Abstract
Glycogen branching enzymes (GBE) or 1,4-α-glucan branching enzymes (EC 2.4.1.18) introduce α-1,6 branching points in α-glucans, e.g., glycogen. To identify structural features in GBEs that determine their branching pattern specificity, the Deinococcus geothermalis and Deinococcus radiodurans GBE (GBEDg and GBEDr, respectively) were characterized. Compared to other GBEs described to date, these Deinococcus GBEs display unique branching patterns, both transferring relatively short side chains. In spite of their high amino acid sequence similarity (88%) the D. geothermalis enzyme had highest activity on amylose while the D. radiodurans enzyme preferred amylopectin. The side chain distributions of the products were clearly different: GBEDg transferred a larger number of smaller side chains; specifically, DP5 chains corresponded to 10% of the total amount of transferred chains, versus 6.5% for GBEDr. GH13-type GBEs are composed of a central (β/α) barrel catalytic domain and an N-terminal and a C-terminal domain. Characterization of hybrid Deinococcus GBEs revealed that the N2 modules of the N domains largely determined substrate specificity and the product branching pattern. The N2 module has recently been annotated as a carbohydrate binding module (CBM48). It appears likely that the distance between the sugar binding subsites in the active site and the CBM48 subdomain determines the average lengths of side chains transferred.
Glycogen is an energy reserve polymer of many animals and microorganisms. It is composed of a backbone of glucose residues linked by α-1,4 glycosidic bonds with α-1,6-linked side chains (7, 31). In bacteria, the linear α-1,4-glucan is synthesized from ADP-glucose by the enzyme glycogen synthase, which is thought to be involved in both initiation and elongation of the chain (40). Side chains are introduced by glycogen branching enzyme (GBE) or 1,4-α-glucan branching enzyme (EC 2.4.1.18). This enzyme catalyzes formation of α-1,6 branch points by cleaving an α-1,4 glycosidic linkage in the donor substrate and transferring the nonreducing end-terminal fragment of the chain to the C-6 hydroxyl position of an internal glucose residue that acts as the acceptor substrate (4). Depending on its source, GBEs have a preference for transferring different lengths of glucan chains (1, 23). Most GBEs are members of subfamily 8 (Eukaryota) or 9 (Bacteria) of glycoside hydrolase family 13 (GH13) (34). Recently, the first GBE from family GH57 was described (28) (http://www.cazy.org).
GH13-type GBEs are composed of three major domains of secondary structure, a central (β/α) barrel catalytic domain or A domain, an N-terminal domain, and a C-terminal domain (1). Domain A is present in all members of family GH13 and consists of a highly symmetrical fold of eight parallel β-strands encircled by eight α-helices. However, some variations occur in GBEs (a missing α-helix 5 and insertion of extra α-helices) (1). Domain A contains the four conserved amino acid regions (I to IV) typical for enzymes of family GH13 (35). In most GH13 enzymes, an extra domain is present, inserted between β-strand 3 and α-helix 3 (domain B), which affects their catalysis and product specificity (16). In GBEs, the length of this loop is only 40 residues, not long enough to be considered a separate domain (1). Domain C is found in most GH13 enzymes and is believed to shield the hydrophobic residues of the catalytic domain from contacts with the solvent. Domain C has also been suggested to be involved in substrate binding (25).
Domain N is typical for GH13 enzymes cleaving or forming endo-α-1,6 linkages (17), namely, isoamylase (EC 3.2.1.68; subfamily 11) (18), pullulanase (EC 3.2.1.41; subfamilies 12 to 14) (27), and both starch (subfamily 8) and glycogen branching enzymes (subfamilies 8 and 9). An exception is the 4-α-d-{(1→4)-α-d-glucano}trehalose trehalohydrolase (EC 3.2.1.141; subfamily 10), which hydrolyzes linear maltooligosaccharide-like substrates (6) (39). The crystal structures of most of these enzymes with their N domains (all or part) have been published previously (1, 6, 18, 27, 39). The exact function of this N domain has remained unclear, and the similarity between the N domains in these different enzymes is low. They vary in length, and some of them consist of two or three modules. However, they all possess one common module that was recently classified as a family 48 carbohydrate-binding-module (CBM48) (19) (http://www.cazy.org/).
In GBEs domain N comprises a module of 150 amino acids, termed the N2 module, that contains the putative CBM48. In some branching enzymes, it is preceded by a module of 100 to 150 amino acids, termed the N1 module. It has been proposed that the N1 module has originated from a DNA duplication of the N2 module (24). Based on the architecture and length of the N domain, GBEs can be divided into group 1, containing both the N1 and the N2 modules, and group 2, containing only the N2 module (12). A 112-amino-acid truncation of the N1 module in E. coli GBE (group 1) resulted in a 40% reduction of enzyme activity (2) and an altered branching pattern (3). Further investigations of this N1 module, by sequential N-terminal deletions, showed that enzymes with the shorter N1 module transferred longer glucan chains (5). No studies have been reported thus far investigating the role of the N2 module (containing the putative CBM48 domain) in GBEs as well as in other GH13 members.
Here, we report a detailed biochemical characterization of two GH13 GBEs from the extremophilic bacteria Deinococcus geothermalis and Deinococcus radiodurans. These two GBEs (GBEDg and GBEDr, respectively) generate unique branching patterns by transferring glucosidic chains that are shorter than those of other GBEs reported to date (9, 36, 38, 41). To investigate the role of the different domains in these enzymes, chimeras of GBEDg and GBEDr were constructed. Their characterization revealed that substrate and chain length specificity in these Deinococcus GH13 GBEs are largely determined by the putative CBM48 part of the N domain.
MATERIALS AND METHODS
Phylogenetic analysis.
Amino acid sequences of all GH13 GBEs reported in the Carbohydrate Active Enzymes database (http://www.cazy.org) were used to construct a phylogenetic tree; these were the following: Agrobacterium tumefaciens (AF033856), Anaerobranca gottschalkii (AM114416), Aquifex aeolicus (AE000704), Bacillus caldolyticus (Z14057), Bacillus cereus (D45838), Bacillus subtilis (AF008220), Butyrivibrio fibrisolvens (M64980), Chlamydophila pneumoniae (AE001632), Cyanobacterium sp. (EF489422 and EF489423), Escherichia coli (AE000419), Geobacillus stearothermophilus (M35089), Magnetospirillum gryphiswaldense (CU459003), Mycobacterium tuberculosis (Z73902), Neisseria denitrificans (AF102867), Rhodothermus obamensis (AB060080), Streptomyces aureofaciens (L11647), Streptomyces coelicolor (AJ001205 and AJ001206), and Yersinia pestis (AJ414159). They were aligned with GBEDg (CP000359) and GBEDr (AE002025) (CLUSTALW interface in MEGA, version 3.1) (20; http://www.megasoftware.com]), followed by bootstrap testing of phylogeny (gap opening, 10; extension penalties, 0.2; 1,000 replicates).
Molecular techniques.
General procedures for DNA manipulation and cloning were done as described previously (32). Restriction endonucleases and T4 DNA ligase were from Fermentas GMBH (St. Leon-Rot, Germany) and were used as recommended by the suppliers. Primers were obtained from Eurogentec (Seraing, Belgium), and sequencing was performed by GATC Biotech (Konstanz, Germany). DNA amplification was performed by PCR on a Thermal Cycler PTC-200 (MJ Research, Waltham, MA) using Expand High Fidelity polymerase (Fermentas, St. Leon-Rot, Germany).
Cloning of Deinococcus GBE genes.
D. geothermalis strain DSM 11300 was a kind gift from Michael J. Daly (Department of Pathology, Uniformed Service University of the Health Sciences, Bethesda, MD). It was grown in TGY (tryptone, glucose, yeast extract) medium at 55°C, and its chromosomal DNA was isolated according to the cetyltrimethylammonium bromide protocol obtained from the Joint Genome Institute. D. radiodurans R1 genomic DNA was a kind gift from Magali Remaud-Siméon (Ingénierie des Systèmes Biologiques et des Procédés, CNRS, INRA, INSA, Toulouse, France). The genes encoding the GBE of D. geothermalis (glgBDg) and D. radiodurans (glgBDr) were amplified from genomic DNA by PCR using the oligonucleotides Dg-FP/Dg-RP and Dr-FP/Dr-RP, respectively (Table 1). Both PCR products were cloned separately into the pET15b vector (Novagen) using NdeI and BglII restriction sites, resulting in plasmids pDg and pDr with N-terminal His tags.
TABLE 1.
Oligonucleotides used to amplify glgBDg and glgBDr genes and to construct truncated and chimeric mutants by PCR
| Primer | Sequence (5′→3′) | Position (nt)a
|
|
|---|---|---|---|
| glgBDg | glgBDr | ||
| Dg-FP | GGAATTCCCATATGGAATTCATGCTTGACCCGTTGCGGGC | 1-20 | |
| Dg-RP | GCTAGCTCTAGAAGATCTTCACCGGTCCTCGCTGCGGGGAC | 1959-1938 | |
| Dr-FP | GATGCATCATATGACGTTTCCCTTGCCCCTCGACCACGAGCACC | 1-34 | |
| Dr-RP | GCTACTAAGATCTTTATCAAGCCTTCTTCCCGCCTTTCTGCTCG | 2124-2099 | |
| CTT-RP | GTAAGGATCCTTACTAGCGGCTGACAAGGCCCTTCACC | 1917-1896 | |
| H1 | GTCATGAATCTGGTGCGGCGCCTGAA | 1531-1556 | 1501-1526 |
| H2 | TTCAGGCGCCGCACCAGATTCATGAC | 1556-1531 | 1526-1501 |
| H3 | GTGACGACGGCTGGTTCCTGAACTAC | 521-546 | 491-516 |
| H4 | GTAGTTCAGGAACCAGCCGTCGTCAC | 546-521 | 516-491 |
| DgNT-FP | CGATCGCATATGGGCGCCGGCCCCATCAGCATCTACGAAGTCCATGTCG | 478-505 | |
| DgCT-RP | GCTACTAAGATCTCATCGCGGTAAAGGCCATTCAGGCGCCGCACCAG | 1571-1540 | |
nt, nucleotide.
Construction of truncated and chimeric genes.
The 3′ 207-nucleotide extension in domain C of glgBDr was truncated (resulting in CTTDr) by PCR using plasmid pDr as a template and the primers Dr-FP and CTT-RP (Table 1). Similarly, the 5′ and 3′ truncated forms of glgBDg (477 nucleotides from 5′ for the N terminus [NTDg] and 387 nucleotides from 3′ for the C terminus [CTDg]) were amplified by PCR using as primers DgNT-FP and Dg-RP for the NTDg chimeric gene and Dg-FP and DgCT-RP for the CTDg chimeric gene. They were all cloned into the NdeI and BglII sites of pET15b vector yielding pDrCTT, pDgNT, and pDgCT plasmids.
Construction of the GGR, RRG, RGG, and GRR chimeric genes was achieved by overlap extension PCR (43). They were named according to the glgB source of their N, A, and C domains (e.g., the GGR gene consists of the glgBDg N domain, the glgBDg A domain, and glgBDr C domain in respective order). Briefly, separate PCRs were performed to yield fragments encompassing the N domain, catalytic A domain, and/or the C-terminal domain for each glgB gene. Appropriate templates and oligonucleotides were combined in subsequent PCRs (Table 2) to yield the respective hybrids. Templates, primers, and amplified fragments are listed in Table 2. The GGR, RRG, RGG, and GRR chimeric genes were cloned into NdeI and BglII sites of the pET15b vector, yielding plasmids pGGR, pRRG, pRGG and pGRR.
TABLE 2.
Templates, primers, and amplified fragments for the construction of chimeric mutants by PCR
| Template(s) | Primers | Amplified fragmenta | Size (bp) |
|---|---|---|---|
| pDr | Dr-FP, CTT-RP | CTTDr | 1,934 |
| pDg | Dg-FP, H2 | NADg | 1,571 |
| pDr | H1, Dr-RP | CDr | 643 |
| NADg, CDr | Dg-FP, Dr-RP | GGR | 2,188 |
| pDg | H2, Dg-RP | CDg | 447 |
| pDr | Dr-FP, H1 | NADr | 1,527 |
| NADr, CDg | Dr-FP, Dg-RP | RRG | 1,948 |
| pDg | Dr-FP, H4 | NDr | 517 |
| pDr | H3, Dg-RP | ACDg | 1,457 |
| NDr, ACDg | Dr-FP, Dg-RP | RGG | 1,948 |
| pDg | Dg-FP, H4 | NDg | 566 |
| pDr | H3, Dr-RP | ACDr | 1,653 |
| NDg, ACDr | Dg-FP, Dr-RP | GRR | 2,193 |
| pDg | Dg-FP, DgCT-RP | CTDg | 1,601 |
| pDg | DgNT-FP, Dg-RP | NTDg | 1,521 |
The N, A, and C domains and combinations of these domains (e.g., NA) are identified by the subscripted abbreviations used in the text for D. geothermalis (Dg) and D. radiodurans (Dr).
Expression and purification.
Overexpression of all proteins, except for NTDg, CTDg, and GRR, was achieved by overnight growth of E. coli BL21(DE3) Star cells containing the corresponding plasmid at 37°C and 210 rpm in Luria-Bertani medium supplemented with 50 μg/ml ampicillin. NTDg, CTDg, and GRR were expressed successfully by induction with 1 mM isopropyl-β-d-thiogalactopyranoside at an optical density at 600 nm of ≈0.6, followed by 5 h of additional growth. For all proteins, the cells were harvested by centrifugation (10 min at 10,000 × g), washed with 50 mM sodium phosphate buffer, pH 8.0, and centrifuged again (10 min at 10,000 × g). The pellets were resuspended in 50 mM sodium phosphate buffer, pH 8.0, and the cells were disrupted by sonication (seven times for 15 s at 7 μm with 30-s intervals) and centrifuged (30 min at 15,000 × g). All proteins were found as soluble proteins in the supernatants except for NTDg, CTDg, and GRR, which were found in the pellets as inclusion bodies. Only GRR was successfully isolated from the inclusion bodies by resuspension of the pellet in 8 M urea, followed by centrifugation (30 min at 15,000 × g). The supernatant containing unfolded GRR was subjected to dialysis against sodium phosphate buffer (25 mM; pH 8), leading to the refolding of GRR, as confirmed by circular dichroism.
All proteins were purified by His tag affinity chromatography using a HiTrap chelating column (Amersham Pharmacia, Uppsala, Sweden) charged with nickel sulfate. The proteins were eluted with a linear gradient of 0 to 500 mM imidazol on an Åkta prime purification system (Amersham Pharmacia). Fractions containing the protein of interest were pooled and dialyzed against sodium phosphate buffer (25 mM; pH 8.0).
Protein determination and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protein concentrations were determined by measuring the absorbance at 280 nm on a Nanodrop ND-1000 spectrophotometer (Isogen Life Science, de Meern, The Netherlands) using the following extinction coefficients (in mg/ml−1 cm−1) estimated by the method of Gill and von Hippel (8): 2.38 for GBEDg, 2.12 for GBEDr, 2.33 for CTTDr, 2.11 for GGR, 2.39 for RRG, 2.41 for RGG, and 2.08 for GRR.
Proteins were analyzed by SDS-PAGE on 10% polyacrylamide gels. The molecular mass was determined using a PageRuler Prestained Protein Ladder (Fermentas) as a standard, and proteins were stained with Bio-Safe Coomassie (Bio-Rad, Germany).
Circular dichroism.
Multiple far-UV spectra (195 to 260 nm) were recorded in sodium phosphate buffer at pH 8.0 and 20°C using a concentration of protein of 0.1 mg/ml on a Jasco J-715 spectropolarimeter (Jasco, Tokyo, Japan) in quartz cells; the following parameters were used: optical path length, 1 mm; data interval, 0.2 nm; bandwidth, 1.0 nm; sensitivity, 20 millidegrees; response time, 0.125 s. All recorded spectra were corrected by subtraction of the buffer spectrum.
Enzyme activity assays. (i) Iodine assay.
The total activity of the enzyme was measured using the iodine assay (10), which is based on formation of a blue complex between iodine-iodide and a linear α-1,4-glucan of a certain length. The sum of the transglycosylation and the hydrolytic activity of the enzyme can be measured by monitoring the decrease in absorbance. The reaction mixtures contained 150 μl of 0.125% (wt/vol) amylose V (Avebe, Foxhol, The Netherlands) or 0.25% potato amylopectin (type III; Sigma) and 50 μl of enzyme at the appropriate concentration (15 to 25 μg/ml). At different time intervals, aliquots of 15 μl were taken, and the reaction was stopped by addition of 1.5 μl of 1 M NaOH and 150 μl of iodine reagent (0.01% [wt/vol] I2, 0.1% [wt/vol] KI). One unit of enzyme activity is defined as the decrease in absorbance of 1.0 per min at 660 nm for amylose and at 530 nm for amylopectin.
To determine the optimal conditions for the activity of the enzymes, the assay was performed at temperatures ranging from 25°C to 45°C and pH 6.5 to 9.5 in Bis-Tris-propane-HCl buffer. For thermostability studies, the enzymes (0.5 mg/ml) were incubated in the absence of substrate for 1 h at different temperatures from 20°C to 70°C, and the residual activity in the supernatant (after centrifugation at 13,000 × g for 1 min) was measured at 34°C.
(ii) Branching assay.
This method allowed determination of the amount of α-1,6 branch points introduced by the action of GBE by measuring the difference in amount of reducing ends before (hydrolytic activity) and after debranching (total activity) of the product by isoamylase (37). The reaction mixtures contained 50 μl of appropriately diluted GBE (1, 2, or 4 μg) in 1,400 μl of 0.125% amylose V (Avebe, Foxhol, The Netherlands) at pH 8.0. Samples (200 μl) were taken at different time points (0 to 20 min), and the reaction was stopped by boiling for 5 min. The numbers of reducing ends before and after debranching were measured at each time point by a 2,2′-bicinchoninate assay (42) using a maltotriose calibration curve. Debranching was performed as follows: 50 μl of reaction mixture was debranched for 20 h at 40°C in 129 μl of 25 mM sodium phosphate buffer (pH 8.0), 20 μl of 1 M acetate buffer (pH 3.8), and 1 μl (0.175 U) of Pseudomonas sp. isoamylase (Megazyme, Wicklow, Ireland). One unit of GBE branching activity is defined as 1 μmol of α-1,6 linkages synthesized per min.
Side chain distribution.
To determine the side chain distribution of the final products, 10 ml of 0.125% (wt/vol) amylose V (Avebe) solution was incubated with 400 units (determined by iodine assay) of GBEDg or GBEDr for 72 h. The reaction was terminated by boiling the mixture for 10 min. The reaction mixture was divided in two equal parts; one part was treated with 10 units of isoamylase from Pseudomonas sp. (Megazyme) after the addition of 750 μl of 1 M citrate buffer, pH 4.0, for 20 h at 40°C to hydrolyze the α-1,6 glucosidic linkages. Both parts of the reaction mixture, before and after debranching, were freeze-dried, dissolved in 90% dimethyl sulfoxide (0.25 mg/ml) and analyzed by high-performance anion-exchange chromatography (HPAEC) analysis. Mixtures of oligosaccharides with degrees of polymerization ranging from 1 to 7 (DP1 to DP7) and a debranched waxy maize starch solution containing a broad mix of oligosaccharides of known composition were used as standards. Separation of oligosaccharides was achieved with a CarboPac PA1 anion exchange column (250 mm by 4 mm; Dionex) coupled to a CarboPac1 guard column (Dionex, Sunnyvale, CA). The following gradient of eluent A (1 ml/min) was used: 95% (10 min), 65% (10 min), 55% (30 min), 35% (4 min), 0% (7 min), and 95% (14 min). Eluent A was sodium hydroxide (0.1 M), and eluent B was sodium acetate (0.6 M) in sodium hydroxide (0.1 M). Detection was performed with an ED40 electrochemical detector (Dionex) equipped with an Au working electrode and an Ag/AgCl reference electrode with a sensitivity of 300 nC. The pulse program used was the following: +0.1 V (0 to 0.41 s), −2.0 V (0.41 to 0.43 s), +0.6 V (0.43 to 0.44 s), and −0.10 V (0.44 to 0.50 s). The integration time was 0.20 to 0.40 s. Data were integrated using a Totalchrom (PerkinElmer, Waltham, MA) data integration system.
RESULTS AND DISCUSSION
Amino acid sequence analysis of the glycogen branching enzymes of Deinococcus.
At present there is no proof for actual synthesis of glycogen in the extremophilic bacteria D. geothermalis and D. radiodurans. However, the key enzymes involved in glycogen metabolism, such as glycogen branching enzyme (including a demonstration of its activity [this paper]), ADP-glucose pyrophosphorylase, and glycogen synthase, have been annotated in both species (28).
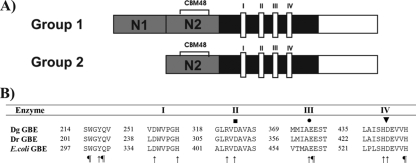
D. geothermalis and D. radiodurans both contain a single gene in their genomes coding for a putative glycogen branching enzyme. The D. geothermalis gene, Dgeo_0981, consists of 1,959 nucleotides and encodes an enzyme of 652 amino acids (GBEDg) that has a predicted molecular mass of 74.4 kDa. The DR1848 gene from D. radiodurans consists of 2,124 nucleotides and encodes an enzyme of 705 amino acids (GBEDr) that has a theoretical molecular mass of 80 kDa. Both enzymes belong to group 2 of GH13 GBEs. Furthermore, they contain the four conserved amino acid regions (I to IV) typical for enzymes of family GH13 (25) plus the four conserved amino acid residues specific for bacterial GBEs, namely W298, Q301, S460, and H530 (E. coli GBE numbering) (Fig. 1) (1). Their N2 modules, containing the CBM48, possess the two consensual aromatic residues crucial for starch or glycogen binding (26) (W77 and Y123 for GBEDg and W61 and Y107 for GBEDr).
FIG. 1.
(A) Domain organization of group 1 and group 2 GH13 GBEs. N domain (gray), catalytic or A domain (black), and C domain (white) are shown. The N domain of group 1 GBEs consists of N1 and N2 modules, while only N2 is present in group 2 GBEs. The N2 module contains a putative CBM48. In domain A the four conserved regions (I to IV) for family GH13 are found. (B) Amino acid alignment of highly conserved regions in the catalytic domains of GBEDg, GBEDr, and E. coli GBE. I to IV are conserved regions in GH13 enzymes. Amino acid positions are given. ▪, putative catalytic nucleophile (25); •, putative acid/base catalyst (25); ▾, putative transition state stabilizer (25); ↑, residue conserved in α-amylase family; ¶, residue conserved in bacterial GBEs (1).
GBEDg and GBEDr show an overall identity in 622 amino acids of 81% (88% similarity). The similarity between both N domains is in the same order (145 to 158 amino acids long, 81% identity, and 88% similarity in 136 amino acids). The identity in their catalytic A domains is relatively high (364 amino acids; 87% identity and 93% similarity). The two enzymes differ most in their C domains (130 amino acids in GBEDg and 195 in GBEDr with only 70% identity and 77% similarity in 113 amino acids). Unique for the GBEDr is a 66-amino-acid extension in the C domain, which is not present in any other GBE known to date. BLAST searches with the 66-residue-long extension did not give a significant result (data not shown).
Characterization of two Deinocccus branching enzymes.
Both Deinococcus GBE genes were cloned, and the enzymes were expressed in E. coli and purified to homogeneity by His tag purification (data not shown), yielding up to 30 mg of purified GBE per liter of E. coli culture. The molecular mass of the proteins as determined by SDS-PAGE was 75 kDa for GBEDg and 80 kDa for GBEDr, in accordance with the calculated values. Both enzymes were active in a relatively broad range of pH (7 to 9) and temperature (30 to 40°C) values and showed maximum activity at pH 8.0 and 34°C, as measured by the iodine assay. Their specific activities on amylose V were 621.3 U/mg for GBEDg and 404.2 U/mg for GBEDr (Table 3). These are relatively high values compared to specific activities previously reported for GH13 GBEs from E. coli, A. aeolicus, G. stearothermophilus, and A. gottschalkii (9, 36, 38, 41). Only the R. obamensis GBE showed a comparable specific activity (33). Specific activities measured by the branching assay were 7.2 U/mg for GBEDg and 4.7 U/mg for GBEDr (values are not directly comparable to iodine assay-specific activities because of a different definition of the unit) (see Materials and Methods). This assay allowed us to discriminate between branching activity and hydrolytic activity; the latter represented less than 1% of total activity (data not shown). Therefore, the iodine assay was used for subsequent characterizations.
TABLE 3.
Specific activity on amylose and amylopectin for the wild-type, the chimeric, and the truncated GBEDg and GBEDr enzymes
| Enzyme | Specific activity (U/mg) on:
|
Amylose/amylopectin activity | |
|---|---|---|---|
| Amylose | Amylopectin | ||
| GBEDg | 621.3 | 474.3 | 1.31 ± 0.08 |
| GBEDr | 404.2 | 538.0 | 0.75 ± 0.06 |
| CTTDr | 442.3 | 556.5 | 0.79 ± 0.05 |
| GGR | 468.8 | 359.2 | 1.30 ± 0.08 |
| RRG | 411.7 | 505.6 | 0.73 ± 0.09 |
| RGG | 624.1 | 342.3 | 1.82 ± 0.09 |
| GRR | <0.5 | <0.5 | NDa |
ND, not determined.
The two enzymes showed clear differences in their thermostability, as expected from their optimal growth temperatures (25 to 30°C for D. radiodurans and 50 to 55°C for D. geothermalis). GBEDg remained fully active after a 1-h incubation at 60°C and lost less than 20% of activity after a 1-h incubation at 65°C. GBEDr exhibited a 10°C lower thermostability than GBEDg and remained fully active at 50°C (1 h incubation).
The two Deinococcus GBEs clearly differ in substrate specificity.
The specific activities of both GBEs were determined on amylopectin using the iodine assay (Table 3) and were compared to their specific activities on amylose as previously measured (see above). In contrast to the results with amylose, the specific activity on amylopectin was higher for GBEDr (538.0 U/mg protein) than for GBEDg (474.3 U/mg protein). The ratio of the specific activities of amylose to amylopectin was 1.31 for GBEDg and 0.75 for GBEDr. E. coli GBE had an amylose/amylopectin ratio of 1.78, which is higher than observed for the two Deinococcus GBEs (2). Thus, in spite of their high overall sequence identity and similarity, GBEDg prefers amylose over amylopectin whereas the opposite is true for GBEDr.
The two Deinococcus GBEs have a preference for transferring short side chains.
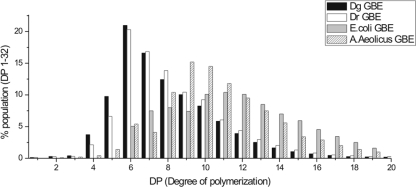
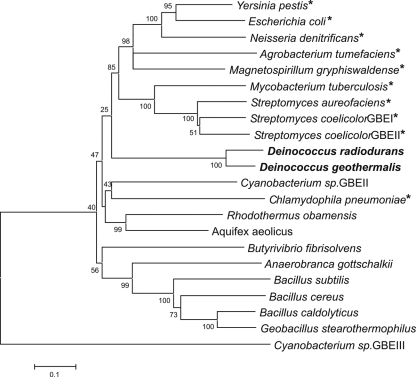
The products obtained after prolonged incubations of both enzymes with amylose showed a similar side chain distribution. Most side chains were between 4 and 17 glucose residues long, with side chains of 6 and 7 glucose residues (∼35% of total) predominantly (Fig. 2). This branching pattern is significantly different from that of the other GH13 GBEs for which a quantitative analysis has been described to date, namely, E. coli GBE (group 1 GBE) wild type and three N1 module-truncated variants (5) and A. aeolicus GBE (group 2 GBE) (41). In these GBEs, the preferred DP of chains transferred (∼40 to 50%) lies between DP10 and DP14, with only 20 to 30% of chains shorter than 10 glucose residues, a value increasing to 70% for Deinococcus GBEs (Fig. 2). Side chain distributions of other GH13 GBEs have been reported, e.g., for G. stearothermophilus GBE (36) and A. gottschalkii GBE (38), both group 2 GBEs. Even though no quantitative data are available for these enzymes, their branching patterns are clearly different from those of the Deinococcus GBEs, with a preference for the transfer of longer chains, as in the case of E. coli and A. aeolicus GBEs. This unique side chain distribution of the Deinococcus GBEs may be a consequence of the fact that they are distantly related to other known GH13 GBEs. The two Deinococcus enzymes form a separate cluster, and although they both belong to group 2 of GH13-type GBEs, they appear equally related to group 1 and group 2 at the amino acid level (Fig. 3).
FIG. 2.
HPAEC-pulsed amperometric detection analysis of the end products obtained by incubation of GBEDg or GBEDr with amylose, followed by debranching with isoamylase. For comparison, the distribution of DPs of the debranched end products of the E. coli GBE (5) and A. aeolicus GBE (41) enzymes are also shown.
FIG. 3.
Neighbor-joining tree of known GH13 GBEs (http://www.cazy.org) constructed using MEGA, version 3.1 (20). Bootstrap values are indicated on the nodes of each branch. The bar indicates 10% amino acid sequence difference. See Materials and Methods for accession numbers. *, GBE from group 1.
Construction of chimeric Deinococcus enzymes showed the importance of the N domain in substrate specificity and branching pattern.
Comparison of the branching patterns of the two Deinococcus GBEs revealed that the branched product of GBEDg contained more short side chains overall, especially of DP5, which corresponded to about 10% of the total (only 6.5% in the case of GBEDr) (Fig. 2). Thus, strikingly, even though both enzymes are highly similar (88% similarity and 81% identity), they exhibit distinct substrate specificities and clear differences in their branching patterns.
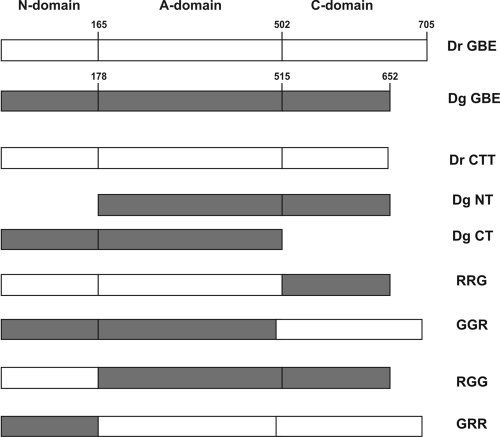
The relatively high overall amino acid sequence identity between GBEDg and GBEDr made them ideal candidates for investigating a possible role of the different domains in causing their differences in substrate preferences and branching patterns. In order to gain more insight into which domain (N, catalytic, or C domain) in the Deinococcus GBEs influenced activity and specificity, a number of chimeric enzymes were constructed and biochemically characterized (Fig. 4).
FIG. 4.
Schematic representation of the GBEDr and GBEDg constructs made for analysis of the N-domain and the C-domain roles in GBE activity.
The characterization of chimeric enzymes has also been used to study the role of the N and C domains in starch branching enzymes, with different and sometimes contradictory results. Domain C was proposed to be involved in substrate specificity (14, 15, 21), in maximal activity (14, 21), and in side chain distribution (15) while domain N was suggested to affect the catalytic efficiency (11, 13, 21) and the branching pattern (13, 21). However, these results cannot be compared directly to GBEs since they show less than 30% identity (less than 50% similarity) to starch branching enzymes, and the differences are even larger in the noncatalytic domains.
Here, we exchanged domain N (N2 module in this case) and domain C between the two Deinococcus GBEs, yielding GGR, RRG, RGG, and GRR chimeric mutants (Fig. 4). Furthermore, the extra 66-amino-acid extension in domain C of GBEDr was deleted (resulting in CTTDr) to analyze its possible effects on enzyme properties. In addition, a truncation of the entire C domain (CTDg) and a truncation of the entire N domain (NTDg) were made in GBEDg (Fig. 4).
The hybrid and truncated enzymes were expressed in E. coli BL21(DE3) Star and purified to homogeneity by His tag affinity chromatography. The GBEDg C- and N-domain-truncated enzymes, as well as the GRR chimeric enzyme, ended up in inclusion bodies. Only the GRR enzyme was successfully purified from the aggregates, and proper folding of this chimera was confirmed by circular dichroism (data not shown).
All chimeras (as well as CTTDr) showed enzyme activity although GRR activity was very low, less than 1% of the wild type (Table 3). An optimum pH of 8.0 and an optimum temperature of 34°C were observed for all chimeric enzymes (data not shown). GRR was not further characterized due to its low activity.
Domain C does not influence substrate specificity or branching pattern.
The specific activity of CTTDr was 442.3 U/mg on amylose and 468.8 U/mg on amylopectin. Both specific activities as well as their ratio of 0.79 were comparable to those of the wild-type GBEDr (Table 3). Furthermore, the side chain distribution produced by CTTDr was very similar to that made by GBEDr (data not shown). The 66-amino-acid extension at the C terminus of GBEDr thus does not affect substrate specificity or branching pattern.
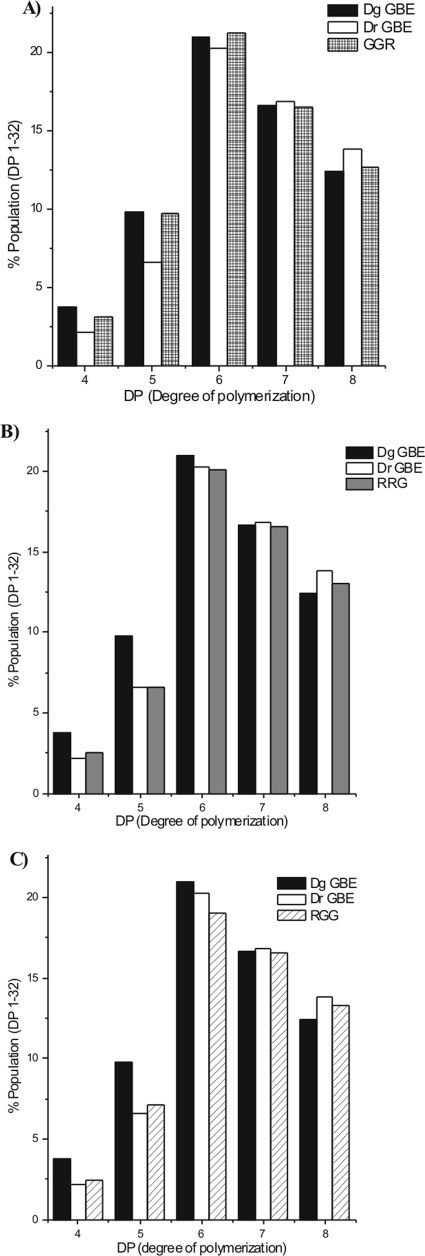
The C-domain-exchanged chimeras GGR and RRG displayed amylose/amylopectin ratios of 1.30 and 0.73, similar to GBEDg and GBEDr, respectively (Table 3). The major difference between their end products was the number of side chains containing five glucose residues. This was close to 10% for GGR (Fig. 5A) and approximately 6.5% for RRG (Fig. 5B), in accordance with their respective wild types (Fig. 5A and B). This finding indicated that the C domain is not involved in determining the substrate specificity or branching pattern of the Deinococcus GBEs.
FIG. 5.
HPAEC-pulsed amperometric detection analysis of the end products (DP4 to DP8) obtained by incubation of the GBEDg, GBEDr or GGR (A), RRG (B), or RGG (C) enzymes with amylose, followed by debranching with isoamylase.
Domain N is involved in determining substrate specificity and branching pattern.
Of the N-domain exchange mutants, only RGG (but not GRR) had enough activity to enable further tests. Therefore, substrate specificity and side chain distribution were measured only for RGG. It exhibited a specific activity on amylose of 624.1 U/mg and on amylopectin of 342.3 U/mg. Its amylose/amylopectin activity ratio was 1.9. Interestingly, this is much higher than the value measured for either parent (Table 3). This result clearly shows that domain N (N2 module) influences the GBE substrate specificity.
The product synthesized by RGG had a branching pattern that was clearly different from the product of GBEDg (Fig. 5C). Interestingly, the side chain distribution of RGG closely resembled that of GBEDr, which was the source of only the N domain of RGG. From this result, it is concluded that N domain plays a critical role in determining the length of the chains transferred by these GBEs.
It has previously been shown that the N1 module of domain N of GBEs of group 1 influences their branching patterns (2, 3, 5). However, no such information is available for GBEs belonging to group 2, which lack the N1 module. The N2 module has recently been annotated as a CBM48 and is most likely involved in binding of the substrate. The CBM48 found in AMP-activated protein kinase has been shown to bind glycogen as well as β-cyclodextrin and oligosaccharides such as maltoheptaose (19, 30). There are clear examples of the importance of CBMs for enzyme activity, e.g., in the case of CGTase, which has a carbohydrate binding site in domain C anchoring the enzyme to the starch granule (22), and the raw starch binding domain E (designated CBM20), which has two carbohydrate binding sites binding raw starch and guiding the linear starch chains toward the active site (29). In the crystal structures of isoamylase and 4-α-d-{(1→4)-α-d-glucano}trehalose trehalohydrolase, the N domain (both with a putative CBM48) is in close proximity to the B domain [loop between the third β-strand and the third α-helix of the (β/α)8 barrel] (25), which is known to be involved in catalysis and product specificity of GH13 enzymes (16). Moreover, it has been suggested that the N domain of D. radiodurans 4-α-d-{(1→4)-α-d-glucano}trehalose trehalohydrolase is involved in the initial steps of sugar binding by contributing to hydrophobic stacking interactions at the entrance of the channel leading to the active site (39). This channel in D. radiodurans 4-α-d-{(1→4)-α-d-glucano}trehalose trehalohydrolase can accommodate seven sugar units in subsites −1 to −7. Since GBE uses substrates longer than DP7, it is indeed possible that the N2 module accommodates the rest of the minus subsites (from subsite −8 on), thus determining the number of glucose units binding in the donor subsites (corresponding to the length of the chains transferred). It is also possible that by interacting with the −7 subsite, N2 influences substrate binding at the −6 subsite. A weaker binding of the donor substrate at the −6 and −7 subsites would result in an increase of transferred chains of DP5.
In the case of group 1 GBEs, the branching patterns are most likely determined by the combined action of both modules N1 and N2 of domain N. A dynamic mechanism in which a flexible N1 would move and restrict exposure of longer chains to the active site has been suggested (5). We propose that the N2 module helps in controlling the length of the chains transferred in these enzymes and plays a main role in determining them in group 2 GBEs. At present, we are studying this region of the protein in more detail to identify amino acids that are crucial for the fine-tuning of the activity of these enzymes.
Conclusions.
Here, we report the first detailed characterization of the highly similar GH13 GBEs from D. geothermalis and D. radiodurans The side chain distributions of their final products are unique and differ significantly from that of other GBEs described thus far in possessing a relatively large percentage of short chains. This exceptional branching pattern makes their products structurally unique. The physicochemical properties of such new, modified products remain to be determined, and they may have new food, cosmetics, and pharmaceutical applications.
Although both enzymes are highly similar (88% similarity and 81% identity), their specificities for amylose and amylopectin differ remarkably. Moreover, they display clear differences in their branching patterns. The variations observed apparently were not caused directly by differences in their catalytic domains (93% similar and 87% identical). Therefore, the influence of their noncatalytic domains was investigated. By constructing chimeric enzyme variants of the two Deinococcus GBEs, we demonstrated that the N domain (N2 module, recently annotated as a CBM48) is involved in substrate specificity and in determining the length of chains transferred by GBEs. Further research will focus on the influence of this putative carbohydrate binding domain on GBE enzyme activity.
Acknowledgments
We thank Peter Sanders for HPAEC analysis and Hans Leemhuis for critically reading the manuscript.
Footnotes
Published ahead of print on 9 January 2009.
REFERENCES
- 1.Abad, M. C., K. Binderup, J. Rios-Steiner, R. K. Arni, J. Preiss, and J. H. Geiger. 2002. The X-ray crystallographic structure of Escherichia coli branching enzyme. J. Biol. Chem. 277:42164-42170. [DOI] [PubMed] [Google Scholar]
- 2.Binderup, K., R. Mikkelsen, and J. Preiss. 2000. Limited proteolysis of branching enzyme from Escherichia coli. Arch. Biochem. Biophys. 377:366-371. [DOI] [PubMed] [Google Scholar]
- 3.Binderup, K., R. Mikkelsen, and J. Preiss. 2002. Truncation of the amino terminus of branching enzyme changes its chain transfer pattern. Arch. Biochem. Biophys. 397:279-285. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, C., and J. Preiss. 1977. Biosynthesis of bacterial glycogen. Purification and properties of the Escherichia coli b alpha-1,4,-glucan:alpha-1,4-glucan 6-glycosyltansferase. Biochemistry 16:3693-3699. [DOI] [PubMed] [Google Scholar]
- 5.Devillers, C. H., M. E. Piper, M. A. Ballicora, and J. Preiss. 2003. Characterization of the branching patterns of glycogen branching enzyme truncated on the N terminus. Arch. Biochem. Biophys. 418:34-38. [DOI] [PubMed] [Google Scholar]
- 6.Feese, M. D., Y. Kato, T. Tamada, M. Kato, T. Komeda, Y. Miura, M. Hirose, K. Hondo, K. Kobayashi, and R. Kuroki. 2000. Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus. J. Mol. Biol. 301:451-464. [DOI] [PubMed] [Google Scholar]
- 7.Francois, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:125-145. [DOI] [PubMed] [Google Scholar]
- 8.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 9.Guan, H., P. Li, J. Imparl-Radosevich, J. Preiss, and P. Keeling. 1997. Comparing the properties of Escherichia coli branching enzyme and maize branching enzyme. Arch. Biochem. Biophys. 342:92-98. [DOI] [PubMed] [Google Scholar]
- 10.Guan, H. P., and J. Preiss. 1993. Differentiation of the properties of the branching isozymes from maize (Zea mays). Plant Physiol. 102:1269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamada, S., H. Ito, H. Ueno, Y. Takeda, and H. Matsui. 2007. The N-terminal region of the starch-branching enzyme from Phaseolus vulgaris L. is essential for optimal catalysis and structural stability. Phytochemistry 68:1367-1375. [DOI] [PubMed] [Google Scholar]
- 12.Hilden, I., L. L. Leggio, S. Larsen, and P. Poulsen. 2000. Characterization and crystallization of an active N-terminally truncated form of the Escherichia coli glycogen branching enzyme. Eur. J. Biochem. 267:2150-2155. [DOI] [PubMed] [Google Scholar]
- 13.Hong, S., R. Mikkelsen, and J. Preiss. 2001. Analysis of the amino terminus of maize branching enzyme II by polymerase chain reaction random mutagenesis. Arch. Biochem. Biophys. 386:62-68. [DOI] [PubMed] [Google Scholar]
- 14.Hong, S., and J. Preiss. 2000. Localization of C-terminal domains required for the maximal activity or for determination of substrate preference of maize branching enzymes. Arch. Biochem. Biophys. 378:349-355. [DOI] [PubMed] [Google Scholar]
- 15.Ito, H., S. Hamada, N. Isono, T. Yoshizaki, H. Ueno, Y. Yoshimoto, Y. Takeda, and H. Matsui. 2004. Functional characteristics of C-terminal regions of starch-branching enzymes from developing seeds of kidney bean (Phaseolus vulgaris L.). Plant Sci. 166:1149-1158. [Google Scholar]
- 16.Janecek, S. 1997. α-Amylase family: molecular biology and evolution. Prog. Biophys. Mol. Biol. 67:67-97. [DOI] [PubMed] [Google Scholar]
- 17.Jespersen, H. M., E. A. MacGregor, M. R. Sierks, and B. Svensson. 1991. Comparison of the domain-level organization of starch hydrolases and related enzymes. Biochem. J. 280:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsuya, Y., Y. Mezaki, M. Kubota, and Y. Matsuura. 1998. Three-dimensional structure of Pseudomonas isoamylase at 2.2 Å resolution. J. Mol. Biol. 281:885-897. [DOI] [PubMed] [Google Scholar]
- 19.Koay, A., K. A. Rimmer, H. D. Mertens, P. R. Gooley, and D. Stapleton. 2007. Oligosaccharide recognition and binding to the carbohydrate binding module of AMP-activated protein kinase. FEBS Lett. 581:5055-5059. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Kuriki, T., D. C. Stewart, and J. Preiss. 1997. Construction of chimeric enzymes out of maize endosperm branching enzymes I and II: activity and properties. J. Biol. Chem. 272:28999-29004. [DOI] [PubMed] [Google Scholar]
- 22.Lawson, C. L., M. R. van, B. Strokopytov, H. J. Rozeboom, K. H. Kalk, G. E. de Vries, D. Penninga, L. Dijkhuizen, and B. W. Dijkstra. 1994. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J. Mol. Biol. 236:590-600. [DOI] [PubMed] [Google Scholar]
- 23.Lerner, L. R., and C. R. Krisman. 1996. Differential action of branching enzymes according to its source. Cell Mol. Biol. (Noisy-le-grand) 42:599-608. [PubMed] [Google Scholar]
- 24.Lo Leggio, L., H. A. Ernst, I. Hilden, and S. Larsen. 2002. A structural model for the N-terminal N1 module of E. coli glycogen branching enzyme. Biologia 57:108-118. [Google Scholar]
- 25.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 26.Machovic, M., and S. Janecek. 2006. The evolution of putative starch-binding domains. FEBS Lett. 580:6349-6356. [DOI] [PubMed] [Google Scholar]
- 27.Mikami, B., H. Iwamoto, D. Malle, H. J. Yoon, E. mirkan-Sarikaya, Y. Mezaki, and Y. Katsuya. 2006. Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site. J. Mol. Biol. 359:690-707. [DOI] [PubMed] [Google Scholar]
- 28.Murakami, T., T. Kanai, H. Takata, T. Kuriki, and T. Imanaka. 2006. A novel branching enzyme of the GH-57 family in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 188:5915-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penninga, D., d. van, V., R. M. Knegtel, S. A. van Hijum, H. J. Rozeboom, K. H. Kalk, B. W. Dijkstra, and L. Dijkhuizen. 1996. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J. Biol. Chem. 271:32777-32784. [DOI] [PubMed] [Google Scholar]
- 30.Polekhina, G., A. Gupta, B. J. van Denderen, S. C. Feil, B. E. Kemp, D. Stapleton, and M. W. Parker. 2005. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure 13:1453-1462. [DOI] [PubMed] [Google Scholar]
- 31.Preiss, J. 1984. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38:419-458. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Shinohara, M. L., M. Ihara, M. Abo, M. Hashida, S. Takagi, and T. C. Beck. 2001. A novel thermostable branching enzyme from an extremely thermophilic bacterial species, Rhodothermus obamensis. Appl. Microbiol. Biotechnol. 57:653-659. [DOI] [PubMed] [Google Scholar]
- 34.Stam, M. R., E. G. Danchin, C. Rancurel, P. M. Coutinho, and B. Henrissat. 2006. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng. Des. Sel. 19:555-562. [DOI] [PubMed] [Google Scholar]
- 35.Svensson, B. 1994. Protein engineering in the alpha-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25:141-157. [DOI] [PubMed] [Google Scholar]
- 36.Takata, H., T. Takaha, T. Kuriki, S. Okada, M. Takagi, and T. Imanaka. 1994. Properties and active center of the thermostable branching enzyme from Bacillus stearothermophilus. Appl. Environ. Microbiol. 60:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda, Y., H. P. Guan, and J. Preiss. 1993. Branching of amylose by the branching isoenzymes of maize endosperm. Carbohydr. Res. 240:253-263. [Google Scholar]
- 38.Thiemann, V., B. Saake, A. Vollstedt, T. Schafer, J. Puls, C. Bertoldo, R. Freudl, and G. Antranikian. 2006. Heterologous expression and characterization of a novel branching enzyme from the thermoalkaliphilic anaerobic bacterium Anaerobranca gottschalkii. Appl. Microbiol. Biotechnol. 72:60-71. [DOI] [PubMed] [Google Scholar]
- 39.Timmins, J., H. K. Leiros, G. Leonard, I. Leiros, and S. McSweeney. 2005. Crystal structure of maltooligosyltrehalose trehalohydrolase from Deinococcus radiodurans in complex with disaccharides. J. Mol. Biol. 347:949-963. [DOI] [PubMed] [Google Scholar]
- 40.Ugalde, J. E., A. J. Parodi, and R. A. Ugalde. 2003. De novo synthesis of bacterial glycogen: Agrobacterium tumefaciens glycogen synthase is involved in glucan initiation and elongation. Proc. Natl. Acad. Sci. USA 100:10659-10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Der Maarel, M. J. E. C., A. Vos, P. Sanders, and L. Dijkhuizen. 2003. Properties of the glucan branching enzyme of the hyperthermophilic bacterium Aquifex aeolicus. Biocatal. Biotransform. 21:199-207. [Google Scholar]
- 42.Waffenschmidt, S., and L. Jaenicke. 1987. Assay of reducing sugars in the nanomole range with 2,2′-bicinchoninate. Anal. Biochem. 165:337-340. [DOI] [PubMed] [Google Scholar]
- 43.Wurch, T., F. Lestienne, and P. J. Pauwels. 1998. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol. Tech. 12:653-657. [Google Scholar]