assistance. Special thanks to Robert E. Mandrell and Michael Cooley (USDA Agricultural Research Service, Western Regional Research Center, Albany, CA) for providing the bacterial strains used in this study.
Footnotes
Published ahead of print on 9 January 2009.
REFERENCES
- 1.Banatvala, N., P. M. Griffin, K. D. Greene, T. J. Barrett, W. F. Bibb, J. H. Green, and J. G. Wells. 2001. The United States National Prospective Hemolytic Uremic Syndrome Study: microbiologic, serologic, clinical, and epidemiologic findings. J. Infect. Dis. 183:1063-1070. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim, K. A., and L. Beutin. 2003. Rapid laboratory identification and characterization of verocytotoxigenic (Shiga toxin producing) Escherichia coli (VTEC/STEC). J. Appl. Microbiol. 95:205-217. [DOI] [PubMed] [Google Scholar]
- 3.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooley, M., D. Carychao, L. Crawford-Miksza, M. T. Jay, C. Myers, C. Rose, C. Keys, J. Farrar, and R. E. Mandrell. 2007. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS ONE 2:e1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 6.Hovde, C. J., S. B. Calderwood, J. J. Mekalanos, and R. J. Collier. 1988. Evidence that glutamic acid 167 is an active-site residue of Shiga-like toxin I. Proc. Natl. Acad. Sci. USA 85:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jelacic, S., C. L. Wobbe, D. R. Boster, M. A. Ciol, S. L. Watkins, P. I. Tarr, and A. E. Stapleton. 2002. ABO and P1 blood group antigen expression and stx genotype and outcome of childhood Escherichia coli O157:H7 infections. J. Infect. Dis. 185:214-219. [DOI] [PubMed] [Google Scholar]
- 8.Kimura, R., R. E. Mandrell, J. C. Galland, D. Hyatt, and L. W. Riley. 2000. Restriction-site-specific PCR as a rapid test to detect enterohemorrhagic Escherichia coli O157:H7 strains in environmental samples. Appl. Environ. Microbiol. 66:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konowalchuk, J., J. I. Speirs, and S. Stavric. 1977. Vero response to a cytotoxin of Escherichia coli. Infect. Immun. 18:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig, K., M. A. Karmali, V. Sarkim, C. Bobrowski, M. Petric, H. Karch, and D. E. Muller-Wiefel. 2001. Antibody response to Shiga toxins Stx2 and Stx1 in children with enteropathic hemolytic-uremic syndrome. J. Clin. Microbiol. 39:2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majoul, I., T. Schmidt, M. Pomasanova, E. Boutkevich, Y. Kozlov, and H. D. Söling. 2002. Differential expression of receptors for Shiga and cholera toxin is regulated by the cell cycle. J. Cell Sci. 115:817-826. [DOI] [PubMed] [Google Scholar]
- 12.Morinaga, N., Y. Iwamaru, K. Yahiro, M. Tagashira, J. Moss, and M. Noda. 2005. Differential activities of plant polyphenols on the binding and internalization of cholera toxin in vero cells. J. Biol. Chem. 280:23303-23309. [DOI] [PubMed] [Google Scholar]
- 13.Neill, M. A. 1998. Treatment of disease due to Shiga toxin-producing Escherichia coli: infectious disease management, p. 357-363. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, DC.
- 14.O'Brien, A. D., and J. D. Kaper. 1998. Shiga toxin-producing Escherichia coli: yesterday, today, and tomorrow, p. 1-11. In J. D. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, DC.
- 15.Obrig, T. G., C. B. Louise, C. A. Lingwood, B. Boyd, L. Barley-Maloney, and T. O. Daniel. 1993. Endothelial heterogeneity in Shiga toxin receptors and responses. J. Biol. Chem. 268:15484-15488. [PubMed] [Google Scholar]
- 16.Oi, H., D. Matsuura, M. Miyake, M. Ueno, I. Takai, T. Yamamoto, M. Kubo, J. Moss, and M. Noda. 2002. Identification in traditional herbal medications and confirmation by synthesis of factors that inhibit cholera toxin-induced fluid accumulation. Proc. Natl. Acad. Sci. USA 99:3042-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostroff, S. M., P. I. Tarr, M. A. Neill, J. H. Lewis, N. Hargrett-Bean, and J. M. Kobayashi. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J. Infect. Dis. 160:994-998. [DOI] [PubMed] [Google Scholar]
- 18.Pastrana, D. V., and D. J. FitzGerald. 2006. A nonradioactive, cell-free method for measuring protein synthesis inhibition by Pseudomonas exotoxin. Anal. Biochem. 353:266-271. [DOI] [PubMed] [Google Scholar]
- 19.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saenz, J. B., T. A. Doggett, and D. B. Haslam. 2007. Identification and characterization of small molecules that inhibit intracellular toxin transport. Infect. Immun. 75:4552-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito, T., M. Miyake, M. Toba, H. Okamatsu, S. Shimizu, and M. Noda. 2002. Inhibition by apple polyphenols of ADP-ribosyltransferase activity of cholera toxin and toxin-induced fluid accumulation in mice. Microbiol. Immunol. 46:249-255. [DOI] [PubMed] [Google Scholar]
- 23.Sandvig, K., I. H. Madshus, and S. Olsnes. 1984. Dimethyl sulphoxide protects cells against polypeptide toxins and poliovirus. Biochem. J. 219:935-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scaglione, P., K. N. Nemec, K. E. Burlingame, A. Grabon, J. Huerta, F. Navarro-Garcia, S. A. Tatulian, and K. Teter. 2008. Structural characteristics of the plasmid-encoded toxin from enteroaggregative Escherichia coli. Biochemistry 47:9582-9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scotland, S. M., G. A. Willshaw, H. R. Smith, and B. Rowe. 1987. Properties of strains of Escherichia coli belonging to serogroup O157 with special reference to production of Vero cytotoxins VT1 and VT2. Epidemiol. Infect. 99:613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serna, A., and E. C. Boedeker. 2008. Pathogenesis and treatment of Shiga toxin-producing Escherichia coli infections. Curr. Opin. Gastroenterol. 24:38-47. [DOI] [PubMed] [Google Scholar]
- 27.Song, S., J. Xue, K. Fan, G. Kou, Q. Zhou, H. Wang, and Y. Guo. 2005. Preparation and characterization of fusion protein truncated Pseudomonas exotoxin A (PE38KDEL) in Escherichia coli. Protein Expr. Purif. 44:52-57. [DOI] [PubMed] [Google Scholar]
- 28.Sugita-Konishi, Y., Y. Hara-Kudo, F. Amano, T. Okubo, N. Aoi, M. Iwaki, and S. Kumagai. 1999. Epigallocatechin gallate and gallocatechin gallate in green tea catechins inhibit extracellular release of Vero toxin from enterohemorrhagic Escherichia coli O157:H7. Biochim. Biophys. Acta 1472:42-50. [DOI] [PubMed] [Google Scholar]
- 29.Yu, M., and D. B. Haslam. 2005. Shiga toxin is transported from the endoplasmic reticulum following interaction with the luminal chaperone HEDJ/ERdj3. Infect. Immun. 73:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, W., M. Bielaszewska, M. Pulz, K. Becker, A. W. Friedrich, H. Karch, and T. Kuczius. 2008. New immuno-PCR assay for detection of low concentrations of Shiga toxin 2 and its variants. J. Clin. Microbiol. 46:1292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao, L., and D. B. Haslam. 2005. A quantitative and highly sensitive luciferase-based assay for bacterial toxins that inhibit protein synthesis. J. Med. Microbiol. 54:1023-1030. [DOI] [PubMed] [Google Scholar]