Abstract
The type III secretion system (T3SS) is a major virulence factor in many gram-negative bacterial pathogens. This secretion system translocates effectors directly into the cytosol of eukaryotic host cells, where the effector proteins facilitate bacterial pathogenesis by interfering with host cell signal transduction and other cellular processes. Plants defend themselves against bacterial pathogens by recognizing either the type 3 effectors or their actions and initiating a cascade of defense responses that often results in programmed cell death of the plant cell being attacked. Here we show that a plant phenolic compound, p-coumaric acid (PCA), represses the expression of T3SS genes of the plant pathogen Dickeya dadantii, suggesting that plants can also defend against bacterial pathogens by manipulating the expression of the T3SS. PCA repressed the expression of T3SS regulatory genes through the HrpX/Y two-component system, a core regulator of the T3SS, rather than through the global regulator GacS/A, which indirectly regulates the T3SS. A further analysis of several PCA analogs suggests that the para positioning of the hydroxyl group in the phenyl ring and the double bond of PCA may be important for its biological activity.
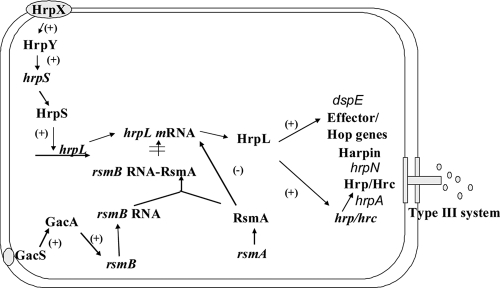
The enterobacterial plant pathogen Dickeya dadantii (formerly named Erwinia chrysanthemi) causes soft-rot, wilt, and blight diseases on a wide range of plant species (6). The family Enterobacteriaceae includes several other genera of plant pathogens, such as Erwinia, Pantoea, and Pectobacterium, and important animal pathogens, such as Escherichia coli, Salmonella spp., and Yersinia spp. Most of the enterobacterial pathogens, including D. dadantii, encode at least one type III secretion system (T3SS) (12), which is a protein secretion system capable of translocating virulence proteins directly into host cells (4, 13). Many bacterial species use a two-component system (TCS) to sense their environment and regulate genes in response to environmental changes. At least two TCSs regulate the D. dadantii 3937 T3SS, including the global regulatory system GacS/A and a TCS encoded by genes in the center of the T3SS gene cluster, HrpX/Y. In the GacS/A-rsmB-RsmA-HrpL regulatory pathway, GacS/A upregulates hrpL mRNA by upregulating rsmB and thereby inactivating RsmA, which would otherwise promote the degradation of hrpL mRNA (1, 22) (Fig. 1). GacS/A also induces the production of pectate lyases in D. dadantii 3937 (22). In the HrpX/Y-HrpS-HrpL regulatory pathway, the TCS HrpX/HrpY activates hrpS, which encodes an enhancer protein (Fig. 1) (25). HrpS interacts with a σ54 RNA polymerase holoenzyme and initiates the transcription of hrpL. HrpL is an alternative sigma factor that is required for the expression of genes encoding the T3SS effectors and structural components such as the units of the needle, the needle extension, and the translocon.
FIG. 1.
Regulatory network controlling the D. dadantii T3SS. The D. dadantii T3SS is regulated by the HrpX/HrpY-HrpS-HrpL and the GacS/GacA-rsmB-HrpL regulatory pathways. The TCS HrpX/HrpY activates hrpS, which encodes a σ54 enhancer. HrpS is required for expression of the alternative sigma factor gene hrpL. HrpL activates expression of genes encoding the T3SS apparatus and its secreted substrates. RsmA is a small RNA-binding protein that acts by lowering the half-life of hrpL mRNA. GacS/GacA upregulates the expression of rsmB, which increases the mRNA level of hrpL by sequestering RsmA. +, positive regulation; −, negative regulation.
The T3SS is an attractive target for development of antimicrobial compounds since it is present mainly in pathogenic gram-negative bacteria and is often required for virulence by these species (5, 18). We recently discovered that the phenolic compounds trans-cinnamic acid (TCA) and o-coumaric acid (OCA) induce the expression of D. dadantii T3SS genes hrpA and hrpN through the rsmB-RsmA pathway (21). To identify potential T3SS repressors, analogs and isomers of TCA and OCA were screened for effects on D. dadantii 3937 hrpA expression. An isomer of OCA, p-coumaric acid (PCA), that repressed the expression of T3SS genes of D. dadantii 3937 is identified in this study. Based on the chemical structures and inhibitory effect of PCA and several analogs of PCA on T3SS gene expression, the structure-activity relationship (SAR) and potential active sites of PCA are identified. To our knowledge, no mechanism for SAR studies on the inhibitory activity of small molecules against T3SS gene expression has ever been reported. Finally, the regulators responsible for the repression of T3SS gene expression by PCA are elucidated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and chemicals.
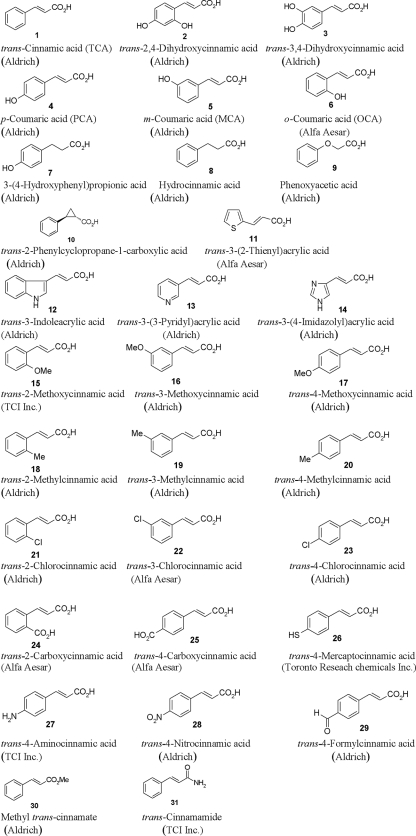
The bacterial strains and plasmids used in this study are listed in Table 1. D. dadantii was grown in Luria-Bertani or T3SS-inducing minimal medium (MM) at 28°C (23). Ampicillin at 100 μg/ml was used in this work. Chemical structures of OCA, PCA, TCA, and related phenolic compounds used in this study are shown in Fig. 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference or source |
|---|---|---|
| D. dadantii strains | ||
| 3937 | Wild type, Saintpaulia sp. (African violet) isolate | N. Hugouvieux-Cotte-Pattat |
| 3937(pAT) | 3937 containing pPROBE-AT | 14 |
| 3937(phrpA) | 3937 containing phrpA; Apr | 22 |
| 3937(phrpN) | 3937 containing phrpN; Apr | 23 |
| 3937(phrpL) | 3937 containing phrpL; Apr | 23 |
| 3937(phrpS) | 3937 containing phrpS; Apr | This work |
| 3937(pmrp) | 3937 containing pmrp; Apr | 14 |
| Plasmids | ||
| pPROBE-AT | Promoter-probe vector; Apr | 11 |
| phrpA | pProbe-AT derivative with PCR fragment containing hrpA promoter region; Apr | 22 |
| phrpN | pProbe-AT derivative with PCR fragment containing hrpN promoter region; Apr | 23 |
| phrpL | pProbe-AT derivative with PCR fragment containing hrpL promoter region; Apr | 23 |
| phrpS | pProbe-AT derivative with PCR fragment containing 709-bp hrpS promoter region; Apr | This work |
| pmrp | pProbe-AT derivative with PCR fragment containing mrp promoter region; Apr | 14 |
| p50HrpN0-117 | pCPP50 derivative with fragment encoding 3937 hrpN lacking nucleotides 50-117 | 24 |
| pCPP50::HrpN | pCPP50 derivative with fragment encoding 3937 hrpN | 24 |
Apr, ampicillin resistance.
FIG. 2.
Chemical structures of OCA, PCA, TCA, and related phenolic compounds.
Flow cytometry analysis.
Promoter activity of hrpA, hrpL, hrpN, and hrpS was determined in a FACSCalibur flow cytometer (BD Biosciences, CA) as described previously (14). The bacterial cells carrying the promoter reporter plasmid were grown in Luria-Bertani broth at 28°C overnight and transferred to appropriate media.
qRT-PCR analysis.
Total RNA from the bacterial cells was isolated by using the Tri reagent method (Sigma, MO) and treated with Turbo DNA-free DNase kits (Ambion, TX). The cDNA levels of target genes in different samples were quantified by quantitative reverse transcription-PCR (qRT-PCR) using Real Master Mix (Eppendorf, Westbury, NY) as described previously (14). qRT-PCR data were analyzed with the Relative Expression Software Tool as described previously (15), with rplU as an endogenous control for data analysis (9).
Western blot analysis.
Wild-type D. dadantii 3937 was grown at 28°C in MM or MM supplemented with different amounts of PCA for 24 h. Proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting. Polyclonal immunoglobulin Y antibody against HrpN prepared from chicken was used as the probe and was preabsorbed with E. coli DH5α and a HrpN mutant. An Immuno-Star AP chemiluminescent substrate (Bio-Rad, CA) was used to detect an anti-chicken secondary antibody (GeneTex, TX).
RESULTS AND DISCUSSION
Screening for T3SS inhibitor.
To identify potential T3SS repressors, 29 analogs and isomers of TCA and OCA were screened for effects on D. dadantii 3937 hrpA expression (Table 2 and Fig. 2). hrpA encodes the T3SS pilus required for protein translocation into plant cells, and this gene is the first in a multigene operon that encodes T3SS structural and secreted proteins. A reporter plasmid, phrpA, which contains a transcriptional fusion of the hrpA promoter controlling expression of green fluorescent protein (GFP) was used to measure the effects of the OCA and TCA analogs and isomers on T3SS gene expression (22). Expression of T3SS genes of phytobacteria is induced in T3SS-inducing MM, which is considered to mimic plant apoplastic conditions, such as a low level of nutrients (7, 14, 17, 19). Bacterial cells containing phrpA were grown in the T3SS-inducing MM supplemented with 100 μM of each compound. GFP intensity, which is a measure of hrpA promoter activity, was assayed by flow cytometry. Among the OCA/TCA analogs screened, PCA (Fig. 2, compound 4) showed the strongest inhibition on T3SS gene expression (Table 2).
TABLE 2.
Expression of D. dadantii 3937 hrpA in MM and MM supplemented with different isomers and analogs of OCA and TCA
| Phenolic compound (compound no.)a | Avg MFI ± SDb at:
|
|
|---|---|---|
| 12 h | 24 h | |
| None | 78.7 ± 6.3 | 92.1 ± 17.1 |
| TCA (1) | 133.9 ± 12.9* | 203.7 ± 16.1* |
| OCA (6) | 115.5 ± 7.9* | 225.8 ± 15.6* |
| MCA (5) | 133.0 ± 38.2 | 203.3 ± 9.6* |
| PCA (4) | 10.2 ± 0.4* | 11.4 ± 1.0* |
| Hydrocinnamic acid (8) | 200.3 ± 35.8* | 213.5 ± 18.9* |
| Phenoxyacetic acid (9) | 222.7 ± 64.3* | 205.7 ± 11.8* |
| trans-2-Phenylcyclopropane-1-carboxylic acid (10) | 67.0 ± 18.4 | 84.0 ± 14.3 |
| trans-3-Indoleacrylic acid (12) | 23.9 ± 1.3* | 121.0 ± 6.2 |
| trans-3-(3-Pyridyl)acrylic acid (13) | 184.9 ± 35.6* | 204.0 ± 16.8* |
| trans-2-Methylcinnamic acid (18) | 157.2 ± 11.7* | 342.5 ± 16.6* |
| trans-2-Chlorocinnamic acid (21) | 166.2 ± 17.8* | 319.8 ± 48.3* |
| Methyl trans-cinnamate (30) | 135.8 ± 8.1* | 219.2 ± 14.5* |
| None | 101.8 ± 3.8 | 134.6 ± 3.3 |
| trans-3-(2-Thienyl)acrylic acid (11) | 201.9 ± 3.7* | 338.2 ± 14.9* |
| trans-3-(4-Imidazolyl)acrylic acid (14) | 118.8 ± 11.1 | 144.1 ± 13.7 |
| trans-2-Methoxycinnamic acid (15) | 213.9 ± 7.4* | 304.3 ± 15.5* |
| trans-2-Carboxycinnamic acid (24) | 109.0 ± 21.5 | 139.7 ± 17.1 |
| 3-(4-Hydroxyphenyl)propionic acid (7) | 138.9 ± 10.5* | 186.9 ± 3.9* |
| trans-Cinnamamide (31) | 126.3 ± 10.2 | 228.5 ± 2.0* |
| trans-2,4-Dihydroxycinnamic acid (2) | 66.6 ± 3.9* | 78.6 ± 5.2* |
| trans-3,4-Dihydroxycinnamic acid (3) | 82.3 ± 9.0 | 111.4 ± 14.6 |
| None | 133.3 ± 4.8 | 176.4 ± 6.1 |
| trans-3-Methoxycinnamic acid (16) | 114.7 ± 12.7 | 143.9 ± 35.9 |
| trans-4-Methoxycinnamic acid (17) | 74.5 ± 7.5* | 100.2 ± 3.7* |
| trans-3-Methylcinnamic acid (19) | 108.0 ± 8.1 | 136.2 ± 21.2 |
| trans-4-Methylcinnamic acid (20) | 58.9 ± 2.7* | 91.1 ± 4.1* |
| trans-3-Chlorocinnamic acid (22) | 113.3 ± 7.5 | 157.0 ± 6.2 |
| trans-4-Chlorocinnamic acid (23) | 54.3 ± 3.3* | 81.3 ± 2.5* |
| trans-4-Carboxycinnamic acid (25) | 114.4 ± 11.9 | 125.2 ± 5.5* |
| trans-4-Mercaptocinnamic acid (26) | 117.4 ± 22.4 | 128.0 ± 9.3* |
| trans-4-Aminocinnamic acid (27) | 125.9 ± 9.4 | 154.6 ± 1.8* |
| trans-4-Nitrocinnamic acid (28) | 100.5 ± 3.0* | 114.2 ± 13.5* |
| trans-4-Formylcinnamic acid (29) | 105.1 ± 4.8* | 137.9 ± 8.1* |
MM was supplemented with 100 μM of the indicated compounds. The compounds were assayed three different times, with MM only (no supplementation) as the control treatment (indicated by “none”) for each set of experiments. The compound numbers are as used in Fig. 2.
D. dadantii 3937 cells carrying GFP reporter phrpA were used in this study. The promoter activities at 12 and 24 h of bacterial growth were determined. GFP mean fluorescence intensity (MFI) was determined for gated populations of bacterial cells by flow cytometry. Values are representative of two or three experiments, and three replicates were used for each experiment. Asterisks indicate statistically significant differences in GFP MFI between bacterial cells grown in MM and MM supplemented with the different compounds (P < 0.01, Student's t test).
SAR of phenolic compound analogs.
OCA (Fig. 2, compound 6) and meta-coumaric acid (MCA) (compound 5) induced hrpA expression (Table 2). PCA, OCA, and MCA are regioisomers, differing only in the position of the hydroxyl group in the phenyl ring (Fig. 2). Two derivatives of PCA, trans-2,4-dihydroxycinnamic acid (compound 2) and trans-3,4-dihydroxycinnamic acid (compound 3), with an additional hydroxyl group in the ortho and meta positions of the phenyl ring, respectively, had reduced inhibitory activity against hrpA expression in comparison to PCA. These results indicated that the para positioning of the hydroxyl group in the phenyl ring is important for the biological activity of PCA. Similarly, chloro-, methyl-, and methoxycinnamic acids demonstrated the same activity pattern, with the para isomers trans-4-chlorocinnamic acid (compound 23), trans-4-methylcinnamic acid (compound 20), and trans-4-methoxycinnamic acid (compound 17) inhibiting hrpA expression but not the ortho isomers (compounds 21, 18, and 15) and the meta isomers (compounds 22, 19, and 16) (Table 2). However, when the hydroxyl group in PCA is replaced by a carboxyl, mercapto, amino, nitro, or formyl group, the resulting cinnamic acid derivatives, trans-4-carboxycinnamic acid (compound 25), trans-4-mercaptocinnamic acid (compound 26), trans-4-aminocinnamic acid (compound 27), trans-4-nitrocinnamic acid (28), and trans-4-formylcinnamic acid (compound 29), respectively, exhibited a reduced inhibitory effect on hrpA (Table 2).
Along with the position of the hydroxyl group in the phenyl ring, the double bond in PCA is essential for its inhibitory activity. For example, if the double bond is reduced, the resulting 3-(4-hydroxyphenyl)propionic acid (compound 7) did not inhibit hrpA expression. Replacing the carboxylic acid group with an amide or methyl ester did not change the activity of TCA (compound 1) to induce T3SS gene expression, as both methyl trans-cinnamate (compound 30) and trans-cinnamamide (compound 31) demonstrated strong inducing activity. Finally, trans-3-indoleacrylic acid (compound 12) had an inhibitory effect on hrpA expression at 12 h of bacterial growth but not at 24 h (Table 2). However, when other heterocyclic rings, such as pyridine, imidazole, and thiophene, are introduced in the same position, the resulting acrylic acids, trans-3-(3-pyridyl)acrylic acid (compound 13), trans-3-(4-imidazolyl)acrylic acid (compound 14), and trans-3-(2-thienyl)acrylic acid (compound 11), respectively, did not have any inhibitory activity.
Inhibitory effect of PCA on T3SS gene expression.
To confirm the inhibitory effect of PCA on the D. dadantii T3SS, the expression of additional T3SS genes was examined. hrpN encodes a T3SS harpin, and hrpN promoter activity was reduced in MM supplemented with 100 μM PCA in comparison to that in unamended MM (Table 3). The promoter of a conserved ATPase gene, mrp, was used as a reference gene (14). Similar levels of mrp expression were observed in D. dadantii 3937(pmrp) when the bacterial cells were grown in MM or MM supplemented with 100 μM PCA, thus showing that the effect of PCA was specific to the T3SS (Table 3).
TABLE 3.
Expression of T3SS genes hrpA, hrpN, hrpS, and hrpL of D. dadantii 3937 in MM and MM supplemented with 100 μM PCA
| Reporter plasmid | Avg MFI ± SD for growth in the indicated mediuma at:
|
|||
|---|---|---|---|---|
| 12 h
|
24 h
|
|||
| MM | MMPCA | MM | MMPCA | |
| phrpA | 64.3 ± 0.9 | 10.3 ± 1.3* | 150.8 ± 4.4 | 18.5 ± 3.5* |
| phrpN | 39.8 ± 5.8 | 6.8 ± 0.8* | 133.3 ± 3.2 | 11.3 ± 3.4* |
| phrpS | 72.7 ± 11.3 | 37.9 ± 1.3* | 95.0 ± 17.7 | 43.6 ± 2.6* |
| phrpL | 12.8 ± 0.1 | 7.8 ± 0.1* | 27.2 ± 1.0 | 11.1 ± 3.3* |
| pmrp | 113.0 ± 7.7 | 124.1 ± 2.7 | 93.4 ± 2.6 | 98.9 ± 1.0 |
| pPROBE-AT | 2.1 ± 0.1 | 2.2 ± 0.2 | 13.4 ± 8.4 | 14.0 ± 10.1 |
The promoter activities were compared at 12 and 24 h of bacterial growth in PCA. GFP mean fluorescence intensity (MFI) was determined for gated populations of bacterial cells by flow cytometry. Values are representative of two experiments, and three replicates were used for each experiment. Asterisks indicate statistically significant differences in GFP intensity between bacterial cells grown in MM and MM supplemented with 100 μM PCA (MMPCA) (P < 0.01, Student's t test).
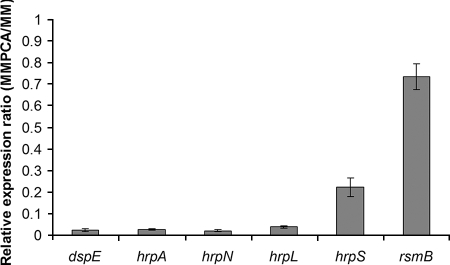
The effect of PCA on T3SS gene transcription was directly measured by qRT-PCR. Compared to that in D. dadantii 3937 grown in unamended MM, there was significantly less hrpA, hrpN, and T3SS effector dspE mRNA in cells grown in MM supplemented with 100 μM PCA (Fig. 3). Since PCA represses the expression of several T3SS genes such as hrpN, we examined the effect of PCA on HrpN protein production. Less HrpN was detected in protein extracts from D. dadantii 3937 grown in MM supplemented with 100 μM PCA than in those from the strain grown in MM supplemented with 10 μM PCA (Fig. 4).
FIG. 3.
Relative mRNA levels of hrpS, hrpL, dspE, hrpA, hrpN, and rsmB of D. dadantii 3937 in MM supplemented with 100 μM PCA compared to mRNA levels in MM without PCA as determined by qRT-PCR. There is no significant difference between MM and MM supplemented with PCA for gene rsmB (P = 0.928), but levels of gene expression of hrpS, hrpL, dspE, hrpA, and hrpN are significantly different between MM and MM supplemented with 100 μM PCA (P < 0.001). Three replicates were used in this experiment. The P value was calculated with the Relative Expression Software Tool as described by Pfaffl et al. (15).
FIG. 4.
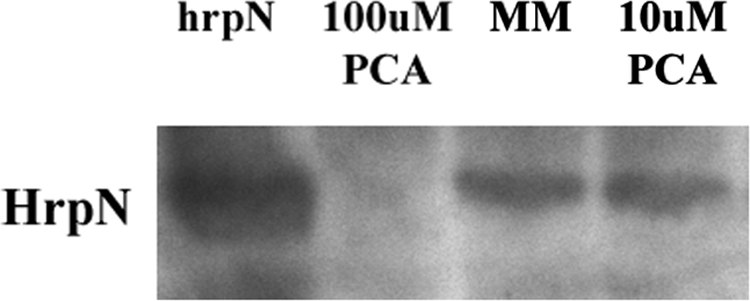
HrpN protein expression of D. dadantii 3937 in MM and MM supplemented with PCA. Lane 1 (left lane), 3937(pCPP50::HrpN); lane 2, D. dadantii 3937 grown in MM supplemented with 100 μM PCA; lane 3, D. dadantii 3937 grown in MM; lane 4, D. dadantii 3937 grown in MM supplemented with 10 μM PCA. Similar amounts of proteins were loaded in each lane.
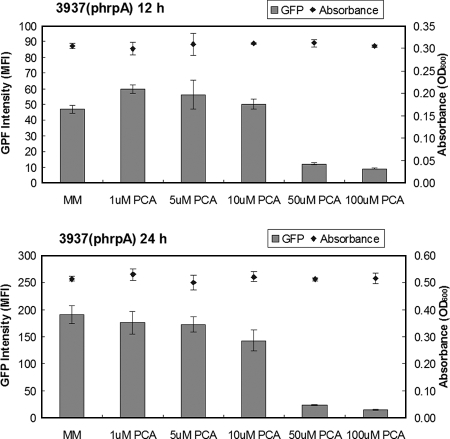
To determine if the repression of T3SS gene expression by PCA was due to toxicity or nutritional status of the phenolic compound, different concentrations of PCA were used to supplement MM to measure the effect of PCA on hrpA expression and bacterial growth. Compared to that for growth in unamended MM, the average GFP fluorescence intensity of D. dadantii 3937(phrpA) cells was reduced more than fourfold when 50 or 100 μM of PCA was added to the medium (Fig. 5). The addition of PCA at concentrations of 1, 5, and 10 μM did not result in a substantial reduction of GFP fluorescence intensity for D. dadantii 3937. No inhibition or promotion of bacterial growth was observed when PCA was added to MM (Fig. 5).
FIG. 5.
Promoter activities of hrpA in D. dadantii 3937 grown in MM and MM supplemented with different amounts of PCA at 12 h and 24 h of growth. To study the effect of PCA on hrpA expression, 50 μl of bacterial suspension (optical density at 600 nm [OD600] = 1.0) was used as the initial inoculum and added into 5 ml MM and MM supplemented with PCA. GFP intensity was determined on gated populations of bacterial cells by flow cytometry and analyzed with the Cell Quest software (BD Biosciences, San Jose, CA). The growth of D. dadantii 3937 in MM supplemented with different concentrations of PCA was recorded. Results from one representative experiment are shown. Three replicates were used in this experiment, and the experiment was repeated twice. MFI, mean fluorescence intensity.
PCA inhibits T3SS through the HrpX/Y-HrpS-HrpL pathway.
Since strong inhibition of hrpA expression by PCA in D. dadantii 3937 was observed, the mechanism of PCA regulation of the T3SS pathway was investigated. To determine whether PCA affects T3SS gene expression through the Gac-Rsm regulatory pathway, rsmB mRNA levels were quantified by qRT-PCR. No significant difference in the level of rsmB mRNA between D. dadantii 3937 cells grown in MM and cells grown in MM supplemented with 100 μM PCA was observed (Fig. 3). Thus, repression of T3SS expression by PCA does not occur through the Gac-Rsm pathway. PCA supplementation did not affect D. dadantii 3937 pectate lyase production, providing further support that PCA does not affect the GacS/A system (data not shown).
To determine if PCA represses T3SS gene expression through the HrpX/Y-HrpS-HrpL pathway, the promoter activities of hrpS and hrpL were examined and found to be reduced in MM supplemented with 100 μM PCA in comparison to those in unamended MM (Table 3). The expression of hrpS and hrpL was also confirmed by qRT-PCR. Our result showed that, compared with those for D. dadantii 3937 grown in MM, significantly smaller amounts of hrpS (relative expression ratio, 0.223; P < 0.001) and hrpL (relative expression ratio, 0.039; (P < 0.001) mRNA were present (Fig. 3). These results demonstrate that PCA inhibits expression of T3SS genes through the HrpX/Y-HrpS-HrpL regulatory pathway.
Phenylpropanoids are a group of secondary metabolites produced by plants from l-phenylalanine. Although the end products vary among plant species, the initial reactions of the phenylpropanoid biosynthesis pathway are conserved (2). These multiple-branch pathways can all be derived from initial steps of the following pathways. (i) TCA is produced by a deamination of l-phenylalanine by phenylalanine ammonia-lyase. (ii) Cinnamic acid 4-hydroxylase catalyzes the addition of the hydroxyl group at the para position of the phenyl ring of TCA, producing PCA. (iii) The carboxyl group of PCA is activated by formation of a thioester bond with coenzyme A (CoA), a process catalyzed by 4-coumarate:CoA ligase, which gives rise to a variety of secondary compounds such as flavonoids, isoflavonoids, stilbenes, and lignin (see Fig. S1 in the supplemental material) (10, 20). Isoflavonoids and stilbenes are phytoalexins which are induced in response to microbial attack and are able to inhibit the growth of pathogens (8), while lignin is a major component of the plant cell wall and provides mechanical strength and impermeability to plant tissues (8). In addition, TCA is an important intermediate in the pathway for salicylic acid biosynthesis involved in the disease resistance of plants (16). In salicylic acid biosynthesis, TCA is converted to OCA through ortho hydroxylation, followed by β-oxidation to produce salicylic acid. Salicylic acid is a plant signaling molecule that triggers basal resistance locally and systemically acquired resistance against a broad spectrum of pathogens including viruses, bacteria, fungi, and oomycetes (3).
It is intriguing to see intermediates in phenylpropanoid biosynthesis either induce or repress the expression of T3SS genes of D. dadantii 3937. Based on the chemical structures and inhibitory effect of PCA and analogs of PCA on T3SS, the SAR and potential active sites of PCA are revealed (Table 2 and Fig. 2). Given the similarity of the T3SS regulatory systems among many plant and animal pathogens, the effects of plant phenolic compounds on D. dadantii 3937 unveiled here provide a new direction for development of novel antimicrobial reagents for agriculture.
Supplementary Material
Acknowledgments
This work is dedicated to Noel T. Keen.
We thank Nicole Perna of the University of Wisconsin for providing access to the annotated D. dadantii genome sequences (https://asap.ahabs.wisc.edu/asap/ASAP1.htm) and M. L. P. Collin and M. J. McBride for critical discussions and reading of the manuscript.
This project is supported by grants from the State Scholarship Fund of the China Scholarship Council awarded to Yan Li, the National Science Foundation (award no. EF-0332163), the Research Growth Initiative of the University of Wisconsin—Milwaukee, and a National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2006-35319-17396.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpinEcc. Mol. Plant-Microbe Interact. 14:516-526. [DOI] [PubMed] [Google Scholar]
- 2.Dixon, R. A., L. Achnine, P. Kota, C. J. Liu, M. S. S. Reddy, and L. Wang. 2002. The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 3:371-390. [DOI] [PubMed] [Google Scholar]
- 3.Grant, M., and C. Lamb. 2006. Systemic immunity. Curr. Opin. Plant Biol. 9:414-420. [DOI] [PubMed] [Google Scholar]
- 4.He, S. Y., K. Nomura, and T. S. Whittam. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181-206. [DOI] [PubMed] [Google Scholar]
- 5.Hudson, D. L., A. N. Layton, T. R. Field, A. J. Bowen, H. Wolf-Watz, M. Elofsson, M. P. Stevens, and E. E. Galyov. 2007. Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51:2631-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugouvieux-Cotte-Pattat, N., G. Condemine, W. Nasser, and S. Reverchon. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213-257. [DOI] [PubMed] [Google Scholar]
- 7.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 8.La Camera, S., G. Gouzerh, S. Dhondt, L. Hoffmann, B. Fritig, M. Legrand, and T. Heitz. 2004. Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 198:267-284. [DOI] [PubMed] [Google Scholar]
- 9.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 10.Metraux, J. P. 2002. Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci. 7:332-334. [DOI] [PubMed] [Google Scholar]
- 11.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 12.Mota, L. J., and G. R. Cornelis. 2005. The bacterial injection kit: type III secretion systems. Ann. Med. 37:234-249. [DOI] [PubMed] [Google Scholar]
- 13.Mota, L. J., I. Sorg, and G. R. Cornelis. 2005. Type III secretion: the bacteria-eukaryotic cell express. FEMS Microbiol. Lett. 252:1-10. [DOI] [PubMed] [Google Scholar]
- 14.Peng, Q., S. Yang, A. O. Charkowski, M. N. Yap, D. A. Steeber, N. T. Keen, and C. H. Yang. 2006. Population behavior analysis of dspE and pelD regulation in Erwinia chrysanthemi 3937. Mol. Plant-Microbe Interact. 19:451-457. [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah, J., P. Kachroo, and D. F. Klessig. 1999. The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11:191-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang, X., Y. Xiao, and J. M. Zhou. 2006. Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant-Microbe Interact. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 18.Wolf, K., H. J. Betts, B. Chellas-Gery, S. Hower, C. N. Linton, and K. A. Fields. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao, Y., L. Lan, C. Yin, X. Deng, D. Baker, J. M. Zhou, and X. Tang. 2007. Two-component sensor RhpS promotes induction of Pseudomonas syringae type III secretion system by repressing negative regulator RhpR. Mol. Plant-Microbe Interact. 20:223-234. [DOI] [PubMed] [Google Scholar]
- 20.Yalpani, N., J. Leon, M. A. Lawton, and I. Raskin. 1993. Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 103:315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, S., Q. Peng, M. San Francisco, Y. Wang, Q. Zeng, and C. H. Yang. 2008. Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS ONE 3:e2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, S., Q. Peng, Q. Zhang, X. Yi, C. J. Choi, R. M. Reedy, A. O. Charkowski, and C. H. Yang. 2008. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937). Mol. Plant-Microbe Interact. 21:133-142. [DOI] [PubMed] [Google Scholar]
- 23.Yang, S., Q. Zhang, J. Guo, A. O. Charkowski, B. R. Glick, A. M. Ibekwe, D. A. Cooksey, and C. H. Yang. 2007. Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Appl. Environ. Microbiol. 73:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yap, M. N., C. M. Rojas, C. H. Yang, and A. O. Charkowski. 2006. Harpin mediates cell aggregation in Erwinia chrysanthemi 3937. J. Bacteriol. 188:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yap, M. N., C. H. Yang, J. D. Barak, C. E. Jahn, and A. O. Charkowski. 2005. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187:639-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.