Abstract
The role of LuxS in Shewanella oneidensis MR-1 has been examined by transcriptomic profiling, biochemical, and physiological experiments. The results indicate that a mutation in luxS alters biofilm development, not by altering quorum-sensing abilities but by disrupting the activated methyl cycle (AMC). The S. oneidensis wild type can produce a luminescence response in the AI-2 reporter strain Vibrio harveyi MM32. This luminescence response is abolished upon the deletion of luxS. The deletion of luxS also alters biofilm formations in static and flowthrough conditions. Genetic complementation restores the mutant biofilm defect, but the addition of synthetic AI-2 has no effect. These results suggest that AI-2 is not used as a quorum-sensing signal to regulate biofilm development in S. oneidensis. Growth on various sulfur sources was examined because of the involvement of LuxS in the AMC. A mutation in luxS produced a reduced ability to grow with methionine as the sole sulfur source. Methionine is a key metabolite used in the AMC to produce a methyl source in the cell and to recycle homocysteine. These data suggest that LuxS is important to metabolizing methionine and the AMC in S. oneidensis.
The AI-2 family of quorum-sensing signals is unique because LuxS, the enzyme responsible for catalyzing the formation of these signals, is conserved in both gram-positive and gram-negative proteobacteria. This broad genetic conservation has led to the conclusion that AI-2 is used for interspecies communication (43). In addition to being implicated with quorum sensing, LuxS is involved in the activated methyl cycle (AMC) (39, 51). This metabolic role of LuxS has resulted in an active debate on whether a mutation in the LuxS gene affects certain phenotypes because of a lack of quorum-sensing abilities or the disruption of a biosynthetic pathway (11, 22, 47, 52).
The AMC generates homocysteine, methionine, adenosine, and S-adenosylmethionine (SAM), a major methyl donor source in the cell (reviewed in reference 47). Homocysteine enters the cycle and is converted to methionine and then to SAM. The conversion of SAM to S-adenosylhomocysteine (SAH) results in the methylation of DNA, RNA, proteins, and metabolites. To complete the cycle, SAH, a toxic metabolite, is then converted to S-ribosylhomocysteine (SRH), and then SRH is converted to homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD) by LuxS. DPD can then spontaneously cyclize into an AI-2 type signal. Two known AI-2 structures are (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran-borate (Vibrio harveyi) (7) and (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (Salmonella enterica serovar Typhimurium) (33). DPD has no known function in the cell besides producing the quorum-sensing signal AI-2.
Recent work proposed that the dissimilatory metal-reducing bacterium Shewanella oneidensis MR-1 has the ability to produce the AI-2 signal (3) and an acyl-homoserine lactone (AHL) (14), a typical gram-negative quorum-sensing signal. It has been suggested that S. oneidensis uses an AHL to enhance hydrogen metabolism, under specific growth conditions (14). Differing from these results, the addition of the supernatant from an S. oneidensis culture (27) or a mutation in luxS (5) was not able to affect iron reduction rates under the experimental conditions of each study.
To date, no physiological traits are known to be regulated by a quorum-sensing signal in S. oneidensis MR-1. In the present study, we report that a mutation in luxS abolishes the ability of S. oneidensis to produce an AI-2-induced reporter strain response. This mutation also disrupts biofilm formation but the addition of the quorum-sensing signal AI-2 does not repair the disruption. The mutation in luxS does create growth defects when grown with methionine, a constituent of the AMC, as the sole sulfur source. This information has thus led to the conclusion that luxS plays a role in the AMC and in that manner influences biofilm formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Agrobacterium tumefaciens strains NTL4(pCF218)(pCF372) and KYC6(pCF218) (18), Pseudomonas aeruginosa PAO1 (23), Pantoea stewartii subsp. stewartii DC283 (10), S. oneidensis MR-1 (34), and Vibrio fischeri strains ES 114 (4) and MJ1 (37) were grown on Luria-Bertani (LB) medium (38) at 30°C, unless stated otherwise. Escherichia coli strains DH5α (Invitrogen), WM3064 (W. Metcalf, personal communication), and MG1655(pJWP01s) (8) were grown on LB medium at 37°C, unless stated otherwise. V. harveyi strains BB120 (2) and MM32 (33) were grown on autoinducer bioassay (AB) medium (21) at 30°C. For growth experiments under sulfur-limiting conditions, M9 medium (38) was prepared and all sulfate salts were exchanged for comparable chloride salt. The medium was supplemented with 20 mM lactate, 15 μM thiamine, and 50 μM cysteine, 100 μM homocysteine, 100 μM methionine, or 200 μM K2SO4. When necessary, supplements or antibiotics were added at the following concentrations (μg/ml): ampicillin, 100; chloramphenicol, 10; diaminopimelic acid (DAP), 100; kanamycin (Km), 50 or 100; and tetracycline (Tc), 10.
Mutagenesis.
All molecular work was performed by standard methods (38). The construction of a luxS gene deletion in S. oneidensis was completed using a mutagenesis procedure described elsewhere (13). Briefly, PCR-amplified luxS fragments 631 bp upstream (primers 5′-GAATTCCCTGTTAATATGGCTGGCAGAG-3′ and 5′-CATATGGTCGCTATCTGTTTATAGGTGC-3′) and 702 bp downstream (primers 5′-CCGCGGCAACAAACGCCGTACGCTAAC-3′ and 5′-GAGCTCGGTAAATTCCGTTTGCTGCG-3′) were inserted into flanking regions of a Km resistance cassette (restriction sites are indicated by the underlines above) in the suicide vector pJK100 (13). The plasmid pJK100 contains both Km and Tc resistance cassettes. The resulting plasmid pluxS:UD was electroporated into a conjugal donor strain, E. coli WM 3064, a strain that requires DAP for growth. The mating experiments involved concentrating 1.0 ml of E. coli WM 3064 pluxS:UD and 0.5 ml of S. oneidensis into 100 μl, and the mixture of cells was spotted onto a LB medium plate with DAP and allowed to grow at 30°C overnight. The cells were then scraped off the plate, suspended in liquid LB medium, and spread onto a LB medium plate with Km and no DAP. Successful double crossover integration of the suicide vector produced S. oneidensis pluxS:UD strains that were Km resistant and Tc sensitive. The chromosomal inserted Km resistance cassette was then removed by conjugating pCM157 (30), a Cre recombinase vector, into S. oneidensis pluxS:UD via E. coli WM 3064. The removal of the resistance cassette was possible because of the loxP sites that flanked the inserted Km resistance gene. The resulting strain S. oneidensis ΔluxS pCM157 was then cured of pCM157. The final in-frame deletion mutant had a 72-bp scar sequence, contained no antibiotic markers, and was named S. oneidensis DL13. The mutant was verified via DNA sequencing. For complementation, S. oneidensis luxS was PCR amplified (5′-GCTTATGAACAGGGAGTGCGTGAATACC-3′ and 5′-GCGACTTGACATACCAAGCTCCCAAAAC-3′), cloned into pGEM-T (Promega), and ligated into pBBR1MCS-3 (26) producing pluxS. The plasmids pluxS and p519ngfp (green fluorescent protein [GFP] plasmid for confocal microscopy) (31) were conjugated into S. oneidensis strains as described above.
Autoinducer assays.
V. harveyi MM32 was used as the qualitative reporter strain to identify the presence of AI-2 (33). The reporter strain was grown in AB medium for 16 h and then diluted 1:5,000. A total of 90 μl of the diluted reporter was then added to a 96-well plate. Cultures of V. harveyi BB120 (AI-2+) and E. coli DH5α (AI-2−) were grown to an optical density at 600 nm (OD600) of 1.5 in AB and LB media, respectively. S. oneidensis cultures were grown on either LB medium to an OD600 of 0.5, 1.5, and 3.0 or overnight on a minimal medium (MM) (20) supplemented with 20 mM lactate (approximate OD600 of 0.5). Then 1 ml of the experimental culture was centrifuged and filter sterilized and 10 μl of the filtered supernatant was added to the reporter stain. The plates were incubated at 30°C and the luminescence of the reporter strain was monitored using a luminometer (LD400; Beckman Coulter) for 18 h.
Two broad-range AHL reporter strains were used to test if S. oneidensis could produce an AHL. The first reporter strain, A. tumefaciens NTL4(pCF218)(pCF372), utilizes the lacZ gene to visualize the presence of an AHL (18, 53). A. tumefaciens NTL4(pCF218)(pCF372) was grown in liquid LB medium overnight and then cross-streaked with LB overnight cultures of a positive control, A. tumefaciens KYC6(pCF218), and S. oneidensis on LB agar supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The second broad-range AHL reporter strain was the GFP reporter strain E. coli MG1655(pJWP01s) (8). E. coli MG1655(pJWP01s) was grown overnight and then diluted and grown to an approximate OD600 of 0.4. Then 1 ml of the filter-sterilized supernatant of the LB overnight experimental cultures (S. oneidensis wild type [WT], V. fischeri ES 114 and MJ1, P. aeruginosa PAO1, and P. stewartii DC283) were added to 4 ml of E. coli MG1655(pJWP01s). Additional experimental cultures of S. oneidensis were grown in LB medium to an OD600 of 1.0 and grown anaerobically in standard M9 medium (supplemented with 50 mg/liter sodium acetate and 20 mg/liter sodium nitrate) to an OD600 of 0.2. The reporter and supernate mix was then incubated at 30°C. Fluorescence was monitored over a period of 4 to 6 h by adding 200 μl of each sample mix to a well in a 96-well plate, and the analysis was done with a fluorometer (Spectrafluor Plus; Tecan).
In vitro AI-2 synthesis.
5′-Methylthioadenosine-S-adenosylhomocysteine nucleosidase (MTA-SAHase) and LuxS were purified and used in a two-step reaction to enzymatically convert SAH (Sigma) to AI-2 (50). Briefly, IPTG (isopropyl-β-d-thiogalactopyranoside) was used to induce the expression of pProEX HT-mtan and pProEX-LuxS (50) in E. coli DH5α for the His-tag purification of MTA-SAHase and LuxS, respectively. The cells were lysed using BugBuster protein extraction solution (Novagen EMD Biosciences), and the desired proteins were retained with Ni-NTA His-Bind resin (Novagen EMD Biosciences). Purified MTA-SAHase (100 μg/ml) was incubated with 2 mM SAH at 37°C for 1 h in an anaerobic chamber. MTA-SAHase was removed by a 10-kDa centrifugal filter (Centricon, Millipore) and the remaining solution was then incubated with LuxS (500 μg/ml) for an additional 2 h. Since homocysteine and AI-2 have a 1:1 stoichiometry in the reaction, the resulting solution was analyzed for homocysteine concentration by adding Ellman's reagent (Sigma) and measuring the absorbance at 412 nm. Additional verification of AI-2 activity came from adding various concentrations of synthetic AI-2 to the reporter strain V. harveyi MM32 (procedure described above).
Microarray analysis.
Overnight LB cultures of the WT and DL13 strains were subcultured into fresh medium and grown to an OD600 of 1.5 in duplicate. The cells were rapidly centrifuged and the cell pellets snap-frozen in liquid nitrogen and stored at −20°C. RNA isolation, microarray construction, hybridization, scanning, image quantification, and data analysis were performed as previously described (6, 19, 48). The experiment had two biological and technical replicates. Statistical analysis was done with JMP Genomics 3.0 software (SAS Institute, Cary, NC). The raw data were log2 transformed and imported into the software for analysis. A distribution analysis and data correlation analysis were done as a quality control step. The overlaid kernel density estimates derived from the distribution analysis allowed the visualization of sources of variation based on the strain, as well as variation attributed to technical factors such as array and dye. The data were subsequently normalized using a standard normalization technique. A mixed model analysis of variance was done to determine the differential expression levels between the WT and DL13 bacterial strains. To control the false discovery rate a testing correction was applied at an α level of 0.05.
Biofilm plate assay.
Ninety-six-well microtiter plate assays were conducted to quantify biofilm growth under static conditions (35, 36). Briefly, cells were grown overnight in MM. Then the cells were diluted to an OD600 of 0.01 and 175 μl was added to each well. If desired, concentrations of AI-2 (0.1, 1, 5, 10 μM) or homocysteine (10 μM) were also added. AI-2 concentration ranges were chosen by comparing the V. harveyi MM32 luminescence response of the S. oneidensis supernatant to known concentrations of AI-2 as described above. The S. oneidensis WT cell-free supernatant and an AI-2 concentration of 10 μM produced a similar luminescence response in the reporter strain V. harveyi MM32. The plates were then incubated at 30°C and monitored over a 3-day period. To process the plates, 10 μl of 0.5% crystal violet was used to stain the cells which adhered to the wells for 10 min. The wells were then washed, and the dye was extracted with methanol. The extract absorbance was quantified at 570 nm.
Flowthrough biofilm assay.
Aerobic biofilms were grown using a standard flowthrough system (9, 45, 46). The system consisted of a MasterFlex L/S pump, MasterFlex silicone tubing, bubble trap, and a three-channel flow chamber (dimensions, 40-mm length by 4-mm width by 1-mm depth; BioCentrum-DTU). The flow chamber was covered with a glass slide (Fisher Brand #1.5, 24 by 50 mm) and sealed with silicon glue. All components were autoclaved except the flow chamber, which was sterilized with 10% H2O2. The system was equilibrated with LB growth medium for at least 5 h before inoculation. WT and DL13 cells were grown in LB medium to an OD600 of 1.0 and then diluted to an OD600 of 0.03. To inoculate the system, medium flow through the system was stopped and 1 to 2 ml of culture was injected into the flow chamber. The chamber was placed glass-side down to increase cell attachment to the glass slide. After 1 h, the chamber was inverted and flow was restored to an approximate flow rate of 112 μl/min. For chemical AI-2 complementation, 5 μM synthetic AI-2 was added to the medium. Images were collected with a Zeiss LSM510 confocal microscope with 40× water immersion and 10× planar objective. Image analysis was performed by the Zeiss image browser.
RESULTS AND DISCUSSION
luxS mutagenesis depletes AI-2 activity.
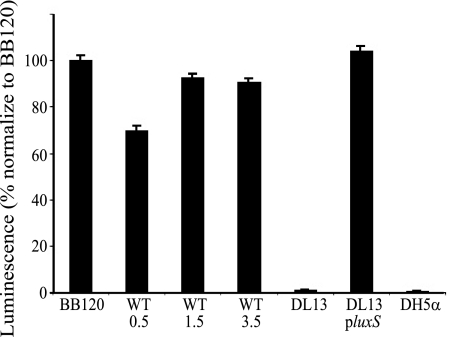
A BLAST search identified a LuxS homologue (SO 1101) in the genome of S. oneidensis with 78% (132/169) amino acid identity and 89% (152/169) similarity to LuxS in V. harveyi. A SO 1101 gene replacement was constructed to produce S. oneidensis DL13 so that the physiological role of the S. oneidensis LuxS could be examined. Supernatants from S. oneidensis WT and DL13 cultures were examined for AI-2 production via the V. harveyi MM32 reporter strain method (Fig. 1). After 15 h, the assays revealed that the supernate from the WT produced an AI-2-induced luminescence response in the reporter strain at levels similar to those of the positive control V. harveyi BB120. The S. oneidensis supernatant from the late exponential to early stationary phases (OD600 of 1.5) had the largest AI-2 luminescence response. DL13 generated near-background levels of luminescence regardless of cell density, similarly to E. coli DH5α (luxS−). DL13 also produced background levels of AI-2 luminescence when grown on MM (data not shown). AI-2 production was restored to WT levels in the mutant strain upon complementation via a plasmid-borne S. oneidensis native luxS, DL13 pluxS (Fig. 1).
FIG. 1.
Luminescence response of V. harveyi MM32 to AI-2 from the filtered supernatant from V. harveyi BB120 (BB120), E. coli DH5α (DH5α), the S. oneidensis WT at an OD600 of 0.5 (WT 0.5), the S. oneidensis WT at an OD600 of 1.5 (WT 1.5), the S. oneidensis WT at an OD600 of 3.5 (WT 3.5), S. oneidensis ΔluxS (DL13), and DL13 pluxS (plasmid-borne luxS). Data are normalized to the average raw luminescence value (9,535) for V. harveyi BB120. The experiment was done in triplicate with eight replicates. Error bars represent the standard errors of the means (n = 8).
AHL quorum sensing in S. oneidensis.
Two broad-range AHL reporter strains were used to identify the physical presence of AHL autoinducer molecules in S. oneidensis supernatant. Both reporter strains A. tumefaciens NTL4(pCF218)(pCF372) (data not shown) and E. coli MG1655(pJWP01s) (Table 1) produced no evidence for the presence of an AHL signal in S. oneidensis supernatant grown aerobically in LB medium or anaerobically in M9 medium. The levels of E. coli MG1655(pJWP01s) GFP production from S. oneidensis supernatant are approximately three orders of magnitude less than those of known AHL producers like V. fischeri MJ1, P. stewartii DC283, and P. aeruginosa PAO1 (Table 1). In addition, an in silico analysis was performed to identify whether S. oneidensis had any LuxR-type AHL-dependent transcription regulatory proteins. S. oneidensis has seven putative transcription regulatory genes that have been labeled as being in the LuxR family of proteins: TIGR loci SO 0351, SO 0864, SO 1860, SO 2648, SO 2725, SO 3305, and SO 4624. A sequence comparison to known LuxR-type homologues (P. aeruginosa LasR and RhlR, P. stewartii EsaR, Aeromonas salmonicida AsaR, and Yersinia enterocolitica YenR) showed only three conserved amino acid residues in the C-terminal region; these residues are also conserved in the helix-turn-helix motif of the broader NarL-FixJ superfamily (18, 20). However, seven amino acid residues spread throughout the N- and C-terminal regions are conserved in LuxR homologues (42). Therefore, the seven putative LuxR family proteins in S. oneidensis are not similar to LuxR-type proteins that are AHL-dependent transcription regulators but are rather members of the NarL-FixJ family of DNA binding proteins.
TABLE 1.
GFP fluorescence production of E. coli MG1655(pJWP01s), an AHL reporter strain, with the addition of filtered culture supernatanta
| Speciesb | Fluorescence/OD600 |
|---|---|
| V. fischeri ES 114 | 13,712 ± 217 |
| V. fischeri MJ1 | 16,018 ± 307 |
| P. aeruginosa PAO1 | 12,149 ± 343 |
| P. stewartii DC283 | 13,917 ± 233 |
| S. oneidensis | 757 ± 15 |
| grown anaerobically to an OD600 of 0.2 | |
| grown aerobically to an OD600 of 1.0 | 770 ± 15 |
| grown aerobically to an OD600 of 3.5 | 798 ± 14 |
| LB blank | 754 ± 14 |
GFP fluorescence/OD600 collected after 4 h of growth. Data are representative of experiments done in triplicate, and one standard deviation is shown.
Each species was grown aerobically on LB medium except one S. oneidensis strain, which was grown anaerobically on M9 salts to an OD600 of 0.2.
Microarray analysis of the luxS mutation.
Gene expression of the WT and DL13 cells were analyzed via microarrays to determine what global affects a luxS mutation may have on transcriptome expression. WT and DL13 cells were harvested at an OD600 of 1.5 because this was when the WT had peak production of AI-2 (Fig. 1). Gene expression profiles with differences of more than a twofold increase can be found in Table 2 and are presented as arithmetic means. A complete list of the microarray data can be found in the supplemental material. Overall, large expression differences were not seen, but the P values show that some differences are statistically significant. The biggest expression difference was only a sixfold increase (SO 3483, small hypothetical protein) in the WT. A luxS mutation did not cause a large difference in global gene expression in S. oneidensis. Depending on the bacterial species, a luxS mutation has been known to cause large (12, 44) or small (17) differences in global gene regulation. The array data produced in this study could suggest that a luxS mutation may be more important for posttranslation modification. This may be a reasonable hypothesis because of the involvement of LuxS in the AMC, which produces a major methyl donor source in the cell.
TABLE 2.
S. oneidensis MR-1 genes showing gene expression compared to DL13 during exponential growth
| Gene category and TIGR locus | Gene name | General role | Induction ratio (fold)a | −log10P valueb |
|---|---|---|---|---|
| Metabolism | ||||
| SO3286 | cydA | Cytochrome d ubiquinol oxidase | 3.7 | 8.3 |
| Transport and binding | ||||
| SO2766 | Degradation of proteins | 4.6 | 5.8 | |
| SO2233 | ATPase transmembrane activity | 2.1 | 3.1 | |
| SO2373 | Bcr/CflA family, resistance transporter | 2.0 | 4.0 | |
| Stress response | ||||
| SO2780 | rpF | Ribosomal protein L32 | 2.6 | 2.8 |
| SO2756 | Antioxidant activity | 2.3 | 4.8 | |
| Hypothetical | ||||
| SO3482 | Hypothetical protein | 6.6 | 7.2 | |
| SO1657 | Hypothetical protein | 2.6 | 4.4 | |
| SO4067 | Hypothetical protein | 2.4 | 3.3 | |
| SO3656 | Hypothetical protein | 2.3 | 7.2 | |
| SO4137 | Hypothetical protein | 2.2 | 4.5 |
Relative gene expression is presented as the mean ratio of the fluorescence intensity of MR-1 cells to that of DL13 cells.
Each gene showed significant differential expression as described in Materials and Methods. P values are represented as −log10 numbers.
The luxS mutation influences biofilm development.
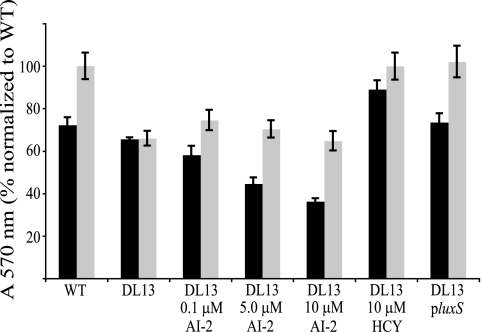
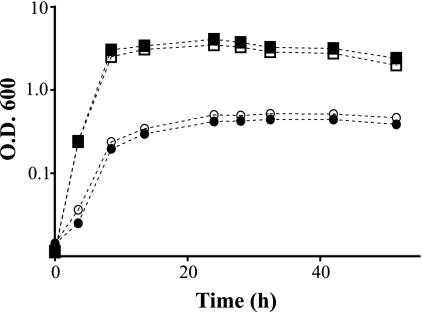
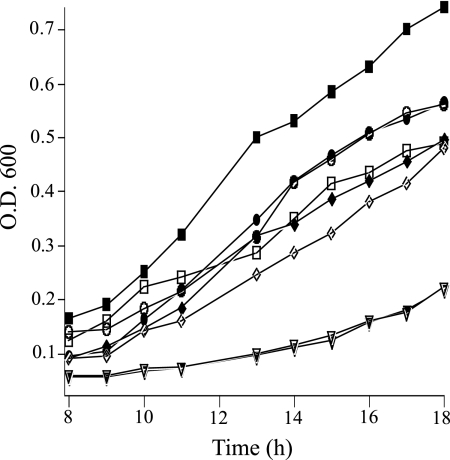
S. oneidensis biofilm assays were conducted under static conditions using a 96-well microtiter plate. After one day of growth, DL13 has a slightly (10%) decreased ability to form a biofilm compared to the WT as measured by crystal violet plate staining (Fig. 2). Upon 3 days of growth, the WT biofilm increased its biomass by about 40%, while the DL13 biofilm remained constant. The DL13 day 3 biofilm had only 66% of the biomass compared to that of the WT. This biofilm defect was absent in DL13 when it was complemented by a plasmid-borne luxS (pluxS). Aerobic planktonic growth curves on MM and LB medium were produced to examine if this luxS mutation specifically alters biofilm formation or causes an overall growth defect. Planktonic growth on either MM or LB medium was not affected by a luxS mutation (Fig. 3). These experiments suggest that a mutation in luxS decreases the ability of S. oneidensis to sustain a biofilm under static conditions in a manner independent of planktonic growth rate.
FIG. 2.
Biofilm microtiter plate assay showing 1 (black bars) and 3 (gray bars) days of S. oneidensis WT, DL13, and DL13 pluxS biofilm growth. Various concentrations of synthetic AI-2 (0, 0.1, 5, and 10 μm) and 10 μM homocysteine (HCY) were added to DL13. The data are normalized to the WT day 3 absorbance at 570 nm. The average raw absorbance value of the WT on day 3 was 0.609. The experiment was completed in duplicate with six replicates. Error bars represent the standard errors of the means (n = 6).
FIG. 3.
Growth curves of the S. oneidensis WT (filled symbols) and DL13 (open symbols) strains on LB medium (squares) and MM (circles). At time zero, the OD600 was 0.02. Data are representative of experiments done in triplicate.
Various concentrations of synthetic AI-2 (0.1 to 10 μm) were added to the biofilm plate assay to test whether the AI-2 signal regulates biofilm formation in S. oneidensis. The concentration ranges of AI-2 were taken from comparing the relative luminescence response of V. harveyi MM32 to the addition of the WT supernatant and known concentrations of AI-2. AI-2 did not restore DL13 biofilm-forming abilities to WT levels after 1 or 3 days of growth (Fig. 2). The affect of homocysteine on static biofilm formation was also examined because homocysteine is a by-product of the reaction that produced synthetic AI-2. Homocysteine had no negative effect on biofilm growth for DL13 or the WT. The addition of homocysteine even seemed to complement the biofilm defect of DL13 after 3 days of growth. The planktonic growth curves of the WT and DL13 with the addition of AI-2 were also produced to rule out the notion that AI-2 was causing an overall growth defect. No aerobic growth differences were seen between DL13 and the WT with the addition of 0.1 to 10 μm AI-2 (data not shown).
These experiments provide evidence that a mutation in luxS can inhibit the ability of S. oneidensis to sustain biofilm growth in a static environment. This biofilm defect could be restored to WT levels by genetic complementation, but the addition of AI-2 had no effect. The inability of AI-2 to restore the biofilm defect suggests that LuxS in S. oneidensis may be involved in the AMC and not quorum sensing. A mutation in luxS would stop the final step in the AMC where LuxS converts SRH into homocysteine and DPD. Adding homocysteine may override the luxS mutation and restore the ability of DL13 to recycle homocysteine within the AMC. Additionally, LuxS is used within the AMC to remove the toxic metabolite SAH. Since these experiments were performed in a static environment, it is possible that this mutation causes a buildup of a toxic metabolite that inhibited biofilm growth.
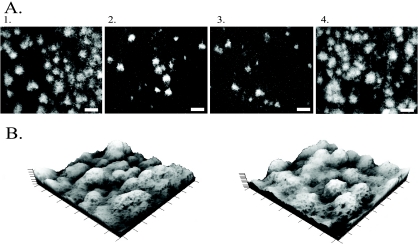
The influence of LuxS on S. oneidensis biofilm development was also evaluated under flow conditions. After 16 h of growth, the WT biofilm covered the majority of the glass slide and had a high density of three-dimensional tower-like structures (Fig. 4A1). Similar structures have also been seen by other studies examining S. oneidensis biofilm formation (45, 46). In comparison, DL13 showed a low density of small microcolonies forming on the surface (Fig. 4A2). The addition of 5 μM AI-2 had no affect on the biofilm of DL13 (Fig. 4A3) but genetic complementation did produce a biofilm similar to that of the WT (Fig. 4A4). The differences in biofilm density only occurred in the early stages of biofilm development, since the DL13 and WT biofilms were similar upon 48 h of growth (Fig. 4B). The addition of homocysteine did not have any affect on WT or DL13 biofilm formation (data not shown). Continuous flow biofilm experiments show that a mutation in luxS inhibited the early stages of S. oneidensis biofilm development. The observation that a luxS mutation can alter initial biofilm development has also been seen in Streptococcus mutans and Klebsiella pneumoniae (1, 32).
FIG. 4.
(A) Two-dimensional confocal images after 16 h of biofilm growth. Panel 1, S. oneidensis WT; panel 2, DL13; panel 3, DL13 supplemented with 5 μM AI-2; panel 4, DL13 pluxS. The scale bars represent 100 μm. (B) Three-dimensional projections of confocal images after 48 h of growth. Left, the S. oneidensis WT; right, DL13. Both three-dimensional projections have the dimensions of 900 μm by 900 μm by 60 μm. All images were taken with a 10× objective lens.
Even though the static and continuous flow biofilm experiments are hard to compare because of the different experimental conditions, one constant finding was that a mutation in luxS seemed to alter biofilm formation for each specific condition. The defect could be restored by genetic complementation but not by the addition of AI-2. This supports the conclusion that the quorum-sensing signal AI-2 is not used to regulate biofilm development in S. oneidensis.
LuxS impacts the activated methyl cycle.
Growth under sulfur-limited conditions was examined to determine if a mutation in luxS disrupts the AMC and homocysteine recycling. Cells were grown in M9 medium with a variety of sole sulfur species: K2SO4, cysteine, homocysteine, or methionine. Growth on K2SO4, cysteine, or homocysteine produced no growth differences between the WT and DL13 (Fig. 5). DL13 showed consistent growth reduction compared to that of the WT when grown with methionine as the sole sulfur source.
FIG. 5.
Growth curves of the S. oneidensis WT (filled symbols) and DL13 (open symbols) on various sources of sulfur. Triangles, cysteine; diamonds, K2SO4; circles, homocysteine; squares, methionine. The data from 0 to 7 h are not shown, because under these conditions, there was long lag phase and little growth was seen. At time zero, the OD600 was 0.03. Data are representative of experiments done in triplicate.
The growth difference seen between the S. oneidensis WT and DL13 strains with methionine as the sole sulfur source is likely to be related to the disruption of the AMC and the mutant's inability to recycle homocysteine and potentially participate in methylation reactions. Similar growth deficiencies in luxS mutants of Bacillus subtilis and Staphylococcus aureus under sulfur-limiting conditions have been linked to the disruption of the AMC (16, 24). The growth difference seen between the WT and DL13 on methionine is an additional link of the role of LuxS in metabolism.
Since a luxS mutation in S. oneidensis can cause problems with methionine metabolism within the AMC, there is a potential for this mutation to also affect the SAM methyltransferase reaction. Altering the cell's ability to participate in methyltransferase reactions could have broad cellular repercussions and could be the reason LuxS plays a role in biofilm development. SAM, the major methyl donor in the cell, has been called the second most-used (the first being ATP) enzyme substrate (28). DNA methylation can alter the cell cycle, gene regulation, virulence, and DNA repair (25, 29). Posttranslational methylation can be used to alter or broaden the function of proteins. One such example is CheR, a chemotaxis methyltransferase that utilizes SAM to transfer methyl residues to methyl-accepting chemotaxis proteins (15, 40, 41).
AI-2 metabolism debate.
More than 60 different bacterial species possess luxS. To date, the seeming ubiquitousness of luxS could be explained because it is an interspecies signal producer and/or a necessary gene in a biosynthetic pathway. The toxic AMC intermediate SAH can be detoxified from the cell by a one-step reaction catalyzed by SAH hydrolase or a two-step process that involves Pfs (MTA-SAHase), LuxS, and the production of AI-2. One could argue that the benefit of utilizing the two-step reaction is that the AI-2 signal is produced. Also, one SAM methyl donation event produces one molecule of AI-2, which makes it a good candidate for a cell density signal and metabolic activity (51, 52). Together this makes a case for AI-2 being a potential signal. Within the Vibrio species, the link between quorum sensing and luxS is well defined. Outside this genus, the debate between the role of LuxS in AI-2 quorum sensing and metabolism still endures. Direct evidence that links the AI-2 signal to luxS-related phenotypes would help resolve this issue (47, 49).
Our findings show that LuxS can alter S. oneidensis biofilm development, but it does not use the AI-2 quorum-sensing signal to regulate biofilm development or act as a global gene regulator. We have also produced evidence that LuxS is important in S. oneidensis for methionine metabolism, most likely through the AMC. This supports the argument that having luxS or even the ability to create a positive AI-2 reporter strain response does not necessarily mean the bacteria have the ability to utilize quorum sensing. We also could not produce any evidence that S. oneidensis can produce an AHL quorum-sensing signal. A question that still remains is why a luxS mutation can alter biofilm development. A metabolic defect could directly influence biofilm growth or in the case of the AMC, it could also disrupt other chemical reactions like posttranslational methylation events.
Supplementary Material
Acknowledgments
We thank Bonnie Bassler, Kim Hardie, Michael Kovach, Christopher Marx, William Metcalf, Jim Tiedje, Paul Williams, and Klaus Winzer for providing strains and plasmids. We thank Kristi DeCourcy for assistance with confocal microscopy. We thank W. Black and S. Melville for helpful discussions and allowing us access to their equipment.
This research was funded by a GAAN fellowship (U.S. Department of Education) and the U.S. Department of Energy's OBES Geosciences program (grant DE-FG02-06ER 15786). This research was funded in part by the U.S. Department of Energy's Office of Science, Biological and Environmental Research, Environmental Remediation Sciences program. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Published ahead of print on 5 January 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Balestrino, D., J. A. Haagensen, C. Rich, and C. Forestier. 2005. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 187:2870-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 3.Bodor, A., B. Elxnat, V. Thiel, S. Schulz, and I. Wagner-Dobler. 2008. Potential for luxS related signalling in marine bacteria and production of autoinducer-2 in the genus Shewanella. BMC Microbiol. 8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretschger, O., A. Obraztsova, C. A. Sturm, I. S. Chang, Y. A. Gorby, S. B. Reed, D. E. Culley, C. L. Reardon, S. Barua, M. F. Romine, J. Zhou, A. S. Beliaev, R. Bouhenni, D. Saffarini, F. Mansfeld, B. H. Kim, J. K. Fredrickson, and K. H. Nealson. 2007. Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 73:7003-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, S. D., M. Martin, S. Deshpande, S. Seal, K. Huang, E. Alm, Y. Yang, L. Wu, T. Yan, X. Liu, A. Arkin, K. Chourey, J. Zhou, and D. K. Thompson. 2006. Cellular response of Shewanella oneidensis to strontium stress. Appl. Environ. Microbiol. 72:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 8.Cho, H., H. Jonsson, K. Campbell, P. Melke, J. W. Williams, B. Jedynak, A. M. Stevens, A. Groisman, and A. Levchenko. 2007. Self-organization in high-density bacterial colonies: efficient crowd control. PLoS Biol. 5:e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 10.Coplin, D. L., R. D. Frederick, D. R. Majerczak, and E. S. Haas. 1986. Molecular cloning of virulence genes from Erwinia stewartii. J. Bacteriol. 168:619-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Keersmaecker, S. C., K. Sonck, and J. Vanderleyden. 2006. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 14:114-119. [DOI] [PubMed] [Google Scholar]
- 12.DeLisa, M. P., J. J. Valdes, and W. E. Bentley. 2001. Quorum signaling via AI-2 communicates the “metabolic burden” associated with heterologous protein production in Escherichia coli. Biotechnol. Bioeng. 75:439-450. [DOI] [PubMed] [Google Scholar]
- 13.Denef, V. J., J. A. Klappenbach, M. A. Patrauchan, C. Florizone, J. L. Rodrigues, T. V. Tsoi, W. Verstraete, L. D. Eltis, and J. M. Tiedje. 2006. Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 72:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Windt, W., N. Boon, S. D. Siciliano, and W. Verstraete. 2003. Cell density related H2 consumption in relation to anoxic Fe(0) corrosion and precipitation of corrosion products by Shewanella oneidensis MR-1. Environ. Microbiol. 5:1192-1202. [DOI] [PubMed] [Google Scholar]
- 15.Djordjevic, S., and A. M. Stock. 1998. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J. Struct. Biol. 124:189-200. [DOI] [PubMed] [Google Scholar]
- 16.Doherty, N., M. T. Holden, S. N. Qazi, P. Williams, and K. Winzer. 2006. Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J. Bacteriol. 188:2885-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dove, J. E., K. Yasukawa, C. R. Tinsley, and X. Nassif. 2003. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology 149:1859-1869. [DOI] [PubMed] [Google Scholar]
- 18.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, H., Y. Wang, X. Liu, T. Yan, L. Wu, E. Alm, A. Arkin, D. K. Thompson, and J. Zhou. 2004. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186:7796-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorby, Y. A., S. Yanina, J. S. McLean, K. M. Rosso, D. Moyles, A. Dohnalkova, T. J. Beveridge, I. S. Chang, B. H. Kim, K. S. Kim, D. E. Culley, S. B. Reed, M. F. Romine, D. A. Saffarini, E. A. Hill, L. Shi, D. A. Elias, D. W. Kennedy, G. Pinchuk, K. Watanabe, S. Ishii, B. Logan, K. H. Nealson, and J. K. Fredrickson. 2006. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. USA 103:11358-11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg, E. P., J. W. Hastings, and S. Ulitzer. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 22.Hardie, K. R., and K. Heurlier. 2008. Establishing bacterial communities by “word of mouth”: LuxS and autoinducer 2 in biofilm development. Nat. Rev. Microbiol. 6:635-643. [DOI] [PubMed] [Google Scholar]
- 23.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hullo, M. F., S. Auger, O. Soutourina, O. Barzu, M. Yvon, A. Danchin, and I. Martin-Verstraete. 2007. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J. Bacteriol. 189:187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeltsch, A., R. Z. Jurkowska, T. P. Jurkowski, K. Liebert, P. Rathert, and M. Schlickenrieder. 2007. Application of DNA methyltransferases in targeted DNA methylation. Appl. Microbiol. Biotechnol. 75:1233-1240. [DOI] [PubMed] [Google Scholar]
- 26.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 27.Lies, D. P., M. E. Hernandez, A. Kappler, R. E. Mielke, J. A. Gralnick, and D. K. Newman. 2005. Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for biofilms. Appl. Environ. Microbiol. 71:4414-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loenen, W. A. 2006. S-Adenosylmethionine: jack of all trades and master of everything? Biochem. Soc. Trans. 34:330-333. [DOI] [PubMed] [Google Scholar]
- 29.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 31.Matthysse, A. G., S. Stretton, C. Dandie, N. C. McClure, and A. E. Goodman. 1996. Construction of GFP vectors for use in gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 145:87-94. [DOI] [PubMed] [Google Scholar]
- 32.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 34.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 37.Ruby, E. G., and K. H. Nealson. 1976. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica; a model of symbiosis based on bacterial studies. Biol. Bull. 151:574-586. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 40.Simms, S. A., and K. Subbaramaiah. 1991. The kinetic mechanism of S-adenosyl-L-methionine: glutamylmethyltransferase from Salmonella typhimurium. J. Biol. Chem. 266:12741-12746. [PubMed] [Google Scholar]
- 41.Springer, W. R., and D. E. Koshland, Jr. 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. USA 74:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens, A. M., and E. P. Greenberg. 1999. Transcriptional activation by LuxR, p. 231-242. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, DC.
- 43.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sztajer, H., A. Lemme, R. Vilchez, S. Schulz, R. Geffers, C. Y. Yip, C. M. Levesque, D. G. Cvitkovitch, and I. Wagner-Dobler. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190:401-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teal, T. K., D. P. Lies, B. J. Wold, and D. K. Newman. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl. Environ. Microbiol. 72:7324-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thormann, K. M., R. M. Saville, S. Shukla, D. A. Pelletier, and A. M. Spormann. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making “sense” of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 48.Wan, X. F., N. C. Verberkmoes, L. A. McCue, D. Stanek, H. Connelly, L. J. Hauser, L. Wu, X. Liu, T. Yan, A. Leaphart, R. L. Hettich, J. Zhou, and D. K. Thompson. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 186:8385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, P., K. Winzer, W. C. Chan, and M. Camara. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B 362:1119-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 51.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53:291-396. [DOI] [PubMed] [Google Scholar]
- 52.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, J., J. W. Beaber, M. I. More, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.