Abstract
Biofilms are composed of bacterial cells encased in a self-synthesized, extracellular polymeric matrix. Poly-β(1,6)-N-acetyl-d-glucosamine (PNAG) is a major biofilm matrix component in phylogenetically diverse bacteria. In this study we investigated the physical and chemical properties of the PNAG matrix in biofilms produced in vitro by the gram-negative porcine respiratory pathogen Actinobacillus pleuropneumoniae and the gram-positive device-associated pathogen Staphylococcus epidermidis. The effect of PNAG on bulk fluid flow was determined by measuring the rate of fluid convection through biofilms cultured in centrifugal filter devices. The rate of fluid convection was significantly higher in biofilms cultured in the presence of the PNAG-degrading enzyme dispersin B than in biofilms cultured without the enzyme, indicating that PNAG decreases bulk fluid flow. PNAG also blocked transport of the quaternary ammonium compound cetylpyridinium chloride (CPC) through the biofilms. Binding of CPC to biofilms further impeded fluid convection and blocked transport of the azo dye Allura red. Bioactive CPC was efficiently eluted from biofilms by treatment with 1 M sodium chloride. Taken together, these findings suggest that CPC reacts directly with the PNAG matrix and alters its physical and chemical properties. Our results indicate that PNAG plays an important role in controlling the physiological state of biofilms and may contribute to additional biofilm-associated processes such as biocide resistance.
Biofilms are composed of bacterial cells encased in a self-synthesized, extracellular polymeric matrix (7). The main function of the biofilm matrix is to provide a structural framework that holds the cells together in a mass and firmly attaches the bacterial mass to the underlying surface. In addition to having a structural role, the matrix provides biofilm cells with a protected microenvironment containing dissolved nutrients, secreted enzymes, DNA, and phages. The matrix may also contribute to the increased antimicrobial resistance exhibited by biofilm cells, either by providing a diffusion barrier or by directly binding to antimicrobial agents and preventing their penetration into the biofilm (19).
Polysaccharides are a major matrix component in most bacterial biofilms (26). Poly-β(1,6)-N-acetyl-d-glucosamine (PNAG) is an extracellular polysaccharide that mediates biofilm cohesion in numerous gram-negative members of the Proteobacteria family, including Escherichia coli, Yersinia pestis, Pseudomonas fluorescens, Bordetella spp., Xenorhabdus nematophila, Aggregatibacter actinomycetemcomitans, and Actinobacillus pleuropneumoniae (4, 8, 15, 22), and in the gram-positive species Staphylococcus aureus and Staphylococcus epidermidis (3, 17). Specific biofilm-related functions ascribed to PNAG include abiotic surface attachment (1), epithelial cell attachment (23, 28), intercellular adhesion (15, 17), and resistance to killing by antibiotics, detergents, antimicrobial peptides, and mammalian phagocytic cells (9, 10, 16, 27, 29).
In the present study we investigated the physical and chemical properties of the PNAG matrix in biofilms produced by the porcine respiratory pathogen A. pleuropneumoniae and the device-associated pathogen S. epidermidis. By using a novel centrifugal filter device assay, we obtained evidence that PNAG significantly inhibits fluid convection and solute transport through A. pleuropneumoniae and S. epidermidis biofilms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. epidermidis strains were passaged weekly on blood agar. A. pleuropneumoniae strains were passaged twice weekly on Tryptic soy agar supplemented with 6 g of yeast extract, 8 g of glucose, and 10 mg of NAD per liter. For biofilm cultures, S. epidermidis was grown in Tryptic soy broth (TSB) supplemented with yeast extract and glucose, and A. pleuropneumoniae was grown in Mueller-Hinton broth (MHB) supplemented with yeast extract, glucose, and NAD, at the concentrations stated above.
TABLE 1.
Bacterial strains
Reagents.
Cetylpyridinium chloride (CPC) (hexadecylpyridinium chloride), benzalkonium chloride (BKC) (predominantly C12 alkyl chain length), rifampin (rifampicin), tetracycline hydrochloride, and vancomycin hydrochloride were purchased from Sigma. Allura red dye solution (red food color; FD&C Red no. 40) was from McCormick (Sparks, MD). Dispersin B, a PNAG-degrading enzyme (13), was obtained from Kane Biotech (Winnipeg, Canada).
Centrifugal filter device assay.
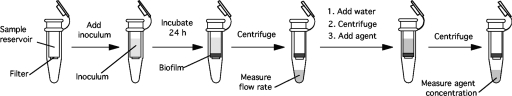
Biofilms were cultured in Microcon centrifugal filter devices (catalog no. YM-100; Millipore). These devices consist of a polycarbonate sample reservoir (6.5-mm inside diameter, 15-mm height, 500-μl capacity) positioned on top of a cellulose membrane with a 100-kDa nominal molecular mass pore size. The sample reservoir snaps into a membrane support base and is separated from the filter by a silicone O ring. The entire sample reservoir assembly fits inside a standard 1.5-ml microcentrifuge tube. These devices are normally used for the concentration and desalting of macromolecular solutions by the use of a centrifuge. A schematic diagram of the centrifugal filter device assay is presented in Fig. 1.
FIG. 1.
Centrifugal filter device assay. Biofilms were cultured in the sample reservoirs of Microcon centrifugal filter devices as described in Materials and Methods. The sample reservoir is positioned on top of a filter with a 100-kDa cutoff. Inocula were transferred directly to the sample reservoir and incubated statically for 24 h. A biofilm formed directly on the filter surface. To measure fluid convection, devices were centrifuged for increasing amounts of time and the flowthrough volume was measured. To measure solute transport, biofilms were first perfused with water by centrifugation to rinse the biofilm and then perfused with the agent. The concentration of the agent in the flowthrough volume was measured.
(i) Inoculation and culturing of filter devices.
Inocula were prepared from agar colonies in fresh TSB or MHB as previously described (9, 14). Aliquots of inocula (300 μl each at 105 to 106 CFU ml−1) were transferred to the sample reservoirs of the centrifugal filter devices. Control devices were inoculated with sterile broth and incubated alongside the bacterium-inoculated devices. In some experiments, media were supplemented with 20 μg of dispersin B ml−1. The devices were incubated statically for 24 h at 37°C in air.
(ii) Measurement of volumetric flow rate of liquid through the biofilms.
After incubation, the devices were centrifuged for 3 min at 4,300 × g in a swinging bucket rotor, and the flowthrough volume was measured. The 3-min centrifugation step was repeated two to three additional times, and the cumulative flowthrough volume was remeasured after each centrifugation step.
(iii) Measurement of solute transport through biofilms.
After incubation, the devices were perfused with 300 μl of water by centrifugation to rinse the biofilms and were then perfused with 300 μl of 0.03% (0.88 mM) CPC, 0.03% (0.88 mM) BKC, 0.5% (vol/vol) Allura red dye solution, 0.75 mM rifampin, 2.8 mM tetracycline, or 1.0 mM vancomycin. The concentration of the agent in the flowthrough volume was measured as follows. For CPC, BKC, and vancomycin, 30 μl of the flowthrough volume was transferred to a well of a 96-well round-bottom microtiter plate containing 270 μl of water. Serial 75% dilutions (225 μl to 75 μl) were made in fresh TSB directly in adjacent wells. For a standard, 30 μl of the agent, at the same concentration used to perfuse the biofilm, was transferred to a well containing 270 μl of water and diluted using the same procedure. Aliquots of S. epidermidis NJ9709 cells (20 μl each at ca. 106 CFU ml−1) were transferred to the wells, and the plate was incubated for 24 h. The concentration of the agent in the flowthrough volume was measured by comparing the MIC of the flowthrough volume to the MIC of the standard, taking into account the dilution factor. For tetracycline, rifampin, and Allura red dye, 200 μl of the flowthrough volume was transferred to a well of a 96-well flat-bottom microtiter plate and the absorbance of the solution (at 440 nm for tetracycline and 490 nm for rifampin and Allura red dye) was measured in a microplate reader. The concentration of the agent in the flowthrough volume was calculated by comparing the absorbance of the flowthrough volume to a standard curve derived from known concentrations of the agent made in the same microtiter plate.
Colony biofilm assay.
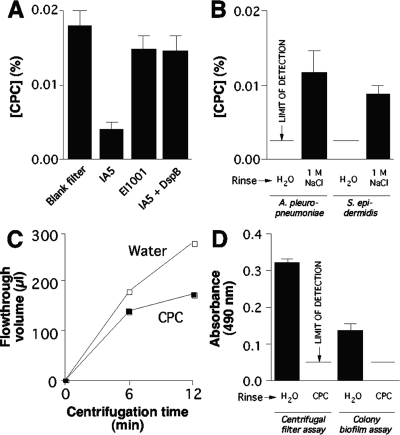
A 47-mm-diameter, 0.45-μm-pore-size polyvinylidene fluoride filter (Millipore catalog no. HVLP04700) was placed on a blood agar plate, and a 10-μl aliquot of inoculum (ca. 106 CFU ml−1) was pipetted onto the filter. After incubation for 18 h, a large colony of bacteria, hereinafter referred to as a colony biofilm, formed on the surface of the membrane. A 10-μl aliquot of 0.03% CPC was pipetted directly onto the colony biofilm. For controls, 10 μl of CPC solution was pipetted directly onto the agar or onto a sterile area of the filter adjacent to the colony biofilm. The CPC was allowed to absorb into the agar for 15 min. The filter was then removed from the plate, and the agar surface was inoculated with a lawn of S. epidermidis NJ9709 cells. After incubation for 24 h, the diameter of the zone of inhibition was measured. To measure the diffusion of Allura red dye through colony biofilms, the filter was removed from the blood agar plate and transferred to a petri dish containing 2% low-gelling-temperature agarose. A 10-μl aliquot of 0.5% Allura red dye solution was pipetted directly onto the colony biofilm or onto a sterile area of the filter adjacent to the colony biofilm. The dye was allowed to absorb into the agarose for 10 min. The filter was then lifted from the plate, and a plug of agarose containing the absorbed dye was removed from the plate by the use of a 5-mm-diameter plastic drinking straw. The agarose plug was transferred to a well of a flat-bottom 96-well microtiter plate, and the plate was placed on a 100°C heating block for 15 min to melt the agarose plug. The absorbance of the dye solution was then measured at 490 nm in a microplate reader.
Cell culture insert biofilm assay.
Biofilms were cultured in polyethylene terephthalate cell culture inserts (BD Biosciences catalog no. 353095) (6.4-mm diameter, 0.4-μm pore size) in the wells of a 24-well microtiter plate. The insert contained 350 μl of inoculum in fresh broth (105 to 106 CFU ml−1), and the well contained 900 μl of sterile broth. After incubation for 24 h, the broth was carefully aspirated from the insert and the insert was placed into a well of a fresh 24-well microtiter plate containing 900 μl of water per well. A total of 300 μl of 0.03% CPC or 0.5% Allura red dye solution was carefully pipetted onto the biofilm, and the plate was incubated at 37°C for 24 h to allow the agent to diffuse from the insert through the biofilm and into the microtiter plate well. The concentration of CPC in the well was measured by a bioassay as described above. The concentration of Allura red dye was measured by spectroscopy at 490 nm as described above.
Scanning electron microscopy.
Biofilms cultured for 24 h in centrifugal filter devices were perfused with water, and the devices were dried for 18 h at 30°C. The filters were removed from the devices and dried for an additional 2 h at 37°C. The top surface of the dried biofilm was examined using a LEO 982 DSM field emission scanning electron microscope (Zeiss). Imaging was done using 1-keV electrons at a working distance of 6 mm. Secondary electrons were collected using an Everhart-Thornley detector. At least two independent areas near the center of the filter were examined in each specimen at magnifications ranging from ×2,000 to ×10,00.
Statistics and reproducibility of results.
All fluid convection and solute transport assays were performed in duplicate. All assays were performed on at least three separate occasions, with nearly identical results. The significance of differences between means was calculated using a Student t test. A P value of <0.05 was considered significant.
RESULTS
PNAG decreases fluid convection through S. epidermidis and A. pleuropneumoniae biofilms.
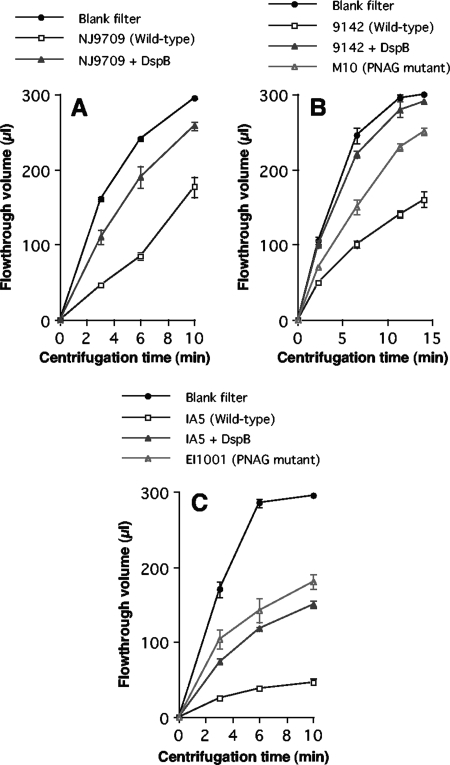
S. epidermidis and A. pleuropneumoniae biofilms were cultured in the sample reservoirs of Microcon centrifugal filter devices (Fig. 1). Biofilms were cultured directly on the filter, which had a 100-kDa nominal molecular mass pore size. This device enabled us to measure the rate of fluid convection through the biofilm by subjecting the devices to low-speed centrifugation and measuring the flowthrough volume after increasing amounts of time (Fig. 2). The rate of fluid convection through S. epidermidis and A. pleuropneumoniae biofilms was significantly less than the rate through control centrifugal filter devices inoculated with sterile broth (Fig. 2). For both species, the rate of fluid convection was significantly increased when biofilms were cultured in the presence of the PNAG-degrading enzyme dispersin B. Biofilms formed by S. epidermidis wild-type strain 9142 exhibited a significantly lower fluid convection rate than biofilms formed by the S. epidermidis PNAG mutant strain M10 (Fig. 2B). Similarly, biofilms formed by A. pleuropneumoniae wild-type strain IA5 exhibited a significantly lower fluid convection rate than biofilms formed by the A. pleuropneumoniae PNAG mutant strain EI1001 (Fig. 2C). These findings indicate that PNAG decreases fluid convection through both S. epidermidis and A. pleuropneumoniae biofilms.
FIG. 2.
Fluid convection through 24-h-old S. epidermidis and A. pleuropneumoniae biofilms cultured in centrifugal filter devices. Devices were subjected to low-speed centrifugation, and the cumulative flowthrough volume was measured after increasing amounts of time. Blank filters were inoculated with sterile broth and incubated alongside the biofilm-containing filters. (A) Fluid convection through S. epidermidis NJ9709 biofilms cultured in the absence or presence of 20 μg of dispersin B (DspB) ml−1. (B) Fluid convection through S. epidermidis 9142 biofilms cultured in the absence or presence of 20 μg ml−1 of dispersin B and through S. epidermidis M10 (PNAG mutant) biofilms. (C) Fluid convection through A. pleuropneumoniae IA5 biofilms cultured in the absence or presence of 20 μg of dispersin B ml−1 and through A. pleuropneumoniae EI1001 (PNAG mutant) biofilms. Values show means and ranges of flowthrough volumes for duplicate centrifugal filter devices.
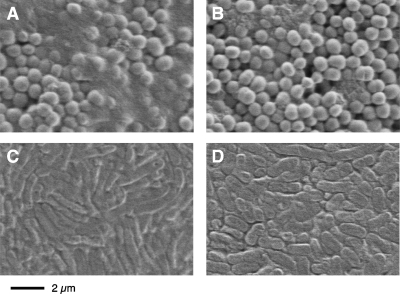
Figure 3 shows scanning electron micrographs of S. epidermidis and A. pleuropneumoniae biofilms cultured in centrifugal filter devices. Note that the biofilms had been dehydrated prior to examination and that such imaging does not directly reflect the original three-dimensional biofilm structure. Biofilms produced by S. epidermidis strain NJ9709 contained an ultrastructurally visible extracellular material (Fig. 3A) that was absent in NJ9709 biofilm cultured in the presence of dispersin B (Fig. 3B). These results are similar to those of previous studies showing the presence of an extracellular fibrous material in S. epidermidis wild-type strains that was absent in PNAG mutant strains (29). A. pleuropneumoniae IA5 biofilms contained a similar extracellular material (Fig. 3C) that was absent in biofilms produced by A. pleuropneumoniae PNAG mutant strain EI1001 (Fig. 3D). A. pleuropneumoniae IA5 biofilms cultured in the presence of dispersin B appeared to be similar to A. pleuropneumoniae EI1001 biofilms (data not shown). These findings suggest that PNAG may block fluid convection by forming a matrix that impedes water flow through the open spaces between the biofilm cells. In addition, wild-type biofilms cultured in the absence of dispersin B appeared wet (smooth and glossy) after centrifugation whereas wild-type biofilms cultured in the presence of dispersin B, as well as PNAG mutant biofilms, appeared dry (textured and mat). These observations suggest that the PNAG matrix can bind a significant amount of water.
FIG. 3.
Scanning electron micrographs of S. epidermidis and A. pleuropneumoniae biofilms cultured in centrifugal filter devices. (A and B) S. epidermidis strain NJ9709 cultured in the absence and presence of 20 μg of dispersin B ml−1, respectively; (C and D) A. pleuropneumoniae wild-type strain IA5 and PNAG mutant strain EI1001, respectively. Images show an area on the top surface of the biofilm near the center of the filter.
PNAG sequesters CPC within the biofilm matrix.
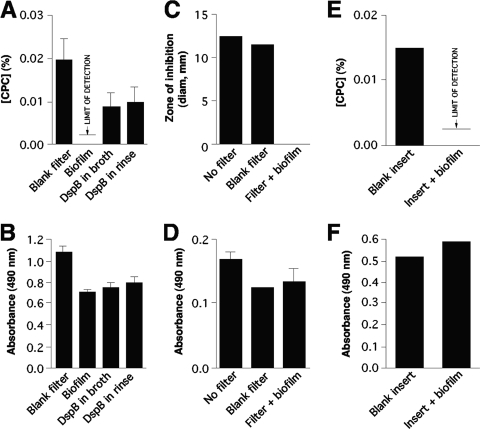
S. epidermidis NJ9709 biofilms cultured in centrifugal filter devices were perfused with 0.03% CPC, and the concentration of CPC in the flowthrough volume was measured (Fig. 4A). No detectable CPC was present in the flowthrough volume (limit of detection = 0.002%). However, when biofilms were cultured in the presence of dispersin B, or when biofilms were perfused with dispersin B prior to perfusion with CPC, a significant amount of CPC was present in the flowthrough volume (Fig. 4A). In contrast, the azo dye Allura red was efficiently transported through S. epidermidis NJ9709 biofilms cultured in both the absence and the presence of dispersin B, and a prerinse with dispersin B did not significantly increase the concentration of dye in the flowthrough volume (Fig. 4B). These finding suggest that PNAG impedes the flow of CPC, but not of Allura red dye, through S. epidermidis biofilms.
FIG. 4.
Transport of CPC (top panels) and Allura red dye (bottom panels) through S. epidermidis NJ9709 biofilms. (A and B) Centrifugal filter device assay. Agents were added to the chambers of filter devices and perfused through the filters by low-speed centrifugation. Values show the concentration of the agent in the flowthrough volume (for CPC) or the absorbance of the flowthrough solution (for Allura red dye) for control filters inoculated with sterile TSB (Blank filter), filters inoculated with NJ9709 (Biofilm), filters inoculated with NJ9709 and cultured in the presence of 20 μg of dispersin B ml−1 (DspB in broth), and filters inoculated with NJ9709 and perfused with 20 μg of dispersin B ml−1 by centrifugation prior to addition of the CPC (DspB in rinse). (C and D) Colony biofilm assay. For panel C, a CPC solution was pipetted directly onto a blood agar plate (No filter), onto a sterile filter placed on the agar surface (Blank filter), or onto a colony biofilm growing on a filter (Filter + biofilm). The CPC was allowed to absorb into the agar for 15 min, after which the filter was removed and the plate was inoculated with a lawn of NJ9709 cells. Values show the diameter of the zone of inhibition after 24 h. For panel D, a filter containing a colony biofilm was transferred to a petri dish containing 2% low-gelling-temperature agarose. Allura red dye was pipetted onto the agarose surface, filter, or colony biofilm as described for panel C and allowed to absorb into the agar for 15 min. Values show the absorbance (at 490 nm) of an agarose plug containing the diffused Allura red dye. (E and F) Cell culture insert assay. Cell culture inserts containing biofilms were rinsed, placed in a fresh microplate containing water, and filled with CPC or Allura red dye. Values show the concentrations of CPC (E) or Allura red dye (F) that diffused into the microplate well after 24 h. As a control, inserts inoculated with sterile TSB (Blank insert) were used. In all panels, values show means and ranges of the results of duplicate assays.
To confirm that CPC and Allura red dye are transported through S. epidermidis biofilms at different rates, we measured the transport of these agents through colony biofilms cultured on filter disks placed on agar surfaces (Fig. 4C and D) and through biofilms cultured in cell culture inserts in broth (Fig. 4E and F). These assays depended on absorption and diffusion for the transport of solutes through the biofilms, which closely resembles the mode of solute transport through natural biofilms (25). In both assays, the rate of CPC transport was significantly less than the rate of dye transport. These findings confirm that the differential data obtained for transport of CPC and Allura red dye through S. epidermidis biofilms cultured in centrifugal filter devices were not the result of an artifact due to centrifugation of the biofilm cells.
We also measured the transport of CPC through A. pleuropneumoniae wild-type IA5 and PNAG mutant EI1001 biofilms cultured in centrifugal filter devices (Fig. 5A). Like the wild-type S. epidermidis biofilms (Fig. 4A), A. pleuropneumoniae IA5 biofilms significantly impeded the transport of CPC, and CPC transport was significantly increased when IA5 biofilms were cultured in the presence of dispersin B. In addition, CPC transport levels were significantly higher in PNAG mutant EI1001 biofilms than in wild-type IA5 biofilms (Fig. 5A). These findings confirm that PNAG impedes transport of CPC through bacterial biofilms.
FIG. 5.
Transport of CPC and Allura red dye through A. pleuropneumoniae and S. epidermidis biofilms. (A) Transport of CPC through A. pleuropneumoniae biofilms cultured in centrifugal filter devices. Values show the concentration of CPC in the flowthrough volume for a control filter inoculated with sterile MHB (Blank filter), a filter inoculated with wild-type strain IA5, a filter inoculated with PNAG mutant strain EI1001, and a filter inoculated with IA5 and cultured in the presence of 20 μg of dispersin B ml−1 (IA5 + DspB). (B) Elution of CPC from A. pleuropneumoniae IA5 and S. epidermidis NJ9709 biofilms in centrifugal filter devices. Biofilms were perfused by centrifugation with 0.03% CPC and then with water. The values show the concentrations of CPC in the flowthrough volume after a third perfusion with water or 1 M NaCl. (C) S. epidermidis NJ9709 biofilms cultured in centrifugal filter devices were perfused with water or CPC, and the rate of water convection was then measured as described for Fig. 1. (D) Transport of Allura red dye through S. epidermidis NJ9709 biofilms cultured in centrifugal filter devices or as colony biofilms. Biofilms were perfused with water or CPC prior to the addition of the Allura red dye. Values show the absorbance of the flowthrough volume (for the centrifugal filter assay) or the absorbance of an agarose plug containing the absorbed dye (for the colony biofilm assay). In all panels, values show means and ranges of the results of duplicate assays.
To investigate the mechanism of CPC binding to A. pleuropneumoniae and S. epidermidis biofilms, wild-type biofilms cultured in centrifugal filter devices were perfused with 0.03% CPC, and the biofilms were then perfused with water or 1 M NaCl. The amount of CPC in the flowthrough volume after the water or NaCl perfusions was then measured (Fig. 5B). For both species, 1 M NaCl caused elution of a significant amount of bioactive CPC from the biofilm. These findings suggest that CPC may react directly with the biofilm matrix by means of electrostatic interactions.
Binding of CPC to S. epidermidis NJ9709 biofilms cultured in centrifugal filter devices significantly impeded fluid convection through the biofilms (Fig. 5C). In addition, binding of CPC to NJ9709 biofilms cultured in centrifugal filter devices, or as colony biofilms on agar, significantly impeded transport of Allura red dye (Fig. 5D). These findings confirm that binding of CPC to S. epidermidis biofilms significantly alters their porosity and filtration rate.
Effect of PNAG on transport of other solutes through S. epidermidis and A. pleuropneumoniae biofilms.
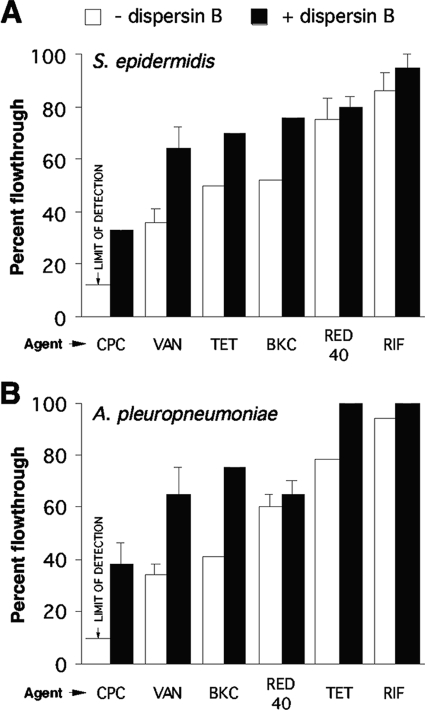
We also measured the transport of various other solutes through wild-type S. epidermidis strain NJ9709 and A. pleuropneumoniae IA5 biofilms by the use of the centrifugal filter device assay (Fig. 6). Transport of the cationic detergent BKC was significantly greater than transport of CPC, with only about half of the BKC being retained by the biofilms. However, like CPC, transport of BKC was significantly increased when biofilms were cultured in the presence of dispersin B. The presence of dispersin B in the culture medium also significantly increased the transport of vancomycin and tetracycline. Rifampin was efficiently transported through both S. epidermidis and A. pleuropneumoniae biofilms.
FIG. 6.
Transport of solutes through S. epidermidis NJ9709 (A) and A. pleuropneumoniae IA5 (B) biofilms cultured in centrifugal filter devices. Biofilms were cultured in the absence or presence of dispersin B. Percentages of flowthrough volume were calculated as follows: (concentration of the agent that flowed through the biofilm/concentration of the agent that flowed through a blank filter device) × 100. The solutes tested were CPC, vancomycin (VAN), tetracycline (TET), BKC, Allura red dye (RED 40), and rifampin (RIF). Values show means and ranges of the results obtained with duplicate filter devices.
DISCUSSION
Our findings indicate that PNAG matrix polysaccharide significantly alters the physical properties of bacterial biofilms. This conclusion is based on the fact that removal of PNAG from the biofilm matrix, either by dispersin B-mediated degradation or by a chromosomal mutation in the PNAG biosynthetic genes, significantly increased the hydraulic conductance and filtration rate across the biofilm (Fig. 2). The low permeability of A. pleuropneumoniae and S. epidermidis biofilms could be due to clogging of the matrix by PNAG polymer, as evidenced by the presence of an ultrastructurally visible extracellular material (Fig. 3), or by the formation of a semiliquid or viscous hydrogel within the matrix, as evidenced by the wet appearance of wild-type biofilms compared to the dry appearance of dispersin B-treated wild-type biofilms and PNAG mutant biofilms. These low-permeability and water-resistant properties of the PNAG matrix are likely to influence the relative effective diffusivity of solutes through the biofilms (25).
CPC is a cationic detergent that is widely used as an antiseptic agent. CPC reacts with phospholipids in the cytoplasmic membrane, causing membrane distortion and cell lysis due to osmotic stress (20). Several studies have shown that biofilm bacteria exhibit significantly increased resistance to CPC compared to the resistance exhibited by planktonic cells (2, 9, 10, 30). Depolymerization of PNAG by dispersin B has been shown to render S. epidermidis and A. actinomycetemcomitans biofilm cells sensitive to killing by CPC (9, 10), indicating that the PNAG matrix contributes to biofilm-mediated CPC resistance. In the present study we showed that CPC binds reversibly to the PNAG-containing matrix of A. pleuropneumoniae and S. epidermidis (Fig. 4 and 5). These findings suggest that PNAG may sequester CPC within the biofilm matrix, thereby preventing it from contacting the biofilm cells. We were unable to utilize dilution plating to determine whether perfusion of biofilms with CPC killed biofilm cells cultured in centrifugal filter devices, because mechanical disruption of perfused biofilms alone caused a dramatic increase in CPC susceptibility (unpublished data).
Bioactive CPC could be eluted from biofilms by treatment with 1 M NaCl, suggesting that CPC binds to biofilms by means of electrostatic interactions and that CPC is not significantly altered as a result of its binding to biofilm cells or biofilm matrix components. Since both CPC and PNAG are cationic, however, it is possible that CPC binds to PNAG by means of its hydrophobic tail. Another interesting possibility is that CPC binds to double-stranded DNA. Previous studies showed that DNA is a component of the extracellular matrix in both S. epidermidis biofilms (24) and A. pleuropneumoniae biofilms (unpublished data). Cationic detergents are known to bind to and precipitate DNA (12), and detergent-DNA complexes can be dissolved by treatment with 1 M NaCl (11). Also, cationic detergents such as cetylpyridinium bromide and cetyl trimethylammonium bromide, both with C16 side chains, bind DNA more efficiently than detergents with shorter side chains (21). This may explain why CPC (C16 side chain) was retained by S. epidermidis biofilms more efficiently than BKC (C12 side chain). If this model is correct, then PNAG may form a stable complex with DNA which is disrupted by dispersin B-mediated depolymerization of PNAG. The formation of a CPC-DNA precipitate could also account for the decreased transport of Allura red dye through S. epidermidis biofilms that were pretreated with CPC (Fig. 5D).
We found that rifampin readily perfused through S. epidermidis biofilms cultured in centrifugal filter devices (Fig. 6). These findings are consistent with those of other studies showing that rifampin diffuses through S. epidermidis biofilms cultured on dialysis membranes (5) and through S. epidermidis colony biofilms cultured on filter disks on agar (31). It is interesting to note that depolymerization of PNAG by dispersin B increased the rate of transport of vancomycin through S. epidermidis biofilms (Fig. 5). These results suggest that vancomycin may react directly with the PNAG-containing matrix. This hypothesis is consistent with the results of previous studies showing that slime extracted from S. epidermidis, which contains PNAG, inhibits the antimicrobial action of glycopeptide antibiotics, including vancomycin (6).
In conclusion, we developed a novel centrifugal filter device assay that can be used to investigate the physical properties of the biofilm matrix. Our findings demonstrate that the biofilm matrix polysaccharide PNAG significantly impedes fluid convection and solute transport through biofilms. Even though this assay relies on the forced transport of water and solutes through the biofilm by centrifugation, the results obtained closely resembled those obtained using colony biofilm and cell culture insert biofilm assays which rely on the more natural processes of adsorption and diffusion for solute transport. Therefore, the centrifugal filter device assay appears to accurately measure the physical properties of the biofilm matrix. The centrifugal filter device assay could be a useful tool for studying the physical, chemical, and biological properties of biofilm matrix components and for testing novel antibiofilm agents that target, destabilize, or destroy the physical integrity of the biofilm matrix.
Acknowledgments
We thank Jared Pienkos, Venkatraman Thulasi, and Era Izano (New Jersey Dental School) for technical assistance, Gerald Pier (Harvard Medical School) for providing some of the S. epidermidis strains used in this study, and Daniel Kadouri (New Jersey Dental School) and Woo Lee (Stevens Institute of Technology) for helpful discussions.
This work was supported in part by Public Health Service award DE15124 (to J.B.K.) and Army Research Office grant W911NF-07-1-0543 (to M.R.L.).
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Agladze, K., X. Wang, and T. Romeo. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 187:8237-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campanac, C., L. Pineau, A. Payard, G. Baziard-Mouysset, and C. Roques. 2002. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob. Agents Chemother. 46:1469-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drace, K., and C. Darby. 2008. The hmsHFRS operon of Xenorhabdus nematophila is required for biofilm attachment to Caenorhabditis elegans. Appl. Environ. Microbiol. 74:4509-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunne, W. M., Jr., E. O. Mason, Jr., and S. L. Kaplan. 1993. Diffusion of rifampicin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother. 37:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farber, B. F., M. H. Kaplan, and A. G. Clogston. 1990. Staphylococcus epidermidis extracted slime inhibits the antimicrobial action of glycopeptide antibiotics. J. Infect. Dis. 161:37-40. [DOI] [PubMed] [Google Scholar]
- 7.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 8.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izano, E. A., I. Sadovskaya, E. Vinogradov, M. H. Mulks, K. Velliyagounder, C. Ragunath, W. B. Kher, N. Ramasubbu, S. Jabbouri, M. B. Perry, and J. B. Kaplan. 2007. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 43:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izano, E. A., I. Sadovskaya, H. Wang, E. Vinogradov, C. Ragunath, N. Ramasubbu, S. Jabbouri, M. B. Perry, and J. B. Kaplan. 2008. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb. Pathog. 44:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, A. S. 1953. Isolation of bacterial nucleic acids using cetyltrimethylammonium bromide (Cetavlon). Biochim. Biophys. Acta 10:607-612. [DOI] [PubMed] [Google Scholar]
- 12.Jones, A. S. 1963. Use of alkyltrimethylammonium bromides for the isolation of ribo- and deoxribonucleic acids. Nature 199:280-282. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan, J. B., K. Velliyagounder, C. Ragunath, H. Rohde, D. Mack, J. K. Knobloch, and N. Ramasubbu. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186:8213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kropec, A., T. Maira-Litran, K. K. Jefferson, M. Grout, S. E. Crampton, F. Götz, D. A. Goldmann, and G. B. Pier. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect. Immun. 73:6868-6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mah, T.-F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 20.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miguel, M. G., A. A. C. C. Pais, R. S. Dias, C. Leal, M. Rosa, and B. Lindman. 2003. DNA-cationic amphiphile interactions. Colloids Surf. A 228:43-55. [Google Scholar]
- 22.Parise, G., M. Mishra, Y. Itoh, T. Romeo, and R. Deora. 2007. Role of a putative polysaccharide locus in Bordetella biofilm development. J. Bacteriol. 189:750-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parise Sloan, G., C. F. Love, N. Sukumar, M. Mishra, and R. Deora. 2007. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J. Bacteriol. 189:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin, Z., Y. Ou, L. Yang, Y. Zhu, T. Tolker-Nielsen, S. Molin, and D. Qu. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083-2092. [DOI] [PubMed] [Google Scholar]
- 25.Stewart, P. S. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 27.Venketaraman, V., A. K. Lin, A. Le, S. C. Kachlany, N. D. Connell, and J. B. Kaplan. 2008. Both leukotoxin and poly-N-acetylglucosamine surface polysaccharide protect Aggregatibacter actinomycetemcomitans cells from macrophage killing. Microb. Pathog. 45:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vuong, C., S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. DeLeo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion and virulence. J. Biol. Chem. 279:54881-54886. [DOI] [PubMed] [Google Scholar]
- 29.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, M., H. Patel, and J. Fletcher. 1996. Susceptibility of biofilm of Streptococcus sanguis to chlorhexidine and cetylpyridinium chloride. Oral Microbiol. Immunol. 11:188-192. [DOI] [PubMed] [Google Scholar]
- 31.Zheng, Z., and P. S. Stewart. 2002. Penetration of rifampicin through Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 46:900-903. [DOI] [PMC free article] [PubMed] [Google Scholar]